Abstract
In relation to emerging multiresistant bacteria, development of antimicrobials and new treatment strategies of infections should be expected to become a high-priority research area. Quorum sensing (QS), a communication system used by pathogenic bacteria like Pseudomonas aeruginosa to synchronize the expression of specific genes involved in pathogenicity, is a possible drug target. Previous in vitro and in vivo studies revealed a significant inhibition of P. aeruginosa QS by crude garlic extract. By bioassay-guided fractionation of garlic extracts, we determined the primary QS inhibitor present in garlic to be ajoene, a sulfur-containing compound with potential as an antipathogenic drug. By comprehensive in vitro and in vivo studies, the effect of synthetic ajoene toward P. aeruginosa was elucidated. DNA microarray studies of ajoene-treated P. aeruginosa cultures revealed a concentration-dependent attenuation of a few but central QS-controlled virulence factors, including rhamnolipid. Furthermore, ajoene treatment of in vitro biofilms demonstrated a clear synergistic, antimicrobial effect with tobramycin on biofilm killing and a cease in lytic necrosis of polymorphonuclear leukocytes. Furthermore, in a mouse model of pulmonary infection, a significant clearing of infecting P. aeruginosa was detected in ajoene-treated mice compared to a nontreated control group. This study adds to the list of examples demonstrating the potential of QS-interfering compounds in the treatment of bacterial infections.
INTRODUCTION
Infections that develop into chronic conditions are a fast-growing problem in the developed world. The underlying biology is thought to be the ability of bacteria to form biofilms (20), which consist of structured and aggregated (often surface-attached) communities of bacteria (18). Multiple studies have documented that such aggregated communities are more resistant to a variety of antibiotics and the action of the immune system compared to their planktonic counterparts (6, 19, 41, 68). Biofilm infections are often connected to patients with medical devices and implants as well as hospitalized patients. Lately, bacterial biofilms have also been associated with nonhealing, chronic wounds (9, 24, 38). There is thus an urgent need for development of new treatment strategies using a combination of drugs targeting a multitude of antimicrobial targets.
Several Gram-negative pathogens use N-acyl homoserine lactone (AHL)-mediated communication systems in a process termed quorum sensing (QS) to coordinate specific gene expression, thereby synchronizing expression of particular phenotypic features between the individual cells (28). QS is thought to play an important role during the initial event of infection for the common opportunistic Gram-negative human pathogen Pseudomonas aeruginosa, which is associated with nosocomial and wound infections, immunocompromise (48, 70), and the genetically inherited disease cystic fibrosis (CF) (22). By employing the QS system to control expression of its virulence factors (many of which are antigenic determinants), P. aeruginosa is able to operate in a stealthy manner until a certain cell density is reached, where the QS systems become activated. Upon activation of the QS systems, a coordinated release of tissue-damaging and immune defense-degrading virulence factors takes place (28, 36). It was recently documented by us that the QS-controlled virulence factor rhamnolipid (also known as heat-stable hemolysin) destroys polymorphonuclear leukocytes (PMNs) by lytic necrosis (36). Besides lysing neutrophils and macrophages, rhamnolipid has also been reported to impair chemotaxis of neutrophils (42, 62). As rhamnolipid is associated with bacteria living in biofilm, it is likely to function as a shield toward important cellular components of the host defense (1, 36, 71). Furthermore, QS promotes increased tolerance of P. aeruginosa biofilms to antibiotic treatments (6) and provides biofilms with structural rigidity through release of extracellular DNA (eDNA) (21, 56).
Two of the P. aeruginosa quorum sensors are based on the LuxRI homologues present in most Gram-negative bacteria with QS systems. The LuxI homologues function as an AHL synthetase producing the required signal molecules, and the LuxR homologues function as transcriptional activators which, upon binding of the cognate signal molecules, activate the transcription of the QS target genes (28). The P. aeruginosa QS system consists of lasRI and rhlRI hierarchically arranged with the las-encoded system, normally at the top level controlling the rhl-encoded system, and a third system that intervenes between the two denoted the pseudomonas quinolone signal (PQS). The three interacting QS systems LasRI, RhlRI, and PQS use the following signal molecules for activation: N-(3-oxododecanoyl)-l-homoserine lactone (3-oxo-C12-HSL), N-butanoyl homoserine lactone (C4-HSL), and 2-heptyl-3-hydroxy-4-quinolone (PQS), respectively (50, 51, 54).
It has been shown that some terrestrial and marine organisms have evolved a system of specific molecules with AHL-antagonistic activity capable of interfering with the bacterial QS system in a possible prevention of colonization (8, 55, 58, 59, 64). Hentzer et al. (33) and Wu et al. (76) demonstrated that the QS system could be used as an effective antimicrobial drug target by altering the tolerance of biofilms to antibiotic treatment and promote a faster clearance of a P. aeruginosa lung infection in mice by using the chemically modified QS inhibitor (QSI) furanone C-30. Crude extracts of garlic (Allium sativum L.) have been shown to inhibit the expression of a large number of QS-controlled genes (58), and Bjarnsholt et al. (8) demonstrated the ability of garlic extracts, similar to C-30 treatments, to promote a rapid clearing of a pulmonary P. aeruginosa infection in mice. Garlic is widely accepted as a herb that through dietary intake can improve human health (61). Epidemiological studies have shown that a daily intake of garlic lowers the risk of certain cancers (25, 63), and several studies have documented an antithrombotic and lipid-lowering cardiovascular effect of some of the constituents in garlic (27).
By means of a bioassay-directed purification procedure, we identified the sulfur-containing compound ajoene (4,5,9-trithiadodeca-1,6,11-triene 9-oxide) to be a QSI present in garlic extract. When garlic is crushed, ajoene and several other organosulfides are produced as degradation products of allicin (diallyl thiosulfinate) (11). Ajoene has been reported to display conventional antimicrobial activities against a number of Gram-positive bacteria and the Gram-negative bacteria Escherichia coli, Klebsiella pneumoniae, and Xanthomonas maltophilia but not P. aeruginosa (45). To further exploit the QSI activity in vitro and in vivo, we employed chemically synthesized ajoene (M. Givskov, 8 December 2010, European patent application no. 10194154.0 and US provisional application no. 61/420,922). The in vitro experiments showed significant inhibition of a subclass of QS-regulated P. aeruginosa genes and a significant synergistic action with tobramycin with respect to the reduction of viability of biofilm cells. Furthermore, a mouse model of pulmonary infection was employed to demonstrate the antimicrobial effect of ajoene on P. aeruginosa infections.
MATERIALS AND METHODS
Bacterial strains.
Sequenced P. aeruginosa PAO1 wild type was obtained from the Pseudomonas Genetic Stock Center (www.pseudomonas.med.ecu.edu; PAO0001). For detection of QSI activity, the following reporter strains were used: a QSI selector 1 strain (QSIS1; E. coli) and monitor strains for lasB-gfp, rhlA-gfp (P. aeruginosa), and luxI-gfp (E. coli), described in references 58, 32, 77, and 4, respectively. Production of AHLs was detected by using reporter strains for lasB-gfp (E. coli) (32) and ahyI-gfp (E. coli) (30). Animal experiments were performed with the wild-type P. aeruginosa strain PAO1, obtained from Barbara Iglewski (University of Rochester Medical Center, Rochester, NY). The strain is QS proficient, except for the reduced production of C4-HSL previously noted for this P. aeruginosa variant (39). The clinical isolate CF438 was obtained from a child with CF and kindly provided by Helle K. Johansen and Oana Ciofu. The mucoid and nonmucoid isogenic strains are described elsewhere (44, 72).
Growth media and conditions for in vitro and in vivo experiments.
ABT minimal medium (B medium [17] plus 2.5 mg thiamine liter−1 and 10% A10 [17]) supplemented with 0.5% (wt/vol) glucose and 0.5% (wt/vol) Casamino Acids was used for growing the monitor strains (overnight cultures) for the QSI indicator screens. All strains were incubated at 30°C with shaking (180 rpm) and supplemented with antibiotics where appropriate. For animal experiments, bacteria from freezer stocks were plated onto blue agar plates (State Serum Institute, Denmark) and incubated at 37°C overnight. Blue agar plates are selective for Gram-negative bacilli (34). One colony was used to inoculate overnight cultures grown in Luria-Bertani (LB) medium at 37°C with shaking.
Extraction and purification of QS-containing fractions from garlic.
Fractions containing QS activity were identified by a QSIS1 bioassay-guided fractionation described previously (58). Garlic cloves were skinned, homogenized in toluene, and stirred overnight with an equal volume of water. The garlic pulp was filtered, and the solvents were separated, dried under vacuum, and tested for QSI activity. Activity was observed only from the toluene extract. The extract was fractionated on a C18 column (125 g, 200 by 50 mm) on a Biotage Isolera flash purification system (Isolera) with a flow rate of 30 ml/min and with sample added as dry load. Samples were collected (without detection) as 100-ml fractions; the first sample was collected at 10% methanol (MeOH) in H2O, the following 10 samples were eluted with a 10 to 100% MeOH gradient (1000 ml), and all subsequent samples were eluted with 100% MeOH (typically, 500 ml). Activity was detected in the 50 to 60% MeOH fractions, which were combined and further purified by semipreparative high-performance liquid chromatography (HPLC) on a Chromolith RP-18e column (100 by 4.6 mm) with a flow rate of 2 ml per min, eluting with 30% acetonitrile (MeCN) and increasing to 37% MeCN over 20 min. A single fraction eluting at 9.5 min was determined to have QSI activity. Positive electrospray (ESI+) high-resolution mass spectrometry (HRMS) gave a mass of 235.0282 Da, corresponding to a formula of C9H15OS3. Comparison of 1H nuclear magnetic resonance (NMR) data identified this fraction as a 60:40 mixture of E- and Z-ajoenes (11).
Chemically synthesized ajoene.
