Abstract
Through functional expression screening, we identified a gene, designated Humanin (HN) cDNA, which encodes a short polypeptide and abolishes death of neuronal cells caused by multiple different types of familial Alzheimer's disease genes and by Aβ amyloid, without effect on death by Q79 or superoxide dismutase-1 mutants. Transfected HN cDNA was transcribed to the corresponding polypeptide and then was secreted into the cultured medium. The rescue action clearly depended on the primary structure of HN. This polypeptide would serve as a molecular clue for the development of new therapeutics for Alzheimer's disease targeting neuroprotection.
Alzheimer's disease (AD) is the most prevalent neurodegenerative disease. To establish curative therapy for this disease, controlling the occurrence of neuronal death is mandatory. Three different kinds of mutant genes cause early-onset familial AD (FAD): mutant amyloid precursor protein (APP), presenilin (PS)1, and PS2 (1). Accumulated evidence (2–10) indicates that all these FAD genes can induce death in neuronal cells or augment their vulnerability to other insults. An important clue in the development of AD therapy, therefore, is the molecules that suppress FAD gene-induced death detectable in neuronal cells in culture. Our strategy was to use a functional screening for antagonistic genes against death insults of interest, termed a death-trap screening, originally developed by D'Adamio and coworkers (11). Here we report a cDNA, encoding a short polypeptide, that suppresses neuronal cell death induced by the three different types of FAD genes and by Aβ, designated Humanin (HN).
Methods
Genes, Polypeptides, and Materials.
V642I-APP cDNA was described (2, 12). Mutant PS cDNAs were subcloned in pcDNA. p Humanin (pHN) was constructed by inserting annealed HN-sense and antisense oligonucleotides into EcoRI and KpnI sites of pFLAG-CMV-5a (pFLAG; Kodak), whose expression produces HN C-terminally fused with FLAG tag GTDYKDDDDK (HN-FLAG; GT is a linker). pHN mutants were constructed by a site-directed mutagenesis kit (Stratagene). Catalytically negative (CN-) procaspase-3 cDNA was described (13). Antibodies against PS N termini were provided by T. Tomita and T. Iwatsubo (University of Tokyo). Anti-Fas antibody (CH-11) and anti-superoxide dismutase-1 (SOD1) antibody were from MBL (Nagoya, Japan). HN peptides were obtained from Peptide Institute (Osaka).
Cell Death Experiments.
F11 cells [7 × 104 cells/well in a 6-well plate cultured in Ham's F-12 plus 18% FBS (HF-18%) for 12–16 h] were transfected with FAD genes and pHN by lipofection (FAD gene, 1 μg; pHN, 1 μg; LipofectAMINE, 4 μl; PLUS Reagent, 8 μl) in the absence of serum for 3 h and were incubated with HF-18%, for 2 h. Culture media were changed to Ham's F-12 plus 10% FBS (HF-10%), and cell mortality was measured at 72 h after transfection. To use HN peptides, cells were transfected with FAD genes in the absence of serum for 3 h, incubated with HF-18% for 2 h, and cultured with HF-10% and HN peptides (with or without inhibitors) for 67 h, and cell mortality and viability were measured. Cell mortality was assessed by Trypan blue and cell viability by WST-8 (Wako, Osaka), as described (12, 14). F11/EcR cell experiments were performed as described (12). Cells were transfected with an ecdysone (EcD)-inducible plasmid with or without pHN, and EcD was added to the media. To use synthetic HN (sHN), cells were transfected in the absence of serum for 3 h, incubated with HF-18% for 12–16 h, and cultured with HF-10% and sHN for 2 h, and EcD was added to the media. Cell mortality was measured 72 h after EcD treatment. Transfection efficiency was consistently 60–70%, as described (12). The primary culture of mouse cortical neurons was performed as described (15). Prepared neurons (1.25 × 105 cells/well) were preincubated with or without 10 nM or 10 μM sHN for 16 h, and treated with 25 μM Aβ1–43 in the presence or absence of 10 nM or 10 μM sHN for 24–72 h. To prevent transient dryness, we changed half volume of medium with prewarmed fresh medium containing 50 μM Aβ1–43 and 10 nM or 10 μM sHN. Lactate dehydrogenase (LDH) assay was performed by using a kit (Wako). Calcein staining was performed as described (16). More than 30 min after 6 μM Calcein-AM (Dojindo, Kumamoto, Japan) treatment, fluorescence was observed by fluorescence microscopy.
Statistics.
All data are presented as mean ± SD of ≥3 independent experiments. Student's t test was used to compare the mean differences between groups. Methods of cDNA library construction, cultured medium experiments, cell-binding experiments, and Northern blot analysis are published as supplemental data on the PNAS web site, www.pnas.org.
Results
Screening of Antagonistic Genes.
