Abstract
In recent years, there has been significant progress in the use of Cre-loxP technology for conditional gene expression in the inner ear. Here, we introduce the basic concepts of this powerful technology, emphasizing the differences between Cre and CreER. We describe the creation and Cre expression pattern of each Cre and CreER mouse line that has been reported to have expression in auditory and vestibular organs. We compare the Cre expression patterns between Atoh1-CreERTM and Atoh1-CreERT2 and report a new line, Fgfr3-iCreERT2, which displays inducible Cre activity in cochlear supporting cells. We also explain how results can vary when transgenic vs. knock-in Cre/CreER alleles are used to alter gene expression. We discuss practical issues that arise when using the Cre-loxP system, such as the use of proper controls, Cre efficiency, reporter expression efficiency, and Cre leakiness. Finally, we introduce other methods for conditional gene expression, including Flp recombinase and the tetracycline-inducible system, which can be combined with Cre-loxP mouse models to investigate conditional expression of more than one gene.
Keywords: cochlea, conditional gene deletion CreER, Cre efficiency, Cre recombinase, ectopic gene expression, Flp recombinase, knock-in, LoxP, reporter lines, Tet-on, Tet-off, transgenic, utricle, vestibular
Introduction
The use of knockout or germline deletion mice has been extremely useful in the past few decades to investigate the role of specific genes in tissues or organs, including the inner ear; however, this approach deletes the gene of interest (GOI) from every cell in the body throughout the life of the mouse and ~15–20 % of germline deletions result in embryonic lethality (Zambrowicz et al. 2003). In addition, germline deletion models identify only the earliest functions of the GOI, with postnatal gene functions often masked by embryonic effects. Germline gene inactivation can also cause pleiotropic effects, where the deletion of a single gene influences multiple phenotypes, again preventing the determination of an individual GOI’s function in a particular organ. Many genes play different roles in various cell types as well as different roles during embryonic development compared to postnatal ages; thus, cell type specificity and temporal control of gene expression (known as conditional gene expression) is needed to fully understand their function. Mouse models that allow conditional gene expression permit the discovery and dissection of GOI functions in a manner that is specific to a chosen cell type and throughout the life of the mouse.
The most common method to alter gene expression in a conditional manner is the use of the Cre-loxP system (Kwan 2002). Here, we discuss the basics of Cre-loxP technology including the differences between Cre and CreER and the generation of knock-in, conventional transgenic, and bacterial artificial chromosome (BAC) transgenic lines. We list each Cre/CreER mouse strain that has been published in the inner ear, organized by when Cre expression occurs (separating embryonic and postnatal ages) and where Cre expression occurs (separating auditory, vestibular, ganglionic, and non-sensory regions). We describe in detail how each mouse line was created, its Cre expression pattern, and the relevant biological discoveries that have been made using each conditional allele. The last section of the review discusses practical issues that arise when using the Cre-loxP system, such as the use of proper controls, Cre efficiency, reporter expression efficiency, and Cre leakiness. We also discuss applications for fate mapping and mosaic Cre expression patterns. Finally, we present the basics of two other methods to control gene expression in a conditional manner—Flp recombinase and the tetracycline-inducible system. This review serves as a thorough introduction to conditional gene deletion and its use in inner ear research as well as a compilation of current information for researchers who routinely use conditional mouse models.
The Cre-loxP system
Cre is an enzyme originally from P1 bacteriophage that acts as a site-specific recombinase, recognizing a short sequence of DNA called a loxP site. A loxP site is a consensus 34-bp DNA sequence that is not present in the mouse genome and has directionality. Cre-mediated recombination of genes flanked by loxP sites (also called a floxed sequence) can result in the excision, inversion, or translocation of DNA depending upon the location and orientation of the loxP sites. The most common use of Cre recombinase is to excise or delete the floxed DNA sequence which occurs when two loxP sites are on the same strand of DNA and are in the same orientation (Nagy 2000). Numerous studies have shown that Cre-mediated recombination can occur in a variety of cell types (Sauer et al. 1989; Kuhn et al. 1995; Kellendonk et al. 1999; Feltri et al. 1999; Nagy et al. 2000) and that only a few Cre molecules per cell are needed to excise the floxed DNA (Nagy 2000). It is important to note that Cre-mediated recombination is a permanent deletion of the floxed DNA, and if cell division occurs after Cre-mediated recombination, all daughter cells will inherit this gene deletion.
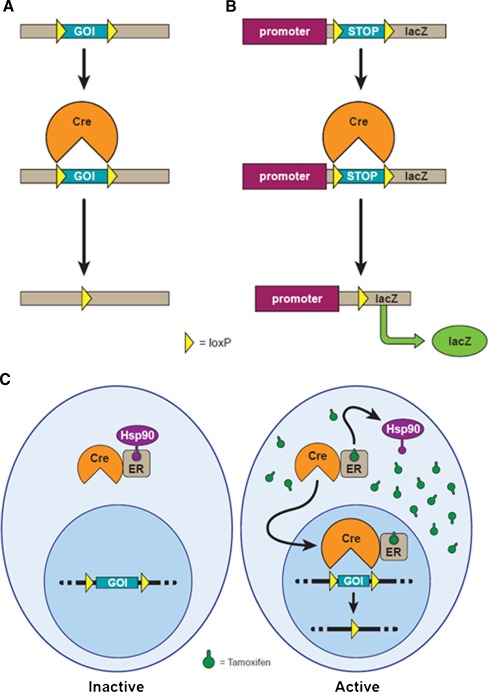
To allow for cell type-specific control of gene deletions, mouse models have been created where Cre expression is controlled by a cell type-specific promoter. These lines can then be crossed with mouse lines containing a relevant part of a GOI that is surrounded by loxP sites in the genome to generate conditional gene deletion (Fig. 1A). The overexpression or ectopic expression of a gene can also be induced using the Cre-loxP system. In this case, a construct containing a promoter and a floxed “stop” sequence upstream of a GOI (i.e., promoter-loxP-stop-loxP-GOI) is inserted into the genome; thus, only Cre+ cells are able to remove the “stop” sequence and overexpress or ectopically express the GOI (Fig. 1B; Zuo 2002; Gao et al. 2004). This strategy is also used for reporter alleles so that lacZ, green fluorescent protein (GFP), or other fluorescent molecules are expressed in Cre+ cells only when the floxed “stop” sequence is removed.
FIG. 1.
Cre-mediated excision and the mechanism of inducible CreER. A Diagram of Cre-mediated deletion of a GOI flanked by loxP sites. B Diagram of a reporter allele where Cre-mediated deletion of a floxed “stop” sequence results in the expression of lacZ. C Diagram of inducible CreER-mediated excision. In the absence of tamoxifen, CreER is sequestered in the cytoplasm by Hsp90. In the presence of tamoxifen, CreER is translocated to the nucleus where it recognizes loxP sequences and cleaves the DNA.
To gain temporal control of gene expression, the ligand-binding domain of an engineered steroid hormone receptor is fused to the Cre enzyme and the Cre molecules are sequestered in the cytoplasm by heat shock protein 90 (Hsp90), keeping Cre inactive. The translocation of the Cre–hormone receptor fusion protein to the nucleus, which allows Cre to become active, only occurs in a ligand-dependent (or inducible) manner (Fig. 1C; Feil et al. 1996; Hayashi and McMahon 2002). To prevent Cre activation by endogenous steroid hormones, the engineered steroid hormone receptor contains specific point mutations that make it insensitive to endogenous steroid hormones while retaining binding affinity for synthetic analogues. The most common use of this strategy is CreER alleles, where an altered ligand-binding domain of the estrogen receptor is fused to Cre and mediates tamoxifen-dependent translocation to the nucleus, but is insensitive to endogenous estradiol (Hayashi and McMahon 2002). Therefore, only exposure to tamoxifen will induce Cre activity allowing temporal control of gene expression. Note that tamoxifen is a prodrug that is hydrolyzed by the liver to its active form, 4-hydroxy-tamoxifen (Furr and Jordan 1984); thus, in vitro induction of CreER requires the use of 4-hydroxy-tamoxifen instead of tamoxifen to activate CreER. Two mutations in the estrogen receptor are commonly used to generate CreER alleles. CreERTM (also called CreERT) contains a single-point mutation, G521R, in the ligand-binding domain of the estrogen receptor, whereas CreERT2 contains a triple mutation (G400V, M543A, and L544A) in this region (Feil et al. 1997; Indra et al. 1999). The triple mutation results in a four to tenfold increase in the sensitivity of CreERT2 to 4-hydroxy-tamoxifen compared to CreERTM; thus, in theory, less tamoxifen is needed to induce Cre activity for CreERT2 alleles (Danielian et al. 1998; Indra et al. 1999; Li et al. 2000). It is important to remember that many other factors may affect Cre activity, such as the activity of the promoter driving Cre expression at the time of induction, so the dose of tamoxifen needed to get optimal results for each CreER line needs to be empirically tested with a reporter line. Other inducible Cre alleles, such as CrePR where the ligand-binding domain of the progesterone receptor is fused to Cre, also exist, but are used less frequently (Wunderlich et al. 2001).
Generation of Cre/CreER lines
Cre/CreER lines are generated by inserting the Cre coding sequence into the endogenous coding region of a gene to “highjack” the promoter of interest (i.e., knock-in) or by creating a transgene by randomly inserting into the genome a fragment of DNA that contains the promoter of interest (and/or enhancer regions) followed by the Cre coding sequence. Conventional transgenic lines use minimal promoter regions (<10 kb), while BAC transgenic lines contain large fragments of DNA (>100 kb). There are pros and cons for each of these methods that affect the expression pattern and expression level of Cre/CreER.
Knock-in alleles use homologous recombination in embryonic stem (ES) cells to insert the Cre coding sequence into the endogenous locus of the GOI. This technique is expensive and time-consuming as hundreds of ES cells need to be screened to find the handful where homologous recombination occurred. However, the knock-in approach is advantageous because it is more likely that the endogenous gene expression pattern will be reproduced by the Cre expression pattern (Tian et al. 2006). If the Cre coding sequence replaces or disrupts the endogenous gene sequence, knock-in alleles are null alleles; thus, only heterozygotes can be used for conditional gene expression. In this case, it is also important to consider that a gene may be haploinsufficient, generating a phenotype when only one allele is present and complicating the conclusions made from the deletion of a GOI. When using such knock-in alleles, it is best to compare results to controls that use the same Cre allele without the floxed GOI to test for potential haploinsufficiency phenotypes.
To prevent a knock-in allele from generating a null allele, an internal ribosome entry site (IRES) can be used to drive Cre expression. The translation of mRNA is generally dependent upon 5′ cap-mediated translation; however, the addition of IRES promotes an internal initiation of translation, allowing a second protein, such as Cre, to be made (Mountford and Smith 1995; Martinez-Salas 1999). In most cases, the IRES-Cre construct is added to the 3′-end of the GOI after the stop codon. It is important to note that the efficiency of IRES-mediated translation varies according to cell type and strain background (Kazadi et al. 2008).
The generation of transgenic lines is much faster and cheaper than knock-in alleles, but is dependent on having a well-defined promoter region. Even then, other regulatory elements outside the promoter region may be needed to recapitulate the endogenous gene expression pattern; thus, many transgenic lines have unexpected Cre expression patterns. In addition, transgenes are randomly inserted into the genome, which can cause variation in Cre expression due to the effects of enhancers or repressors in the local genomic region (called positional effects). Thus, multiple founders need to be analyzed for each transgenic line created (Zuo 2002; Tian et al. 2006). The addition of insulators (i.e., an intron region of the chicken beta-globin gene) in front of the transgenic promoter may lessen these positional effects (Burgess-Beusse et al. 2002). Sometimes the insertion of a transgene into the genome can inactivate an endogenous gene, so it is best to use transgenic Cre lines as heterozygotes. Since conventional transgenes are relatively small, multiple copies of the transgene are commonly inserted into the genome at the same site or at multiple insertion sites. This can increase the level of Cre expression and thus affect the pattern of Cre expression. In addition, multiple transgene copies can be reduced after successive generations and cause genetic drift of Cre expression.
In contrast, BAC transgenic lines contain very large segments of DNA that should include all regulatory elements necessary to recapitulate the endogenous gene expression pattern driven by the promoter of interest. Although BAC transgenes are not subject to strong positional effects, which often occur in conventional transgenic lines, they are well known to rearrange or break apart when integrated into the host genome. In fact, rearrangements can even occur in offspring after germline transmission is obtained; thus, Cre expression patterns may be different between founder lines and successive generations. BAC transgene insertion into the genome can also inactivate endogenous genes, so it is also wise to use these alleles as heterozygotes. It is also important to note that BAC vectors may contain additional loxP sites. If just one loxP sequence is retained in the BAC cloning vector, it can be transferred into the mouse genome and can affect the outcome of Cre-mediated excision. In addition, the large size of BAC transgenes increases the likelihood that other genes and/or promoter elements are within these transgenes (Tian et al. 2006; Yu and Zuo 2009).
Reporter lines
To determine the pattern of Cre activity and to quantify the percentage of Cre+ cells, reporter mouse lines are used. The most commonly used line is the ROSA26LacZ reporter (also called ROSA26R or R26R), where lacZ, encoding β-galactosidase (βgal), is expressed in Cre+ cells after the removal of the floxed “stop” sequence (Soriano 1999). βgal expression can only be visualized in the presence of its substrate, X-gal, or using βgal antibodies. Many other reporter lines exist that use the ROSA26 locus or the CAG promoter (a combination of the CMV enhancer with the chicken β-actin promoter) such as ROSA26eGFP (Giel-Moloney et al. 2007), ROSA26eYFP (Srinivas et al. 2001), CAGlacZ (Akagi et al. 1997), and CAGeGFP (Kawamoto et al. 2000). In theory, a reporter line driven by the ROSA26 locus or a CAG promoter is ubiquitously expressed, and recent work has demonstrated that the CAG promoter is eight to tenfold stronger than the ROSA26 promoter (Nyabi et al. 2009; Chen et al. 2011).
xA more complex reporter line that is commonly used is Z/EG. Here, the CAG promoter is used to express lacZ in all cells until Cre-mediated recombination deletes the floxed lacZ gene, allowing the expression of a second reporter, enhanced GFP (eGFP) (Novak et al. 2000). Since eGFP has endogenous fluorescence, Cre activity can be monitored in live samples and Cre+ live cells can be isolated using FACS. Other reporter lines that use the same strategy are the Z/AP line, where human alkaline phosphatase is expressed after the deletion of the floxed lacZ sequence (Lobe et al. 1999), and the mT/mG line, where membrane-targeted GFP is expressed after the deletion of a floxed membrane-targeted tdTomato (Muzumdar et al. 2007).
Imaging the fluorescent reporter molecule does not reflect the current expression of Cre, but instead provides a history of Cre expression since the progeny of Cre+ cells will permanently express the reporter regardless of whether they currently express Cre. In addition, it is important to consider that it takes time for the Cre enzyme to be translated and translocated into the nucleus for recombination to occur (Nagy 2000). It takes additional time for the reporter protein to be synthesized at a level high enough for detection. Several groups have estimated this to take approximately 12 h by comparing the expression of the cell type-specific promoter driving Cre with reporter expression (Bouchard et al. 2004; Matei et al. 2005). The delay for inducible CreER alleles occurs for another reason. After injection, it takes time for tamoxifen to enter the blood stream, be hydrolyzed to its active form, reach cells in the inner ear, and activate the CreER molecule for translocation to the nucleus. Cre-mediated recombination has been detected at the level of the genome 24–30 h after tamoxifen injection (Weber et al. 2008); however, detection using different reporter lines may vary depending on the strength of the fluorescent reporter molecule (Madisen et al. 2010). For this reason, most labs wait 5–10 days after tamoxifen injection to analyze reporter samples.
Cre/CreER lines for the inner ear
In recent years, there has been a significant expansion of Cre and CreER mouse lines that are useful for inner ear research. We will continue to have additional strains in the future due to the NIH Neuroscience Blueprint Cre-Driver Network whose goal is to provide the Neuroscience Community with mouse strains suitable for the cell type-specific perturbation of gene function in the nervous system (http://www.credrivermice.org/index). In addition, the NIH Knockout Mouse Project (KOMP) and the European Conditional Mouse Mutant Program have initiated plans to create floxed alleles for every gene in the mouse genome (Austin et al. 2004; Auwerx et al. 2004). The combination of these resources offers the unique opportunity to study the role of any GOI in a particular cell type of the inner ear at a particular age. Here, we discuss conditional gene expression relevant to research in the inner ear with a review of Cre and CreER mouse lines that have Cre activity in specific cell types (Tables 1 and 2), as well as a discussion of practical issues that arise when using conditional gene expression. In the following section, several Cre/CreER alleles are presented more than once as their Cre expression pattern has been described in multiple inner ear cell types and at different stages of development. It is important to note that the majority of the Cre/CreER alleles that we discuss have Cre expression in other tissues and organs in addition to the inner ear.
TABLE 1.
