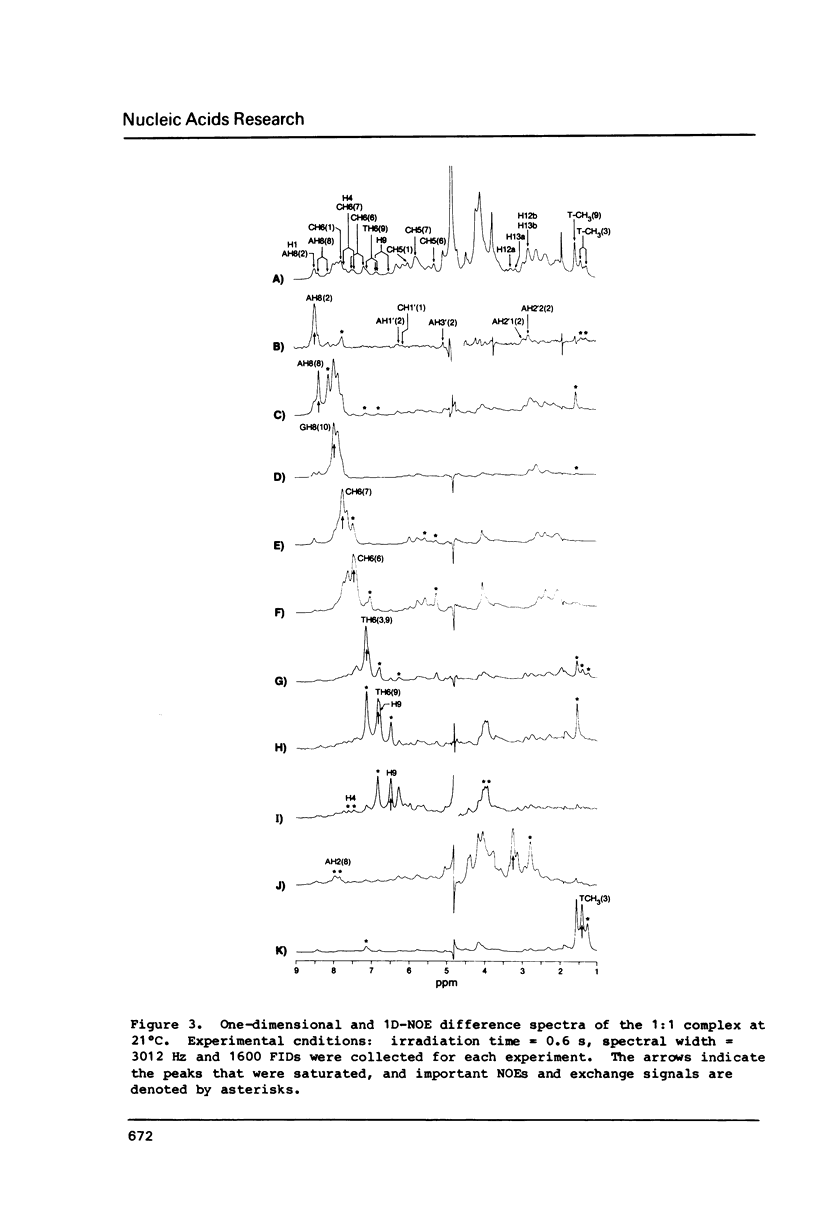
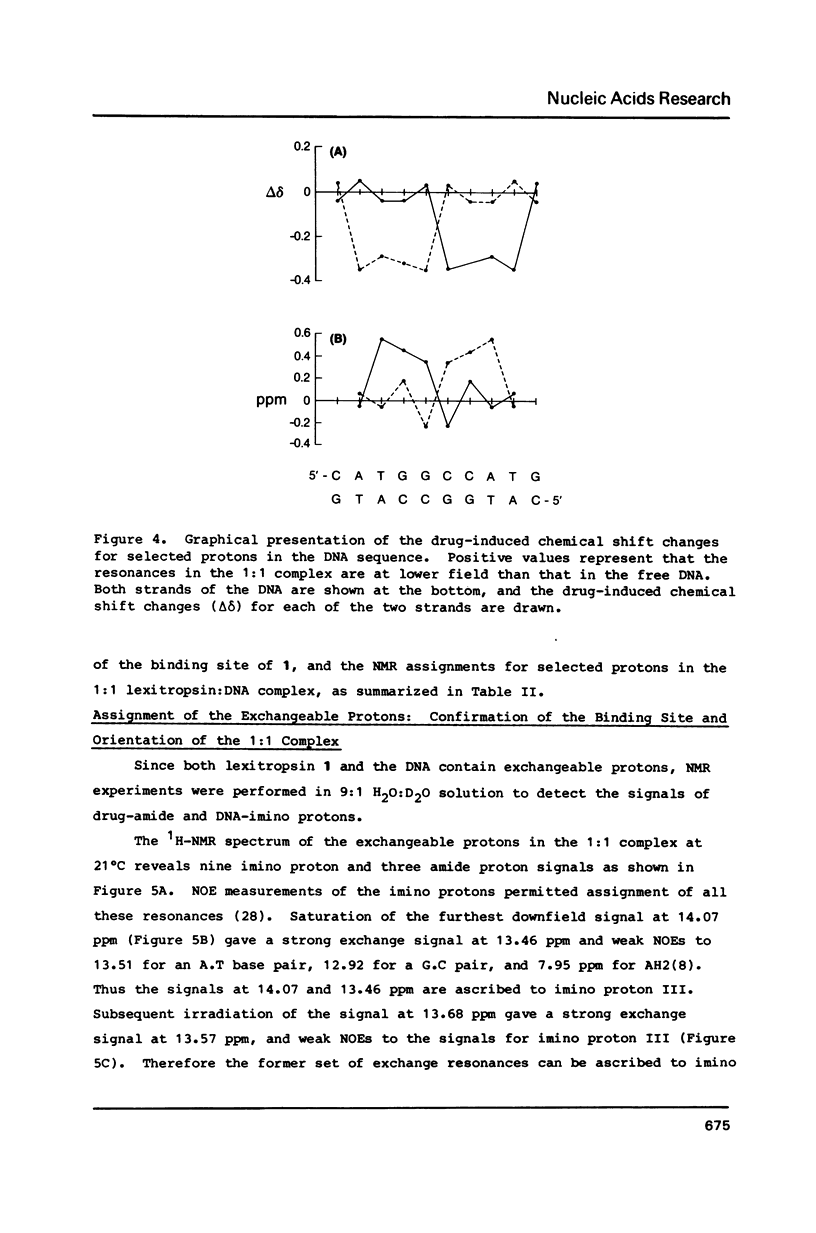
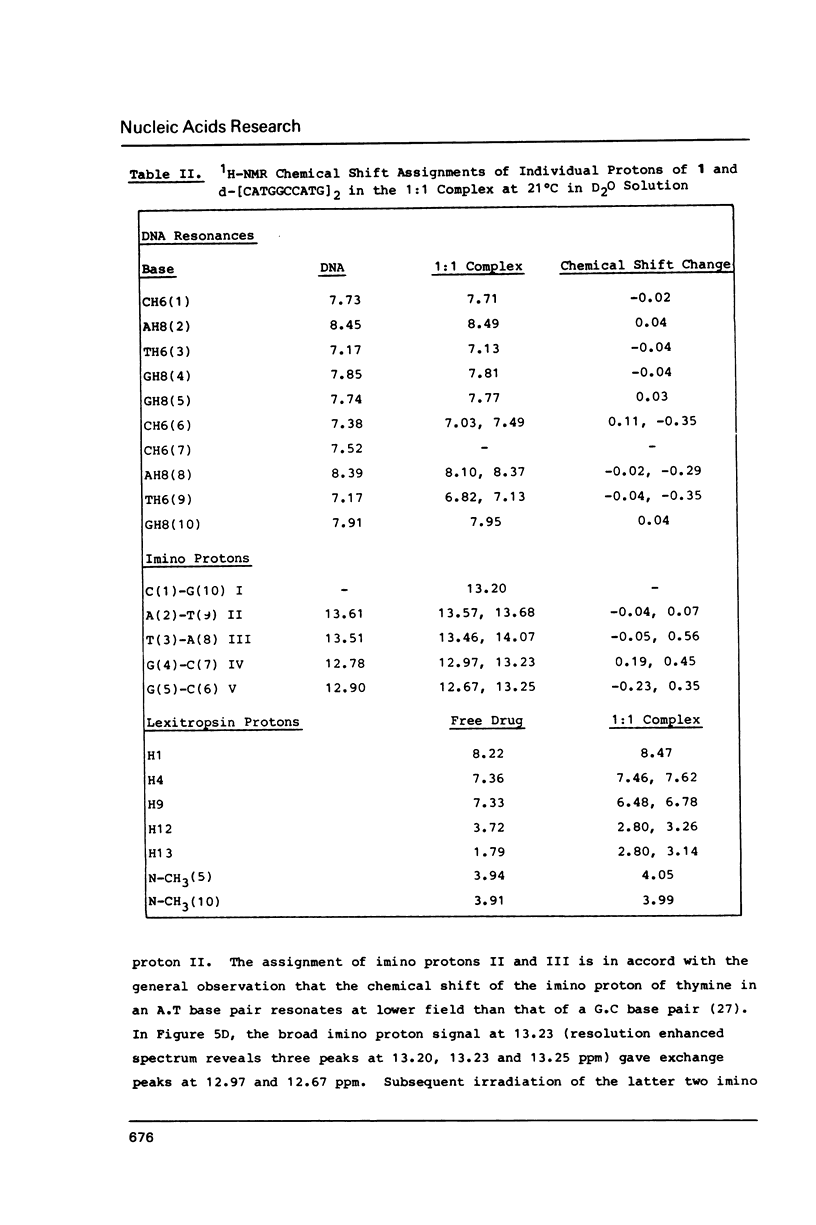
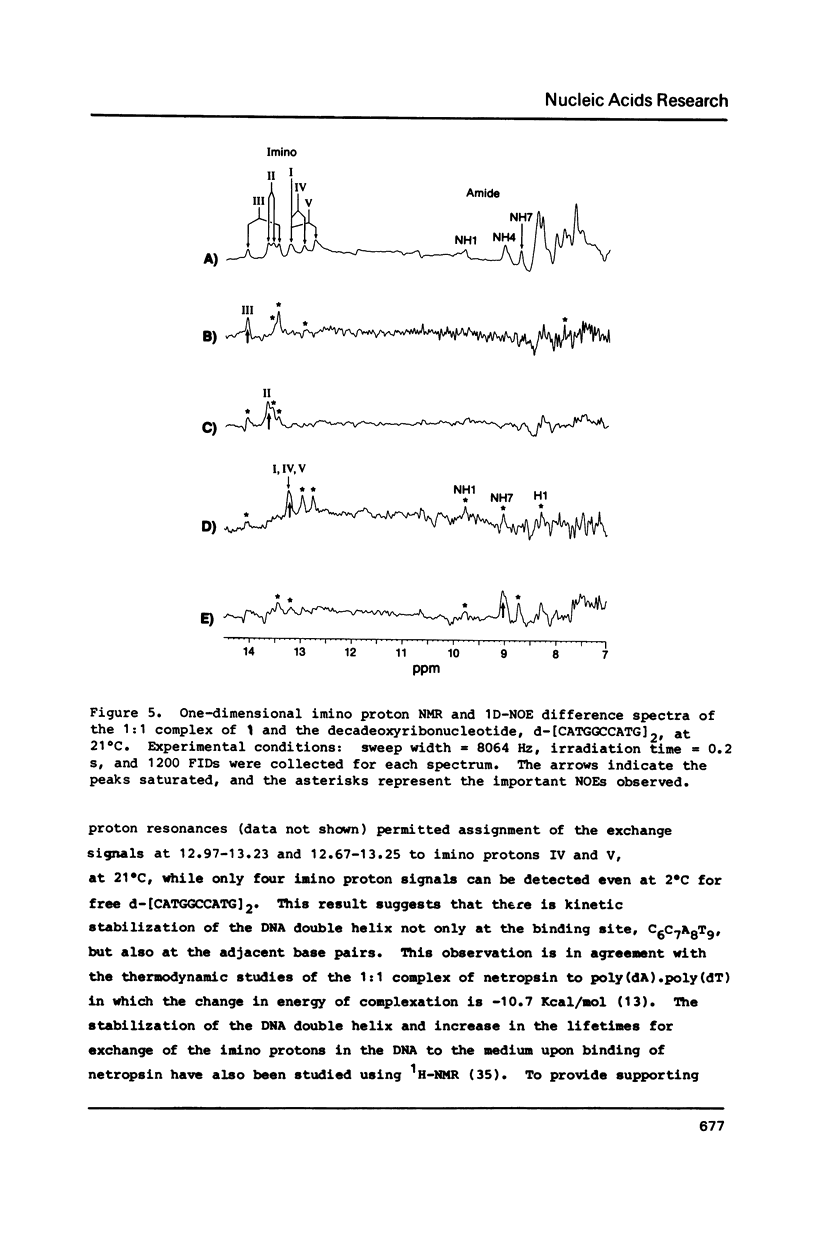
Abstract
All 1H-NMR resonances of d-[CATGGCCATG]2 and the 1:1 complex of lexitropsin 1 and the DNA were assigned by the NOE difference, COSY and NOESY methods. Addition of 1 causes the base and imino protons for the sequence 5'-CCAT to undergo the most marked drug-induced chemical shift changes, thereby indicating that 1 is located in this base pair sequence. NOEs confirmed the location and orientation of the drug in the 1:1 complex, with the amino terminus oriented to C(6). The van der Waals interaction between H12a,b of 1 and AH2(8) may be responsible for reading of the 3' A.T base pair in the 5'-CCAT sequence. Exchange NMR effects allow an estimate of approximately equal to 62 s-1 for the intramolecular "slide-swing" exchange of the lexitropsin between two equivalent binding sites with delta G = 58 +/- 5 kJ mol-1 at 301 degrees K.
Full text
PDF










Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berman H. M., Neidle S., Zimmer C., Thrum H. Netropsin, a DNA-binding oligopeptide structural and binding studies. Biochim Biophys Acta. 1979 Jan 26;561(1):124–131. doi: 10.1016/0005-2787(79)90496-9. [DOI] [PubMed] [Google Scholar]