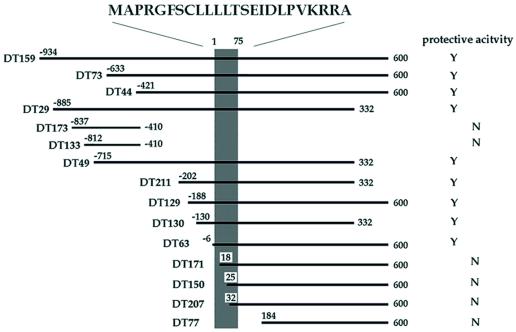
We applied death-trap screening to a V642I-APP-inducible F11 neuronal clone (F11/EcR/V642I) (F11 cells are hybrid cells of E13 rat primary neuron with mouse neuroblastoma NTG18). On EcD treatment, F11/EcR/V642I cells express V642I-APP, resulting in death in most cells within a few days (17). We transfected these cells with a cDNA library in pEF-BOS constructed from the occipital cortex of an AD patient brain, treated cells with EcD for 72 h, and recovered the plasmids from survived cells. This procedure was repeated for four rounds in total, and we picked 250 clones of plasmids. Using dot blot hybridization with randomly selected clones, we classified them into 36 crosshybridizable groups. The largest group consisted of 28 clones. This group totally encoded a cDNA consisting of 1,567 bases. We assayed whether transfection of each DNA fragment resulted in suppression of EcD-induced death of F11/EcR cells cotransfected with EcD-inducible pIND encoding V642I-APP (pIND-V642I-APP) (Fig. 1). The comparison of each sequence with each activity revealed that the V642I-APP-antagonizing activity was in a 75-base ORF that encodes a 24-residue peptide MAPRGFSCLLLLTSEIDLPVKRRA. We designated this molecule Humanin.
Figure 1.
Effects of DT clones on V642I-APP-induced death of neuronal cells. The region encoding the activity suppressing F11/EcR cell death by V642I-APP. The cDNA fragments are aligned within the longest clone [from −934 to 600; No. 1 base corresponds to the first base of the HN ORF (shaded area), one base before which is numbered −1], and their activities against V642I-APP-induced death of F11/EcR cells are indicated. F11/EcR cells were transfected with pIND-V642I-APP (1 μg) with 1 μg of either pEF-BOS or each cDNA fragment-coding pEF-BOS, followed by 72 h treatment with EcD, and then cell mortality was measured. When there was a statistically significant difference (P < 0.01) in cell mortality between pEF-BOS-transfected cells and each cDNA fragment-transfected cells, the protective activity was assessed to be present in the cDNA fragment and was indicated as Y; N indicates the absence of significant activity. All of the DT cDNAs with significant activity, shown as Y, caused almost complete suppression of V642I-APP-induced F11/EcR cell death. Nonprotective HN cDNAs such as DT171 were selected for the following reason: in death-trap screening, one way to augment the recovery of the plasmids from survived cells is to culture insult-treated cells for a short period and to harvest the cells at the time point when 70–80% of total cells are dead. By repeating the recovery and the retransfection, the antagonizing clones are concentrated. As in the text, we repeated this selection for a total of four rounds, picked out the final clones, and classified them into crosshybridizable groups by using dot blot hybridization with randomly selected clones. Therefore, some of the obtained clones were false positive, and each crosshybridizable group included inactive fragments, more or less.
The deduced longest HN-containing cDNA sequence, described in Fig. 6 (which is published as supplemental data on the PNAS web site, www.pnas.org), is considered to be identical to human FLJ22981 fis cDNA [gi 10439530 dbj AK026634.1 AK026634); identities = 1545/1553 (99%)]. The homology to other registered DNAs is shown in the supplemental text. No protein was found to be homologous to HN among registered molecules thus far, whereas some proteins, in their signal sequences, contain regions <10 amino acids in length, somewhat similar to portions in HN. Effects of some death-trap (DT) clones, some containing and some not containing the ORF, are provided by Fig. 6.
Suppression by HN cDNA of Neuronal Cell Death by FAD Genes and Extracellular Secretion of HN.
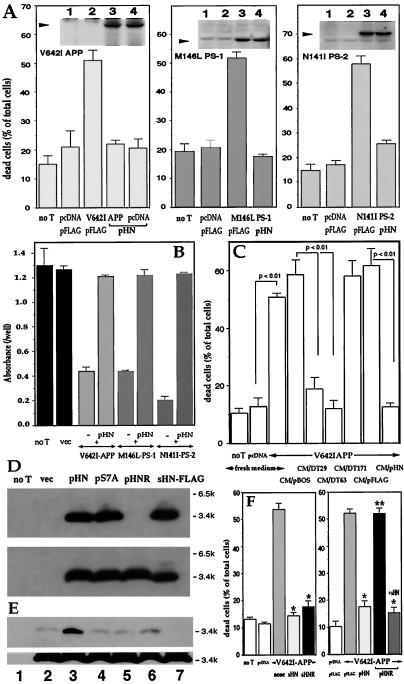
We subcloned the ORF of HN into pFLAG (pHN). Transfection of pHN to F11 cells suppressed toxicity by either V642I-APP, M146L-PS1, or N141I-PS2 cDNA, without altering basal death rates (Fig. 2A). Fig. 2 Insets indicate that, (i) each FAD protein was overexpressed; and (ii) pHN cotransfection did not affect the expression level of each FAD protein. Antagonism of pHN against these FAD genes was confirmed by cell viability assay (Fig. 2B).