Ajoene was synthesized from commercially available distilled allyl disulfide as a 1:4 mixture of E and Z isomers as described by Givskov (European patent application no. 10194154.0 and US provisional application no. 61/420,922). Synthetic ajoene was purified by silica gel chromatography and characterized by 1H NMR, 13C NMR, and HRMS. The purity was greater than 98%. Synthetic ajoene was used in all experiments conducted in the study described in this paper.
Determination of inhibitor strength.
The following bioassays were used to determine inhibitor activity of ajoene. ABT medium (150 μl) supplemented with 0.5% (wt/vol) glucose and 0.5% (wt/vol) Casamino Acids was added to all wells in a 96-well microtiter dish (Black Isoplate; Perkin Elmer). To the first column, ajoene was added to a final concentration of 200 μg/ml and a 2-fold serial dilution was made. No ajoene was added to the last column, which was used as a reference. At last, 150-μl overnight cultures of the QSI monitors (lasB-gfp [32], rhlA-gfp [77], or luxI-gfp [4]) were added to all the wells to a final optical density at 450 nm (OD450) of 0.1. Additionally, the signal molecule N-(3-oxohexanoyl)-l-homoserine lactone was added to the luxI-gfp reporter screen in a final concentration of 100 nM. The growth of the bacterial cells (OD450) and green fluorescent protein (GFP) expression (excitation wavelength, 485 nm; emission wavelength, 535 nm) were measured on a multilabel plate reader (Wallac 1420 VICTOR2; Perkin Elmer) every 15 min over 14 h. The temperature was held constant at 34°C.
Production of QS signal molecules.
Production of C4-HSL and 3-oxo-C12-HSL was detected in the supernatant from an overnight culture of a clinical isolate (CF438) by using the AHL-specific reporter strains and the method described by Hentzer et al. (32).
RNA preparation for DNA microarray analysis.
P. aeruginosa PAO1 exponentially growing (OD600, 0.5) at 37°C and 180 rpm in AB medium (B medium plus 10% A10) supplemented with 0.5% Casamino Acids was diluted to an OD600 of 0.05. When an OD600 of 0.5 was reached, the culture was divided into 5 cultures of 50 ml and the following four concentrations of ajoene were added: 10 μg/ml, 20 μg/ml, 40 μg/ml, and 80 μg/ml. No ajoene was added to one culture. At an OD600 of 2.0, samples were retrieved and 2 volumes of RNAlater (Ambion) was added. Isolation of RNA was performed using an RNeasy minipurification kit (Qiagen) with on-column DNase treatment. The following synthesis of cDNA and hybridization were performed by the microarray core unit at Rigshospitalet, Denmark. The gene expressions were analyzed by the use of the software ArrayStar (version 3.0; DNAStar). DNA microarray analysis of global gene expression was performed according to guidelines provided by Affymetrix and repeated three times with RNA from three individual growth experiments.
RT-PCR.
The purified RNA used for DNA microarray analysis was also used for real-time PCR (RT-PCR). cDNA was made from 1 μg of RNA using high-capacity RNA-to-cDNA master mix (Applied Biosystems). For quantitative real-time PCR, amplification was performed with Power SYBR green master mix in a Step One Plus thermal cycler (Applied Biosystems). The primers were designed using Primer Express software (version 3.0; Applied Biosystems). Forty cycles were run with denaturation at 95°C for 15 s, annealing at 55°C for 30 s, and extension at 60°C for 45 s. The gene rpoD was used as a control for standardization. Primer sequences were as follows: rhlA forward, 5′-GGCGATCGGCCATCT-3′; rhlA reverse, 5′-AGCGAAGCCATGTGCTGAT-3′; lasB forward, 5′-CGACAACGCGTCGCAGTA-3′; lasB reverse, 5′-AGGTAGAACGCACGGTTGTACA-3′; rpoD forward, 5′-ACAAGATCCGCAAGGTACTGAAG-3′; and rpoD reverse, 5′-CGCCCAGGTGCGAATC-3′.
Measurements of total rhamnolipid production.
Samples for measurements of total rhamnolipid concentration were retrieved from the cultures grown for DNA microarray analysis and RT-PCR at OD600s of 1.5 and 2.0 and kept at −80°C until further examinations. HPLC with ESI+ HRMS detection (47) was used to quantify rhamnolipids as their [M + NH4]+ (peak area) on the basis of external standard quantification of a NMR-validated rhamnolipid B standard. A series of diluted standard was analyzed before and after the samples, in order to minimize potential differences in ionization levels of rhamnolipid between the samples. Other rhamnolipids were assumed to give the same ionization efficiency as rhamnolipid B. Total rhamnolipid concentration was derived from six major rhamnolipids with the following masses [M + NH4]+: 668.4 (rhamnolipid B [C10-C10-rha-rha]), 694.4 (C10-C12D-rha-rha), 696.4 (C10-C12-rha-rha), 522.4 (C10-C10-rha), 548.4 (C10-C12D-rha), and 550.4 (C10-C12-rha).
Stability of ajoene.
Overnight cultures of P. aeruginosa strain PAO1 and the ΔlasR-ΔrhlR and ΔlasI-ΔrhlI mutants were diluted to an OD600 of 0.2 and incubated with 100 μg/ml ajoene at 37°C and 4°C for 18 h. The samples were sterile filtrated (pore size, 0.22 μm), and to measure the QSI activity of ajoene, the supernatants were tested in the P. aeruginosa QSI screen using the lasB-gfp monitor strain (see “Determination of inhibitor strength” above).
Effect of ajoene on P. aeruginosa QS signal molecule content.
Production of AHL was quantified by HPLC with tandem mass spectrometry (MS/MS) as described in reference 60. P. aeruginosa was grown as described for DNA array analysis and RT-PCR, with the same concentrations of ajoene added. At OD600s of 1.5, 1.8, and 2.0, samples were retrieved and sterile filtrated (pore size, 0.22 μm). Acidified ethyl acetate was added to the supernatant in a 1:1 ratio and left at room temperature overnight. The top phase was withdrawn, concentrated under nitrogen gas, and resuspended in 500 μl ethanol, with 1 μl analyzed by HPLC-MS/MS (60). In this case, external standard quantification was done. The method detects C4-HSL, open lactone-C4-HSL, 3-oxo-C12-HSL, and open lactone-3-oxo-C12-HSL. Detection limits were in the 10 to 30 nM range.
Effect of serum albumin on QSI activity of ajoene.
The P. aeruginosa QSI screen (with the lasB-gfp monitor strain) was used to test the effect of serum albumin on ajoene activity. Bovine serum albumin was dissolved in ABT medium to a concentration of 100 mg/ml. Three hundred microliters was added to the first row of a 96-well microtiter dish (Black Isoplate; Perkin Elmer). To the rest of the rows, 150 μl medium without serum albumin was added. A 2-fold serial dilution of serum albumin was made, and ajoene was added to the following final concentrations: 12.5 μg/ml and 25 μg/ml. Finally, 150 μl of the lasB-gfp monitor strain was added to all the wells (for a more detailed description, see “Determination of inhibitor strength” above).
In vitro biofilms.
Biofilms were grown at 37°C in continuous-culture, once-through, three-channel flow cells with individual channel dimensions of 1 by 4 by 40 mm perfused with sterile AB trace minimal medium containing 0.3 mM glucose as described by Christensen et al. (15) and Pamp and Tolker-Nielsen (49). Overnight cultures were diluted to an OD600 of 0.1 in 0.9% NaCl, and 250 μl per channel was used for inoculation. All microscopic observations and image acquisitions were performed using a confocal laser scanning microscope (Leica TCS SP5; Leica Microsystems, Germany). Images were obtained with a ×40 dry objective and ×100 oil objective. To visualize dead bacterial cells and lysed PMNs, propidium iodide (PI; P-4170; Sigma, Steinheim, Germany) was used, whereas expression of GFP was used as a measure for live bacterial cells. Image scanning was carried out at 488-nm (green) and 543-nm (red) laser lines from an Ar/Kr laser. An Imaris software package (Bitplane AG) was used to generate pictures of the biofilm. Tobramycin was diluted in 0.9% NaCl. The medium containing ajoene was kept on ice during the experiment.
Preparation of PMNs.
Isolation of PMNs was performed as described by Bjarnsholt et al. (6), with modifications. Human blood was collected from healthy volunteers in BD Vacutainer tubes containing 0.129 M sodium citrate. PMNs were resuspended in RPMI 1640 with NaHCO3 to obtain a concentration of 1.5× 107 PMNs/ml.
PMN exposure of biofilms.
The exposure experiment was performed as described by Bjarnsholt et al. (6). We evaluated the biofilm and PMN interactions every 30 min for 2 h. Necrotic PMNs were demonstrated as PMNs with increased red fluorescence from the supplemented DNA stain PI.
Animals.
Female BALB/c mice were purchased from Taconic M&B A/S (Ry, Denmark) at 9 to 11 weeks of age and were maintained on standard mouse chow and water ad libitum for a minimum period of 1 week before the challenge.
The animal studies were carried out in accordance with the European convention and directive for the protection of vertebrate animals used for experimental and other scientific purposes and the Danish law on animal experimentation. All experiments were authorized and approved by the National Animal Ethics Committee, Denmark (the Animal Experiments Inspectorate, dyreforsoegstilsynet.dk) and given permit number 2008/561-1466. All surgery was performed using fentanyl (Hypnorm)-midazolam, and pentobarbital was used to euthanize the mice at the termination of the experiments. All efforts were made to minimize suffering.
Pulmonary infection model.
The pulmonary infection model in mice was prepared and performed as described previously (10). The ajoene solution used for the treatment group was prepared as follows: ajoene was dissolved in 96% ethanol to a concentration of 100 mg/ml and diluted 40 times in a 20% vehicle solution [(2-hydroxypropyl)-β-cyclodextrin (catalog no. C0926; Sigma) dissolved in 0.9% NaCl] to a concentration of 2.5 mg/ml, which reduced the concentration of ethanol to 2.4%. Each mouse was treated with 25 mg ajoene kg−1 body weight (BW) subcutaneously (s.c.) once a day as prophylactic treatment for 2 days, right after infection, and subsequently at 2 days postinfection. The placebo group received 96% ethanol diluted in the vehicle corresponding to the amount of ethanol that the ajoene-treated group received.