- Borah B., Roy S., Zon G., Cohen J. S. Unique pyrimidine 2D-COSY aromatic cross-peaks as monitors of pyrimidine environments and mobility in oligo- and polynucleotides. Biochem Biophys Res Commun. 1985 Dec 17;133(2):380–388. doi: 10.1016/0006-291x(85)90917-9. [DOI] [PubMed] [Google Scholar]
- Debart F., Rayner B., Imbach J. L., Chang D. K., Lown J. W. Structure and conformation of the duplex consensus acceptor exon:intron junction d[(CpTpApCpApGpGpT). (ApCpCpTpGpTpApG)] deduced from high-field 1H-NMR of non-exchangeable and imino protons. J Biomol Struct Dyn. 1986 Dec;4(3):343–363. doi: 10.1080/07391102.1986.10506354. [DOI] [PubMed] [Google Scholar]
- Debart F., Rayner B., Imbach J. L., Lee M., Chang D. K., Pon R. T., Lown J. W. Structure and conformation of the duplex consensus 5'-splice site d[(CpApGpGpTpApApGpT).(ApCpTpTpApCpCpTpG)] deduced from high field 1H-NMR of the non-exchangeable and imino protons. J Biomol Struct Dyn. 1987 Aug;5(1):47–65. doi: 10.1080/07391102.1987.10506374. [DOI] [PubMed] [Google Scholar]
- Frederick C. A., Grable J., Melia M., Samudzi C., Jen-Jacobson L., Wang B. C., Greene P., Boyer H. W., Rosenberg J. M. Kinked DNA in crystalline complex with EcoRI endonuclease. Nature. 1984 May 24;309(5966):327–331. doi: 10.1038/309327a0. [DOI] [PubMed] [Google Scholar]