Figure 2.
Effect of pHN and secretion of the transcribed HN into cultured media. (A and B) F11 cells were transfected with pcDNA or either V642I-APP, M146L-PS1, or N141I-PS2 cDNA with pFLAG or pHN for 72 h. In A and B, cell mortality and viability were measured, respectively. Negative controls without transfection (no T) or with empty plasmid transfection (vec) were also examined. (A Insets) Expression of cognate FAD proteins by transfection with FAD genes. Arrows indicate the cognate holoproteins (1, no T; 2, vec; 3, FAD gene; 4, FAD gene + pHN). (C) F11 cells were transfected with pcDNA or V642I-APP cDNA in the absence of serum for 3 h, incubated with HF-18% for 2 h, and cultured with CM/pHN, other transfected CM, or fresh media (fresh HF-10%) for 67 h. Cell mortality was measured 72 h after transfection. (D) F11 cells were transfected with pHN, pS7A (pHN mutant with S7A), or pHNR for 72 h, and the cell lysates (Lower) and media (Upper) were analyzed by M2 antibody. (E) F11 cells were transfected with pHN for 24 h and treated with or without secretagogues or an inhibitor for 6 h. CM and cell lysates were submitted to immunoblot analysis with M2 antibody [1, no transfection; 2, pHN transfection alone; 3, pHN + 48 mM KCl; 4, pHN + 1 μM forskolin; 5, pHN + 0.1 mM 3-isobutyl-1-methylxanthine (IBMX); 6, pHN + 1 μM forskolin + 0.1 mM IBMX; 7, pHN + 20 ng/ml (+)Brefeldin A]. (F) F11 cells were transfected with pcDNA or V642I-APP cDNA with cotransfection of pFLAG or pHN or with treatment of 10 μM HN peptides. After 72-h culture, cell mortality was measured. *, significant suppression (P < 0.01); **, no significant suppression vs. V642I-APP-induced cell death.
The cultured medium of F11 cells transfected with pHN (CM/pHN) carried protective activity. When F11 cells were transfected with V642I-APP cDNA and then cultured in CM/pHN, cell mortality drastically decreased (Fig. 2C). Also, CM/DT29 and CM/DT63, but not CM/DT171, suppressed V642I-APP-induced cell death. CM/pHN as well as lysates of cells transfected with pHN contained bands of the HN immunoreactivity at 3–4 kDa, consistent with the deduced size of sHN-FLAG (Fig. 2D). Titration by using sHN-FLAG assessed that HN was at 8–9 μM in CM/pHN (data not shown). HN was thus transcribed from coding DT clones and pHN and secreted into the medium.
Secretory Activity Is Encoded in the HN Sequence.
The N-terminal 23 or entire 24 residues in HN partially satisfy the provisions for signal sequences (18, 19). The HN sequence consists of a hydrophobic core region GFSCLLLLTSEIDL flanked by a C-terminal polar region PVKRRA and an N-terminal region MAPR. Because (i) intracellularly expressed HN was secreted into the medium at micromolar levels; (ii) the FLAG tag was located at the C terminus; and (iii) secreted HN retained the deduced molecular weight, the entire sequence of HN would have a signal sequence-like activity, and after membrane insertion, whole HN could be released into the endoplasmic reticulum lumen without being cleaved. If a short signal sequence has few charged residues in its N-terminal region, the signal peptide or its portion could be released into the endoplasmic reticulum lumen (18). Such a structure is retained by HN.
Secretion of HN was affected by L9R, which disqualifies HN from satisfying provisions for signal sequences (www.cbs.dtu.dk/services/SignalP). The L9R mutant of HN (HNR) was expressed inside the cells by the cognate cDNA (pHNR) transfection but was not secreted to the medium (Fig. 2D). These data demonstrate that the primary sequence of HN encodes the mechanism, whereby newly synthesized HN is extracellularly secreted. The release of HN was stimulated strongly by high K+ and weakly by forskolin plus 3-isobutyl-1-methylxanthine (Fig. 2E). These are secretory stimuli through intracellular activities of Ca2+ and cAMP, respectively. The release of HN was blocked by (+)Brefeldin A, an inhibitor of endoplasmic reticulum-Golgi transport. Thus, HN release is regulated by intracellular secretory mechanisms.
Where HN acts was assessed by using HNR. When F11 cells were transfected with V642I-APP and HNR cDNAs, cells underwent death as strongly as observed for V642I-APP-transfected cells (Fig. 2F Right), whereas death of cells transfected with V642I-APP and HNR cDNAs was suppressed by extracellularly added sHN. Extracellularly added sHNR did inhibit death by V642I-APP (Fig. 2F Left). Because intracellularly expressed HNR was not secreted (Fig. 2E), the rescue action of HN cDNA requires extracellular secretion of the corresponding polypeptide, not its presence inside the cell, indicating that HN acts on cells from the outside.