Summary of Cre lines described in the review
| Conditional allele | Type | Expression pattern | Level of expression | Original citation | Source |
|---|---|---|---|---|---|
| Atoh1-Cre | Knock-in | E13.5: vestibular sensory regions; E14.5: basal turn of the cochlea; P0: HCs and SCs in cochlea and vestibular organs | Cochlear and vestibular HCs (90 %), cochlear SCs (60 %), utricle SCs (6 %), saccule SCs (15 %), cristae SCs (42 %) | Yang et al. (2010a) | Not commercially available |
| Atoh1-Cre | Transgenic | E11: otocyst; E18.5: HCs and SCs in cochlea and vestibular organs, spiral and vestibular ganglion neurons | Cre expression not quantified | Matei et al. (2005) | Jax stock #11104 |
| CAG-Cre | Transgenic | Ubiquitous expression | Cre expression not quantified | Sakai and Miyazaki (1997) | Not commercially available |
| Col1A1-Cre | Transgenic | Not characterized with a reporter line | Liu et al. (2004) | MMRRC stock #15398-UCD | |
| Col2A1-Cre | Transgenic | E10.5: non-sensory regions of the otocyst and mesenchymal cells | Cre expression not quantified | Ovchinnikov et al. (2000) | Jax stock #3554 |
| Fgf16-Cre | Knock-in | E10.5: otic vesicle; P1: semicircular canal cristae, stria vascularis, cochlear spiral prominence epithelium | Cre expression not quantified | Hatch et al. (2009) | Not commercially available |
| Foxg1-Cre | Knock-in | E8.5: otic placode; E13.5: cochlea, vestibular organs, spiral and vestibular ganglia | Cre expression not quantified | Hebert and McConnell (2000) | Jax stock #4337 |
| hGFAP-Cre | Transgenic | E13.5: utricle, cristae, non-sensory cells around vestibular organs, postnatal cochlear SCs | Cre expression not quantified | Zhuo et al. (2001) | Jax stock #4600 |
| Gfi1-Cre | Knock-in | E13.5: vestibular HCs; E15.5: cochlear HCs in basal turn; E18.5: cochlear and vestibular HCs | Cochlear HCs (93 %), HCs in utricle, saccule, and cristae (90 %) | Yang et al. (2010b) | Not commercially available |
| Hoxb2-r4-Cre | Transgenic | E8.5: otic placode; P0: cochlea, vestibular organs, spiral and vestibular ganglia | Cre expression not quantified | Szeto et al. (2009) | Not commercially available |
| Otog-Cre | Transgenic | Not characterized with a reporter line | Cohen-Salmon et al. (2002) | Not commercially available | |
| Pax2-Cre | BAC transgenic | E8.5: otic placode; P0: cochlea, vestibular organs, spiral and vestibular ganglia | Cre expression not quantified | Ohyama and Groves (2004) | MMRRC stock #10569-UNC |
| Pax3-Cre | Knock-in | E9: otic vesicle and developing spiral and vestibular ganglion; E11.5 to E17.5: cochlea, utricle and saccule | Cre expression not quantified | Engleka et al. (2005) | Jax stock #5549 |
| Pax8-Cre | Knock-in | E8.5: otic placode; E16.5: epithelial components of the inner ear and spiral and vestibular ganglia | Cre expression not quantified | Bouchard et al. (2002) | EMMA stock #EM:00141 |
| Pou3f4-Cre | Transgenic | E14.5: otic mesenchyme, adult: temporal bone, spiral ligament, spiral limbus, tympanic border cells, Reissner’s membrane, mesenchymal cells, non-sensory cells in utricle | Cre expression not quantified | Ahn et al. (2009) | Not commercially available |
| Pou4f3-Cre | Transgenic | E12.5: utricle sensory epithelium; E13.5: zone of non-proliferating cells in cochlea; P6: cochlear HCs and SCs, vestibular HCs and stroma | Cre expression not quantified | Sage et al. (2006) | Not commercially available |
| Prestin-Cre | BAC transgenic | P6: cochlear and vestibular HCs, spiral ganglia region | Cre expression not quantified | Tian et al. (2004) | Not commercially available |
| Prestin-Cre | Transgenic | P14: cochlear inner HCs, vestibular HCs, spiral ganglia region; P50: last row of cochlear outer HCs | Cre expression not quantified | Li et al. (2004) | Not commercially available |
| Prox1-eGFP/Cre | Knock-in | P23: cochlear HCs, pillar cells, Deiters’ cells, GER and LER | Outer HCs (30 %), inner HCs (4 %), pillar cells and Deiters’ cells (almost 100 %) | Liu et al. (2012) | Not commercially available |
| SHH-EGFP/Cre | Knock-in | P0: spiral ganglion neurons, peripheral nerve fibers surrounding HCs and SCs | Spiral ganglion neurons (100 %) | Harfe et al. (2004) | Jax stock #5622 |
| Sox2-Cre | Transgenic | Ubiquitous expression | Cre expression not quantified | Hayashi et al. (2002) | Jax stock #4783 |
| Wnt1-Cre | Transgenic | E9: otic vesicle, developing spiral and vestibular ganglion; E11.5 to E17.5: cochlea, utricle and saccule | Cre expression not quantified | Danielian et al. (1998) | Jax stock #3829 |
TABLE 2.
Summary of CreER lines described in the review
| Conditional allele | Type | Tamoxifen induction | Expression pattern | Original citation | Source |
|---|---|---|---|---|---|
| Atoh1-CreERTM | Transgenic | E16 only | Cochlear HCs (not quantified) | Chow et al. (2006) | MMRRC stock #29581-UNC |
| P0 only | Cochlear inner HCs (40 %), outer HCs (50 %), utricle and saccule HCs (40 %), cristae HCs (10 %) | ||||
| P0 and P1 | Cochlear inner HCs (80 %), outer HCs (90 %), utricle and saccule HCs (60 %), cristae HCs (15 %) | ||||
| P0, P1, and P2 | Cochlear inner HCs (80 %), outer HCs (90 %), utricle and saccule HCs (60 %), cristae HCs (20 %) | ||||
| Atoh1-CreERT2 | Transgenic | E12.75 | Cochlear inner HCs (40 %), outer HCs (44 %) | Machold and Fishell (2005) | Jax stock #7684 |
| E12.75 and E13.75 | Cochlear inner HCs (90 %), outer HCs (97 %) | ||||
| P0 and P1 | Cochlear HCs (10–20 % depending on turn), utricle HCs (not quantified) | ||||
| CAG-CreERTM | Transgenic | Varies | Ubiquitous expression | Hayashi and McMahon (2002) | Jax stock #4453 |
| Fgfr3-iCreERT2 | PAC transgenic | P0 and P1 | Pillar and Deiters’ cells (100 %), cochlear outer HCs (25–75 % depending on turn), Hensen or Claudius cells (not quantified), spiral lamina | Rivers et al. (2008) | Not commercially available |
| P2 and P3 | Pillar and Deiters’ cells (100 %), cochlear outer HCs (>30 % depending on turn), Hensen or Claudius cells (not quantified), spiral lamina | ||||
| P6 and P7 | Pillar and Deiters’ cells (100 %), cochlear outer HCs (>20 % depending on turn), Hensen or Claudius cells (not quantified), spiral lamina | ||||
| P12 and P13 or P30 | Pillar and Deiters’ cells (100 %), Hensen or Claudius cells (not quantified), spiral lamina | ||||
| Ngn1-CreERT2 | BAC transgenic | E8.5 to E13.5 | HCs, SCs and non-sensory epithelium of vestibular organs, spiral and vestibular ganglia, GER (no quantifications were done) | Raft et al. (2007) | Jax stock #8529 |
| E8.5 only | Vestibular ganglion neurons (not quantified) | ||||
| E12.5 only | Spiral ganglion neurons (not quantified) | ||||
| Plp-CreERT2 | Transgenic | P0–P7, P3–P9, or P10-P16 | Inner phalangeal cells, Schwann cells in the spiral lamina, vestibular SCs, vestibular Schwann cells and satellite cells (no quantifications were done) | Doerflinger et al., (2003) | Jax stock #5975 |
| P0 and P1 | Inner phalangeal cells (50–80 % depending on turn), pillar and Deiters’ cells (5–10 %), utricle HCs and SCs (not quantified) | ||||
| Prestin-CreERT2 | Knock-in | P0, P1, and P2 | Cochlear outer HCs (15–60 % depending on turn) | Fang et al. (2012) | Not commercially available |
| P2 and older | Cochlear outer HCs (100 %) | ||||
| Prox1-CreERT2 | Knock-in | E16 | Deiters’ cells (72 %), outer pillar cells (18 %), inner pillar cells (3 %), cochlear outer HCs (7 %) | Srinivasan et al. (2007) | Not commercially available |
| P0 and P1 | Pillar cells (5 %), Deiters’ cells (5–10 %) | ||||
| ROSA26-CreER | Knock-in | Varies | Ubiquitous expression | Vooijs et al. (2001) | Not commercially available |
| ROSA26-CreER | Knock-in | Varies | Ubiquitous expression | Badea et al. (2003) | Jax stock #4847 |
Cre/CreER lines for the developing otic vesicle and otocyst
This section will summarize mouse lines with Cre expression occurring in the developing structures of the inner ear during early embryogenesis. While reporter activity persists to postnatal ages, the onset of Cre expression began embryonically.
Foxg1-Cre is a knock-in mouse line that is a modified version of the Foxg1-lacZ mouse (Xuan et al. 1995) where the Cre coding sequence was inserted into the Foxg1 locus and disrupts endogenous Foxg1 expression. Using ROSA26LacZ reporter mice (Hebert and McConnell 2000) and Z/AP reporter mice (Pirvola et al. 2002), Cre+ cells were first detected at embryonic day (E) 8.5 in the otic placode. By E13.5, reporter expression was found throughout the otic epithelium, including the cochlea, vestibular organs, and both spiral and vestibular ganglia, while the surrounding mesenchyme was Cre-negative. Foxg1-Cre has been used in numerous studies to investigate the deletion or overexpression of various genes (Pirvola et al. 2002; Arnold et al. 2006; Zelarayan et al. 2007; Barrionuevo et al. 2008; Jones et al. 2008; Rickheit et al. 2008; Grimsley-Myers et al. 2009; Schultz et al. 2009; Wang et al. 2009; Yamamoto et al. 2009; Deng et al. 2010; Freyer and Morrow 2010; Haugas et al. 2010; Hurd et al. 2010; Hwang et al. 2010; Sipe and Lu 2011). We highlight a few studies here. Yamamoto et al. (2011) challenged the established dogma that Notch signaling is required to specify cochlear sensory progenitor cells using Foxg1-Cre-mediated deletion of RBP-J. Three other papers used Foxg1-Cre to delete Notch receptors (Delta1, Jagged1, and Notch1) to demonstrate a role for Notch signaling in lateral induction and lateral inhibition in the inner ear (Kiernan et al. 2005, 2006; Brooker et al. 2006). Foxg1-Cre was also used to ectopically express the Notch intracellular domain (NICD) which showed that Notch signaling is sufficient to generate ectopic sensory patches (Hartman et al. 2010; Pan et al. 2010). The role of N-myc in the development of the otic vesicle and its control over proliferation in the otic epithelium were discovered using both Foxg1-Cre and Pax2-Cre lines (Dominguez-Frutos et al. 2011; Kopecky et al. 2011). Comparison of the germline knockout of Sonic hedgehog (SHH) with Foxg1-Cre-mediated deletion of Smoothened demonstrated SHH’s direct role in the formation of ventral otic structures (cochlea and saccule) and SHH’s indirect role in the formation of dorsal structures (utricle, semicircular canal cristae, and the endolymphatic duct). Conditional deletion of Smoothened also revealed that SHH signaling regulates the proliferation of neurogenic progenitors which give rise to spiral and vestibular ganglion neurons (Brown and Epstein 2011). Fgf8’s role in regulating the development of cochlear pillar cells was also discovered using the Foxg1-Cre allele (Jacques et al. 2007). Heterozygous Foxg1-Cre mice have only one copy of the Foxg1 gene (Hebert and McConnell 2000), which has been reported to cause haploinsufficiency phenotypes that include proliferation in other organs (Shen et al. 2006; Eagleson et al. 2007; Siegenthaler et al. 2008). However, no change in proliferation in the inner ear has been reported in several papers where proper controls of Foxg1-Cre mice (without the floxed allele) were used (Yamamoto et al. 2009, 2011; Hartman et al. 2010; Brown and Epstein 2011).
The Pax2-Cre transgenic allele was created using a BAC that contains a 101-kb region 5′ to the mouse Pax2 gene as well as 20 kb of the Pax2 coding region, which includes the first three exons of the Pax2 gene. An IRES followed by the Cre coding sequence was inserted into exon 2 of this Pax2 sequence. Cre expression patterns were characterized with ROSA26LacZ and Z/EG reporter mice and first detected at E8.5 in the otic placode. By postnatal day (P) 0, most cells in the cochlea and vestibular organs were Cre+, including cells in the organ of Corti, stria vascularis, Reissner’s membrane, sensory epithelia of the vestibular organs, and both the spiral and vestibular ganglia (Ohyama and Groves 2004). Pax2-Cre has also been used extensively (Arnold et al. 2006; Rocha-Sanchez et al. 2007; Doetzlhofer et al. 2009; Grimsley-Myers et al. 2009; Soukup et al. 2009; Wang et al. 2009; Abraira et al. 2010; Fritzsch et al. 2010; Jahan et al. 2010a; Basch et al. 2011; Dominguez-Frutos et al. 2011; Kopecky et al. 2011; Pan et al. 2011). Neurod1 deletion using Pax2-Cre disrupted the basal-to-apical gradient of hair cell (HC) differentiation and produced ectopic HCs in the spiral and vestibular ganglia regions. These results suggest an antagonistic relationship between Neurod1 and Atoh1 during inner ear development (Jahan et al. 2010b). Pax2-Cre was also used to demonstrate that bone morphogenetic protein (BMP) signaling promotes the formation of the abneural side of the cochlea (i.e., organ of Corti and lesser epithelial ridge (LER)) while repressing the formation of the neural side (i.e., greater epithelial ridge (GER); Ohyama et al. 2010). Pax2-Cre-mediated deletion and activation of canonical Wnt signaling demonstrated Wnt’s role in mediating the otic placode–cranial epidermis fate decision by promoting an otic placode fate (Ohyama et al. 2006). The Pax2-Cre allele was also used to show the interaction between Notch signaling and Wnt signaling in the otic placode (Jayasena et al. 2008).
Pax3-Cre is a knock-in mouse line where the Cre coding sequence was inserted into the first exon of the Pax3 gene, creating a null allele of Pax3. Homozygous Pax3-Cre mice die embryonically (Engleka et al. 2005). Cre expression was characterized in the developing otic epithelium using the RCE:loxP reporter where the CAG promoter was inserted after the ROSA26 locus followed by a floxed “stop” sequence and eGFP (Sousa et al. 2009). Cre+ cells were first detected at E9 in the prosensory epithelium of the otic vesicle, as well as within the developing spiral and vestibular ganglion. In later stages of development (E11.5 to E17.5), Pax3-Cre activity is present in HCs and supporting cells (SCs) of the cochlea, utricle, and saccule. There was also Cre activity detected in the cells of the GER, stria vascularis, endolymphatic duct, and periotic mesenchyme. No Cre activity was detected in the semicircular canals cristae at any age. This use of Pax3-Cre (together with Wnt1-Cre described below) provided the novel and unexpected finding that embryonic neural tube cells contribute to the formation of the otic vesicle and can develop into both sensory and non-sensory cells (Freyer et al. 2011).
The transgenic Wnt1-Cre line uses the Wnt1 enhancer to drive the expression of Cre (Danielian et al. 1998). Using the RCE:loxP reporter, Cre expression was very similar to the Pax3-Cre line, but with fewer Cre+ cells in each of the locations (Freyer et al. 2011).
Pax8-Cre is a knock-in mouse line where the Cre coding sequence replaced exon 3 of the Pax8 gene, creating a null allele of Pax8 (Bouchard et al. 2002). Using the Z/AP reporter line, Cre activity was first detected at E8.5 in the otic placode, and by E10.5, most cells in the otic vesicle were Cre+. At E16.5, most epithelial components of the inner ear had Cre activity, as well as the spiral and vestibular ganglia; however, reporter expression in the semicircular canal cristae was patchy (Bouchard et al. 2004).
The transgenic Hoxb2-r4-Cre line uses the Hoxb2 r4 enhancer, a 1.4-kb fragment of the 5′ region of the Hoxb2 gene, to drive the expression of Cre. Using the ROSA26LacZ reporter line, βgal expression was first detected in the pre-otic field at the five-somite stage (~E8.5); thus, when the otic placode and otic vesicle develop, these cells are Cre+ as well. Accordingly, at P0, most cells are Cre+ in the cochlea and vestibular organs, as well as in the spiral and vestibular ganglia, stria vascularis, and Reissner’s membrane (Szeto et al. 2009).