Statistical analysis.
The number of mice in each group was calculated to provide a power of 0.8 or higher for continuous data. For analyzing quantitative data, the Mann-Whitney U test was used for calculating P values in the statistical program GraphPad Prism (version 5.0; GraphPad Software, Inc., San Diego, CA). P values of ≤0.05 were considered significant.
Apoptosis assay.
Apoptosis tests in the lung epithelial cell line A549 (purchased from the German Collection of Microorganisms and Cell Lines [DMSZ], Braunschweig, Germany) was performed by flow cytometry using a fluorescein isothiocyanate (FITC)-conjugated antibody to cleaved poly(ADP-ribose) polymerase (PARP; Cell Signaling Technology, Denver, CO). Samples of 5 × 105 cells were incubated in the presence of various concentrations of ajoene in six-well plates for 4 h. Subsequently, cells were detached with trypsin-EDTA, washed twice, fixed in 2% formaldehyde for 15 min, permeabilized by 0.1% saponin (Roth GmbH, Karlsruhe, Germany) in PBS for 60 min, and incubated with anti-cleaved PARP according to the manufacturer's protocol for 30 min. For positive control, cells were cultured in the presence of apoptosis inducer tetrandrine (Sigma-Aldrich, Steinheim, Germany) for 4 h (12).
Proliferation assay.
Interference of ajoene with A549 cell proliferation was measured employing MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt] proliferation assays (Promega, Mannheim, Germany) following the manufacturer's protocol.
Toxicity assay.
Cytotoxicity testing was performed using a lactate dehydrogenase (LDH) release-based assay kit purchased from Roche Applied Science (Mannheim, Germany). A549 lung epithelial cells were exposed to concentration series of test substances for 24 h before LDH release was determined according to the manufacturer's instructions.
RESULTS
Ajoene is the major bioactive QSI compound in garlic extract.
Previously, demonstrations of inhibition of QS in P. aeruginosa with crude and partially purified garlic extracts (8) encouraged us to further identify and assess the efficacy of the pure QS inhibitor. In our hands, Spanish garlic appeared to contain a higher level of bioactivity, with respect to QS inhibition, than garlic obtained from other countries, including China and Argentina (data not shown). The compounds present in garlic bulb extracts were stable to time, protease activity, and various evaporation techniques at room temperature, and it was only at high temperatures that degradation was observed. An iterative process of fractionation and assaying was applied to crude garlic extracts to determine the presence of any potential QSI compounds (bioassay-guided fractionation).
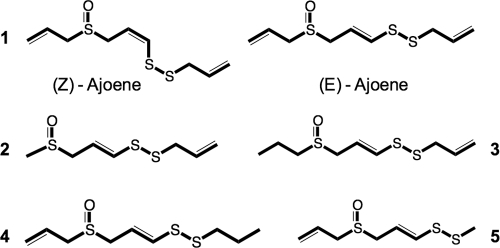
Several different water extracts were investigated, with several different columns for fractionation and purification initially tested. These columns included matrices consisting of C18, Sephadex G10 and LH20, and hydrophilic interaction chromatography (HILIC) matrix; however, in the end, ajoene was isolated on C18 material, as described in the Materials and Methods section. During examination of the initial water extract, the activity was lost. Investigations showed that the active compound was most likely degraded or adsorbed to sodium sulfate (used as a drying agent). Consequently, the focus was switched to the toluene phase. The toluene extracts continuously showed a high level of QSI activity. The treatment of these extracts was similar to that of the water extracts; however, all drying agents were avoided in case of absorption of the active compounds. In this case, the primary column matrices were silica based (SI, DIOL) or C18. Bioassay-guided fractionation based on QSI screens (with QSIS1 and the lasB-gfp and rhlA-gfp reporter strains) of these extracts isolated a single primary compound responsible for the in vitro activity. This compound was isolated, examined by MS and NMR (data not shown), and identified as ajoene (Fig. 1). In addition, several ajoene derivatives (Fig. 1) were shown to be present. Ajoene is a lipid-soluble allyl sulfide formed from allicin, which is converted from alliin by an enzymatic process when garlic is crushed (11). A range of different organosulfur compounds is formed in this process, with ajoene being among the most abundant (37). To further investigate the QSI bioactivity, we used chemically synthesized ajoene prepared by a recently published method (Givskov, European patent application no. 10194154.0 and US provisional application no. 61/420,922). Both naturally occurring and chemically synthesized ajoene exist as two isomers, the Z and E isomers, in different ratios dependent on the preparation method (Fig. 1).
Fig 1.
Ajoene and derivatives. Compound 1, ajoene, present in two isomers, E and Z; compounds 2 to 5, ajoene derivatives.
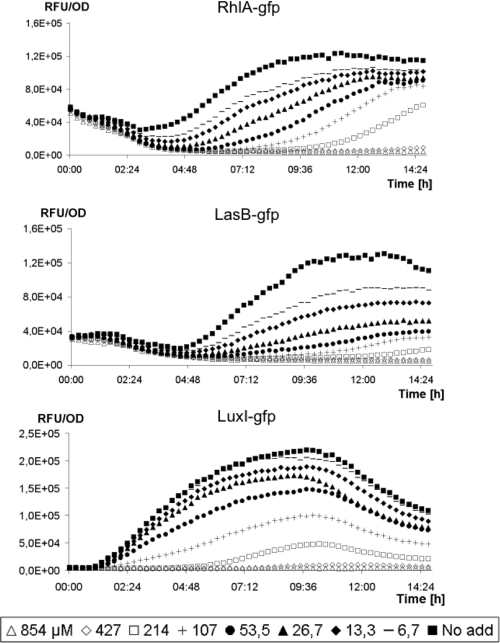
To determine the QSI activity of ajoene, dose-response curves were created using two QSI reporter systems which contain fusions of the QS-controlled lasB promoter and rhlA promoter to gfp (ASV), encoding an unstable GFP variant in a P. aeruginosa background (32, 77). We also used a QS reporter system harbored in an E. coli background where the luxR gene and the promoter region of the luxI are fused to gfp (ASV) (4). In all three reporter systems, induction of the QS system can be measured as increasing fluorescence. The presence of an antagonist decreases GFP expression, and thus, fluorescence is proportional to the concentration or effectiveness of the QSI present. Growth of the reporter strains was monitored to make sure that the concentrations of added ajoene were not affecting primary metabolic functions and thereby altering growth rate. Fifty percent inhibitory concentrations (IC50s) were calculated from the curves expressing the specific fluorescence (GFP expression/cell density) (Fig. 2), giving the following values: lasB-gfp reporter, 15 μM; rhlA-gfp reporter, 50 μM; and luxI-gfp reporter, 100 μM. The calculations were performed by plotting the maximal slopes from the curves obtained with the different reporter strains as a function of the concentrations of ajoene. The slope represents the synthesis rate (change in the number of relative fluorescence units/OD450/change in time [ΔRFU/OD450/Δtime]).
Fig 2.
Expression of QS-controlled specific fluorescence (GFP expression/cell density). The QS bioassays used were with P. aeruginosa harboring either the rhlA-gfp or the lasB-gfp fusion and E. coli harboring the luxI-gfp fusion incubated with synthesized ajoene.
Target gene specificity.
DNA microarray analysis was used to identify the target gene specificity of ajoene. As a reference, we have used the QS regulon previously identified by Hentzer et al. (33). In the past, this data set has been used to validate the target specificity of putative QSI compounds. The study by Hentzer et al. (33) defines QS-regulated genes to be those genes for which the expression is altered more than 5-fold in a ΔlasI-ΔrhlI mutant in response to the addition of exogenous C4-HSL and 3-oxo-C12-HSL. Genes with a less than 5-fold alteration in expression between treated and nontreated cultures were not included in this study. Exponentially growing P. aeruginosa cultures were treated with the following four concentrations of ajoene: 10 μg/ml (42.7 μM), 20 μg/ml (85.4 μM), 40 μg/ml (170.8 μM), and 80 μg/ml (341.6 μM). None of these concentrations affected growth (see Fig. S1 in the supplemental material). The samples were retrieved at an OD600 of 2.0, as previous investigations have shown that the highest activity among the QS genes is at this particular cell density.
Only a small number of genes were significantly (P < 0.05) downregulated more than 5-fold by the four different concentrations of ajoene: 0 at 10 μg/ml, 0 at 20 μg/ml, 2 at 40 μg/ml, and 11 at 80 μg/ml. According to the QS regulon defined by Hentzer et al. (33), 10 of the 11 genes and, according to Rasmussen et al. (59), all the genes downregulated more than 5-fold by ajoene are defined as QS regulated. There is a clear relationship between the increasing concentration of ajoene used for treatment and the degree of regulation of the target genes. The transcriptomic analysis indicated that the optimum concentration for repression of the target genes is close to 80 μg/ml ajoene. Attempts to repress more than those 11 genes (out of a total of 5,570 P. aeruginosa genes) were not possible without supplementing cultures with concentrations that would also affect growth. This means that ajoene administered at this optimum concentration exhibits a high degree of target specificity toward a small subgroup of the QS regulon. Five genes were significantly (P < 0.05) upregulated more than 5-fold in response to treatment with 80 μg/ml ajoene, and with the three lower ajoene concentrations, there were only a few genes for which expression was significantly altered. Three of the genes encode components of a type VI secretion system (tagQ1, PA0070; tssB1, PA0083; hcp1, PA0085), and the other two genes are exaC (PA1984) and PA0182, which is a probable short-chain dehydrogenase. Among the genes significantly downregulated by ajoene were the following QS-regulated important virulence factors: LasA protease (lasA, PA1871); chitinase (chiC, PA2300); the cytotoxic galactophilic lectin (lecA, PA2570); the rhamnosyltransferase AB operon (rhlA, PA3478; rhlB, PA3479); the PvdS-regulated endoprotease (prpL, PA4175) that degrades casein, elastin, lactoferrin, transferrin, and decorin (75); and the associated chitin-binding protein cbpD (PA0852), which mediates attachment to chitin-containing substrates and presumably assists in biofilm formation (26) (Table 1). None of the treatments seemed to affect transcription of the genes encoding central regulatory genes of the QS circuit. This is similar to other previously published QSI compounds, including furanone C-30 (33), patulin, and penicillic acid (59), and indicates that interaction of the inhibitor with its target may occur at the posttranscriptional level.