Effect of HN Polypeptide and Its Structural Derivatives.
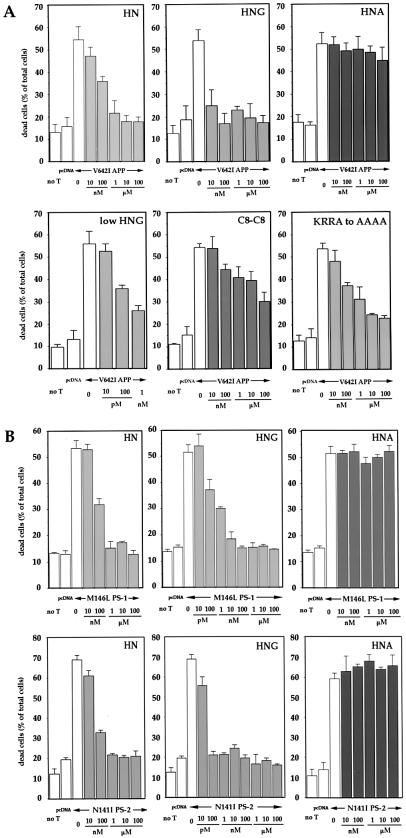
Ten micromolars sHN completely protected V642I-APP-transfected F11 cells from death (Fig. 3A). Although 10 nM sHN exerted little suppression, the inhibitory action reached complete suppression at 1–10 μM. HN did not affect the basal death rate. Complete protection was also observed at ≥10 nM synthetic S14G HN (sHNG). In contrast, sHN with C8A (sHNA) failed to suppress V642I-APP-induced cell death at 100 μM. The importance of Cys8 was further suggested by a dimer form through Cys8, which indicated an intermediate potency between those of HN and HNA. The action potency of the HN with C-terminal AAAA substitution for KRRA was marginally altered, suggesting that C-terminal residues were not important. sHN suppressed cell death by M146L-PS1 or N141I-PS2 cDNA with similar dose-response profiles (Fig. 3B). sHNA failed to suppress death by either FAD gene, whereas sHNG was fully protective at 10 nM against either FAD gene without altering the basal death rates. The cell viability assay confirmed that 10 μM sHN and 10 nM sHNG, but not 10 μM sHNA, blocked cytotoxicity by V642I-APP, M146L-PS1, and N141I-PS2 cDNA (Fig. 7A, which is published as supplemental data on the PNAS web site, www.pnas.org). Treatment of V642I-APP-, M146L-PS1-, or N141I-PS2-transfected cells with 10 μM sHN, sHNG, or sHNA did not alter the expression of these FAD gene products (Fig. 7B). These results demonstrate an essential role of the primary structure in the action of HN.
Figure 3.
Effect of HN polypeptides on neuronal cell death by FAD genes. (A) F11 cells were transfected with V642I-APP cDNA and were treated with various concentrations of sHN, sHNG, sHNA, the dimer form of sHN through C8 (C8-C8), and sHN whose C-terminal KRRA was replaced with AAAA (KRRA to AAAA). Seventy-two hours after transfection, cell mortality was measured by Trypan blue exclusion assay. (B) F11 cells were transfected with M146L-PS1 or N141I-PS2 cDNA, and were treated with various concentrations of sHN, sHNG (S14G), or sHNA (C8A). Seventy-two hours after transfection, cell mortality was similarly measured.
HN Inhibits Aβ-Induced Death in Primary Neurons.
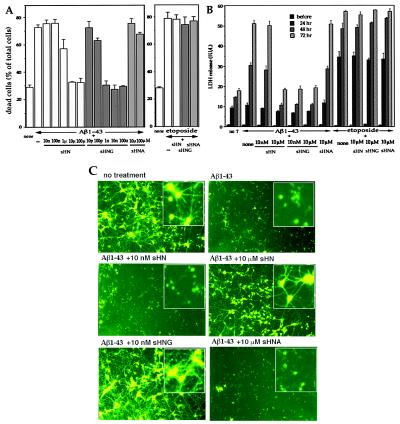
Aβ is a major component of senile plaques, a pathological hallmark deposition in the AD brain. Secreted Aβ may contribute to the pathogenesis of AD and, in fact, Aβ treatment kills neurons (20). Treatment of primary cortical neurons with 25 μM Aβ1–43 caused massive cell death with dystrophic neuritic changes. The microscopic views of treated neurons are provided by Fig. 8A (which is published as supplemental data on the PNAS web site, www.pnas.org) In this system, 25 μM of a similar Aβ peptide Aβ1–40, which provides a less aggregative Aβ control, caused no neurotoxicity (21), supporting for the specific neurotoxic action of Aβ1–43. When neurons were pretreated with 10 μM sHN, Aβ1–43-induced death as well as dystrophic changes of neurites were suppressed. Although cell damage, assessed by cell mortality (Fig. 4A) and released LDH (Fig. 4B), occurred by Aβ1–43, 10 μM sHN recovered these indices to those observed in a basal state. In contrast, 10 μM sHN or sHNG did not protect neurons from toxicity by etoposide (Fig. 4 A and B and Fig. 8B). Ten nanomolars sHNG protected neurons from the Aβ toxicity, whereas neither 10 nM sHN nor 10 μM sHNA did so (Fig. 4 A–C), assessed by multiple assays.