There are several Cre lines that use a collagen promoter to drive the expression of Cre recombinase. Col1A1-Cre is a transgenic line that uses a 3.6-kb fragment of the rat alpha1(I) collagen (Col1A1) promoter (Liu et al. 2004). This line was not characterized in the developing otocyst using a reporter allele; however, endogenous Col1A1 was detected by in situ hybridization at E11.5 with ubiquitous expression in the otocyst. Col1A1-Cre was used to delete the retinoblastoma protein (Rb) in the developing inner ear, which resulted in an increase in HCs and SCs in both the cochlea and utricle. Rb-null HCs had stereocilia bundles, were innervated, and continued to divide at late embryonic ages. Thus, cell fate determination and differentiation into HCs and SCs does not require the presence of Rb and can occur even while cells are proliferating (Sage et al. 2005). There is a second Col1A1-Cre allele that uses a truncated 2.3-kb fragment of the rat Col1A1 promoter (Liu et al. 2004), but this line has not been described in the inner ear.
The type II collagen promoter (Col2A1) has also been used to generate a transgenic Cre mouse line. Col2A1-Cre uses 3 kb of the Col2A1 promoter, the first exon of Col2A1, with a mutated initiation codon, and a 3.02-kb fragment of intron 1 followed by IRES to drive the expression of Cre (Ovchinnikov et al. 2000). Using the ROSA26LacZ reporter line, βgal expression was first detected at E10.5 as small clusters of cells in the non-sensory regions of the otocyst, as well as in surrounding mesenchymal cells. Col2A1-Cre has been used to ectopically express the NICD in a transient manner by combining it with the tetracycline-inducible system. The expression of NICD induced ectopic HCs and SCs in the non-sensory regions of the cochlea and vestibular system, demonstrating that Notch signaling is sufficient for the initiation of sensory cell fate in the developing inner ear (Pan et al. 2010).
Otogelin-Cre or Otog-Cre is a transgenic BAC mouse line where Cre is under the control of the murine Otog promoter. A reporter line was not used to characterize Otog-Cre; however, endogenous Otog expression was detected at E10 in the otic vesicle and at E18 in the non-sensory cells of the sensory epithelium of the cochlea and vestibular organs. Otog-Cre-mediated deletion of connexin26 resulted in normal development of the inner ear, but by hearing onset, progressive cell death was described beginning with inner phalangeal and border cells and continuing to outer HCs and other SC subtypes. Inner HCs remained intact and the vestibular system was not affected. These results demonstrate that connexin26 and the gap junction network are required for the survival of cochlear HCs and SCs and, thus, auditory function (Cohen-Salmon et al. 2002).
The BAC transgenic line, Neurogenin1-CreERT2 (also called Ngn1-CreERT2 or Neurog1-CreERT2), contains 113 kb of the 5′ sequence, the Ngn1 coding sequence, and 71 kb of the 3′ sequence. The CreERT2 coding sequence replaced the Ngn1 coding sequence in the BAC. Using the Z/EG reporter line and tamoxifen (1 mg/40 g) given twice daily by gavage from E8.5 to E13.5, analysis at E14.5 revealed many Cre+ HCs and SCs in the utricle and saccule, while only a few Cre+ SCs were found in the cristae. There were also many Cre+ cells in the surrounding non-sensory epithelium of the utricle and saccule as well as in the spiral and vestibular ganglia. In the cochlea, Cre+ cells were found only in the GER. Similar results were described when tamoxifen was given at E8.5 and E9.5. These results indicate that the sensory regions of the utricle and saccule, but not the cristae and cochlea, are derived from the neurogenic region of the otocyst (Raft et al. 2007). More fate mapping studies using the Ngn1-CreERT2 line were conducted to determine the specification of vestibular and spiral ganglion neurons. Early tamoxifen injections (E8.5) primarily labeled vestibular ganglion neurons, while tamoxifen given at E12.5 primarily labeled spiral ganglion neurons; thus, Ngn1+ precursor cells change with age to generate these two cell populations (Koundakjian et al. 2007). Ngn1-CreERT2-mediated deletion of ephrin-B2 with tamoxifen given by gavage at E9.5–E10.5 demonstrated ephrin-B2’s role in the fasciculation of spiral ganglion neurons (Coate et al. 2012). There is also a non-inducible Ngn1-Cre allele that used the same BAC construct, but it has only been described in the brain (Lundell et al. 2009).
Fgf16-Cre is a knock-in mouse line with an IRES-Cre cassette inserted into the first coding exon of Fgf16. Even though this line is a null allele, the inner ears of homozygotes are structurally and functionally normal. Using the ROSA26LacZ reporter line, βgal expression was first apparent in the otic vesicle at E10.5. By P1, Cre+ cells were detected in the sensory and non-sensory regions of the semicircular canal cristae, at the base of the stria vascularis, and in the cochlear spiral prominence epithelium (Hatch et al. 2009).
Cre/CreER lines for the developing organ of Corti
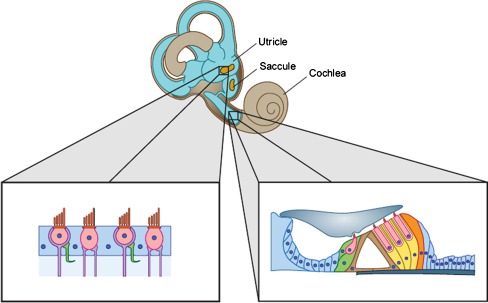
This section will focus on Cre lines with expression occurring in the developing sensory epithelium of the cochlea (Fig. 2) during late embryogenesis, when prosensory cells are in the process of committing to either a HC or SC fate.
FIG. 2.
Diagram of the inner ear and cell types in the sensory epithelium of the cochlea and utricle. In the magnified image of the utricle, HCs are in pink and SCs are in blue. The magnified image of the cochlea focuses on the organ of Corti, with HCs in pink. Green SCs underneath the inner HC are inner phalangeal cells/border cells. Brown SCs between the inner and outer HCs are pillar cells. Yellow SCs underneath the three outer HCs are Deiters’ cells. The orange SC lateral to the outer HCs is a Hensen cell.
Pou4f3-Cre is a transgenic allele which uses a 9-kb region 5′ to the Pou4f3 gene as the promoter to drive Cre expression. Using the ROSA26LacZ reporter, βgal expression was first detected at E13.5 in the zone of non-proliferating cells of the developing cochlea and by P6 in both HCs and SCs. Quantification of Cre+ cells was not reported (Sage et al. 2006). Pou4f3-Cre was used to delete Rb, which extended HC and SC proliferation to neonatal ages while preserving the initial development of mechanoelectrical transduction; however, degeneration occurred in the adult organ of Corti that led to severe hearing loss. These experiments suggest that transient regulation of Rb might be a strategy to achieve cochlear HC regeneration (Sage et al. 2006). Pou4f3-Cre-mediated deletion of Dicer1 caused malformations in HC stereocilia bundles, degeneration of HCs, and subsequent hearing loss. Interestingly, vestibular HCs were less effected and only a mild vestibular phenotype was detected (Friedman et al. 2009) The Pou4f3-Cre line has also been used to ablate HCs in a mosaic but reproducible manner by the expression of a drug-inducible, dimerizable caspase 3 (iCsp3) that leads to hearing loss after a week of drug administration. In the cochlea, ~60 % of HCs expressed iCsp3 (Fujioka et al. 2011).
There are two Atoh1-Cre lines, a knock-in allele (Yang et al. 2010a) and a transgenic allele (Matei et al. 2005). The Atoh1-Cre knock-in line replaced the entire Atoh1 coding sequence with the Cre coding sequence. Homozygous Atoh1-Cre mice die soon after birth, whereas heterozygous Atoh1-Cre mice are viable and display no visible defects. Using the ROSA26LacZ reporter, βgal expression was first detected in the basal turn of the cochlea at E14.5 and progressed to middle and apical turns over the next few days. By P0, βgal expression was observed in all turns of the cochlea in both HCs and SC, including Deiters’ cells, pillar cells, and inner phalangeal cells. Approximately 90 % of HCs and 60 % of SCs were Cre+. These findings prompted the conclusion that Atoh1 is expressed in prosensory progenitor cells before HC and SC fates are specified (Yang et al. 2010a). The Atoh1-Cre transgenic line used the same Atoh1 enhancer fragment used to create the Atoh1-eGFP mouse (Chen et al. 2002) and was also characterized with the ROSA26LacZ reporter line. In contrast to the Atoh1-Cre knock-in line, βgal expression was first detected in the Atoh1-Cre transgenic line at E11 in areas that correspond to the future sensory epithelium. At E18.5, almost all cochlear HCs expressed βgal, except for those in the most apical tip. The Atoh1-Cre transgenic line also had Cre expression in SCs. Quantification of Cre+ cells was not reported (Matei et al. 2005). The Atoh1-Cre transgenic line has been used to delete Dicer1, showing the important roles of microRNAs in cochlear gene expression profiles and maintaining the apex to base gradient of gene expression. This model also demonstrated that microRNAs are required for HC survival (Weston et al. 2011). Transgenic Atoh1-Cre-mediated deletion of beta or gamma actin isoforms showed that only one of these genes is needed for the normal development of HC stereocilia bundles, while each gene plays a different role in the maintenance of stereocilia during aging (Perrin et al. 2010). Comparison of Atoh1-Cre-mediated Neuro1d deletion with Pax2-Cre-mediated Neuro1d deletion demonstrated that Neurod1’s roles in HC differentiation and maturation are relevant only at very early embryonic ages (Jahan et al. 2010b).
There are also two inducible Cre alleles which use the Atoh1 enhancer to drive the expression of CreER: Atoh1-CreERTM (Chow et al. 2006) and Atoh1-CreERT2 (Machold and Fishell 2005). The transgenic Atoh1-CreERTM allele was induced with tamoxifen (100 μg/g, IP) at E16 and analyzed at E19 using a Rosa26-loxP-stop-loxP-NICD-IRES-eGFP line (which expresses both NICD and eGFP after Cre-mediated excision of the floxed stop sequence). eGFP expression was only detected in HCs; however, quantification was not performed. The eGFP+ HCs in this model also had ectopic expression of NICD, but appeared normal (Liu et al. 2012b). The transgenic Atoh1-CreERT2 line was characterized with the ROSA26eYFP reporter and embryonic tamoxifen induction (the tamoxifen dose and route of administration were not specified). After a single tamoxifen dose at E12.75, eYFP expression was detected in 40 % of inner HCs and 44 % of outer HCs at E18.5. These percentages increased to 90 % of inner HCs and 97 % of outer HCs when a second tamoxifen dose was given at E13.75. Cre activity was not observed in SCs with either induction paradigm. The Atoh1-CreERT2 allele was used to delete Eya1 and Six1, demonstrating that these genes are required for HC development and Atoh1 expression (Ahmed et al. 2012).
Gfi1-Cre is a knock-in allele where the Cre coding sequence replaced the endogenous Gfi1 gene; thus, homozygous Gfi1-Cre mice cannot be used because of their severe phenotype where HCs do not form. This line was characterized with the ROSA26LacZ reporter where βgal expression was first seen in inner HCs of the basal turn at E15.5 that progressed to outer HCs and other turns of the cochlea over the following days. By E18.5, nearly all HCs (~93 %) in the entire cochlea were labeled by βgal. No other cell types in the cochlea were labeled (Yang et al. 2010b). Gfi1-Cre-mediated deletion of BMP2 demonstrated that this gene is not required for normal HC formation or hearing ability (Hwang et al. 2010).
The Prox1-CreERT2 knock-in allele was created by inserting IRES-CreERT2 into the mouse Prox1 locus (Srinivasan et al. 2007). Heterozygous Prox1-CreERT2 mice have normal Prox1 expression, normal morphology of the organ of Corti, and normal hearing. Homozygous mice were not used since the Cre coding sequence is followed by a Neo-cassette which may affect the expression levels of Prox1 (Yu et al. 2010). The Cre expression pattern was characterized using the ROSA26eYFP reporter line and tamoxifen (100 μg/g, IP) given at E16. When the cochlea was analyzed at E19, Cre expression was detected in ~72 % of Deiters’ cells, ~18 % outer pillar cells, ~3 % inner pillar cells, and ~7 % of outer HCs. No Cre+ inner HCs were found (Liu et al. 2012b).
Cre/CreER lines for the developing vestibular organs
This section will focus on Cre lines with expression occurring in the developing sensory epithelium of vestibular organs: the utricle, saccule, and semicircular canal cristae (Fig. 2).
Cre activity from the transgenic Pou4f3-Cre mouse line was first seen using the ROSA26LacZ reporter in the sensory epithelium of the utricle at E12.5. By E13.5, βgal expression was detected in vestibular HCs. At P6, the only βgal+ cells in the sensory epithelium were HCs, but there were also βgal+ cells in the stroma (containing nerve fibers) beneath the epithelium. Quantification of Cre+ cells was not reported. Pou4f3-Cre-mediated deletion of Rb was also studied in the utricle where HCs continued to divide until 6 weeks of age and partial vestibular function was maintained at 6 months. Vestibular Rb-null HCs died at a slow rate, which contrasts with the rapid cell death seen in cochlear HCs when Rb was deleted using the same Cre line. Thus, Rb plays different roles in the survival of these two types of HCs (Sage et al. 2006).
ROSA26LacZ reporter expression was also detected in the vestibular system with the two Atoh1-Cre lines. Cre+ cells in the Atoh1-Cre knock-in allele were first detected in vestibular sensory regions at E13.5 and increased to P0 where both HCs and SCs in the utricle, saccule, and cristae expressed βgal. Specifically, there were ~90 % Cre+ HCs in each of these organs, and the number of Cre+ SCs varied from 6 % in the utricle to 15 % in the saccule and 42 % in the cristae. These data also suggest that, as in the cochlea, Atoh1 is expressed in vestibular progenitor cells before HC and SC fates are specified (Yang et al. 2010a). Cre activity in vestibular organs of the Atoh1-Cre transgenic line was detected at E11 and by E18.5 was found in vestibular HCs and SCs. Quantification of Cre+ cells was not reported (Matei et al. 2005). The Atoh1-CreERTM and Atoh1-CreERT2 alleles were not characterized in the embryonic vestibular system.
The knock-in Gfi1-Cre allele also has Cre expression in vestibular HCs beginning at E13.5 and progressively increasing to P0, where ~90 % of HCs in the saccule, utricle, and cristae are βgal+. No vestibular SCs were Cre+ (Yang et al. 2010b).
Cre/CreER lines for the postnatal organ of Corti
This section will focus on Cre lines with expression occurring in the sensory epithelium of the cochlea after birth.
The transgenic Atoh1-CreERTM line also has Cre activity in the postnatal cochlea After tamoxifen injection(s) (3 mg/40 g, IP) at birth, the only Cre+ cells found in the organ of Corti were HCs (as revealed using the ROSA26LacZ reporter line). One tamoxifen injection at P0 resulted in 40 % Cre+ inner HCs and 50 % Cre+ outer HCs. These percentages increased to 80 % Cre+ inner HCs and 90 % Cre+ outer HCs when tamoxifen was given once at P0 and again 24 h later at P1. No further increase was seen with a third injection given at P2 (Chow et al. 2006; Weber et al. 2008). In contrast to the results obtained with embryonic Rb deletion using the Col1A1-Cre (Sage et al. 2005) and Pou4f3-Cre lines (Sage et al. 2006), deletion of Rb in neonatal HCs using Atoh1-CreERTM produced S phase reentry followed by cell death. Thus, Rb plays an age-dependent role in HC proliferation (Weber et al. 2008). Atoh1-CreERTM-mediated deletion of Pkd1 ruled out the involvement of this protein as the major component in the mechanoelectrical transduction channel complex or in planar cell polarity mechanisms, but demonstrated its requirement for normal stereocilia number and structure (Steigelman et al. 2011). This line has also been used to drive the expression of DTA as a method of damaging HCs in the neonatal cochlea, in vivo (Cox et al. 2010). Ectopic NICD expression in neonatal HCs using the Atoh1-CreERTM allele resulted in the reactivation of Sox2 in inner and outer HCs and of Prox1 only in outer HCs. Despite the expression of NICD, Sox2, and Prox1, HCs continued to develop normally, demonstrating that once a HC fate is committed activation of the Notch pathway will not impact their development (Liu et al. 2012b).
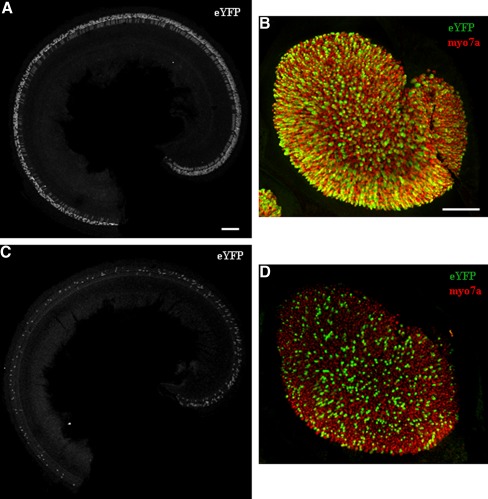
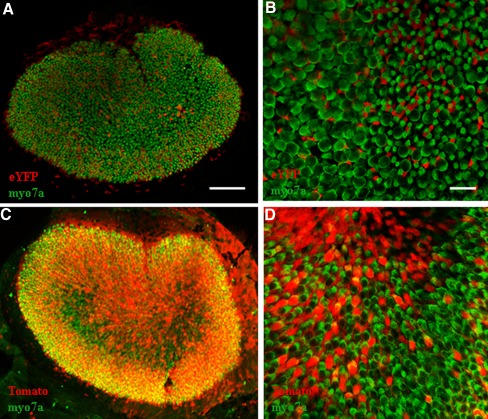
We also investigated the Cre expression pattern of Atoh1-CreERT2 mice (Machold and Fishell 2005) with tamoxifen induction after birth. We bred this line with the ROSA26eYFP reporter and gave tamoxifen injections (3 mg/40 g, IP) once a day at P0 and P1. Analysis at P6 revealed only Cre+ HCs, but a much lower number than what was observed with the Atoh1-CreERTM line under the identical conditions. We found ~20 % Cre+ inner and outer HCs in the apical turn of the cochlea and <10 % in middle and basal turns (Fig. 3). Both Atoh1-CreER strains are transgenics; thus, positional effects and the copy number of the transgene likely play a role in the difference of Cre activity observed.