Table 1.
Alterations in gene expression by ajoenea
| Gene no. | Gene | Description | Fold change in gene expression with ajoene concn (μg/ml) ofb: |
|||
|---|---|---|---|---|---|---|
| 10 | 20 | 40 | 80 | |||
| PA0852 | cbpD | Chitin-binding protein | −2.8 | −2.5 | −3.9* | −6.9* |
| PA1871 | lasA | LasA protease precurser | −2.6 | −2.2 | −3** | −8.7** |
| PA2069 | Probable carbamoyl transferase | −2.3 | −2.4 | −4 | −5.3* | |
| PA2146 | Conserved hypothetical protein | −1.3 | −1.8* | −2.6* | −7.3** | |
| PA2300 | chiC | Chitinase | −2.5 | −2.1* | −5.1** | −24.6** |
| PA2570 | pa1L | LecA | −1.8 | −2 | −3.3 | −6.3* |
| PA3478 | rhlB | Rhamnosyltransferase chain B | −2.6 | −2 | −3.3** | −8.7** |
| PA3479 | rhlA | Rhamnosyltransferase chain A | −2.2 | −1.5 | −2.6 | −8.8** |
| PA4141 | Hypothetical protein | −1 | 1.1 | −1.3 | −5.4** | |
| PA4142 | Probable secretion protein | −2 | −2.2 | −2.7 | −5.1* | |
| PA4175 | prpL | Pvds-regulated endoprotease | −3.7 | −3.3 | −5.3* | −6.8* |
Genes included are >5 times downregulated by 80 μg/ml ajoene treatment.
Fold change in gene expression compared to an untreated control. Data represent the averages of three individual experiments.
, P < 0.05, Student's t test;
, P < 0.01, Student's t test.
To verify the microarray data, RT-PCR was performed with the two stringently QS-regulated genes lasB and rhlA (Fig. 3). When comparing the two experimental methods, repression of the two genes followed the same trend, with a slightly stronger reduction observed with the RT-PCR-based method. The RT-PCR data showed that a concentration of 80 μg/ml ajoene lowered expression of rhlA almost 12-fold and that of lasB almost 5-fold. According to Rasmussen et al. (59), the genes listed in Table 1 (except for prpL, which is exclusively regulated by the Las QS system) are subject to regulation by both the Las and Rhl QS systems. This suggests that ajoene may primarily target the Rhl system.
Fig 3.
Fold change in gene expression of rhlA and lasB measured by RT-PCR (dark gray bars) and DNA microarray (light gray bars). Data represent the average of three individual experiments. *, P < 0.05, Student's t test. Error bars are means ± SDs.
Attenuation of rhamnolipid production by ajoene.
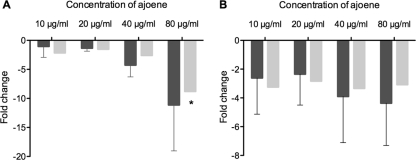
To exemplify the actual efficacy of ajoene in downregulating one of the important virulence factors, the concentration of rhamnolipid present in the cultures grown for DNA array analysis and RT-PCR was directly quantified by liquid chromatography (LC)-MS. The production of rhamnolipids encoded by the rhlA, rhlB, and rhlC genes (PA3479, PA3478, and PA1131, respectively) is initiated in early stationary phase and coordinately regulated by the Rhl and the PQS systems (57, 73). Samples where therefore retrieved at OD600s of 1.5 and 2.0 to monitor rhamnolipid production before and after the synthesis was fully induced. The data showed clearly that there was an increase in rhamnolipid production from an OD600 of 1.5 to one of 2.0. The concentrations of rhamnolipid in the samples correlated inversely with increasing concentrations of ajoene. When treated with 20 μg/ml ajoene at an OD600 of 2.0, the rhamnolipid content was reduced to approximately 1/3 of that for the untreated culture, and there was almost no detectable rhamnolipid present in the sample when the cells were treated with 80 μg/ml ajoene (Fig. 4).
Fig 4.
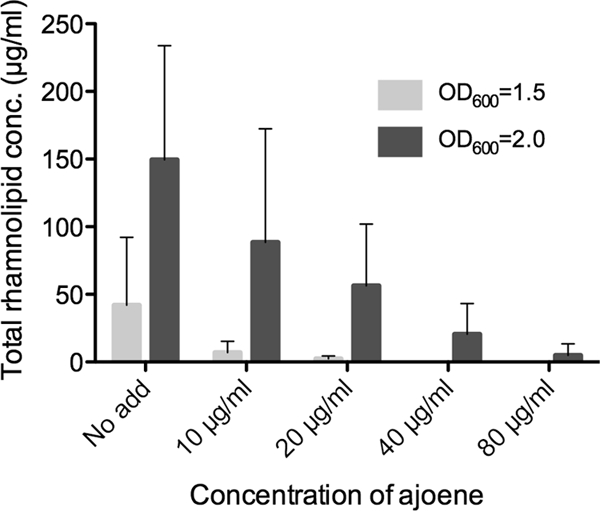
Total rhamnolipid concentration in untreated (no added ajoene [no add]) and ajoene-treated planktonic P. aeruginosa. The cultures were grown in medium supplemented with 10 μg/ml, 20 μg/ml, 40 μg/ml, and 80 μg/ml of ajoene (rhamnolipid is below the detection level for the 40-μg/ml and 80-μg/ml ajoene treatments at an OD of 1.5). Samples were retrieved at an OD600 of 1.5 (light gray bars) and at an OD600 of 2.0 (dark gray bars). Data represent the average of three individual experiments. Error bars are means ± SDs.
Effect of ajoene on P. aeruginosa QS signal molecule production.
According to the transcriptomic analysis, the genes inhibited by ajoene treatment could indicate that a posttranscriptional effect on gene products is responsible for generation of C4-HSL. To test this, the concentrations of C4-HSL and 3-oxo-C12-HSL in untreated and ajoene-treated cultures were determined by HPLC-MS/MS at three different cell densities (OD600s, 1.5, 1.8, and 2.0). The concentration of C4-HSL was found to inversely correlate with increasing concentration of ajoene, and at a concentration of 80 μg/ml of ajoene, the level was reduced almost 3-fold compared to the untreated control. With respect to the concentration of 3-oxo-C12-HSL, there was no consistent effect with increasing concentrations of ajoene (Fig. 5).
Fig 5.
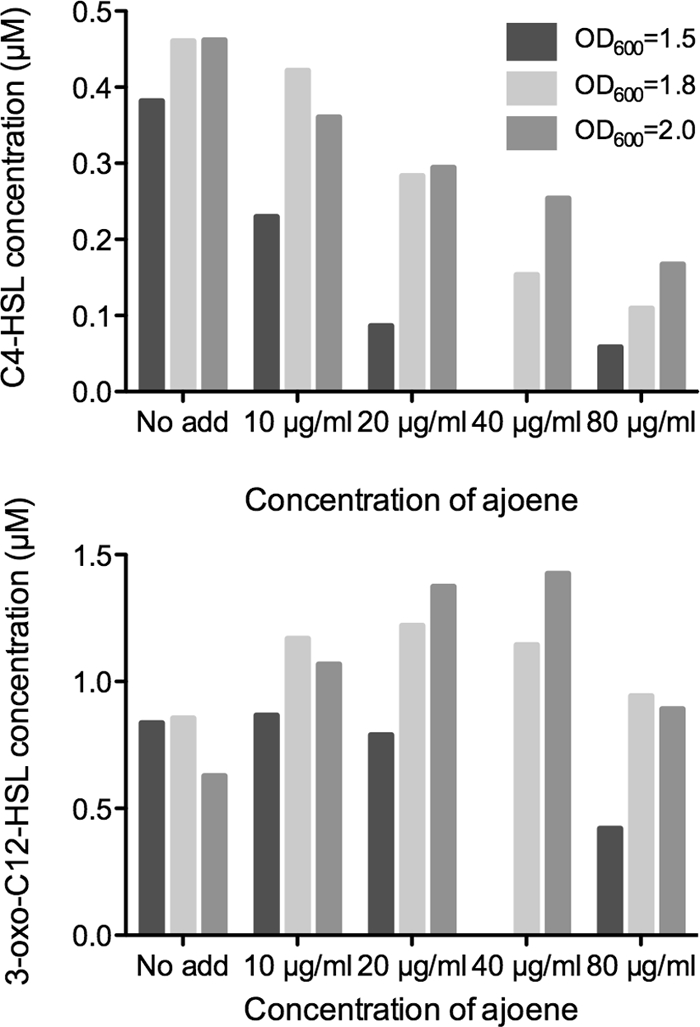
Concentrations of C4-HSL and 3-oxo-C12-HSL in untreated (no added ajoene [no add]) and ajoene-treated planktonic cultures of P. aeruginosa. The cultures were grown in medium supplemented with 10 μg/ml, 20 μg/ml, 40 μg/ml, and 80 μg/ml of ajoene. Samples were retrieved at an OD600 of 1.5 (dark gray bars), at an OD600 of 1.8 (light gray bars), and at an OD600 of 2.0 (medium gray bars).
Ajoene treatment renders in vitro biofilms rhamnolipid deficient and thereby prevents the killing of PMNs.
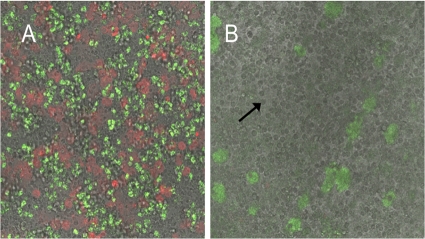
The effect of ajoene on rhamnolipid production prompted us to investigate whether this would in fact inhibit lysis of PMNs. P. aeruginosa biofilms were grown for 4 days in either the presence or absence of 100 μg/ml ajoene. When freshly isolated PMNs were subsequently introduced into the flow chambers of the biofilms grown in the absence of ajoene, PI staining indicated extensive necrosis of the PMNs (Fig. 6A). In contrast, when the biofilms were grown in the presence of ajoene prior to PMN exposure, no necrosis of the PMNs was observed (Fig. 6B).