- Hare D. R., Wemmer D. E., Chou S. H., Drobny G., Reid B. R. Assignment of the non-exchangeable proton resonances of d(C-G-C-G-A-A-T-T-C-G-C-G) using two-dimensional nuclear magnetic resonance methods. J Mol Biol. 1983 Dec 15;171(3):319–336. doi: 10.1016/0022-2836(83)90096-7. [DOI] [PubMed] [Google Scholar]
- Kearns D. R. NMR studies of conformational states and dynamics of DNA. CRC Crit Rev Biochem. 1984;15(3):237–290. doi: 10.3109/10409238409102803. [DOI] [PubMed] [Google Scholar]
- Kissinger K., Krowicki K., Dabrowiak J. C., Lown J. W. Molecular recognition between oligopeptides and nucleic acids. Monocationic imidazole lexitropsins that display enhanced GC sequence dependent DNA binding. Biochemistry. 1987 Sep 8;26(18):5590–5595. doi: 10.1021/bi00392a002. [DOI] [PubMed] [Google Scholar]
- Klevit R. E., Wemmer D. E., Reid B. R. 1H NMR studies on the interaction between distamycin A and a symmetrical DNA dodecamer. Biochemistry. 1986 Jun 3;25(11):3296–3303. doi: 10.1021/bi00359a032. [DOI] [PubMed] [Google Scholar]
- Kopka M. L., Yoon C., Goodsell D., Pjura P., Dickerson R. E. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1376–1380. doi: 10.1073/pnas.82.5.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LePecq J. B., Paoletti C. A fluorescent complex between ethidium bromide and nucleic acids. Physical-chemical characterization. J Mol Biol. 1967 Jul 14;27(1):87–106. doi: 10.1016/0022-2836(67)90353-1. [DOI] [PubMed] [Google Scholar]