FIG. 3.
Comparison of the transgenic lines, Atoh1-CreERTM and Atoh1-CreERT2. Representative confocal projection images of the apical turn of the cochlea (A) and utricle (B) from Atoh1-CreERTM;ROSA26eYFP mice, induced with tamoxifen at P0 and P1. Representative confocal projection images of the apical turn of the cochlea (C) and utricle (D) from Atoh1-CreERT2;ROSA26eYFP mice, induced with tamoxifen at P0 and P1. In A and C, eYFP is in white. In B and D, eYFP is in green and the HC marker, myosin VIIa (myo7a), is in red. Scale bars, 100 μm.
There are three mouse strains that use some form of the prestin locus to drive the expression of Cre or CreER: Prestin-Cre (transgenic), Prestin-Cre (BAC transgenic), and Prestin-CreERT2. The Prestin-Cre (transgenic) allele uses a 9-kb fragment of mouse prestin, which contains the putative thyroid hormone-responsive element, the first coding exon (exon 3), and part of intron 3, but not the promoter region upstream of exon 1, to drive the expression of Cre. Using the ROSA26LacZ reporter line, Cre activity was first detected at P14 in inner HCs, and this increased so that by P135 most inner HCs were Cre+. In addition, faint βgal expression was observed in the last row of outer HCs in the apical and middle turns of the cochlea starting at P50 (Li et al. 2004). However, analysis of successive generations of this line with the ROSA26eYFP and Ai6 (where deletion of the floxed “stop” sequence drives the expression of ZsGreen; Madisen et al. 2010) reporter lines did not reveal detectable reporter signals in adult HCs, likely due to genetic drift of transgenic expression (Dearman and Zuo, unpublished).
The Prestin-Cre (BAC transgenic) mouse line used a modified BAC containing ~150 kb of genomic DNA which includes the coding region of prestin (~50 kb) followed by an IRES-Cre cassette after the stop codon in exon 20. This line has only a single copy of the transgenic BAC; however, it was not determined how much of the prestin promoter region is contained within the BAC, which may be the reason why Cre expression is different from endogenous prestin expression. When this line was characterized with the ROSA26LacZ reporter line, βgal expression was first detected at P6 in the majority of outer HCs in the middle and basal turns of the cochlea, while only a few outer HCs in the apical turn were Cre+. Some inner HCs also had βgal expression, and this expression level increased between P6 and P60 (Tian et al. 2004). Conditional deletion of the adherence junction protein, vezatin, using the Prestin-Cre (BAC transgenic) line showed no effects on hearing development or function at early ages, but an increased sensitivity to noise and age-related hearing loss. These findings highlight the role of HC–SC junctions in hearing (Bahloul et al. 2009). Deletion of thyroid hormone receptor beta using this Cre line clarified that malformation of the tectorial membrane (cochlear cause) and not delayed BK channel expression in HCs (retro-cochlear cause) produced hearing loss in the absence of this receptor (Winter et al. 2009).
Most recently, Prestin-CreERT2 was generated using a knock-in method which resulted in a Cre expression pattern that more accurately recapitulates the endogenous expression of prestin. IRES-CreERT2 was inserted into the prestin locus after the stop codon in exon 20. Unlike most knock-in alleles, endogenous prestin was not affected and homozygous Prestin-CreERT2 mice have normal hearing, as tested by auditory brainstem response (ABR). Using the CAGeGFP and Ai6 (where deletion of the floxed “stop” sequence drives expression of ZsGreen; Madisen et al. 2010) reporters, Cre expression patterns were investigated after different tamoxifen induction paradigms. Cre activity was very specific to the outer HCs of the cochlea with all induction paradigms. Early tamoxifen injections (3 mg/40 g, IP) once a day at P0, P1, and P2 produced a gradient of Cre+ outer HCs with 60 % in the base, 35 % in the middle, and 15 % in the apex. In contrast, tamoxifen injections given once a day for 2 days beginning at P2 or any time after resulted in close to a 100 % of Cre+ outer HCs throughout the length of the cochlea (Fang et al. 2012).
The Prox1-CreERT2 knock-in allele (Srinivasan et al. 2007) also has Cre expression in the postnatal cochlea. Using several reporter lines and tamoxifen (3 mg/40 g, IP) injections given once a day at P0 and P1, only pillar cells and Deiters’ cells were Cre+ in the organ of Corti. Specifically, 5 % of pillar cells were Cre+ throughout the length of the cochlea, and the percentage of Cre+ Deiters’ cells varied among turns with 10 % in the apex and 5 % in the middle and base. Deletion of Rb using the Prox1-CreERT2 allele resulted in cell cycle reentry of both pillar cells and Deiters’ cells, while only pillar cells were able to complete the cell cycle and increase in number. This finding highlights the heterogeneity in the role of Rb between these two SC subtypes (Yu et al. 2010).
There is a second allele which uses the Prox1 locus to drive Cre expression. Prox1-eGFP/Cre is a knock-in line where an eGFP/Cre fusion protein was inserted downstream of the Prox1 translation start site (Srinivasan et al. 2010). This allele was recently characterized in the cochlea using the ROSA26eYFP reporter line. At P23, Cre expression was detected in almost all pillar cells and Deiters’ cells, ~30 % outer HCs, and ~4 % inner HCs. There was also Cre activity detected in the GER and LER (Liu et al. 2012b).
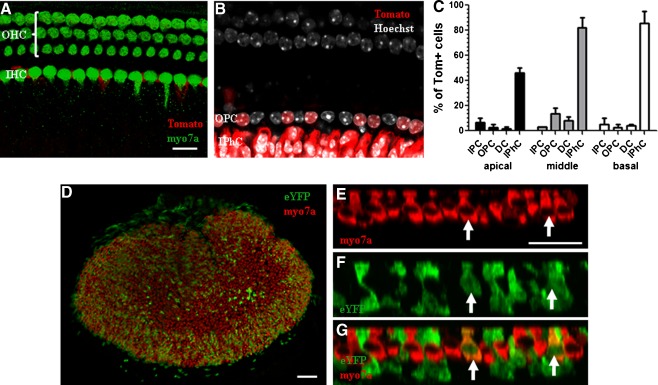
Fgfr3-iCreERT2 is a phage artificial chromosome (PAC) transgenic mouse line that expresses a “codon-improved” version of Cre (iCre) driven by the Fgfr3 promoter. Specifically, iCreERT2 was inserted into the first exon of the Fgfr3 gene in the PAC allele (Rivers et al. 2008; Young et al. 2010). Using the Ai14 reporter line (where deletion of the floxed “stop” sequence drives expression of tdTomato; Madisen et al. 2010), we tested tamoxifen induction at several ages where the majority of Cre+ cells in the cochlea were pillar cells and Deiters’ cells. After tamoxifen (3 mg/40 g, IP) injection once a day at P0 and P1, tdTomato expression was detected in ~100 % of pillar and Deiters’ cells, 25–75 % of outer HCs depending on the turn of the cochlea, and a small fraction of Hensen or Claudius cells (Fig. 4A–C). When we gave tamoxifen (3 mg/40 g, IP) once a day at P2 and P3 (Fig. 4D) or at P6 and P7 (Fig. 4E), the percentage of Cre+ outer HCs decreased to <30 %, while Cre+ pillar and Deiters’ cells remained at 100 %. There were still some Cre+ Hensen or Claudius cells with both induction paradigms. When we gave tamoxifen (3 mg/40 g, IP) once a day at P12 and P13 (Fig. 4F) or one injection of tamoxifen (9 mg/40 g, IP) at P30 (data not shown), we no longer observed Cre+ outer HCs, while Cre+ pillar and Deiters’ cells remained at 100 %. A low percentage of Cre+ Hensen and Claudius cells also remained. Cre+ inner HCs were never found with any induction paradigm. Cre+ cells were also found in the spiral lamina (data not shown).
FIG. 4.
Fgfr3-iCreERT2 expression pattern in the cochlea. Representative confocal images of the organ of Corti from Fgfr3-iCreERT2;Ai14 mice, induced with tamoxifen at P0 and P1. A Slice image taken at the HC layer with tdTomato expression (red) and HCs labeled by myosin VIIA (myo7a, green). B Slice image at the SC nuclear layer with tdTomato expression (red) and SC nuclei labeled by Hoechst (white). Scale bar, 10 μm. Quantification of tdTomato+ cells expressed as a percentage of total cells by type after tamoxifen induction at P0/P1 (C), P2/P3 (D), P6/P7 (E), and P12/P13 (F). DCs Deiters’ cells, PCs pillar cells, OHC outer hair cells.
Plp-CreERT2 is a transgenic mouse line that uses 2.4 kb of the 5′ region, exon 1, and intron 1 of the mouse proteolipid protein 1 gene to drive CreER expression (Doerflinger et al. 2003). Several tamoxifen induction paradigms have been characterized with either the ROSA26LacZ or ROSA26eYFP reporter lines. Tamoxifen (33 mg/kg, IP) given once a day from P0 to P7, P3 to P9, or P10 to P16 results in Cre+ inner phalangeal cells in the organ of Corti and Cre+ Schwann cells in the spiral lamina. The amount of Cre+ cells was not quantified (Gomez-Casati et al. 2010). We also characterized this line using the Ai14 reporter line (where deletion of the floxed “stop” sequence drives expression of tdTomato; Madisen et al. 2010). Tamoxifen (3 mg/40 g, IP) given once a day at P0 and P1 results in 50 % tdTomato+ inner phalangeal cells in the apical turn and 80 % tdTomato+ inner phalangeal cells in the middle and basal turns of the cochlea. We also found 5–10 % Tomato+ pillar and Deiters’ cells. No Cre+ HCs were found (Fig. 5A–C). We also found a large number of Cre+ Schwann cells in the spiral lamina (data not shown). There was no effect on morphology or cellular organization of the inner ear when BDNF was deleted using the Plp-CreERT2 allele and tamoxifen injections were given once a day from P5 to P11 (Gomez-Casati et al. 2010).
FIG. 5.
Plp-CreERT2 expression pattern in the cochlea and utricle. Representative confocal images of the organ of Corti from Plp-CreERT2;Ai14 mice, induced with tamoxifen at P0 and P1. A Slice image taken at the HC layer with tdTomato expression (red) and HCs labeled by myosin VIIA (myo7a, green). B Slice image at the SC nuclear layer with tdTomato expression (red) and SC nuclei labeled by Hoechst (white). Scale bar, 20 μm. C Quantification of tdTomato+ cells expressed as a percentage of total cells by type. IPC inner pillar cells, OPC outer pillar cells, DC Deiters’ cells, IPhC inner phalangeal cells. D–G Representative confocal images of the utricle from Plp-CreERT2; ROSA26eYFP mice, induced with tamoxifen at P0 and P1. E–G Artificial slice images of HCs labeled by myosin VIIA (myo7A, red) and eYFP expression (green). Cre activity is expressed in both HCs (arrows) and SCs. Scale bars, 50 μm (D) and 20 μm (E–G).
There are at least eight lines that use either the mouse or human glial fibrillary acidic protein (GFAP) promoter to drive the expression of Cre or CreER, of which only one allele has been used in the inner ear. The transgenic hGFAP-Cre line used 2.2 kb of the 5′ region from the human GFAP gene with the Gfa2 promoter to drive the expression of Cre (Zhuo et al. 2001). Using the mT/mG and ROSA26eYFP reporter lines, Cre expression was detected in some SCs postnatally, but the amount was not quantified (Hartman et al. 2010).
Cre/CreER lines for the postnatal vestibular system
Several of the alleles that have Cre activity in the postnatal cochlea are also expressed in vestibular organs; however, less is known. This section will focus on alleles where Cre activity occurs in the sensory epithelia of vestibular organs after birth.
The transgenic Atoh1-CreERTM allele also has Cre expression in vestibular HCs when tamoxifen is given at birth. Again, the percentage of Cre+ HCs increased with multiple tamoxifen injections (3 mg/40 g, IP). One tamoxifen injection at P0 resulted in 40 % Cre+ HCs in the utricle and saccule, but only 10 % Cre+ HCs in the cristae. With tamoxifen at P0 and P1, 55–60 % of HCs in the utricle and saccule were Cre+ and Cre+ HCs in the cristae increased slightly to 15 %. No increase in Cre+ HCs in the utricle and saccule was seen with a third injection given at P2, but it did increase Cre+ HCs in the cristae to 20 % (Chow et al. 2006).
We also found Cre activity in the utricle of the transgenic Atoh1-CreERT2 allele using the ROSA26eYFP reporter line. Similar to the cochlea, Cre activity was specific to HCs, but there were fewer Cre+ HCs in the utricle after tamoxifen injection at P0 and P1 (Fig. 3). We did not characterize the other vestibular organs.
Using the ROSA26LacZ reporter line, Cre activity was detected in vestibular HCs in both the Prestin-Cre (transgenic) line (Li et al. 2004) and the Prestin-Cre (BAC transgenic) allele (Tian et al. 2004). No Cre+ cells were found in vestibular organs using the Prestin-CreERT2 knock-in allele (Fang et al. 2012).
After tamoxifen (33 mg/kg, IP) injection once a day from P0 to P7, P3 to P9, or P10 to P16, the transgenic Plp-CreERT2 line showed Cre activity in most SCs in the utricle and saccule, while only some SCs were Cre+ in the cristae. Cre activity was also observed in vestibular Schwann cells and satellite cells (Gomez-Casati et al. 2010). We also characterized this line using the ROSA26eYFP reporter line. When tamoxifen (3 mg/40 g, IP) was given once each at P0 and P1, both Cre+ HCs and SCs were found in the utricle (Fig. 5D–G). Other vestibular organs were not characterized.
Cre activity in the vestibular system has only been described in one of the transgenic hGFAP-Cre lines (Zhuo et al. 2001). Reporter expression was first detected at E13.5 in cells within the utricle and cristae as well as in non-sensory cells in the regions around the vestibular organs. Ectopic expression of NICD using the hGFAP-Cre line resulted in ectopic sensory patches that contained both HCs and SCs. Interestingly, ectopic HCs were not Cre+, which suggests that they were induced by neighboring NICD+ cells. This finding is consistent with the known role of Notch in lateral inhibition. In addition, there was a marked decrease in the number of HCs found in the ectopic sensory patches of adult mice where NICD remained permanently expressed (Hartman et al. 2010).
Cre/CreER lines for the spiral and vestibular ganglia
Similar to the vestibular organs, the spiral and vestibular ganglia regions are less studied in the reviewed Cre alleles. Here, we list what is known about Cre activity in these regions; however, with the exception of the SHH-eGFP/Cre line, very little detail has been reported.
The Atoh1-Cre transgenic line showed Cre activity in both the spiral ganglion and vestibular ganglion neurons (Matei et al. 2005); however, this was not detected in the Atoh1-Cre knock-in allele (Yang et al. 2010a).
Cre activity was detected in the majority of cells in the spiral ganglia region in the Prestin-Cre (transgenic) line (Li et al. 2004), while only a small fraction were Cre+ in the Prestin-Cre (BAC transgenic) allele (Tian et al. 2004). In addition, the Prestin-Cre (BAC transgenic) allele had Cre activity in the vestibular ganglia (Tian et al. 2004). No Cre+ cells were found in these neuronal regions using the Prestin-CreERT2 allele (Fang et al. 2012).
The SHH-eGFP/Cre knock-in allele was created by inserting an eGFP/Cre fusion protein into the SHH locus, replacing the first 12 amino acids of SHH and creating a null allele. Heterozygous mice exhibited no noticeable phenotypes (Harfe et al. 2004). The expression of eGFP was used to detect SHH expression in spiral ganglion neurons between E13.5 and E17.5. All spiral ganglion neurons expressed eGFP at E13.5, followed by the loss of eGFP expression progressing from the base to the apex. Using the ROSA26eYFP reporter line, Cre expression at P0 was detected in almost 100 % of spiral ganglion neurons and in some peripheral nerve fibers surrounding HCs and SCs. Surrounding glial cells were Cre-negative. Vestibular ganglia were not investigated. The basal-to-apical decline in SHH-eGFP expression in spiral ganglion neurons occurs at a similar time as the basal-to-apical gradient of cochlear HC differentiation, which suggests that SHH signaling may inhibit HC differentiation (Liu et al. 2010).