Fig 6.
Combined fluorescence and light microscopic investigations at day 4 of biofilms of P. aeruginosa exposed to PMNs (arrow) for 180 min at 37°C and then subsequently stained with the DNA stain PI. (A) Biofilm grown without ajoene in the medium; (B) biofilm grown in the presence of 100 μg/ml ajoene in the medium. Red fluorescence indicates lysed PMNs, and green fluorescence indicates the P. aeruginosa biofilm.
Ajoene enhanced tobramycin effect on P. aeruginosa biofilm.
We have previously published the results of an in vitro treatment study of biofilms grown in the presence of QSI bioactivities, including those associated with garlic extract and furanone C-30, where QS inhibition was found to greatly enhance the antimicrobial effect of tobramycin (8, 33). Biofilms of a P. aeruginosa strain were grown in either the presence or absence of 100 μg/ml ajoene. At day 3, the biofilms were treated with 10 μg/ml tobramycin for 24 h. A pilot study on biofilms grown in the absence of ajoene indicated that treatment with 10 μg/ml, 100 μg/ml, or 340 μg/ml tobramycin showed no difference in the extent of killing, as judged from live-dead staining and inspection by means of confocal scanning laser microscopy. Our analysis showed a more than 90% killing of cells when the biofilms were grown in the presence of ajoene and subsequently treated with 10 μg/ml tobramycin (Fig. 7A). The synergistic effect was also evaluated on the clinical isolate from a patient with CF, CF438 (the first isolate from a child diagnosed with CF), which possesses functional QS systems, and once again, extensive killing of the biofilm was recorded (Fig. 7B).
Fig 7.
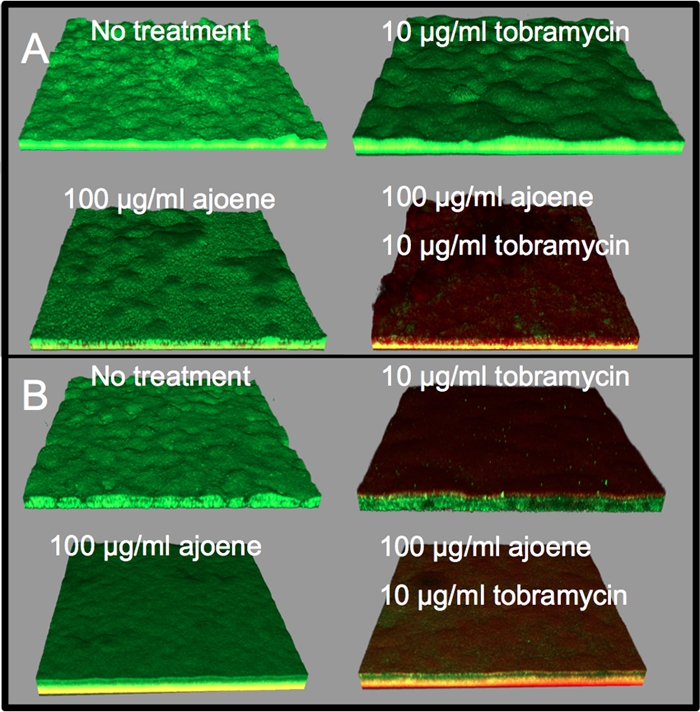
Biofilms of P. aeruginosa PAO1 (green) (A) and clinical P. aeruginosa isolate CF438 (green; stained with Syto 9) (B) at day 4 after no treatment, treatment with 100 μg/ml ajoene for 4 days, treatment with 10 μg/ml tobramycin for the final 24 h, or treatment with a combination of tobramycin-ajoene. Dead cells are stained with the DNA stain PI (red). The yellow color reflects a mixture of live and dead cells. The biofilms were visualized with a confocal scanning laser microscope.
Antimicrobial effects in vivo.
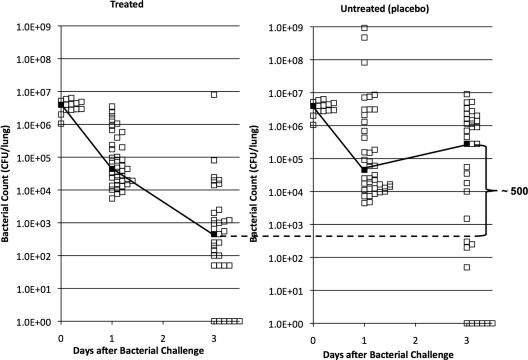
We performed three individual in vivo treatment experiments with ajoene in a pulmonary infection model in mice using 25 μg ajoene g−1 BW. When combining the experiments, a significant difference was seen on day 3 (P < 0.002), with a more than 500-fold difference in clearance recorded between the groups (Fig. 8). Experiments with a lasR rhlR double mutant showed that this is the maximum obtainable difference in clearance that can be obtained in this infection model (6).
Fig 8.
Combined results of three separate experiments of ajoene treatment versus no treatment (placebo) using the mouse model of pulmonary infection. The BALB/c mice were intratracheally challenged (at day 0) with alginate beads containing 1.5 × 108 CFU/ml P. aeruginosa. The two groups of mice were either untreated (placebo) or treated with ajoene at 25 μg/g BW once a day. The mice were given 2 days of prophylactic treatment or placebo. Mice were sacrificed on day 1 or day 3 postinfection, and the contents of bacteria in the lungs were determined. The median values are indicated with filled black squares. The statistical significance of the difference in clearance was tested by a Mann-Whitney U test (analysis of nonparametric data), and P values for the difference at day 1 and day 3 were 0.9 and 0.002, respectively.
Treatment of mice infected with clinical CF isolates.
In CF patients, the appearance of the mucoid phenotype is considered a clinical marker for the onset of the chronic infection, which correlates with a poor prognosis for the infected CF patients. To test the efficacy of ajoene treatment on a QS-proficient mucoid P. aeruginosa strain, we adopted a clinical isolate from a patient who had had a chronic infection with P. aeruginosa for 16 years (44, 72). After administration of 25 μg/g ajoene, we noted an induction of abscesses, which resulted in an increase in the bacterial load in the lungs. This phenomenon was not observed with the placebo group. The experiment was performed twice with similar results. We then repeated the experiment with 12.5-μg/g and 6.25-μg/g ajoene treatments. The concentration of 12.5 μg/g gave the best results with respect to bacterial clearance and apparent well-being of the mice, but the clearance in the treatment group was not significantly different from that in the placebo group (P < 0.5). Treatment with 6.25 μg/g of ajoene did not have any significant effect on clearance compared with the placebo group (data not shown). An isogenic, nonmucoid derivative of the clinical isolate was also investigated in a treatment study using 12.5 μg/g of ajoene. This isolate cleared very rapidly from the lungs of the mice. Therefore, the experiment was evaluated at day 1 after infection. The difference between the treated group and the placebo group showed a nonsignificant (P < 0.1) 2-orders-of-magnitude reduction (data not shown). Both isolates have a functional QS system (44). To investigate the efficacy of ajoene on an early clinical P. aeruginosa isolate, an experiment with the first isolate of P. aeruginosa from a CF patient was conducted by treating the infected mice with 12.5 μg/g of ajoene. The experiment was evaluated at day 1, since the strain is highly virulent and therefore the mice would not survive for a 3-day period. There was a significant 20-fold difference (P < 0.05) between the treated group and the placebo group (data not shown). The isolate tested positive for a functional QS system (data not shown).
Stability of ajoene under experimental conditions.
To test the stability in the presence of bacteria and whether ajoene could interact directly with the QS signal molecules, 50 μg/ml of ajoene was incubated overnight at 37°C and 4°C with P. aeruginosa (PAO1) or a QS-deficient ΔlasI rhlI or ΔlasR rhlR mutant and pure C4-HSL, pure 3-oxo-C12-HSL, or no addition. Following the incubations, ajoene activity in the supernatant was assessed by means of a lasB-gfp dose-response bioassay. The activity in the supernatant disappeared after incubation with bacteria at 37°C overnight, whereas there was no decrease in ajoene activity when incubated with bacteria at 4°C or incubated in the absence of bacteria at 4°C and 37°C. There was no change in ajoene activity when incubated with pure C4-HSL and 3-oxo-C12-HSL, which indicates that ajoene is not likely to chemically react with the signal molecules and cause their inactivation (data not shown).
Effect of BSA on the bioactivity of ajoene.
Because of putative annihilating effects of serum proteins on the bioactivity of ajoene, concentrations ranging from 12.5 μg/ml to 50 μg/ml of ajoene were incubated with bovine serum albumin (BSA). There was no notable reduction in the QSI activity of ajoene over the range of concentrations tested with serum albumin at concentrations up to 50 mg/ml, i.e., the concentration which corresponds to the content in adult serum (see Fig. S2 in the supplemental material).
Effects of ajoene on human lung epithelial cells.
A549 human lung epithelial cells were employed as a model system to quantify potential apoptosis-inducing effects. According to cytometric determination of cleaved PARP as a marker, ajoene promotes apoptosis with a 50% effective concentration (EC50) of about 100 μM (23 μg/ml). The PARP-activating effect of ajoene on lung epithelial cells was about 10-fold weaker than that of the positive control, tetrandrine, a natural compound used in Chinese medicine for the treatment of lung disorders such as silicosis (12). Consistent with these findings, ajoene inhibited proliferation of A549 cells with an IC50 of about 100 μM and is about 1 order of magnitude less potent in impairment of proliferation than the control compound, tetrandrine. General cytotoxicity in terms of induced cytolysis was determined by the release of LDH and, hence, LDH activity from damaged A549 cells. Ajoene induced LDH release in a dose-dependent manner, with an EC50 of 200 μM. It was found to be approximately 10-fold less cytotoxic than the established furanone QS inhibitor compound C-30 (see Fig. S3A to D in the supplemental material). Taken together, results from three independent assays coherently characterize ajoene as a compound with relatively weak interference with the cell physiology of the lung epithelium.