Figure 4.
Effect of HN on Aβ-induced cell death in primary neurons. (A and B) Primary cortical neurons were treated with 25 μM Aβ1–43 in the presence or absence of sHN or its derivatives. In A, cell mortality was measured 72 h after Aβ treatment with or without HN peptides. Neurons were similarly treated, as a positive control, with 20 μM etoposide in the presence or absence of 10 μM HN peptides for 72 h. In B, the LDH activity in the culture media was monitored by sampling 6 μl of the media culturing neurons treated with 25 μM Aβ1–43 in the presence or absence of HN peptides at 24, 48, or 72 h after the onset of Aβ treatment. The LDH release was also measured in neurons treated with 20 μM etoposide in the presence or absence of HN peptides at 24, 48, or 72 h after the onset of etoposide treatment. These experiments were performed independently for both assays. (C) Fluorescence microscopic views by Calcein-AM staining for neuronal viability. Seventy-two hours after treatment with 25 μM Aβ1–43, in the presence or absence of HN peptides, neurons were stained with Calcein-AM. Insets indicate magnified views of similarly treated independent cultures. Representative views are indicated.
No Effect of HN on Neuronal Cell Death by Q79 or SOD1 Mutants.
Expression of Q79, implicated in Huntington's disease and spinocerebellar ataxia, caused death in F11/EcR cells, which was not inhibited by cotransfection of pHN or pHNG or by treatment with HN peptides (Fig. 9 A and B, www.pnas.org). Familial amyotrophic lateral sclerosis-linked mutants of SOD1 also caused death when the cognate cDNAs were transfected to F11 cells, and the increased cell death rates were not inhibited by cotransfected pHN or by treatment with HN peptides (Fig. 9 C and D, www.pnas.org). pHN cotransfection or sHN treatment little affected expression of Q79 or mutant SOD1. Thus, HN had no effect on cell death by polyQ or SOD1 mutants.
Analysis of the Action of HN.
Using HPLC-purified radiolabeled sHNG, we performed the experiment shown in Fig. 5A, in which we incubated F11 cells with a high picomolar concentration (estimated to be 0.36–0.91 nM) of radiolabeled sHNG in the presence or absence of 2 μM unlabeled sHNG or sHNA, crosslinked cells with radiolabeled sHNG, precipitated the cells, and counted the radioactivity of the precipitated cells. As a result, F11 cells potently associated sHNG, and this association was displaced significantly by unlabeled sHNG but marginally by unlabeled sHNA, indicating there is a specific binding site(s) on the cell surface.
Figure 5.
Analysis of the action of HN. (A) F11 cells were incubated with radiolabeled sHNG in the presence or absence (1) of 2 μM unlabeled sHNG (2) or sHNA (3) for 2 h at 4°C and then treated with 200 μM BS3 for 20 min at 4°C. After adding Tris⋅HCl buffer and washing cells with PBS four times, cells were resuspended in PBS and subjected to radiocounting. The total input of radioactivity was 590,990 cpm/sample. (B) F11 cells were transfected with pcDNA or V642I-APP cDNA and incubated with or without 10 μM sHN in the presence or absence of 10 nM wortmannin (W), 100 μM genistein (G), or 50 μM PD98059 (PD). Seventy-two hours after the onset of transfection, cell mortality was similarly measured. (Inset) Jurkat cells were incubated with or without 100 ng/ml CH-11 (anti-Fas; MBL) in the presence or absence of 10 μM sHN (anti-Fas + sHN) for various periods, and cell mortality was similarly measured. (C) F11 cells were transfected with CN-procaspase-3 cDNA with pcDNA or V642I-APP cDNA by lipofection [0.2 μg CN-procaspase-3 cDNA, 0.8 μg V642I-APP cDNA, LipofectAMINE 2 μl, PLUS Reagent 4 μl (Upper); 0.4 μg CN-procaspase-3 cDNA, 0.6 μg V642I-APP cDNA, LipofectAMINE 2 μl, PLUS Reagent 4 μl (Lower)] in the presence or absence of 10 μM sHN or 100 μM Ac-DEVD-CHO [no transfection (lanes 1, 6); pcDNA transfection (lanes 2, 7); CN-procaspase-3 transfection (lanes 3, 8); CN-procaspase-3 + V642I-APP (lanes 4, 9); CN-procaspase-3 + V642I-APP + sHN (lanes 5, 10); CN-procaspase-3 + V642I-APP + Ac-DEVD-CHO (lane 11)]. Forty-eight hours after onset of transfection, cell lysates were submitted to immunoblot analysis with anti-caspase-3, anti-APP, or anti-tubulin antibody. Arrows, procaspase-3.