Cre/CreER lines in non-sensory regions of the inner ear
Another Cre line from the POU domain family of transcription factors, Pou3f4-Cre, has a very different Cre expression pattern compared to the Pou4f3-Cre allele. The Pou3f4 enhancer was fused to the herpes thymidine kinase promoter to drive Cre expression in the Pou3f4-Cre transgenic allele. When analyzed with the ROSA26LacZ reporter, βgal expression was detected in the otic mesenchyme as early as E14.5. In adult mice, Cre+ cells were found in structures derived from otic mesenchyme, including the temporal bone, spiral ligament, spiral limbus, tympanic border cells, Reissner’s membrane, and mesenchymal cells that lie underneath the sensory epithelia of the cochlea and vestibular organs. The non-sensory epithelium of the utricle was also Cre+, as well as a few cells in the spiral ganglion region. Cre activity was not detected in structures derived from the embryonic otic epithelium, such as the organ of Corti, the sensory epithelia of the vestibular organs, and the stria vascularis (Ahn et al. 2009).
Cre/CreER lines with ubiquitous expression
There are two knock-in alleles that use the ROSA26 locus to drive the expression of CreERTM. Both have ubiquitous expression in all tissues of the body; however, the number of Cre+ cells varies depending on the tissue and tamoxifen induction paradigm used. Deletion of connexin26 using the ROSA26-CreERTM allele (Vooijs et al. 2001) and tamoxifen injection at E19 caused outer HC and SC death beginning at the onset of hearing (P14). In addition, the tunnel of Corti and spaces of Nuel never opened. These results demonstrate that connexin26 plays a role in the postnatal maturation and survival of cells in the organ of Corti (Sun et al. 2009; Wang et al. 2009). The second ROSA26-CreERTM allele (Badea et al. 2003) was used to delete Kif3a, a ciliary protein, at E10.5. The HCs in Kif3a conditional knockout mice had stereocilia bundles with planar cell polarity defects that were similar to a mutant mouse model of another ciliary protein, Ift88 (also called Polaris). Together, these results demonstrate the requirement of ciliary genes for the regulation of planar cell polarity in HC stereocilia bundles (Jones et al. 2008).
The CAG promoter has also been used to generate mouse lines with ubiquitous expression of Cre/CreER. The transgenic CAG-Cre allele generates Cre-mediated recombination before the two-cell stage of the embryo (Sakai and Miyazaki 1997). The transgenic CAG-CreERTM allele is likely more useful since Cre expression can be induced at any age. There is a tamoxifen dose-dependent level of Cre recombination in this line, as well as a low percentage of cells that undergo spontaneous Cre-mediated recombination in the absence of tamoxifen (called “leakiness”; Hayashi and McMahon 2002; Oesterle et al. 2011). Ectopic expression of NICD induced by CAG-CreERTM at E10.5 resulted in the formation of ectopic HC/SC patches in the non-sensory regions of both the cochlea and utricle; however, when tamoxifen was given at E13, ectopic HC/SC patches were only found in utricular non-sensory regions (Liu et al. 2012a). Similarly, ectopic expression of NICD induced by CAG-CreERTM in cultured cochlear explants at E13.5 did not result in the formation of ectopic HCs or SCs (Basch et al. 2011). When tamoxifen was given once a day at P0 and P1, no ectopic HCs were observed in either the cochlea or utricle (Liu et al. 2012a). Taken together, these results suggest that the ability of Notch signaling to produce HCs and SCs is age-dependent and differs between the cochlea and vestibular system. CAG-CreERTM-mediated deletion of the transcription factor, Tbx1, with tamoxifen injections at E10.5 and E11.5 resulted in a smaller utricle and saccule organs and a “cochlea” that lacked the spiral shape, but still had some HCs while the semicircular canal cristae failed to form. When Tbx1 deletion occurred with a later tamoxifen induction (E14.5), the cochlea appeared normal and the semicircular canal cristae were underdeveloped (Xu et al. 2007). This CreER line was also used to delete the cell cycle inhibitor, p27Kip1, with tamoxifen induction at P3 that resulted in the proliferation of cochlear SCs. SCs also proliferated when tamoxifen was administered at 6 weeks of age, but to a lesser extent. These findings demonstrate that p27Kip1 is required to maintain the quiescence of postnatal SCs and that dependence on p27Kip1 declines with age (Oesterle et al. 2011).
The transgenic Sox2-Cre allele is another ubiquitously expressed Cre line that uses a 12.5-kb enhancer/promoter element from Sox2 to drive the expression of Cre. Using the ROSA26LacZ reporter, βgal expression was observed at E7.5 in all three germ layers of the mouse embryo as well as in epiblast-derived extra-embryonic membranes (Hayashi et al. 2002).
Potentially useful Cre/CreER lines for inner ear research
There are hundreds of Cre/CreER lines that have been made for research in various organs of the body and may also be useful for studies in the inner ear (Table 3). For example, the knock-in Shh-CreERT2 allele (Harfe et al. 2004), the transgenic Sox2-CreER line (Favaro et al. 2009; Arnold et al. 2011), and the knock-in Lgr5-eGFP-IRES-CreERT2 line (Barker et al. 2007) are all driven by promoters with known expression in the inner ear. In addition, the NIH Neuroscience Blueprint Cre-Driver Network (http://www.credrivermice.org/index) has generated many new Cre/CreER alleles that may be useful for research in the inner ear (i.e., Otoferlin-Cre, http://www.informatics.jax.org/javawi2/servlet/WIFetch?page=alleleDetail&key=667536; Otoferlin-CreERT2, http://www.informatics.jax.org/javawi2/servlet/WIFetch?page=alleleDetail&key=667535; and Calbindin2-CreERT2 (calbindin2 is also known as calretinin), http://www.informatics.jax.org/searches/accession_report.cgi?id=MGI:4880758).
TABLE 3.
Potentially useful Cre/CreER lines for inner ear research
| Conditional allele | Type | Original citation | Source |
|---|---|---|---|
| Atoh1-CrePR | Knock-in | Rose et al. (2009) | Jax stock #13594 |
| Calretinin-CreER | Knock-in | Taniguchi et al. (2011) | Jax stock #13730 |
| Calretinin-IRES-Cre | Knock-in | Taniguchi et al. (2011) | Jax stock #10774 |
| Dlx5-CreER | Knock-in | Taniguchi et al. (2011) | Jax stock #10705 |
| Gli1-CreER | Knock-in | (Ahn et al. 2004) | Jax stock #7913 |
| Hes1-CreER | Knock-in | Kopinke et al. (2011) | Not commercially available |
| Id2-CreER | Knock-in | Rawlins et al. (2009) | Jax stock #16222 |
| Isl1-IRES-Cre | Knock-in | Srinivas et al. (2001) | Not commercially available |
| Isl1-mER-Cre-mER | Knock-in | Laugwitz et al. (2005) | Not commercially available |
| Lgr5-eGFP-IRES-CreERT2 | Knock-in | Barker et al. (2007) | Jax stock #8875 |
| Notch1-Cre | Knock-in | Vooijs et al. (2007) | Jax stock #6953 |
| Otoferlin-Cre | Knock-in | N/A | MMRRC stock #032781-MU |
| Otoferlin-CreER | Knock-in | N/A | MMRRC stock #032782-MU |
| Parvalbumin-2A-Cre | Knock-in | Madisen et al. (2010) | Jax stock #12358 |
| Parvalbumin-CreER | Knock-in | Taniguchi et al. (2011) | Jax stock #10777 |
| Shh-CreERT2 | Knock-in | Harfe et al. (2004) | Jax stock #5623 |
| Sox2-CreER | Knock-in | Arnold et al. (2011) | Not commercially available |
| Sox2-CreER | Transgenic | Favaro et al. (2009) | Not commercially available |
| Sox9-CreER | Transgenic | Kopp et al. (2011) | Not commercially available |
Practical issues related to conditional gene expression
Nature of conditional alleles
In general, the Cre expression pattern is the most important factor when deciding which Cre/CreER allele to use in your research. It is also important to consider the type of conditional allele (knock-in, transgenic, or BAC transgenic). Comparison of the three strains that use the prestin promoter to drive Cre expression can illustrate the differences between the different types of conditional alleles. The Prestin-Cre (transgenic) allele and Prestin-Cre (BAC transgenic) allele both show Cre activity in cell types where the endogenous prestin protein is not detected, such as inner HCs, vestibular HCs, and spiral ganglion neurons (Li et al. 2004; Tian et al. 2004). However, the knock-in Prestin-CreERT2 line has Cre activity that is very specific to the outer HCs of the cochlea, where endogenous prestin is found (Fang et al. 2012). The most likely explanation is that the regulatory elements which regulate the prestin promoter are quite far from the promoter region and thus are not part of either transgene. In addition, unlike most knock-in alleles, the Prestin-CreERT2 line is not a null allele of prestin. Instead, it uses IRES to drive the expression of CreERT2; thus, endogenous prestin is still made and homozygous Prestin-CreERT2 mice have normal expression of prestin (Fang et al. 2012).
Comparison of the two Atoh1-Cre alleles provides another example that illustrates the difference between knock-in and transgenic lines. Cre expression from the knock-in Atoh1-Cre line was first detected in the basal turn of the cochlea at E14.5 which corresponds with the Atoh1 in situ results that were first detected in the basal turn at E13.5 (Yang et al. 2010a). In contrast, Cre expression from the transgenic Atoh1-Cre allele was detected at E11 in the developing sensory epithelium (Matei et al. 2005). The two transgenic Atoh1-CreER alleles also have varying Cre expression patterns. With the same tamoxifen induction paradigm and reporter line, the Atoh1-CreERTM allele has Cre activity in 80–90 % of HCs (Chow et al. 2006; Weber et al. 2008), while the Atoh1-CreERT2 line has Cre activity in 20 % or less of HCs depending on the turn of the cochlea (Fig. 3). Given that both lines use the Atoh1 enhancer to drive Cre expression, it is tempting to think of them as equal and assume that the Atoh1-CreERT2 line would be better since CreERT2 has a higher affinity for tamoxifen. However, these strains are transgenics; thus, positional effects and the copy number of the transgene likely play a role in the difference of Cre activity observed. Therefore, it is very important to empirically test each Cre line with reporter lines to correctly interpret the effects of specific gene manipulations.
Cre efficiency
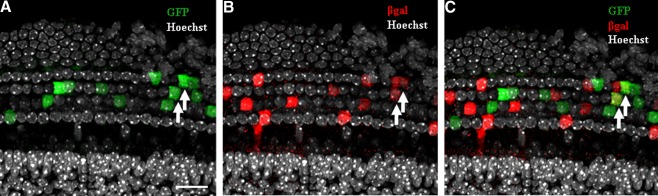
There are many other factors to consider when making conclusions from data obtained using Cre/CreER alleles, such as Cre efficiency, reporter expression efficiency, and Cre leakiness. The ability of the Cre recombinase enzyme to excise DNA between loxP sites is referred to as Cre efficiency or recombination frequency. Both cellular Cre and nuclear CreER levels and the number of loxP sites in the genome can be important for Cre efficiency. Even though only a few Cre molecules per cell are needed to excise floxed DNA (Nagy 2000), the expression level of the promoter driving Cre can affect Cre efficiency (Araki et al. 1997). In addition, Cre recombination efficiency can be affected by the location of loxP sites within the GOI and the distance between loxP sites (Zheng et al. 2000; Vooijs et al. 2001). For CreER alleles, the dose and timing of tamoxifen injections can affect the level of CreER activity (Vooijs et al. 2001). When CreER levels are relatively low, Cre efficiency can be variable or mosaic for more than one floxed locus. To illustrate this point, we bred the Prox1-CreERT2 allele with both the CAGeGFP and ROSA26lacZ reporter lines. In the cochlea of Prox1-CreERT2;CAGeGFP;ROSA26lacZ mice, the majority of cells are either GFP+ or βgal+, while only a few cells express both reporter molecules (Fig. 6). Thus, the majority of cells in this model only have enough Cre activity to excise one floxed allele and only a few cells display Cre activity for both floxed alleles. This may have significant implications when using two independent floxed genes (i.e., one floxed GOI and one reporter for tracing purposes). Moreover, it is well known that homozygous floxed alleles of the same gene may exhibit differential excision efficiency; therefore, it is highly recommended that instead of using homozygous floxed alleles, a deleted (delta) allele and a floxed allele of the same gene should be used to ensure complete deletion when a weak Cre line is used, such as Prox1-CreERT2.
FIG. 6.
Illustration of Cre efficiency using a low efficiency Cre and two reporter lines. A–C Representative confocal images of the organ of Corti of Prox1-CreERT2;CAGeGFP;ROSA26lacZ mice, induced with tamoxifen at P0 and P1. Many cells expressed either GFP (green) (A) or lacZ (βgal antibody labeling in red) (B), while only two cells were both GFP+ and lacZ+ (arrows). Nuclei are labeled with Hoechst in white. Scale bar, 20 μm.
Another application of a Cre line with low efficiency is to generate a mosaic of Cre+ and Cre-negative cells in the same tissue. Mosaic Cre expression patterns can also be created using an inducible CreER allele by giving lower doses of tamoxifen to activate CreER in fewer cells. The use of mosaic Cre expression to delete a GOI provides the opportunity to determine whether the effects of GOI inactivation are cell autonomous or non-cell autonomous (Weber et al. 2008; Yu et al. 2010).
Reporter expression efficiency
Reporter expression efficiency is dependent upon Cre efficiency, but also relies on the expression level of the promoter driving the reporter gene and the detection sensitivity of the reporter molecule. A good reporter line should express reporter genes ubiquitously in all tissues, and the reporter molecule should have a high level of sensitivity. The ROSA26 locus was thought to be ubiquitously expressed in every cell of the mouse; however, recent evidence suggests that it is downregulated in the adult. Thus, cells with no detectable reporter expression may still have Cre activity. To address this issue, new Cre reporter lines were created where the ROSA26 locus was modified by the insertion of a construct containing the CAG promoter followed by a floxed “stop” sequence and the brightest fluorescent proteins available, ZsGreen or tdTomato. When directly compared to the ROSA26eYFP reporter line using several different Cre lines in the brain, these reporter lines, termed Ai6 for ZsGreen and Ai9 or Ai14 for tdTomato, had substantially stronger fluorescence and labeled more Cre+ cells than expected from previous reports (Madisen et al. 2010).
We performed similar studies in the utricle using Plp-CreERT2;ROSA26eYFP mice compared to Plp-CreERT2;Ai14 mice. After tamoxifen (3 mg/40 g, IP) induction at P0 and P1, we found a much larger number of Cre+ HCs and SCs using the Ai14 reporter compared to the eYFP reporter (Fig. 7). Since most of the Cre/CreER alleles described in this review were not characterized with one of these new Ai reporter lines, it is likely that current reports are an underestimate of their Cre activity. It is also worth noting that the ZsGreen fluorescence in the Ai6 line has uneven distribution at adult ages with punctate staining in the cytoplasm that affects the quality of the image.
FIG. 7.
Comparison of the ROSA26eYFP and the Ai14 reporter lines in the utricle. Representative confocal images of the utricle of Plp-CreERT2;ROSA26eYFP mice (A, B) and Plp-CreERT2;Ai14 mice (C, D) both induced with tamoxifen at P0 and P1. HCs labeled by myosin VIIA (myo7a, green), while YFP and tdTomato are in red. Scale bars, 100 μm (A, C) and 20 μm (B, D).
Similar efficiency issues apply when deleting the floxed region in a GOI, and one should not assume that the percentage of reporter+ cells will be the same as the percentage of GOI-deleted cells. If possible, it is best to perform immunostaining of the GOI to confirm the extent of the deletion. In addition, when using inducible alleles to delete genes postnatally, it is important to consider the protein half-life of the GOI since it may be very stable in a mature cell (Hayashi and McMahon 2002). Cre-mediated excision occurs at the level of the genome; thus, it takes time for the protein that was made prior to tamoxifen induction to be degraded to see a phenotype caused by deletion of the GOI.
Cre leakage
Another important issue to consider with inducible Cre lines is “leakage.” It is possible that some CreER molecules are translocated to the nucleus in the absence of tamoxifen. Although Cre “leakage” effect is uncommon, it needs to be empirically tested for each CreER line (Zuo 2002). This can be easily done with controls using a reporter allele without tamoxifen injection. In fact, the CAG-CreERTM allele has been reported to have a low level of Cre leakiness (Hayashi and McMahon 2002; Oesterle et al. 2011).
Strain background
Strain background effects also need to be considered when making conclusions. Due to high efficiency for gene targeting, 129 ES cells are commonly used to generate knock-in alleles and offspring after germline transmission are often mixed with the C57Bl/6 background. Both of these strains are not ideal for hearing research due to the Ahl loci which causes early onset of age-related hearing loss (Johnson and Zheng 2002; Ohlemiller 2004); however, these strains have normal hearing before 2 months of age (Zheng et al. 1999). If animals older than 2 months are needed for a study, it would be best to back-cross the desired lines to the CBA/CaJ strain, which is the “gold standard” for auditory research (Erway et al. 1996; Zheng et al. 1999; Johnson et al. 2000; Yoshida et al. 2000; Johnson and Zheng 2002; Willott et al. 2003). The recent KOMP effort was designed to create null and floxed alleles for every gene in the genome using the C57Bl/6 background because it is the most widely used strain and the complete genome sequence is available (Austin et al. 2004). Thus, strain effects with C57Bl/6 in hearing research should be taken into consideration when taking advantage of these valuable resources from KOMP or other consortiums. In addition, strain background can have an effect on Cre efficiency and the pattern of Cre expression (Hebert and McConnell 2000).