DISCUSSION
Intriguingly, worldwide emerging problems of infection control parallel a marked slowdown in the development of new antibiotics. Many pharmaceutical companies no longer have antibiotic drugs in the pipeline or research activities in the field. There are both strategic and economic reasons for this, but there have been profound scientific difficulties for the pharmaceutical industry in this context. One major limiting factor is that the study of free-living, planktonic bacteria has provided the basis for our general understanding of microbial life and, in particular, infectious diseases. Consequently, in the traditional design of antibiotic drugs, it is not appreciated that the biofilm habitat may dominate in chronic infections just as it does in the environment. Antimicrobial treatment of biofilms is a challenge, in particular when it comes to heterogeneity, which is likely one of the special features that provide biofilm bacteria with such remarkable resilience.
The archetypical biofilm disease has for a long time been considered to be P. aeruginosa infection in CF patients. Furthermore, a recent investigation by us pointed out that the early colonizers of children with CF have functional QS systems. The first system lost is the Las system, but the strains are still able to express rhamnolipid and other PQS- and C4-HSL-controlled genes (7). Therefore, strategies that help in disabling the protective mechanisms of P. aeruginosa, in particular, the rhamnolipid shield and eDNA production, are likely to be employed as a worthwhile addition to conventional antimicrobial chemotherapy. Different approaches can be used to identify and harness the QS inhibitors obtained from natural sources. Extraction of natural products has provided several positive QSI molecules, among them the Delisea pulchra furanones (32) and patulin from Penicillium coprobium (59). It is interesting that sulfur-containing compounds appear to be a new class of molecules capable of inhibiting QS (3, 5). Li et al. (40) found two sulfur-containing compounds from an in silico-based virtual screening to target the AI-2 QS system from Vibrio harveyi, and several analogues were identified to exert bioactivity (52). These molecules contain a sulfone group, in contrast to ajoene, which contains disulfide and sulfinyl groups. For a more detailed description of identified QSIs, see the work of Galloway et al. (29).
We have shown that approximately 80 μg/ml (341.6 μM) ajoene efficiently switches off the expression of the rhlA-gfp fusion in our dose-response bioassays and extensively downregulates rhlA gene expression, which corresponds to the decrease in concentration of rhamnolipid directly measured by HPLC. The gene rhlA encodes a rhamnosyltransferase which catalyzes a glycosyl transfer reaction in rhamnolipid synthesis. Rhamnolipids are glycolipids that have strong surfactant abilities. One rhamnolipid in particular, rhamnolipid B, is one of the two more abundant rhamnolipids and has been shown to cause necrosis of PMNs (36). Previously published data with a ΔrhlA mutant (71) (which is defective only in rhamnolipid synthesis) make us strongly believe that rhamnolipid is responsible for the lytic killing of the PMNs, also in vivo. We have shown that ajoene treatment of in vitro biofilms prevents the killing of PMNs. These experiments furthermore suggest that ajoene treatment is capable of attenuating the production of rhamnolipids. In addition, ajoene treatment is capable of rescuing the PMNs and is likely able to restore the action of the PMNs. The same results were observed with a ΔrhlA mutant (71): almost no necrotic PMNs were detected when exposed to biofilms of the rhamnolipid-deficient mutant.
One important issue in the treatment of bacterial biofilm infections is the lowered effectiveness of administered antibiotics. An infection in the airways of CF patients results in high concentrations of anionic polyelectrolytes like DNA (14) released from lysed inflammatory cells, such as the PMNs and bacteria. It has been shown that anionic polyelectrolytes, in particular, DNA, bind to and reduce the activity of cationic antibiotics like tobramycin (74, 78), which can lead to a decrease in the biological availability of tobramycin to as low as 5% of the existing dose (43). This suggests that by blocking the production of eDNA, it is possible to attenuate the otherwise subsequent inactivation of tobramycin. We have shown in vitro that addition of 100 μg/ml ajoene to a biofilm followed by addition of 10 μg/ml tobramycin kills more than 90% of the biofilm bacteria, whereas the presence of only tobramycin or ajoene had no effect. It is documented that the release of bacterial eDNA is controlled by QS (2), which, taking into consideration and combined with our results, points to a possible attenuation of the release of eDNA by ajoene.
This synergistic effect is also relevant in vivo. It has been demonstrated that treatment of an in vivo P. aeruginosa foreign-body biofilm infection with a combination of a QSI and tobramycin likewise showed a synergistic clearing effect on the bacteria (16). The results could be obtained with the use of either of the QSIs: furanone C-30, ajoene, or horseradish juice extract. In addition, rhamnolipid-mediated lysis of attacking PMNs may significantly contribute to the tobramycin annihilating effects in vivo. Our data indicate that this chain of neutralizing events may be obstructed by treatment with QSIs, including ajoene. A recent study showed similar promising results with treatment of a Burkholderia cenocepacia infection with a combination of tobramycin and the QSI baicalin hydrate in a mouse pulmonary model of infection (13). Several published papers by our group have demonstrated that QS deficiency (either by mutation or by QSI treatments) leads to faster clearing of the bacteria than of bacteria with functional QS in a mouse model of pulmonary infection (6, 8, 33). In this study, ajoene was administered prophylactically and was continued after infection. Enumeration by plate counts showed a significant difference between the treated group and the control group on day 3. This is concordant with the results obtained by Bjarnsholt et al. (8) in a study in which raw garlic extract was used as treatment. In addition, in a study by Harjai et al. (31), where garlic extract was given to mice orally, mice showed significantly reduced renal bacterial content of P. aeruginosa at day 5 postinfection.
Three different clinical isolates were tested in the lung model to demonstrate the efficacy of ajoene toward different isolates retrieved from CF patients. Ajoene treatment of a QS-proficient mucoid strain obtained from a CF patient chronically infected for 16 years and the isogenic nonmucoid strain did not have any significant effect compared to that on the placebo group, whereas ajoene treatment of a first CF isolate showed a significant difference. This increased susceptibility of the early clinical isolate to the QSI induced by ajoene is in line with our demonstration of the predominance of intact QS in early isolates from CF patients (7). These data were obtained at 1 day postinfection because of difficulties in keeping the mice in a proper healthy condition. Furthermore, the ajoene concentration was 2-fold lower than that in studies with wild type. These modifications (which were taken to comply with ethical constraints) in the experimental procedure might be the reason for the lower effectiveness of ajoene that we obtained with the clinical strains compared to wild type. When comparing the present studies with ajoene to earlier studies with garlic extracts, the present study offers a convincing indication of ajoene being the major active component in garlic able to reduce a P. aeruginosa infection. Two other sulfur-containing molecules previously isolated by us from garlic were found to possess QS-inhibitory activity toward the Vibrio fischeri LuxR QS system but not against the P. aeruginosa QS systems (53).
The question is whether it would be possible to obtain the promising treatment results in clinical trials performed on patients suffering from P. aeruginosa infections. The ajoene content in garlic is found to be concentrations of up to 172 μg/g of E-ajoene and 476 μg/g of Z-ajoene, as judged from rice oil-heated (80°C) freshly prepared garlic extracts (46). To match this relatively low herbal content of ajoene with the dosages required for the present animal treatments, individuals would be required to intake approximately 5 kg of raw garlic per day. Despite this, a recently published pilot study investigating the effect of garlic capsules orally administered to CF patients reported a nonsignificant but nevertheless reduced decline in lung function in 1 s (FEV1) in the treated group compared with the corresponding placebo group (65). The exact amount of ajoene present in the capsules was not determined. However, water extracts made directly on the content of the capsules showed bioactivity directed against our lasB-gfp reporter (data not shown). It therefore remains unknown if ajoene was present in biologically relevant amounts or whether the capsules contained ajoene-enhancing components that would increase the effects of small amounts of ajoene. In fact, upon subsequent purification close to 100% purity, as determined by LC-diode array detection-MS, we found that synthesized ajoene actually lost activity in the in vivo infectious models (data not shown). For example, fresh garlic extract shows a much more pronounced effect than synthesized ajoene on the transcriptome of P. aeruginosa (58). In contrast to our garlic extracts used previously, our transcriptome analysis revealed that synthetic ajoene affected only a few but nevertheless important number of QS-controlled genes, including lasA, chiC, and rhlAB, whereas lasB was not downregulated more than 5-fold. The effect on primarily QS-controlled genes and the small amount of genes affected suggest that synthetic ajoene, in contrast to our previous garlic extracts, inhibits expression of only a minor part of the QS regulon. In comparison, with more than 80% of the QS-controlled genes being downregulated by furanone C-30 treatment and no effect on signal generation, it is unlikely that ajoene targets both LasR and RhlR.
Sonnleitner et al. (66) have investigated the influence of the SM-like RNA-binding protein Hfq on the QS system. They documented a decrease in elastase, catalase, and pyocyanin production in an hfq-knockout mutant (66) and confirmed by transcriptome analysis that the effect on this subset of QS-regulated virulence factors was mediated by reduced expression of the corresponding genes (67). The authors suggested the following interactive path of Hfq and the QS system: Hfq binds to and stabilizes the regulatory RNA RsmY, which subsequently binds to the RsmA protein, which, in turn, negatively regulates RhlI messenger translation. Furthermore, they showed a decrease in the concentration of C4-HSL in both a PAO1 ΔrsmY mutant and a PAO1 hfq mutant strain compared to the wild type (67), which corresponds to the results of our investigations, where the amount of C4-HSL in cultures decreases with increasing ajoene concentrations. Furthermore, when comparing our transcriptome analysis with the transcriptome analysis of the PAO1 hfq mutant strain, there is a compelling correlation with genes which are significantly downregulated by ajoene treatment. It is, however, not trivial to compare the two studies. For example, the PAO1 hfq mutant strain showed a reduced growth rate compared to the wild type (66). Furthermore, the authors grew their samples in LB medium and isolated RNA from cultures grown to an OD600 of 2.5. In comparison, our samples, which were retrieved at an OD600 of 2.0, were grown in AB minimal medium supplemented with 0.5% Casamino Acids. Despite this, we suggest that either the Hfq protein or the RsmY RNA may constitute a possible target of ajoene.