Protection by HN of V642I-APP-induced cell death was not affected by 10 nM wortmannin or 50 μM PD98059 but was totally suppressed by 100 μM genistein, suggesting that HN exerts cytoprotection through certain tyrosine kinase pathways not activating either PI-3 kinases or MAP kinases (Fig. 5B). We also examined whether HN affects caspase activation by V642I-APP in F11 cells. Consistent with induction of apoptosis by V642I-APP in F11 cells (2, 17), V642I-APP expression reduced the amount of CN-procaspase-3 without affecting that of tubulin. This reduction was recovered by acetyl-l-aspartyl-l-glutamyl-l-valyl-l-aspart-1-al (Ac-DEVD-CHO), suggesting that V642I-APP activates procaspase-3. There, HN protected CN-procaspase-3 from V642I-APP-induced cleavage (Fig. 5C). At least one target of HN action should thus be a proapoptotic mechanism.
The most established death pathway involving caspase-3 is the Fas system. Treatment with CH-11, an anti-Fas antibody, caused massive death in Jurkat cells in the presence or absence of 10 μM sHN (Fig. 5B Inset), indicating that HN cannot impair cell death by Fas. Although this result added support to the notion that the cytoprotective action of HN is not omnipotent, whether this failure was attributable to deficiency of HN-activated mechanisms in Jurkat cells or to inactivity of the HN signal on Fas-induced cell death remained unclear.
Expression of HN poly(A)+ RNA.
Northern blot analysis of human tissues indicated that HN poly(A)+ RNA was detected in the heart, skeletal muscles, kidney, and liver (Fig. 10, www.pnas.org). Lesser but significant expression was observed in the brain and the gastrointestinal tract. Little HN poly(A)+ RNA was in the immune system. The size of the expressed major message was ≈1.6 kb, which corresponds closely to the deduced size of DT159. The other messages were of different sizes, ≈3 and ≈1 kb. Because similar results were obtained by probing with DT77, coding for the 3′ untranslated region (UTR) of HN (Fig. 1) and with the antisense primer for the 5′UTR −432 to −414 of the DT clone, these RNAs likely represented the full-length HN message and its spliced variants. Similar results with minor differences were noted in the mouse tissues. Regional investigation of the human brain indicated that the cerebellum and the occipital lobe expressed relatively more messages than other regions.
Discussion
HN protects neuronal cells from death by three types of FAD genes and Aβ. HN is a unique secretory polypeptide, the sequence of which encodes activities for both secretion and cytoprotection. Such a factor, acting extracellularly with the antagonistic capability for this wide spectrum of the AD-relevant insults, has, to our best knowledge, never before been identified. Even focusing on the Aβ antagonism, only activity-dependent neurotrophic factor (22), basic fibroblast growth factor (23), and insulin-like growth factor-I (24) are known, among neurotropic factors, to antagonize neuronal death. In addition, HN did not inhibit cytotoxicity by Q79, SOD1 mutants, etoposide, or Fas, or basally occurring death, indicating that HN has a clear action specificity. As augmented Aβ42 secretion from cells expressing these FAD genes has been established, a simple interpretation is that HN commonly antagonizes cytotoxicity of FAD genes in F11 cells by suppressing Aβ42-forming APP processing or an Aβ42-triggered toxic mechanism. However, this is unlikely, based on multiple lines of evidence, including that, (i) Aβ peptides do not cause death in F11 cells (2), and (ii) mutant V642I-APP lacking the 41st and 42nd residues in the Aβ region can induce cytotoxicity as strong as V642I-APP in F11 cells (2). We also observe that HN inhibits death of F11 cells transfected with K595N/M596L-APP cDNA without affecting augmented Aβ secretion (25). In addition, it is quite unlikely that HN peptides suppress Aβ toxicity through direct interaction, because the effective concentration of HNG was lower by three to four orders than the toxic concentration of Aβ. Thus, the action of HN in suppressing Aβ neurotoxicity would not account for the action of HN on neuronal cell death by FAD genes but provides evidence suggesting that HN could also attenuate neuronal death in sporadic AD, in which Aβ neurotoxicity may contribute to neuronal loss.
There is an alternative possibility that FAD genes exert neurotoxicity not through Aβ secretion and that intracellular mechanisms triggered by each FAD gene and Aβ are commonly suppressed by HN. For instance, FAD genes promote Aβ-forming processing of APP, resulting in increased Aβ secretion on the one hand and accumulation of the cytoplasmic domains of APP on the other, which can potentially induce neurotoxicity (12, 21, 26, 27). Also, extracellular Aβ binds to cell-surface APP (28), and ligand binding to cell-surface APP generates cytotoxic signals via the cytoplasmic region of APP (25), suggesting that extracellular Aβ may induce cell death through APP and its cytoplasmic domain. Thus, HN might block cytotoxic signals from the cytoplasmic region of APP to commonly attenuate neurotoxicity by FAD genes and Aβ.