Toxicity
The scientific community has recently expressed concern that the expression of Cre might have toxic effects to cells, which is unrelated to the deletion of the GOI. The most appropriate control to measure potential Cre toxicity is to use the same Cre allele without the floxed GOI (Editorial 2007). There is also concern that tamoxifen treatment may have toxic effects. While tamoxifen does impair reproductive development (Gill-Sharma et al. 2001; Mehasseb et al. 2009), no effects on hearing sensitivity, as measured by ABR, have been reported (Li et al. 2004; Tian et al. 2004; Chow et al. 2006; Yu et al. 2010).
Fate mapping
Fate mapping (or lineage tracing) takes advantage of the permanent gene deletion caused by Cre-mediated recombination. Immunostaining of the Cre protein cannot be used for fate mapping because the Cre enzyme is only expressed while the cell type-specific promoter is active. The use of cell type-specific Cre/CreER alleles in combination with a reporter allele will permanently label Cre+ cells regardless of whether they change cell fate or if the promoter driving Cre is downregulated. Therefore, daughter cells (progeny) from Cre+ cells will continue to have the reporter label since the deletion of the floxed DNA sequence occurs at the level of the genome and is passed on (Nagy 2000). This concept is advantageous when trying to investigate the cell of origin for a structure, tumor, or regenerated cell population since cell-specific markers may be turned off with time or with cell fate changes. Many use the terms fate mapping and lineage tracing interchangeably, and while the basic concepts hold true for both, lineage tracing is fate mapping performed at the single-cell level. It is also important to note that if the goal is to determine the cell of origin during development, it is best to use a knock-in Cre/CreER line so that the Cre expression pattern faithfully recapitulates endogenous expression of the promoter driving Cre/CreER. However, if the goal is to trace cells when there is a cell fate change (i.e., after ectopic expression NICD or in HC regeneration studies), a knock-in Cre/CreER line is not required. Control samples of the same age without the gene manipulation or drug treatment can be used to show what cell types normally have Cre expression. For example, the Ngn1-CreERT2 line was used to trace the progeny of Ngn1-expressing cells after the sensory epithelia were formed (Raft et al. 2007), and the Atoh1-Cre (knock-in) line was used to conclude that Atoh1 expression occurs in the common progenitor cells that give rise to both HCs and SCs (Yang et al. 2010a).
Complex uses of Cre technology
As discussed previously, Cre-mediated recombination will excise DNA between floxed regions when two loxP sites are on the same strand of DNA and are in the same orientation. Mutations in the loxP sequence have been created to generate incompatible or mismatched loxP sites where the efficiency of recombination is reduced (Langer et al. 2002). This concept was used with loxP and loxH to generate a mosaic expression of iCsp3 (Fujioka et al. 2011). In addition, Brainbow 1.0 reporter lines use two loxP sites and two mutated loxP sites, allowing two possible recombination outcomes to occur in equal proportion. Brainbow 1.0 cells are red and in the presence of Cre become either yellow or cyan, depending on which segment of DNA is deleted. Brainbow 1.1 lines added to this complexity with an additional set of mutated loxP sites and a third reporter gene. Additional colors are possible when more than one copy of the Brainbow transgene is inserted into the genome because each construct recombines independently, allowing more than one fluorescent protein to be expressed in a single cell (Livet et al. 2007).
Cre-mediated recombination can also result in the inversion of DNA between loxP sites when the loxP sites are in opposite directions (i.e., loxP sites face either other). Brainbow 2.0 utilizes this concept with a construct where red fluorescent protein (RFP) is in the sense direction, cyan fluorescent protein (CFP) is in the antisense orientation, and both are surrounded by loxP sites facing each other. Thus, RFP is normally expressed, and when Cre-mediated recombination occurs, cells stop making RFP and begin making CFP. Brainbow 2.1 mice have two invertible DNA segments positioned in tandem to generate three different possible recombination outcomes which occur stochastically. Both Brainbow transgenic lines produce neurons in a variety of colors, which allow tracing of neuronal processes at the individual level and visualization of their synaptic interactions (Livet et al. 2007). These lines are driven by the Thy1 promoter and may be useful for neuronal tracing in the inner ear; however, none of the Brainbow lines had reporter expression in cochlear HCs or SCs when appropriate Cre lines (Atoh1-CreERTM and Prox1-CreERT2) were used (Fang et al. 2009).
Recently, a knock-in R26R-Confetti mouse line was generated and first used in the intestine. Instead of the Thy1 promoter, the Brainbow 2.1 cassette was inserted into the ROSA26 locus with the addition of a strong CAG promoter and a floxed “stop” codon. Since there are five loxP sites, there are ten recombination outcomes, six of which include the “stop” codon and four of which produce a fluorescent protein (CFP, GFP, YFP, or RFP; Snippert et al. 2010). Since the ROSA26 locus is known to be active in the inner ear, this line is expected to give reporter expression in various cell types, including HCs and SCs.
The Brainbow and R26-Confetti mouse lines offer advantages over single-color reporter lines because tracing at the single-cell level (lineage tracing) can be performed when CreER lines are used. This was recently done in the intestine where Lgr5+ cells were labeled with the R26-Confetti mouse. Soon after tamoxifen induction, cells of various colors were observed due to the random recombination of the R26-Confetti allele; however, after several weeks, each intestinal crypt was labeled by a single color, demonstrating that it was populated by a single Lgr5+ cell (Snippert et al. 2010). Similar studies could be performed in the inner ear. For example, prosensory progenitor cells could be labeled with the R26-Confetti line to show how many HCs and SCs are derived from a single progenitor cell. In addition, the fluorescent proteins in Brainbow mice are also present in neuronal processes and synapses; thus, this line could be used with a spiral/vestibular ganglion CreER allele to visualize the innervation pattern of a single neuron.
Other methods for conditional gene expression
In addition to Cre recombinase, Flp recombinase from yeast has been used to generate conditional alleles in mice. Similar to recombination between loxP sites by Cre, Flp recognizes FRT sites and can excise, invert, or translocate the fragment of DNA between FRT sites in a permanent manner (Dymecki 1996). To optimize Flp recombinase activity, a more efficient Flpe enzyme was made that has higher activity at physiological temperatures (Buchholz et al. 1998). Flpe was further modified with the addition of a nuclear localization sequence which improved its performance in mammalian cells (Schaft et al. 2001). In 2007, a codon-optimized version of Flpe, termed Flpo, was generated. In mammalian cells, Flpo is reported to have five times more activity than Flpe (Raymond and Soriano 2007; Kranz et al. 2010). Currently, no Flp mouse lines have been reported with specific activity in the inner ear, although several attempts were made to express Flp specifically in cochlear HCs (Dearman and Zuo 2010). We are also not aware of any inducible Flp mouse lines.
To transiently alter gene expression, the tetracycline-inducible system is used to either repress or activate gene transcription. The tetracycline transactivator (tTA) is a fusion protein that combines the tet repressor-binding domain and an activation domain of VP16, a transcription factor from herpes simplex virus (Gossen and Bujard 1992). It binds to and activates the promoter of a second transgene which contains a tetracycline response element (TRE). When tetracycline or doxycycline is given, it binds to tTA and prevents tTA from binding to the TRE sequence, therefore blocking transcription (also called Tet-off system). Mutations in four amino acids of the tet repressor-binding domain of tTA generate a reverse tetracycline transactivator (rtTA) which works in the opposite manner to activate transcription in the presence of tetracycline or doxycycline (also called Tet-on system; Gossen et al. 1995). Both tTA and rtTA have been shown to work in transgenic mice, where doxycycline is given in the food and/or drinking water (Kistner et al. 1996). The cell type specificity comes from the use of cell type-specific promoters to express the tTA or rtTA protein. An important hallmark of the Tet-on/off systems is that the expression of the GOI can be transiently activated or inactivated only when doxycycline is present. When doxycycline is withdrawn, expression of the GOI will be reversed. Doxycycline will effectively cross the blood–labyrinth barrier, as demonstrated by the use of the Tet-on system in the cochlea to study ectopic expression of NICD (Pan et al. 2010). For gene functional studies in the inner ear, it often becomes essential, albeit challenging, to combine two independent methods (i.e., Cre-loxP and Tet-on/off) in a single mouse.
Concluding remarks
Many significant advances have been made in the inner ear using conditional gene expression and Cre-loxP technology. In addition to fate mapping with reporter lines to determine the cell origin of various inner ear cell types, temporal control of gene expression has led to the discovery of age-dependent changes in the function of a particular gene (i.e., Rb and Notch signaling). Here, we summarize 32 Cre/CreER lines with expression in the developing or postnatal inner ear to provide a resource for auditory and vestibular researchers. For postnatal studies, there are many good alleles to study specific cell types in the cochlea; however, this is not the case for vestibular organs. Better characterization, as well as more Cre/CreER lines, is needed. In addition, the majority of the Cre/CreER lines we summarized were characterized using the ROSA26LacZ reporter, which likely underestimated their Cre activity. It would be wise to repeat Cre expression pattern analysis in future studies using one of the Ai reporter lines.
Since conditional gene expression is a growing field and a young one for inner ear research, we have also provided a table of 19 Cre/CreER alleles that may be useful in auditory and vestibular organs, but have not been characterized yet. The NIH Neuroscience Blueprint Cre-Driver Network will continue to create additional strains in the future. We anticipate that Flp and tetracycline-inducible strains expressed in specific cell types of the inner ear will also be created in the future and can be combined with the Cre-loxP system for more complex approaches. In summary, conditional gene expression offers much opportunity to expand our knowledge of the inner ear and build upon the findings obtained using germline knockout mice.
Acknowledgments
We thank Betsy Williford in the Biomedical Communications Department of St. Jude Children’s Research Hospital for her assistance in creating the artwork for figures. This work was supported in part by grants from the National Institute of Health (DC006471 (J.Z.), DC008800 (J.Z.), DC010310 (B.C.C.) and CA21765); the Office of Naval Research (N000140911014); the Sir Henry Wellcome Trust fellowship (M.M.M.L.); and the American Lebanese Syrian Associated Charities (ALSAC) of St. Jude Children’s Research Hospital. J. Zuo is a recipient of The Hartwell Individual Biomedical Research Award.
Glossary
- ABR
Auditory brainstem response
- BAC
Bacterial artificial chromosome
- βgal
Beta-galactosidase
- BMP
Bone morphogenetic protein
- BDNF
Brain-derived neurotrophic factor
- CFP
Cyan fluorescent protein
- Col1A1
Alpha (1) collagen promoter
- Col2A1
Type II collagen promoter
- DTA
Diphtheria toxin fragment A
- E
Embryonic day
- eGFP
Enhanced green fluorescent protein
- ES
Embryonic stem cells
- GER
Greater epithelial ridge
- GFAP
Glial fibrillary acidic protein
- GFP
Green fluorescent protein
- GOI
Gene of interest
- Hsp90
Heat shock protein 90
- iCsp3
Drug-inducible dimerizable caspase 3
- IRES
Internal ribosome entry site
- KOMP
NIH knockout mouse project
- LER
Lesser epithelial ridge
- myo7a
Myosin VIIa
- NICD
Notch intracellular domain
- P
Postnatal day
- PAC
Phage artificial chromosome
- Pkd1
Polycystic kidney disease 1
- Rb
Retinoblastoma protein
- RFP
Red fluorescent protein
- rtTA
Reverse tetracycline transactivator
- SHH
Sonic hedgehog
- TRE
Tetracycline responsive element
- tTA
Tetracycline transactivator
References
- Editorial (2007) Toxic alert. Nature 449:378 [DOI] [PubMed]
- Abraira VE, Satoh T, Fekete DM, Goodrich LV. Vertebrate Lrig3–ErbB interactions occur in vitro but are unlikely to play a role in Lrig3-dependent inner ear morphogenesis. PLoS One. 2010;5:e8981. doi: 10.1371/journal.pone.0008981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed M, Wong EY, Sun J, Xu J, Wang F, Xu PX. Eya1–Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Dev Cell. 2012;22:377–390. doi: 10.1016/j.devcel.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn S, Joyner AL. Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell. 2004;118:505–516. doi: 10.1016/j.cell.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Ahn KJ, Passero F, Jr, Crenshaw EB., 3rd Otic mesenchyme expression of Cre recombinase directed by the inner ear enhancer of the Brn4/Pou3f4 gene. Genesis. 2009;47:137–141. doi: 10.1002/dvg.20454. [DOI] [PubMed] [Google Scholar]
- Akagi K, Sandig V, Vooijs M, Valk M, Giovannini M, Strauss M, Berns A. Cre-mediated somatic site-specific recombination in mice. Nucleic Acids Res. 1997;25:1766–1773. doi: 10.1093/nar/25.9.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, Imaizumi T, Okuyama K, Oike Y, Yamamura K. Efficiency of recombination by Cre transient expression in embryonic stem cells: comparison of various promoters. J Biochem. 1997;122:977–982. doi: 10.1093/oxfordjournals.jbchem.a021860. [DOI] [PubMed] [Google Scholar]
- Arnold JS, Braunstein EM, Ohyama T, Groves AK, Adams JC, Brown MC, Morrow BE. Tissue-specific roles of Tbx1 in the development of the outer, middle and inner ear, defective in 22q11DS patients. Hum Mol Genet. 2006;15:1629–1639. doi: 10.1093/hmg/ddl084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R, Sengupta S, Seandel M, Geijsen N, Hochedlinger K. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell. 2011;9:317–329. doi: 10.1016/j.stem.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin CP, et al. The Knockout Mouse Project. Nat Genet. 2004;36:921–924. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auwerx J, et al. The European dimension for the mouse genome mutagenesis program. Nat Genet. 2004;36:925–927. doi: 10.1038/ng0904-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea TC, Wang Y, Nathans J. A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J Neurosci. 2003;23:2314–2322. doi: 10.1523/JNEUROSCI.23-06-02314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahloul A, Simmler MC, Michel V, Leibovici M, Perfettini I, Roux I, Weil D, Nouaille S, Zuo J, Zadro C, Licastro D, Gasparini P, Avan P, Hardelin JP, Petit C. Vezatin, an integral membrane protein of adherens junctions, is required for the sound resilience of cochlear hair cells. EMBO Mol Med. 2009;1:125–138. doi: 10.1002/emmm.200900015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, Es JH, Kuipers J, Kujala P, Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Barrionuevo F, Naumann A, Bagheri-Fam S, Speth V, Taketo MM, Scherer G, Neubuser A. Sox9 is required for invagination of the otic placode in mice. Dev Biol. 2008;317:213–224. doi: 10.1016/j.ydbio.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Basch ML, Ohyama T, Segil N, Groves AK. Canonical Notch signaling is not necessary for prosensory induction in the mouse cochlea: insights from a conditional mutant of RBPjkappa. J Neurosci. 2011;31:8046–8058. doi: 10.1523/JNEUROSCI.6671-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard M, Souabni A, Mandler M, Neubuser A, Busslinger M. Nephric lineage specification by Pax2 and Pax8. Genes Dev. 2002;16:2958–2970. doi: 10.1101/gad.240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard M, Souabni A, Busslinger M. Tissue-specific expression of Cre recombinase from the Pax8 locus. Genesis. 2004;38:105–109. doi: 10.1002/gene.20008. [DOI] [PubMed] [Google Scholar]
- Brooker R, Hozumi K, Lewis J. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development. 2006;133:1277–1286. doi: 10.1242/dev.02284. [DOI] [PubMed] [Google Scholar]