The effects of ajoene on lasB transcription, monitored by RT-PCR and DNA array analysis, were only minor, in contrast to the observed effects on lasB-gfp expression. We see a strong reduction of fluorescence with the lasB-gfp reporter with increasing ajoene concentrations. The reporter strain is a translational fusion and reflects the reported effect of the small regulatory RNA molecules on posttranscriptional levels (as previously reported as a reduction in elastase production [66]). This is in support of our hypothesis that ajoene targets Hfq and the regulatory RNAs and that the effect of the lower parts of the QS hierarchy on transcription is mediated by a reduction in the C4-HSL concentration. Furthermore, no effect on the transcription of lasI and rhlI is found on the transcriptomic analysis, which supports the view that the effect of ajoene on reducing RhlI expression is posttranscriptional. The much more pronounced effect of garlic extracts on a multitude of QS-controlled gene expression previously reported by us (58), taken together with our shortcomings in the extraction of hydrophilic compounds, suggests that garlic may in fact contain a multitude of QSI compounds or stabilizing agents that may act in synergy and thereby in concert cover a much larger spectrum of QS-controlled virulence gene expression. If true, quantities of garlic attainable in the food diet may in fact contribute to a natural prophylaxis against bacterial infections.
To address the question of whether synthesized ajoene constitutes a pharmaceutically relevant drug candidate, we investigated toxicity effects on human epithelium cells. Ajoene exerts proapoptotic, antiproliferative, and cytotoxic effects on A549 lung epithelial cells. The concentrations showing half-maximal effects in our assays were in the range of 23 to 46 μg/ml (100 to 200 μM). Compared to tetrandrine, a substance used as a lung therapeutic agent in Chinese medicine, ajoene is clearly less toxic (by a factor of 10) toward respiratory epithelial cells. Interestingly, ajoene is less toxic for A549 cells than for HL-60 leukemia cells, which already respond by intense apoptosis at concentrations of about 4.7 μg/ml (20 μM) (23). In comparison, the concentration that we used in the mouse experiments was 25 μg/g (107 μM). To more thoroughly evaluate the potential of ajoene as a putative component of future CF medicine, it will be important to compare its cytotoxic effects with those induced by antibiotics administered in the treatment of the CF syndrome.
In conclusion, we have demonstrated the use of synthetic ajoene to attenuate the virulence of P. aeruginosa by lowering expression of important QS-controlled virulence genes in P. aeruginosa. It is shown for the first time that successful antimicrobial treatments with the QS systems as targets can be obtained by inhibiting only a few but important virulence genes and not the entire QS regulon (mediated through LasR and RhlR). This new finding leads us to suggest that within the framework of QS inhibition as an antimicrobial strategy, small regulatory RNA molecules operating in the lower part of the QS hierarchy may constitute a new, functional antimicrobial drug target. At present, this possible novel antimicrobial target needs to be extensively pursued and confirmed by molecular approaches. Interestingly, small regulatory RNAs or microRNAs and their cognate targets are strongly implicated in cancer as either oncogenes or tumor and metastasis suppressors. Targeting of small regulatory RNAs to therapeutic antimicrobial ends would therefore parallel future developments in anticancer therapy, with cancer-specific microRNAs to be exploited not only to produce a direct anticancer effect but also to improve the response of tumor cells to conventional treatments (35, 69). Similarly, the biofilm-weakening properties of ajoene with respect to enhancing the effect of conventional antibiotics such as tobramycin may become instrumental for the future development of combinatory treatments.
It is worth acknowledging that QS inhibition does not remove the P. aeruginosa ability to form a biofilm. However, all available data indicate that a QS-deficient biofilm is more fragile than a QS-proficient biofilm (6). Since, for example, the matrix component DNA is missing (matrix production is C4-HSL-RhlR controlled), the biofilm is sensitive to shear forces and can slough off, depending on the hydrodynamic forces. In addition, since rhamnolipid is not formed, the biofilm becomes sensitive to the action of PMNs (because the PMNs are not killed when they get in contact with the biofilm). We have previously shown that in vitro biofilms of QS-deficient bacteria can be phagocytosed by freshly isolated PMNs, in contrast to QS-proficient biofilms (6). Central in our model for biofilm tolerance of PMNs is that rhamnolipid production forms a protective shield against the incoming PMNs, and we have several data supporting this (1, 36, 71). The use of QSIs should therefore greatly enhance the antimicrobial properties of the PMNs and allow them to efficiently eradicate biofilm-forming bacteria. Furthermore, rhamnolipid lyses the PMNs, which subsequently spill out their content of DNA, hydrolytic enzymes, and oxygen radicals. This creates an “evil circle,” particularly with respect to tissue damage, increasing inflammation and induction of mutations in P. aeruginosa that result in the appearance of the mucoid phenotype, which significantly contributes to exacerbations. The key thing is that this should never happen, and a QS-inhibitory drug should prevent this from happening. Our animal experiments with clinical isolates suggest that late mucoid isolates are not sensitive to the blocking of QS-controlled phenotypes, whereas early isolates are likely to show sensitivity. Ajoene might be able to prevent initial adherence and colonization of P. aeruginosa, and this treatment strategy might prevent the chronic lung infection by mucoid strains of P. aeruginosa in CF patients. The decrease in infection in the mouse experiments, the removal of in vitro biofilms in a combinatorial experiment with tobramycin, and the initial toxicity test with ajoene suggest the potential of using ajoene as an antipathogenic drug for treatment of chronic P. aeruginosa infections in the future.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the German Mukoviszidose Ve, the Danish Strategic Research Council, and the Novo Nordisk Foundation to M.G.
Footnotes
Published ahead of print 6 February 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Alhede M, et al. 2009. Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology 155:3500–3508 [DOI] [PubMed] [Google Scholar]
- 2. Allesen-Holm M, et al. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59:1114–1128 [DOI] [PubMed] [Google Scholar]
- 3. Amara N, et al. 2009. Covalent inhibition of bacterial quorum sensing. J. Am. Chem. Soc. 131:10610–10619 [DOI] [PubMed] [Google Scholar]
- 4. Andersen JB, et al. 2001. gfp-based N-acyl homoserine-lactone sensor systems for detection of bacterial communication. Appl. Environ. Microbiol. 67:575–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Binghe Wang NN, et al. March 2009. Compositions for regulating or modulating quorum sensing in bacteria, methods of using the compounds, and methods of regulating or modulating quorum sensing in bacteria. International publication number WO 2009/029317 A2. Patent Cooperation Treaty US2008/066028
- 6. Bjarnsholt T, et al. 2005. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology 151:373–383 [DOI] [PubMed] [Google Scholar]
- 7. Bjarnsholt T, et al. 2010. Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PLoS One 5:e10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bjarnsholt T, et al. 2005. Garlic blocks quorum sensing and promotes rapid clearing of pulmonary Pseudomonas aeruginosa infections. Microbiology 151:3873–3880 [DOI] [PubMed] [Google Scholar]
- 9. Bjarnsholt T, et al. 2008. Why chronic wounds will not heal: a novel hypothesis. Wound Repair Regen. 16:2–10 [DOI] [PubMed] [Google Scholar]
- 10. Bjarnsholt T, et al. In vitro screens for quorum sensing inhibitors and in vivo confirmation of their effect. Nat. Protoc. 5:282–293 [DOI] [PubMed] [Google Scholar]
- 11. Block E, et al. 1984. The chemistry of alkyl thiosulfate esters. 8. (E,Z)-Ajoene: a potent antithrombotic agent from garlic. J. Am. Chem. Soc. 106:8295–8296 [Google Scholar]
- 12. Borowski A, et al. 2008. Interleukin-13 acts as an apoptotic effector on lung epithelial cells and induces pro-fibrotic gene expression in lung fibroblasts. Clin. Exp. Allergy 38:619–628 [DOI] [PubMed] [Google Scholar]
- 13. Brackman G, Cos P, Maes L, Nelis HJ, Coenye T. 2011. Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob. Agents Chemother. 55:2655–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brandt T, Breitenstein S, von der Hardt H, Tummler B. 1995. DNA concentration and length in sputum of patients with cystic fibrosis during inhalation with recombinant human DNase. Thorax 50:880–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Christensen BB, et al. 1999. Molecular tools for study of biofilm physiology. Methods Enzymol. 310:20–42 [DOI] [PubMed] [Google Scholar]
- 16. Christensen LD, et al. 2012. Synergistic antibacterial efficacy of early combination treatment with tobramycin and quorum-sensing inhibitors against Pseudomonas aeruginosa in an intraperitoneal foreign-body infection mouse model. J. Antimicrob. Chemother. [Epub ahead of print.] doi:10.1093/jac/dks002 [DOI] [PubMed] [Google Scholar]
- 17. Clark DJ, Maaloe O. 1967. DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 23:99–112 [Google Scholar]
- 18. Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711–745 [DOI] [PubMed] [Google Scholar]
- 19. Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322 [DOI] [PubMed] [Google Scholar]
- 20. Costerton W, et al. 2003. The application of biofilm science to the study and control of chronic bacterial infections. J. Clin. Invest. 112:1466–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Davies DG, et al. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295–298 [DOI] [PubMed] [Google Scholar]