Although it should be investigated whether HN suppresses neuronal loss by FAD genes in vivo, no mouse model causing massive neuronal loss has been established by transgenic expression of FAD genes. However, the presence of an AD-specific neuroprotective factor, as suggested by HN, provides an important clue to generate such mouse models. Also, it remains uncertain why the S14G mutant indicates higher potency than HN, despite the fact that HN is encoded by a native DNA, whereas it is reasonable to assume that posttranslational S14 modification may allow HN to become as potent as HNG. Although HN poly(A)+ RNA was found to be present in various tissues, there is a possibility that HN is not produced in vivo at functional levels. There is also the possibility that HN also functions as a domain in other macromolecules. The unique combination of unbiased death-trap screening with a cDNA library from an intact lobe of an AD patient brain may provide a novel approach, termed disease-based death-trap screening, for efficient screening of protective genes against a wide spectrum of neurodegenerative diseases.
In summary, HN has several properties that suggest that HN becomes a useful clue for protecting neurons from death in AD: (i) HN completely protects neurons from death by Aβ with the effective concentration of HNG being low nM; (ii) HN and HNG inhibit death of neuronal cells by FAD mutants of APP, PS1, and PS2; (iii) HN cDNA encodes a chemically synthesizable short polypeptide that acts on cells from the outside; (iv) HN or HNG has no effect on death by genes linked to Huntington's disease/spinocerebellar ataxia or familial amyotrophic lateral sclerosis. Not only the high potency of the observed neuroprotection but also its comprehensiveness and specificity for AD-relevant insults are notable. Although future studies are required to examine toxicity, stability, and effectiveness in vivo, our data suggest that HN and its derivatives are appropriate starting points for future efforts to generate new AD therapeutics targeting neuroprotection.
Supplementary Material
Acknowledgments
We thank T. Tomita and T. Iwatsubo for antibodies; P. St. George-Hyslop, L. D'Adamio, and A. Kakizuka for plasmids; S. Koyasu for Jurkat cells; Y. and Y. Tamai for indispensable support; E. Ogata and J. T. Potts, Jr. for essential help; and T. Hiraki, S. Narumi, D. Wylie, and K. Nishihara for expert assistance. This work was supported by grants from the Japan Medical Association, Ono Medical Research Foundation, Ministry of Education, Science, and Culture of Japan, and the Organization for Pharmaceutical Safety and Research.
Abbreviations
- AD
Alzheimer's disease
- FAD
familial AD
- APP
amyloid precursor protein
- PS
Presenilin
- HN
Humanin
- pHN
a plasmid encoding HN cDNA
- s
synthetic
- EcD
ecdysone
- CM
cultured medium
- sHNA
sHN with C8A
- sHNG
synthetic S14G HN
- SOD1
superoxide dismutase-1
- CN
catalytically negative
- DT
death-trap
- HNR
L9R mutant of HN
- EcR
ecdysone receptor
- F11/EcR
F11 cells stably overexpressing EcR
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. YA029066).
References
- 1.Shastry B S, Giblin F J. Brain Res Bull. 1999;48:121–127. doi: 10.1016/s0361-9230(98)00156-7. [DOI] [PubMed] [Google Scholar]
- 2.Yamatsuji T, Okamoto T, Takeda S, Fukumoto H, Iwatsubo T, Suzuki N, Asami-Odaka A, Ireland S, Kinane T B, Nishimoto I. Science. 1996;272:1349–1352. doi: 10.1126/science.272.5266.1349. [DOI] [PubMed] [Google Scholar]
- 3.Zhao B, Chrest F J, Horton W E, Jr, Sisodia S S, Kusiak J W. J Neurosci Res. 1997;47:253–263. [PubMed] [Google Scholar]
- 4.Luo J J, Wallace W, Riccioni T, Ingram D K, Roth G S, Kusiak J W. J Neurosci Res. 1999;55:629–642. doi: 10.1002/(SICI)1097-4547(19990301)55:5<629::AID-JNR10>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 5.Wolozin B, Iwasaki K, Vito P, Ganjei J K, Lacana E, Sunderland T, Zhao B, Kusiak J W, Wasco W, D'Adamio L. Science. 1996;274:1710–1713. doi: 10.1126/science.274.5293.1710. [DOI] [PubMed] [Google Scholar]