- Brown AS, Epstein DJ. Otic ablation of smoothened reveals direct and indirect requirements for Hedgehog signaling in inner ear development. Development. 2011;138:3967–3976. doi: 10.1242/dev.066126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz F, Angrand PO, Stewart AF. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat Biotechnol. 1998;16:657–662. doi: 10.1038/nbt0798-657. [DOI] [PubMed] [Google Scholar]
- Burgess-Beusse B, Farrell C, Gaszner M, Litt M, Mutskov V, Recillas-Targa F, Simpson M, West A, Felsenfeld G. The insulation of genes from external enhancers and silencing chromatin. Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16433–16437. doi: 10.1073/pnas.162342499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Johnson JE, Zoghbi HY, Segil N. The role of Math1 in inner ear development: uncoupling the establishment of the sensory primordium from hair cell fate determination. Development. 2002;129:2495–2505. doi: 10.1242/dev.00114. [DOI] [PubMed] [Google Scholar]
- Chen CM, Krohn J, Bhattacharya S, Davies B. A comparison of exogenous promoter activity at the ROSA26 locus using a PhiC31 integrase mediated cassette exchange approach in mouse ES cells. PLoS One. 2011;6:e23376. doi: 10.1371/journal.pone.0023376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow LM, Tian Y, Weber T, Corbett M, Zuo J, Baker SJ. Inducible Cre recombinase activity in mouse cerebellar granule cell precursors and inner ear hair cells. Dev Dyn. 2006;235:2991–2998. doi: 10.1002/dvdy.20948. [DOI] [PubMed] [Google Scholar]
- Coate TM, Raft S, Zhao X, Ryan AK, Crenshaw EB, 3rd, Kelley MW. Otic mesenchyme cells regulate spiral ganglion axon fasciculation through a Pou3f4/EphA4 signaling pathway. Neuron. 2012;73:49–63. doi: 10.1016/j.neuron.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Salmon M, Ott T, Michel V, Hardelin JP, Perfettini I, Eybalin M, Wu T, Marcus DC, Wangemann P, Willecke K, Petit C. Targeted ablation of connexin26 in the inner ear epithelial gap junction network causes hearing impairment and cell death. Curr Biol. 2002;12:1106–1111. doi: 10.1016/S0960-9822(02)00904-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox BC, Lenoir A, Zhang L, Steigelman KA, Zuo J (2010) Hair cell damage in the neonatal mouse cochlea using forced expression of diphtheria toxin. MidWinter Research Meeting of the Association for Research in Otolaryngology, Anaheim, CA
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/S0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Dearman J, Zuo J (2010) Creating a hair cell specific Flp transgenic mouse. MidWinter Research Meeting of the Association for Research in Otolaryngology, Anaheim, CA
- Deng M, Pan L, Xie X, Gan L. Requirement for Lmo4 in the vestibular morphogenesis of mouse inner ear. Dev Biol. 2010;338:38–49. doi: 10.1016/j.ydbio.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerflinger NH, Macklin WB, Popko B. Inducible site-specific recombination in myelinating cells. Genesis. 2003;35:63–72. doi: 10.1002/gene.10154. [DOI] [PubMed] [Google Scholar]
- Doetzlhofer A, Basch ML, Ohyama T, Gessler M, Groves AK, Segil N. Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev cell. 2009;16:58–69. doi: 10.1016/j.devcel.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Frutos E, Lopez-Hernandez I, Vendrell V, Neves J, Gallozzi M, Gutsche K, Quintana L, Sharpe J, Knoepfler PS, Eisenman RN, Trumpp A, Giraldez F, Schimmang T. N-myc controls proliferation, morphogenesis, and patterning of the inner ear. J Neurosci. 2011;31:7178–7189. doi: 10.1523/JNEUROSCI.0785-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymecki SM. Flp recombinase promotes site-specific DNA recombination in embryonic stem cells and transgenic mice. Proc Natl Acad Sci U S A. 1996;93:6191–6196. doi: 10.1073/pnas.93.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagleson KL, Schlueter McFadyen-Ketchum LJ, Ahrens ET, Mills PH, Does MD, Nickols J, Levitt P. Disruption of Foxg1 expression by knock-in of Cre recombinase: effects on the development of the mouse telencephalon. Neuroscience. 2007;148:385–399. doi: 10.1016/j.neuroscience.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engleka KA, Gitler AD, Zhang M, Zhou DD, High FA, Epstein JA. Insertion of Cre into the Pax3 locus creates a new allele of Splotch and identifies unexpected Pax3 derivatives. Dev Biol. 2005;280:396–406. doi: 10.1016/j.ydbio.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Erway LC, Shiau YW, Davis RR, Krieg EF. Genetics of age-related hearing loss in mice. III. Susceptibility of inbred and F1 hybrid strains to noise-induced hearing loss. Hear Res. 1996;93:181–187. doi: 10.1016/0378-5955(95)00226-X. [DOI] [PubMed] [Google Scholar]
- Fang J, Dearman J, Zuo J (2009) Characterization of the Brainbow transgenic labeling system in the mouse cochlea. Midwinter Research Meeting of the Association for Research in Otolaryngology, Baltimore, MD
- Fang J, Zhang WC, Yamashita T, Gao J, Zhu MS, Zuo J. Outer hair cell-specific prestin-CreER(T2) knockin mouse lines. Genesis. 2012;50:124–131. doi: 10.1002/dvg.20810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro R, Valotta M, Ferri AL, Latorre E, Mariani J, Giachino C, Lancini C, Tosetti V, Ottolenghi S, Taylor V, Nicolis SK. Hippocampal development and neural stem cell maintenance require Sox2-dependent regulation of Shh. Nat Neurosci. 2009;12:1248–1256. doi: 10.1038/nn.2397. [DOI] [PubMed] [Google Scholar]
- Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P (1996) Ligand-activated site-specific recombination in mice. Proc Natl Acad Sci U S A 93:10887–10890 [DOI] [PMC free article] [PubMed]
- Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- Feltri ML, D'Antonio M, Previtali S, Fasolini M, Messing A, Wrabetz L (1999) P0-Cre transgenic mice for inactivation of adhesion molecules in Schwann cells. Ann N Y Acad Sci 883:116–123 [PubMed]
- Freyer L, Morrow BE. Canonical Wnt signaling modulates Tbx1, Eya1, and Six1 expression, restricting neurogenesis in the otic vesicle. Dev Dyn. 2010;239:1708–1722. doi: 10.1002/dvdy.22308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyer L, Aggarwal V, Morrow BE. Dual embryonic origin of the mammalian otic vesicle forming the inner ear. Development. 2011;138:5403–5414. doi: 10.1242/dev.069849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman LM, Dror AA, Mor E, Tenne T, Toren G, Satoh T, Biesemeier DJ, Shomron N, Fekete DM, Hornstein E, Avraham KB. MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proc Natl Acad Sci U S A. 2009;106:7915–7920. doi: 10.1073/pnas.0812446106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Dillard M, Lavado A, Harvey NL, Jahan I. Canal cristae growth and fiber extension to the outer hair cells of the mouse ear require Prox1 activity. PLoS One. 2010;5:e9377. doi: 10.1371/journal.pone.0009377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Tokano H, Fujioka KS, Okano H, Edge AS. Generating mouse models of degenerative diseases using Cre/lox-mediated in vivo mosaic cell ablation. J Clin Invest. 2011;121:2462–2469. doi: 10.1172/JCI45081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furr BJ, Jordan VC. The pharmacology and clinical uses of tamoxifen. Pharmacol Ther. 1984;25:127–205. doi: 10.1016/0163-7258(84)90043-3. [DOI] [PubMed] [Google Scholar]
- Gao J, Wu X, Zuo J. Targeting hearing genes in mice. Brain Res Mol Brain Res. 2004;132:192–207. doi: 10.1016/j.molbrainres.2004.06.035. [DOI] [PubMed] [Google Scholar]
- Giel-Moloney M, Krause DS, Chen G, Etten RA, Leiter AB. Ubiquitous and uniform in vivo fluorescence in ROSA26-EGFP BAC transgenic mice. Genesis. 2007;45:83–89. doi: 10.1002/dvg.20269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill-Sharma MK, Balasinor N, Parte P. Effect of intermittent treatment with tamoxifen on reproduction in male rats. Asian J Androl. 2001;3:115–119. [PubMed] [Google Scholar]
- Gomez-Casati ME, Murtie J, Taylor B, Corfas G. Cell-specific inducible gene recombination in postnatal inner ear supporting cells and glia. J Assoc Res Otolaryngol. 2010;11:19–26. doi: 10.1007/s10162-009-0191-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- Grimsley-Myers CM, Sipe CW, Geleoc GS, Lu X. The small GTPase Rac1 regulates auditory hair cell morphogenesis. J Neurosci. 2009;29:15859–15869. doi: 10.1523/JNEUROSCI.3998-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, Tabin CJ. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Hartman BH, Reh TA, Bermingham-McDonogh O. Notch signaling specifies prosensory domains via lateral induction in the developing mammalian inner ear. Proc Natl Acad Sci U S A. 2010;107:15792–15797. doi: 10.1073/pnas.1002827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EP, Urness LD, Mansour SL. Fgf16(IRESCre) mice: a tool to inactivate genes expressed in inner ear cristae and spiral prominence epithelium. Dev Dyn. 2009;238:358–366. doi: 10.1002/dvdy.21681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugas M, Lillevali K, Hakanen J, Salminen M. Gata2 is required for the development of inner ear semicircular ducts and the surrounding perilymphatic space. Dev Dyn. 2010;239:2452–2469. doi: 10.1002/dvdy.22373. [DOI] [PubMed] [Google Scholar]
- Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Lewis P, Pevny L, McMahon AP. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech Dev. 2002;119(Suppl 1):S97–S101. doi: 10.1016/S0925-4773(03)00099-6. [DOI] [PubMed] [Google Scholar]
- Hebert JM, McConnell SK. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev Biol. 2000;222:296–306. doi: 10.1006/dbio.2000.9732. [DOI] [PubMed] [Google Scholar]
- Hurd EA, Poucher HK, Cheng K, Raphael Y, Martin DM. The ATP-dependent chromatin remodeling enzyme CHD7 regulates pro-neural gene expression and neurogenesis in the inner ear. Development. 2010;137:3139–3150. doi: 10.1242/dev.047894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CH, Guo D, Harris MA, Howard O, Mishina Y, Gan L, Harris SE, Wu DK. Role of bone morphogenetic proteins on cochlear hair cell formation: analyses of Noggin and Bmp2 mutant mice. Dev Dyn. 2010;239:505–513. doi: 10.1002/dvdy.22200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, Metzger D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques BE, Montcouquiol ME, Layman EM, Lewandoski M, Kelley MW. Fgf8 induces pillar cell fate and regulates cellular patterning in the mammalian cochlea. Development. 2007;134:3021–3029. doi: 10.1242/dev.02874. [DOI] [PubMed] [Google Scholar]
- Jahan I, Kersigo J, Pan N, Fritzsch B. Neurod1 regulates survival and formation of connections in mouse ear and brain. Cell Tissue Res. 2010;341:95–110. doi: 10.1007/s00441-010-0984-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan I, Pan N, Kersigo J, Fritzsch B. Neurod1 suppresses hair cell differentiation in ear ganglia and regulates hair cell subtype development in the cochlea. PLoS One. 2010;5:e11661. doi: 10.1371/journal.pone.0011661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasena CS, Ohyama T, Segil N, Groves AK. Notch signaling augments the canonical Wnt pathway to specify the size of the otic placode. Development. 2008;135:2251–2261. doi: 10.1242/dev.017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KR, Zheng QY. Ahl2, a second locus affecting age-related hearing loss in mice. Genomics. 2002;80:461–464. doi: 10.1006/geno.2002.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KR, Zheng QY, Erway LC. A major gene affecting age-related hearing loss is common to at least ten inbred strains of mice. Genomics. 2000;70:171–180. doi: 10.1006/geno.2000.6377. [DOI] [PubMed] [Google Scholar]
- Jones C, Roper VC, Foucher I, Qian D, Banizs B, Petit C, Yoder BK, Chen P. Ciliary proteins link basal body polarization to planar cell polarity regulation. Nat Genet. 2008;40:69–77. doi: 10.1038/ng.2007.54. [DOI] [PubMed] [Google Scholar]
- Kawamoto S, Niwa H, Tashiro F, Sano S, Kondoh G, Takeda J, Tabayashi K, Miyazaki J. A novel reporter mouse strain that expresses enhanced green fluorescent protein upon Cre-mediated recombination. FEBS Lett. 2000;470:263–268. doi: 10.1016/S0014-5793(00)01338-7. [DOI] [PubMed] [Google Scholar]
- Kazadi K, Loeuillet C, Deutsch S, Ciuffi A, Munoz M, Beckmann JS, Moradpour D, Antonarakis SE, Telenti A. Genomic determinants of the efficiency of internal ribosomal entry sites of viral and cellular origin. Nucleic Acids Res. 2008;36:6918–6925. doi: 10.1093/nar/gkn812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan AE, Cordes R, Kopan R, Gossler A, Gridley T. The Notch ligands DLL1 and JAG2 act synergistically to regulate hair cell development in the mammalian inner ear. Development. 2005;132:4353–4362. doi: 10.1242/dev.02002. [DOI] [PubMed] [Google Scholar]
- Kellendonk C, Tronche F, Casanova E, Anlag K, Opherk C, Schutz G (1999) Inducible site-specific recombination in the brain. J Molec Biol 285:175–182 [DOI] [PubMed]
- Kiernan AE, Xu J, Gridley T. The Notch ligand JAG1 is required for sensory progenitor development in the mammalian inner ear. PLoS Genet. 2006;2:e4. doi: 10.1371/journal.pgen.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistner A, Gossen M, Zimmermann F, Jerecic J, Ullmer C, Lubbert H, Bujard H. Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proc Natl Acad Sci U S A. 1996;93:10933–10938. doi: 10.1073/pnas.93.20.10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky B, Santi P, Johnson S, Schmitz H, Fritzsch B. Conditional deletion of N-Myc disrupts neurosensory and non-sensory development of the ear. Dev Dyn. 2011;240:1373–1390. doi: 10.1002/dvdy.22620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopinke D, Brailsford M, Shea JE, Leavitt R, Scaife CL, Murtaugh LC. Lineage tracing reveals the dynamic contribution of Hes1+ cells to the developing and adult pancreas. Development. 2011;138:431–441. doi: 10.1242/dev.053843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp JL, Dubois CL, Schaffer AE, Hao E, Shih HP, Seymour PA, Ma J, Sander M. Sox9+ ductal cells are multipotent progenitors throughout development but do not produce new endocrine cells in the normal or injured adult pancreas. Development. 2011;138:653–665. doi: 10.1242/dev.056499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koundakjian EJ, Appler JL, Goodrich LV. Auditory neurons make stereotyped wiring decisions before maturation of their targets. J Neurosci. 2007;27:14078–14088. doi: 10.1523/JNEUROSCI.3765-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz A, Fu J, Duerschke K, Weidlich S, Naumann R, Stewart AF, Anastassiadis K. An improved Flp deleter mouse in C57Bl/6 based on Flpo recombinase. Genesis. 2010;48:512–520. doi: 10.1002/dvg.20641. [DOI] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K (1995) Inducible gene targeting in mice. Science 269:1427–1429 [DOI] [PubMed]
- Kwan KM. Conditional alleles in mice: practical considerations for tissue-specific knockouts. Genesis. 2002;32:49–62. doi: 10.1002/gene.10068. [DOI] [PubMed] [Google Scholar]
- Langer SJ, Ghafoori AP, Byrd M, Leinwand L. A genetic screen identifies novel non-compatible loxP sites. Nucleic Acids Res. 2002;30:3067–3077. doi: 10.1093/nar/gkf421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin LZ, Cai CL, Lu MM, Reth M, Platoshyn O, Yuan JX, Evans S, Chien KR. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Indra AK, Warot X, Brocard J, Messaddeq N, Kato S, Metzger D, Chambon P. Skin abnormalities generated by temporally controlled RXRalpha mutations in mouse epidermis. Nature. 2000;407:633–636. doi: 10.1038/35036595. [DOI] [PubMed] [Google Scholar]
- Li M, Tian Y, Fritzsch B, Gao J, Wu X, Zuo J. Inner hair cell Cre-expressing transgenic mouse. Genesis. 2004;39:173–177. doi: 10.1002/gene.20040. [DOI] [PubMed] [Google Scholar]
- Liu F, Woitge HW, Braut A, Kronenberg MS, Lichtler AC, Mina M, Kream BE. Expression and activity of osteoblast-targeted Cre recombinase transgenes in murine skeletal tissues. Int J Dev Biol. 2004;48:645–653. doi: 10.1387/ijdb.041816fl. [DOI] [PubMed] [Google Scholar]
- Liu Z, Owen T, Zhang L, Zuo J. Dynamic expression pattern of Sonic hedgehog in developing cochlear spiral ganglion neurons. Dev Dyn. 2010;239:1674–1683. doi: 10.1002/dvdy.22302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Owen T, Fang J, Zuo J. Overactivation of Notch1 signaling induces ectopic hair cells in the mouse inner ear in an age-dependent manner. PloS One. 2012a;7:e34123. doi: 10.1371/journal.pone.0034123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Owen T, Fang J, Srinivasan RS, Zuo J. In vivo notch reactivation in differentiating cochlear hair cells induces sox2 and prox1 expression but does not disrupt hair cell maturation. Dev Dyn. 2012;241:684–696. doi: 10.1002/dvdy.23754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- Lobe CG, Koop KE, Kreppner W, Lomeli H, Gertsenstein M, Nagy A. Z/AP, a double reporter for Cre-mediated recombination. Dev Biol. 1999;208:281–292. doi: 10.1006/dbio.1999.9209. [DOI] [PubMed] [Google Scholar]
- Lundell TG, Zhou Q, Doughty ML. Neurogenin1 expression in cell lineages of the cerebellar cortex in embryonic and postnatal mice. Dev Dyn. 2009;238:3310–3325. doi: 10.1002/dvdy.22165. [DOI] [PubMed] [Google Scholar]