- 22. Davies JC. 2002. Pseudomonas aeruginosa in cystic fibrosis: pathogenesis and persistence. Paediatr. Respir. Rev. 3:128–134 [DOI] [PubMed] [Google Scholar]
- 23. Dirsch VM, Gerbes AL, Vollmar AM. 1998. Ajoene, a compound of garlic, induces apoptosis in human promyeloleukemic cells, accompanied by generation of reactive oxygen species and activation of nuclear factor kappaB. Mol. Pharmacol. 53:402–407 [DOI] [PubMed] [Google Scholar]
- 24. Fazli M, et al. 2009. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J. Clin. Microbiol. 47:4084–4089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fleischauer AT, Arab L. 2001. Garlic and cancer: a critical review of the epidemiologic literature. J. Nutr. 131:1032S–1040S [DOI] [PubMed] [Google Scholar]
- 26. Folders J, Tommassen J, van Loon LC, Bitter W. 2000. Identification of a chitin-binding protein secreted by Pseudomonas aeruginosa. J. Bacteriol. 182:1257–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fukao H, Yoshida H, Tazawa Y, Hada T. 2007. Antithrombotic effects of odorless garlic powder both in vitro and in vivo. Biosci. Biotechnol. Biochem. 71:84–90 [DOI] [PubMed] [Google Scholar]
- 28. Fuqua WC, Winans SC, Greenberg EP. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Galloway WR, Hodgkinson JT, Bowden SD, Welch M, Spring DR. 2011. Quorum sensing in Gram-negative bacteria: small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem. Rev. 111:28–67 [DOI] [PubMed] [Google Scholar]
- 30. Garde C, et al. 2010. Quorum sensing regulation in Aeromonas hydrophila. J. Mol. Biol. 396:849–857 [DOI] [PubMed] [Google Scholar]
- 31. Harjai K, Kumar R, Singh S. Garlic blocks quorum sensing and attenuates the virulence of Pseudomonas aeruginosa. FEMS Immunol. Med. Microbiol. 58:161–168 [DOI] [PubMed] [Google Scholar]
- 32. Hentzer M, et al. 2002. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 148:87–102 [DOI] [PubMed] [Google Scholar]
- 33. Hentzer M, et al. 2003. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 22:3803–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Høiby N. 1974. Epidemiological investigations of the respiratory tract bacteriology in patients with cystic fibrosis. Acta Pathol. Microbiol. Scand. B Microbiol. Immunol. 82:541–550 [PubMed] [Google Scholar]
- 35. Izumiya M, Tsuchiya N, Okamoto K, Nakagama H. 2011. Systematic exploration of cancer-associated microRNA through functional screening assays. Cancer Sci. 102:1615–1621 [DOI] [PubMed] [Google Scholar]
- 36. Jensen PO, et al. 2007. Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology 153:1329–1338 [DOI] [PubMed] [Google Scholar]
- 37. Kaschula CH, Hunter R, Parker MI. Garlic-derived anticancer agents: structure and biological activity of ajoene. Biofactors 36:78–85 [DOI] [PubMed] [Google Scholar]
- 38. Kirketerp-Moller K, et al. 2008. Distribution, organization, and ecology of bacteria in chronic wounds. J. Clin. Microbiol. 46:2717–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kohler T, van Delden C, Curty LK, Hamzehpour MM, Pechere JC. 2001. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J. Bacteriol. 183:5213–5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li M, et al. 2008. Structure-based discovery and experimental verification of novel AI-2 quorum sensing inhibitors against Vibrio harveyi. ChemMedChem 3:1242–1249 [DOI] [PubMed] [Google Scholar]
- 41. Mah TF, et al. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306–310 [DOI] [PubMed] [Google Scholar]
- 42. McClure CD, Schiller NL. 1992. Effects of Pseudomonas aeruginosa rhamnolipids on human monocyte-derived macrophages. J. Leukoc. Biol. 51:97–102 [DOI] [PubMed] [Google Scholar]
- 43. Mendelman PM, et al. 1985. Aminoglycoside penetration, inactivation, and efficacy in cystic fibrosis sputum. Am. Rev. Respir. Dis. 132:761–765 [DOI] [PubMed] [Google Scholar]
- 44. Moser C, et al. 2009. Novel experimental Pseudomonas aeruginosa lung infection model mimicking long-term host-pathogen interactions in cystic fibrosis. APMIS 117:95–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Naganawa R, et al. 1996. Inhibition of microbial growth by ajoene, a sulfur-containing compound derived from garlic. Appl. Environ. Microbiol. 62:4238–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Naznin MT, Akagawa M, Okukawa K, Maeda T, Morita N. 2008. Characterization of E- and Z-ajoene obtained from different varieties of garlics. Food Chem. 106:1113–1119 [Google Scholar]
- 47. Nielsen KF, Dalsgaard PW, Smedsgaard J, Larsen TO. 2005. Andrastins A-D, Penicillium roqueforti metabolites consistently produced in blue-mold-ripened cheese. J. Agric. Food Chem. 53:2908–2913 [DOI] [PubMed] [Google Scholar]
- 48. Obritsch MD, Fish DN, MacLaren R, Jung R. 2005. Nosocomial infections due to multidrug-resistant Pseudomonas aeruginosa: epidemiology and treatment options. Pharmacotherapy 25:1353–1364 [DOI] [PubMed] [Google Scholar]
- 49. Pamp SJ, Tolker-Nielsen T. 2007. Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 189:2531–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pearson JP, et al. 1994. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. U. S. A. 91:197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pearson JP, Passador L, Iglewski BH, Greenberg EP. 1995. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 92:1490–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Peng H, et al. 2009. Synthesis and evaluation of new antagonists of bacterial quorum sensing in Vibrio harveyi. ChemMedChem 4:1457–1468 [DOI] [PubMed] [Google Scholar]
- 53. Persson T, et al. 2005. Rational design and synthesis of new quorum-sensing inhibitors derived from acylated homoserine lactones and natural products from garlic. Org. Biomol. Chem. 3:253–262 [DOI] [PubMed] [Google Scholar]
- 54. Pesci EC, et al. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 96:11229–11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Peters L, et al. 2003. Secondary metabolites of Flustra foliacea and their influence on bacteria. Appl. Environ. Microbiol. 69:3469–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Purdy Drew KR, Sanders LK, Culumber ZW, Zribi O, Wong GC. 2009. Cationic amphiphiles increase activity of aminoglycoside antibiotic tobramycin in the presence of airway polyelectrolytes. J. Am. Chem. Soc. 131:486–493 [DOI] [PubMed] [Google Scholar]
- 57. Rahim R, et al. 2001. Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol. Microbiol. 40:708–718 [DOI] [PubMed] [Google Scholar]
- 58. Rasmussen TB, et al. 2005. Screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J. Bacteriol. 187:1799–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rasmussen TB, et al. 2005. Identity and effects of quorum-sensing inhibitors produced by Penicillium species. Microbiology 151:1325–1340 [DOI] [PubMed] [Google Scholar]
- 60. Rau MH, et al. 2010. Early adaptive developments of Pseudomonas aeruginosa after the transition from life in the environment to persistent colonization in the airways of human cystic fibrosis hosts. Environ. Microbiol. 12:1643–1658 [DOI] [PubMed] [Google Scholar]
- 61. Rivlin RS. 2001. Historical perspective on the use of garlic. J. Nutr. 131:951S–954S [DOI] [PubMed] [Google Scholar]
- 62. Shryock TR, Banschbach SSMW, Kramer JC. 1984. Effect of Pseudomonas aeruginosa rhamnolipid on human neutrophil migration. Curr. Microbiol. 10:323–328 [Google Scholar]
- 63. Shukla Y, Kalra N. 2007. Cancer chemoprevention with garlic and its constituents. Cancer Lett. 247:167–181 [DOI] [PubMed] [Google Scholar]
- 64. Skindersoe ME, et al. 2008. Quorum sensing antagonism from marine organisms. Mar. Biotechnol. (NY) 10:56–63 [DOI] [PubMed] [Google Scholar]
- 65. Smyth AR, et al. Garlic as an inhibitor of Pseudomonas aeruginosa quorum sensing in cystic fibrosis—a pilot randomized controlled trial. Pediatr. Pulmonol. 45:356–362 [DOI] [PubMed] [Google Scholar]
- 66. Sonnleitner E, et al. 2003. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb. Pathog. 35:217–228 [DOI] [PubMed] [Google Scholar]
- 67. Sonnleitner E, Schuster M, Sorger-Domenigg T, Greenberg EP, Blasi U. 2006. Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Mol. Microbiol. 59:1542–1558 [DOI] [PubMed] [Google Scholar]
- 68. Stewart PS, Costerton JW. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135–138 [DOI] [PubMed] [Google Scholar]
- 69. Thomas M, Lieberman J, Lal A. 2010. Desperately seeking microRNA targets. Nat. Struct. Mol. Biol. 17:1169–1174 [DOI] [PubMed] [Google Scholar]
- 70. van Delden C, Iglewski BH. 1998. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg. Infect. Dis. 4:551–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Van Gennip M, et al. 2009. Inactivation of the rhlA gene in Pseudomonas aeruginosa prevents rhamnolipid production, disabling the protection against polymorphonuclear leukocytes. APMIS 117:537–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. van Gennip M, et al. 2009. Augmented effect of early antibiotic treatment in mice with experimental lung infections due to sequentially adapted mucoid strains of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 64:1241–1250 [DOI] [PubMed] [Google Scholar]
- 73. Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Weiner DJ, Bucki R, Janmey PA. 2003. The antimicrobial activity of the cathelicidin LL37 is inhibited by F-actin bundles and restored by gelsolin. Am. J. Respir. Cell Mol. Biol. 28:738–745 [DOI] [PubMed] [Google Scholar]
- 75. Wilderman PJ, et al. 2001. Characterization of an endoprotease (PrpL) encoded by a PvdS-regulated gene in Pseudomonas aeruginosa. Infect. Immun. 69:5385–5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wu H, et al. 2004. Synthetic furanones inhibit quorum-sensing and enhance bacterial clearance in Pseudomonas aeruginosa lung infection in mice. J. Antimicrob. Chemother. 53:1054–1061 [DOI] [PubMed] [Google Scholar]
- 77. Yang L, et al. 2009. Computer-aided identification of recognized drugs as Pseudomonas aeruginosa quorum-sensing inhibitors. Antimicrob. Agents Chemother. 53:2432–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zribi OV, Kyung H, Golestanian R, Liverpool TB, Wong GC. 2006. Condensation of DNA-actin polyelectrolyte mixtures driven by ions of different valences. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 73:031911 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.