- 6.Weihl C C, Ghadge GD, Kennedy S G, Hay N, Miller R J, Roos R P. J Neurosci. 1999;19:5360–5369. doi: 10.1523/JNEUROSCI.19-13-05360.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo Q, Furukawa K, Sopher B L, Pham D G, Xie J, Robinson N, Martin G M, Mattson M P. NeuroReport. 1996;8:379–383. doi: 10.1097/00001756-199612200-00074. [DOI] [PubMed] [Google Scholar]
- 8.Guo Q, Sebastian L, Sopher B L, Miller M W, Glazner G W, Ware C B, Martin G M, Mattson M P. Proc Natl Acad Sci USA. 1999;96:4125–4130. doi: 10.1073/pnas.96.7.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Z, Hartmann H, Do V M, Abramowski D, Sturchler-Pierrat C, Staufenbiel M, Sommer B, van de Wetering M, Clevers H, Saftig P, et al. Nature (London) 1998;395:698–702. doi: 10.1038/27208. [DOI] [PubMed] [Google Scholar]
- 10.Czech C, Lesort M, Tremp G, Terro F, Blanchard V, Schombert B, Carpentier N, Dreisler S, Bonici B, Takashima A, et al. Neuroscience. 1998;87:325–336. doi: 10.1016/s0306-4522(98)00162-6. [DOI] [PubMed] [Google Scholar]
- 11.Vito P, Lacana E, D'Adamio L. Science. 1996;271:521–525. doi: 10.1126/science.271.5248.521. [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto Y, Niikura T, Ito Y, Nishimoto I. J Biol Chem. 2000;275:34541–34551. doi: 10.1074/jbc.M005332200. [DOI] [PubMed] [Google Scholar]
- 13.Niikura T, Hashimoto Y, Okamoto T, Abe Y, Yasukawa T, Kawasumi M, Hiraki T, Kita Y, Terashita K, Kouyama K, et al. J Neurosci. 2001;21:1902–1910. doi: 10.1523/JNEUROSCI.21-06-01902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hashimoto Y, Jiang H, Niikura T, Ito Y, Hagiwara A, Umezawa K, Abe Y, Murayama Y, Nishimoto I. J Neurosci. 2000;20:8401–8409. doi: 10.1523/JNEUROSCI.20-22-08401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sudo H, Jiang H, Yasukawa T, Hashimoto Y, Niikura T, Kawasumi M, Matsuda S, Takeuchi Y, Aiso S, Matsuoka M, et al. Mol Cell Neurosci. 2000;16:708–723. doi: 10.1006/mcne.2000.0910. [DOI] [PubMed] [Google Scholar]
- 16.Bozyczko-Coyne D, McKenna B W, Connors T J, Neff N T. J Neurosci Methods. 1993;50:205–216. doi: 10.1016/0165-0270(93)90009-g. [DOI] [PubMed] [Google Scholar]
- 17.Niikura T, Murayama N, Hashimoto Y, Ito Y, Yamagishi Y, Matsuoka M, Takeuchi Y, Aiso S, Nishimoto I. Biochem Biophys Res Commun. 2000;274:445–454. doi: 10.1006/bbrc.2000.3143. [DOI] [PubMed] [Google Scholar]
- 18.Martoglio B, Dobberstein B. Trends Cell Biol. 1998;8:410–415. doi: 10.1016/s0962-8924(98)01360-9. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen H, Brunak S, von Heijne G. Protein Eng. 1999;12:3–9. doi: 10.1093/protein/12.1.3. [DOI] [PubMed] [Google Scholar]
- 20.Loo D T, Copani A, Pike C J, Whittemore E R, Walencewicz A J, Cotman C W. Proc Natl Acad Sci USA. 1993;90:7951–7955. doi: 10.1073/pnas.90.17.7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sudo H, Hashimoto Y, Niikura T, Shao Z, Yasukawa T, Ito Y, Yamada M, Hata M, Hiraki T, Kawasumi M, et al. Biochem Biophys Res Commun. 2001;282:548–556. doi: 10.1006/bbrc.2001.4604. [DOI] [PubMed] [Google Scholar]
- 22.Brenneman D E, Hauser J, Neale E, Rubinraut S, Fridkin M, Davidson A, Gozes I. J Pharmacol Exp Ther. 1998;285:619–627. [PubMed] [Google Scholar]
- 23.Mark R J, Keller J N, Kruman I, Mattson M P. Brain Res. 1997;756:205–214. doi: 10.1016/s0006-8993(97)00196-0. [DOI] [PubMed] [Google Scholar]
- 24.Dore S, Kar S, Quirion R. Proc Natl Acad Sci USA. 1997;94:4772–4777. doi: 10.1073/pnas.94.9.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hashimoto Y, Ito Y, Niikura T, Shao Z, Hata M, Oyana F, Nishimoto I. Biochem Biophys Res Commun. 2001;283:460–468. doi: 10.1006/bbrc.2001.4765. [DOI] [PubMed] [Google Scholar]
- 26.Lu D C, Rabizadeh S, Chandra S, Shayya R F, Ellerby L M, Ye X, Salvesen G S, Koo E H, Bredesen D E. Nat Med. 2000;6:397–404. doi: 10.1038/74656. [DOI] [PubMed] [Google Scholar]
- 27.Passer B, Pellegrini L, Russo C, Siegel R M, Lenardo M J, Schettini G, Bachmann M, Tabaton M, D'Adamio L. J Alzheimers Dis. 2000;2:289–301. doi: 10.3233/jad-2000-23-408. [DOI] [PubMed] [Google Scholar]
- 28.Lorenzo A, Yuan M, Zhang Z, Paganetti P A, Sturchler-Pierrat C, Staufenbiel M, Mautino J, Vigo F S, Sommer B, Yonkner B A. Nat Neurosci. 2000;3:460–464. doi: 10.1038/74833. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.