- Machold R, Fishell G. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005;48:17–24. doi: 10.1016/j.neuron.2005.08.028. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Salas E. Internal ribosome entry site biology and its use in expression vectors. Curr Opin Biotechnol. 1999;10:458–464. doi: 10.1016/S0958-1669(99)00010-5. [DOI] [PubMed] [Google Scholar]
- Matei V, Pauley S, Kaing S, Rowitch D, Beisel KW, Morris K, Feng F, Jones K, Lee J, Fritzsch B. Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Dev Dyn. 2005;234:633–650. doi: 10.1002/dvdy.20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehasseb MK, Bell SC, Habiba MA. The effects of tamoxifen and estradiol on myometrial differentiation and organization during early uterine development in the CD1 mouse. Reproduction. 2009;138:341–350. doi: 10.1530/REP-09-0054. [DOI] [PubMed] [Google Scholar]
- Mountford PS, Smith AG. Internal ribosome entry sites and dicistronic RNAs in mammalian transgenesis. Trends Genet : TIG. 1995;11:179–184. doi: 10.1016/S0168-9525(00)89040-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26:99–109. doi: 10.1002/(SICI)1526-968X(200002)26:2<99::AID-GENE1>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. doi: 10.1002/1526-968X(200011/12)28:3/4<147::AID-GENE90>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Nyabi O, Naessens M, Haigh K, Gembarska A, Goossens S, Maetens M, Clercq S, Drogat B, Haenebalcke L, Bartunkova S, Vos I, Craene B, Karimi M, Berx G, Nagy A, Hilson P, Marine JC, Haigh JJ. Efficient mouse transgenesis using Gateway-compatible ROSA26 locus targeting vectors and F1 hybrid ES cells. Nucleic Acids Res. 2009;37:e55. doi: 10.1093/nar/gkp112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterle EC, Chien WM, Campbell S, Nellimarla P, Fero ML. p27(Kip1) is required to maintain proliferative quiescence in the adult cochlea and pituitary. Cell cycle. 2011;10:1237–1248. doi: 10.4161/cc.10.8.15301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlemiller KK. Age-related hearing loss: the status of Schuknecht's typology. Curr Opin Otolaryngol Head Neck Surg. 2004;12:439–443. doi: 10.1097/01.moo.0000134450.99615.22. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Groves AK. Generation of Pax2-Cre mice by modification of a Pax2 bacterial artificial chromosome. Genesis. 2004;38:195–199. doi: 10.1002/gene.20017. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Mohamed OA, Taketo MM, Dufort D, Groves AK. Wnt signals mediate a fate decision between otic placode and epidermis. Development. 2006;133:865–875. doi: 10.1242/dev.02271. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Basch ML, Mishina Y, Lyons KM, Segil N, Groves AK. BMP signaling is necessary for patterning the sensory and nonsensory regions of the developing mammalian cochlea. J Neurosci. 2010;30:15044–15051. doi: 10.1523/JNEUROSCI.3547-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov DA, Deng JM, Ogunrinu G, Behringer RR. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis. 2000;26:145–146. doi: 10.1002/(SICI)1526-968X(200002)26:2<145::AID-GENE14>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Pan W, Jin Y, Stanger B, Kiernan AE. Notch signaling is required for the generation of hair cells and supporting cells in the mammalian inner ear. Proc Natl Acad Sci U S A. 2010;107:15798–15803. doi: 10.1073/pnas.1003089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan N, Jahan I, Kersigo J, Kopecky B, Santi P, Johnson S, Schmitz H, Fritzsch B. Conditional deletion of Atoh1 using Pax2-Cre results in viable mice without differentiated cochlear hair cells that have lost most of the organ of Corti. Hear Res. 2011;275:66–80. doi: 10.1016/j.heares.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin BJ, Sonnemann KJ, Ervasti JM. beta-actin and gamma-actin are each dispensable for auditory hair cell development but required for Stereocilia maintenance. PLoS Genet. 2010;6:e1001158. doi: 10.1371/journal.pgen.1001158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirvola U, Ylikoski J, Trokovic R, Hebert JM, McConnell SK, Partanen J. FGFR1 is required for the development of the auditory sensory epithelium. Neuron. 2002;35:671–680. doi: 10.1016/S0896-6273(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Raft S, Koundakjian EJ, Quinones H, Jayasena CS, Goodrich LV, Johnson JE, Segil N, Groves AK. Cross-regulation of Ngn1 and Math1 coordinates the production of neurons and sensory hair cells during inner ear development. Development. 2007;134:4405–4415. doi: 10.1242/dev.009118. [DOI] [PubMed] [Google Scholar]
- Rawlins EL, Clark CP, Xue Y, Hogan BL. The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development. 2009;136:3741–3745. doi: 10.1242/dev.037317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CS, Soriano P. High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PLoS One. 2007;2:e162. doi: 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickheit G, Maier H, Strenzke N, Andreescu CE, Zeeuw CI, Muenscher A, Zdebik AA, Jentsch TJ. Endocochlear potential depends on Cl− channels: mechanism underlying deafness in Bartter syndrome IV. EMBO J. 2008;27:2907–2917. doi: 10.1038/emboj.2008.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers LE, Young KM, Rizzi M, Jamen F, Psachoulia K, Wade A, Kessaris N, Richardson WD. PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nat Neurosci. 2008;11:1392–1401. doi: 10.1038/nn.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha-Sanchez SM, Morris KA, Kachar B, Nichols D, Fritzsch B, Beisel KW. Developmental expression of Kcnq4 in vestibular neurons and neurosensory epithelia. Brain Res. 2007;1139:117–125. doi: 10.1016/j.brainres.2006.12.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MF, Ahmad KA, Thaller C, Zoghbi HY. Excitatory neurons of the proprioceptive, interoceptive, and arousal hindbrain networks share a developmental requirement for Math1. Proc Natl Acad Sci U S A. 2009;106:22462–22467. doi: 10.1073/pnas.0911579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage C, Huang M, Karimi K, Gutierrez G, Vollrath MA, Zhang DS, Garcia-Anoveros J, Hinds PW, Corwin JT, Corey DP, Chen ZY. Proliferation of functional hair cells in vivo in the absence of the retinoblastoma protein. Science. 2005;307:1114–1118. doi: 10.1126/science.1106642. [DOI] [PubMed] [Google Scholar]
- Sage C, Huang M, Vollrath MA, Brown MC, Hinds PW, Corey DP, Vetter DE, Chen ZY. Essential role of retinoblastoma protein in mammalian hair cell development and hearing. Proc Natl Acad Sci U S A. 2006;103:7345–7350. doi: 10.1073/pnas.0510631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai K, Miyazaki J. A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem Biophys Res Commun. 1997;237:318–324. doi: 10.1006/bbrc.1997.7111. [DOI] [PubMed] [Google Scholar]
- Sauer B, Henderson N (1989) Cre-stimulated recombination at loxP-containing DNA sequences placed into the mammalian genome. Nucleic Acids Res 17:147–161 [DOI] [PMC free article] [PubMed]
- Schaft J, Ashery-Padan R, Hoeven F, Gruss P, Stewart AF. Efficient FLP recombination in mouse ES cells and oocytes. Genesis. 2001;31:6–10. doi: 10.1002/gene.1076. [DOI] [PubMed] [Google Scholar]
- Schultz JM, et al. Noncoding mutations of HGF are associated with nonsyndromic hearing loss, DFNB39. Am J Hum Genet. 2009;85:25–39. doi: 10.1016/j.ajhg.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Nam HS, Song P, Moore H, Anderson SA. FoxG1 haploinsufficiency results in impaired neurogenesis in the postnatal hippocampus and contextual memory deficits. Hippocampus. 2006;16:875–890. doi: 10.1002/hipo.20218. [DOI] [PubMed] [Google Scholar]
- Siegenthaler JA, Tremper-Wells BA, Miller MW. Foxg1 haploinsufficiency reduces the population of cortical intermediate progenitor cells: effect of increased p21 expression. Cerebr Cortex. 2008;18:1865–1875. doi: 10.1093/cercor/bhm209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipe CW, Lu X. Kif3a regulates planar polarization of auditory hair cells through both ciliary and non-ciliary mechanisms. Development. 2011;138:3441–3449. doi: 10.1242/dev.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, Flier LG, Sato T, Es JH, Born M, Kroon-Veenboer C, Barker N, Klein AM, Rheenen J, Simons BD, Clevers H. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Soukup GA, Fritzsch B, Pierce ML, Weston MD, Jahan I, McManus MT, Harfe BD. Residual microRNA expression dictates the extent of inner ear development in conditional Dicer knockout mice. Dev Biol. 2009;328:328–341. doi: 10.1016/j.ydbio.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa VH, Miyoshi G, Hjerling-Leffler J, Karayannis T, Fishell G. Characterization of Nkx6-2-derived neocortical interneuron lineages. Cerebr Cortex. 2009;19(Suppl 1):i1–10. doi: 10.1093/cercor/bhp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan RS, Dillard ME, Lagutin OV, Lin FJ, Tsai S, Tsai MJ, Samokhvalov IM, Oliver G. Lineage tracing demonstrates the venous origin of the mammalian lymphatic vasculature. Genes Dev. 2007;21:2422–2432. doi: 10.1101/gad.1588407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan RS, Geng X, Yang Y, Wang Y, Mukatira S, Studer M, Porto MP, Lagutin O, Oliver G. The nuclear hormone receptor Coup-TFII is required for the initiation and early maintenance of Prox1 expression in lymphatic endothelial cells. Genes Dev. 2010;24:696–707. doi: 10.1101/gad.1859310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steigelman KA, Lelli A, Wu X, Gao J, Lin S, Piontek K, Wodarczyk C, Boletta A, Kim H, Qian F, Germino G, Geleoc GS, Holt JR, Zuo J. Polycystin-1 Is required for stereocilia structure but not for mechanotransduction in inner ear hair cells. J Neurosci. 2011;31:12241–12250. doi: 10.1523/JNEUROSCI.6531-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Tang W, Chang Q, Wang Y, Kong W, Lin X. Connexin30 null and conditional connexin26 null mice display distinct pattern and time course of cellular degeneration in the cochlea. J Comp Neurol. 2009;516:569–579. doi: 10.1002/cne.22117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto IY, Leung KK, Sham MH, Cheah KS. Utility of HoxB2 enhancer-mediated Cre activity for functional studies in the developing inner ear. Genesis. 2009;47:361–365. doi: 10.1002/dvg.20507. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsani D, Fu Y, Lu J, Lin Y, Miyoshi G, Shima Y, Fishell G, Nelson SB, Huang ZJ. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Li M, Fritzsch B, Zuo J. Creation of a transgenic mouse for hair-cell gene targeting by using a modified bacterial artificial chromosome containing Prestin. Dev Dyn. 2004;231:199–203. doi: 10.1002/dvdy.20106. [DOI] [PubMed] [Google Scholar]
- Tian Y, James S, Zuo J, Fritzsch B, Beisel KW. Conditional and inducible gene recombineering in the mouse inner ear. Brain Res. 2006;1091:243–254. doi: 10.1016/j.brainres.2006.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vooijs M, Jonkers J, Berns A. A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep. 2001;2:292–297. doi: 10.1093/embo-reports/kve064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vooijs M, Ong CT, Hadland B, Huppert S, Liu Z, Korving J, Born M, Stappenbeck T, Wu Y, Clevers H, Kopan R. Mapping the consequence of Notch1 proteolysis in vivo with NIP-CRE. Development. 2007;134:535–544. doi: 10.1242/dev.02733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chang Q, Tang W, Sun Y, Zhou B, Li H, Lin X. Targeted connexin26 ablation arrests postnatal development of the organ of Corti. Biochem Biophys Res Commun. 2009;385:33–37. doi: 10.1016/j.bbrc.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T, Corbett MK, Chow LM, Valentine MB, Baker SJ, Zuo J. Rapid cell-cycle reentry and cell death after acute inactivation of the retinoblastoma gene product in postnatal cochlear hair cells. Proc Natl Acad Sci U S A. 2008;105:781–785. doi: 10.1073/pnas.0708061105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston MD, Pierce ML, Jensen-Smith HC, Fritzsch B, Rocha-Sanchez S, Beisel KW, Soukup GA. MicroRNA-183 family expression in hair cell development and requirement of microRNAs for hair cell maintenance and survival. Dev Dyn. 2011;240:808–819. doi: 10.1002/dvdy.22591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willott JF, Tanner L, O'Steen J, Johnson KR, Bogue MA, Gagnon L. Acoustic startle and prepulse inhibition in 40 inbred strains of mice. Behav Neurosci. 2003;117:716–727. doi: 10.1037/0735-7044.117.4.716. [DOI] [PubMed] [Google Scholar]
- Winter H, Ruttiger L, Muller M, Kuhn S, Brandt N, Zimmermann U, Hirt B, Bress A, Sausbier M, Conscience A, Flamant F, Tian Y, Zuo J, Pfister M, Ruth P, Lowenheim H, Samarut J, Engel J, Knipper M. Deafness in TRbeta mutants is caused by malformation of the tectorial membrane. J Neurosci. 2009;29:2581–2587. doi: 10.1523/JNEUROSCI.3557-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich FT, Wildner H, Rajewsky K, Edenhofer F. New variants of inducible Cre recombinase: a novel mutant of Cre-PR fusion protein exhibits enhanced sensitivity and an expanded range of inducibility. Nucleic Acids Res. 2001;29:E47. doi: 10.1093/nar/29.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Chen L, Baldini A. In vivo genetic ablation of the periotic mesoderm affects cell proliferation survival and differentiation in the cochlea. Dev Biol. 2007;310:329–340. doi: 10.1016/j.ydbio.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan S, Baptista CA, Balas G, Tao W, Soares VC, Lai E. Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron. 1995;14:1141–1152. doi: 10.1016/0896-6273(95)90262-7. [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Okano T, Ma X, Adelstein RS, Kelley MW. Myosin II regulates extension, growth and patterning in the mammalian cochlear duct. Development. 2009;136:1977–1986. doi: 10.1242/dev.030718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N, Chang W, Kelley MW. Rbpj regulates development of prosensory cells in the mammalian inner ear. Dev Biol. 2011;353:367–379. doi: 10.1016/j.ydbio.2011.03.016. [DOI] [PubMed] [Google Scholar]
- Yang H, Xie X, Deng M, Chen X, Gan L. Generation and characterization of Atoh1-Cre knock-in mouse line. Genesis. 2010;48:407–413. doi: 10.1002/dvg.20633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Gan J, Xie X, Deng M, Feng L, Chen X, Gao Z, Gan L. Gfi1-Cre knock-in mouse line: a tool for inner ear hair cell-specific gene deletion. Genesis. 2010;48:400–406. doi: 10.1002/dvg.20632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida N, Hequembourg SJ, Atencio CA, Rosowski JJ, Liberman MC. Acoustic injury in mice: 129/SvEv is exceptionally resistant to noise-induced hearing loss. Hear Res. 2000;141:97–106. doi: 10.1016/S0378-5955(99)00210-5. [DOI] [PubMed] [Google Scholar]
- Young KM, Mitsumori T, Pringle N, Grist M, Kessaris N, Richardson WD. An Fgfr3-iCreER(T2) transgenic mouse line for studies of neural stem cells and astrocytes. Glia. 2010;58:943–953. doi: 10.1002/glia.20976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Zuo J. The practical use of Cre and loxP technologies in mouse auditory research. Meth Mol Biol. 2009;493:87–102. doi: 10.1007/978-1-59745-523-7_6. [DOI] [PubMed] [Google Scholar]
- Yu Y, Weber T, Yamashita T, Liu Z, Valentine MB, Cox BC, Zuo J. In vivo proliferation of postmitotic cochlear supporting cells by acute ablation of the retinoblastoma protein in neonatal mice. J Neurosci. 2010;30:5927–5936. doi: 10.1523/JNEUROSCI.5989-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrowicz BP, et al. Wnk1 kinase deficiency lowers blood pressure in mice: a gene-trap screen to identify potential targets for therapeutic intervention. Proc Natl Acad Sci U S A. 2003;100:14109–14114. doi: 10.1073/pnas.2336103100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelarayan LC, Vendrell V, Alvarez Y, Dominguez-Frutos E, Theil T, Alonso MT, Maconochie M, Schimmang T. Differential requirements for FGF3, FGF8 and FGF10 during inner ear development. Dev Biol. 2007;308:379–391. doi: 10.1016/j.ydbio.2007.05.033. [DOI] [PubMed] [Google Scholar]
- Zheng QY, Johnson KR, Erway LC. Assessment of hearing in 80 inbred strains of mice by ABR threshold analyses. Hear Res. 1999;130:94–107. doi: 10.1016/S0378-5955(99)00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Sage M, Sheppeard EA, Jurecic V, Bradley A. Engineering mouse chromosomes with Cre-loxP: range, efficiency, and somatic applications. Mol Cell Biol. 2000;20:648–655. doi: 10.1128/MCB.20.2.648-655.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo L, Theis M, Alvarez-Maya I, Brenner M, Willecke K, Messing A. hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis. 2001;31:85–94. doi: 10.1002/gene.10008. [DOI] [PubMed] [Google Scholar]
- Zuo J. Transgenic and gene targeting studies of hair cell function in mouse inner ear. J Neurobiol. 2002;53:286–305. doi: 10.1002/neu.10128. [DOI] [PubMed] [Google Scholar]