- Lown J. W., Krowicki K., Bhat U. G., Skorobogaty A., Ward B., Dabrowiak J. C. Molecular recognition between oligopeptides and nucleic acids: novel imidazole-containing oligopeptides related to netropsin that exhibit altered DNA sequence specificity. Biochemistry. 1986 Nov 18;25(23):7408–7416. doi: 10.1021/bi00371a024. [DOI] [PubMed] [Google Scholar]
- Marky L. A., Snyder J. G., Remeta D. P., Breslauer K. J. Thermodynamics of drug-DNA interactions. J Biomol Struct Dyn. 1983 Oct;1(2):487–507. doi: 10.1080/07391102.1983.10507457. [DOI] [PubMed] [Google Scholar]
- Morgan A. R., Lee J. S., Pulleyblank D. E., Murray N. L., Evans D. H. Review: ethidium fluorescence assays. Part 1. Physicochemical studies. Nucleic Acids Res. 1979 Oct 10;7(3):547–569. doi: 10.1093/nar/7.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi A., Morden K. M., Patel D. J., Tinoco I., Jr Kinetics for exchange of the imino protons of the d(C-G-C-G-A-A-T-T-C-G-C-G) double helix in complexes with the antibiotics netropsin and/or actinomycin. Biochemistry. 1983 Mar 1;22(5):1107–1113. doi: 10.1021/bi00274a018. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Shapiro L. Sequence-dependent recognition of DNA duplexes. Netropsin complexation to the AATT site of the d(G-G-A-A-T-T-C-C) duplex in aqueous solution. J Biol Chem. 1986 Jan 25;261(3):1230–1240. [PubMed] [Google Scholar]
- Roy S., Redfield A. G. Nuclear Overhauser effect study and assignment of D stem and reverse-Hoogsteen base pair proton resonances in yeast tRNAAsp. Nucleic Acids Res. 1981 Dec 21;9(24):7073–7083. doi: 10.1093/nar/9.24.7073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda Y., Ohlendorf D. H., Anderson W. F., Matthews B. W. DNA-binding proteins. Science. 1983 Sep 9;221(4615):1020–1026. doi: 10.1126/science.6308768. [DOI] [PubMed] [Google Scholar]
- Wartell R. M., Larson J. E., Wells R. D. Netropsin. A specific probe for A-T regions of duplex deoxyribonucleic acid. J Biol Chem. 1974 Nov 10;249(21):6719–6731. [PubMed] [Google Scholar]
- Zakrzewska K., Lavery R., Pullman B. Theoretical studies of the selective binding to DNA of two non-intercalating ligands: netropsin and SN 18071. Nucleic Acids Res. 1983 Dec 20;11(24):8825–8839. doi: 10.1093/nar/11.24.8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer C. Effects of the antibiotics netropsin and distamycin A on the structure and function of nucleic acids. Prog Nucleic Acid Res Mol Biol. 1975;15(0):285–318. doi: 10.1016/s0079-6603(08)60122-1. [DOI] [PubMed] [Google Scholar]
- Zimmer C., Wähnert U. Nonintercalating DNA-binding ligands: specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material. Prog Biophys Mol Biol. 1986;47(1):31–112. doi: 10.1016/0079-6107(86)90005-2. [DOI] [PubMed] [Google Scholar]


