Abstract
RNase III is a key enzyme in the pathways of RNA degradation and processing in bacteria and has been suggested as a global regulator of antibiotic production in Streptomyces coelicolor. Using RNA-Seq, we have examined the transcriptomes of S. coelicolor M145 and an RNase III (rnc)-null mutant of that strain. RNA preparations with reduced levels of structural RNAs were prepared by subtractive hybridization prior to RNA-Seq analysis. We initially identified 7,800 transcripts of known and putative protein-coding genes in M145 and the null mutant, JSE1880, along with transcripts of 21 rRNA genes and 65 tRNA genes. Approximately 3,100 of the protein-coding transcripts were categorized as low-abundance transcripts. For further analysis, we selected those transcripts of known and putative protein-coding genes whose levels changed by ≥2-fold between the two S. coelicolor strains and organized those transcripts into 16 functional categories. We refined our analysis by performing RNA immunoprecipitation of the mRNA preparation from JSE1880 using a mutant RNase III protein that binds to transcripts but does not cleave them. This analysis identified ca. 800 transcripts that were enriched in the RNA immunoprecipitates, including 28 transcripts whose levels also changed by ≥2-fold in the RNA-Seq analysis. We compare our results with those obtained by microarray analysis of the S. coelicolor transcriptome and with studies describing the characterization of small noncoding RNAs. We have also used the RNA immunoprecipitation results to identify new substrates for RNase III cleavage.
INTRODUCTION
The double-strand-specific endonuclease RNase III is found in bacteria and eukaryotes (12, 25, 33). In addition to its general role in RNA processing, RNase III is involved in the regulation of gene expression in Escherichia coli and other bacteria. Thus, RNase III autoregulates its own expression in E. coli and is also involved in the regulation of the expression of the polynucleotide phosphorylase (PNPase) gene, pnp (3, 40). There are a number of other examples of the regulation of gene expression by RNase III in E. coli and other bacteria (reviewed in references 12, 25, and 33).
Streptomyces are Gram-positive soil bacteria notable for their ability to sporulate and for the production of antibiotics (6, 7, 11). Members of the genus Streptomyces produce nearly 70% of all antibiotics used in clinical and veterinary medicine worldwide (5). Much is known about the regulation of antibiotic production in the paradigm for the study of antibiotic production in streptomycetes, Streptomyces coelicolor (6, 7). Of particular relevance to the present report are the studies of Champness and coworkers (1, 2) on the absB locus of S. coelicolor. absB was shown to encode a homolog of E. coli RNase III, and the function of the absB product as a double-strand-specific endoribonuclease has been subsequently verified (1, 2, 10). Mutations in the absB locus reduce or abolish the production of all four of the antibiotics normally synthesized by S. coelicolor (2). Moreover, Aceti and Champness demonstrated decreased levels of the actII-orf4 and redD transcripts, encoding the pathway-specific regulators or Streptomyces antibiotic regulatory proteins (SARPs) for actinorhodin and undecylprodigiosin biosynthesis, respectively, in the absB mutant (1). These data strongly suggest that RNase III functions as a global regulator of antibiotic production in S. coelicolor.
A number of published studies have examined the levels of gene expression under various conditions and by utilizing various experimental approaches in S. coelicolor. These studies include the use of proteome analysis to examine the response of S. coelicolor to phosphate limitation (37) and to examine overall levels of gene expression in that organism (43) and the use of microarrays to examine the expression of genes involved in differentiation and antibiotic production under various conditions (17, 18, 22–24, 27, 37, 43). Recently, high-throughput cDNA sequencing (RNA-Seq) was used to examine the expression of small noncoding RNAs in S. coelicolor (42).
To facilitate the analysis of genes that serve as targets for RNase III regulation of antibiotic production and to obtain more general information about the role of RNase III in the control of gene expression in S. coelicolor, we have performed an RNA-Seq analysis of the transcriptomes of S. coelicolor M145, a parental strain, and JSE1880 (16), an S. coelicolor mutant lacking RNase III. This analysis was extended by using RNA immunoprecipitation to identify specific transcripts that are cleaved by RNase III and to which RNase III may bind without cleavage. Our studies represent the first application of the RNA-Seq approach coupled with RNA immunoprecipitation to the analysis of a Streptomyces transcriptome.
MATERIALS AND METHODS
Strains and growth conditions.
The S. coelicolor strains used in this study were M145, a prototrophic strain derived from S. coelicolor A3 (2) and lacking the plasmids SCP1 and SCP2 (29), and JSE1880, derived from M145 and containing a null mutation in the RNase III (rnc) gene in which the entire rnc coding sequence was replaced with a viomycin resistance cassette (16). Strains were grown on R2YE plates with viomycin (30 μg/ml) as necessary, atop cellophane membranes, and plates were inoculated with ca. 106 viable spores, which were not pregerminated. Plates were grown at 30°C. Mycelium was scraped off the plates at mid-log phase (ca. 20 h postinoculation), prior to the initiation of antibiotic synthesis, for the preparation of RNA. The RNAs used in the experiments described below were derived from mycelium harvested at the single mid-log-phase time point indicated above.
RNA preparation, RNA immunoprecipitation and cDNA synthesis.
Total RNA was prepared from two R2YE plates each of S. coelicolor M145 and JSE1880 using the modified Kirby procedure (30) as described previously (21). RNA preparations were treated twice with DNase I to remove contaminating DNA. Total RNAs, usually 10-μg portions, were enriched in mRNAs by two rounds of treatment with the MICROBExpress kit (Ambion). In this procedure, total RNA was mixed with a set of oligonucleotides that bind to the 16S and 23S rRNAs. Next, the rRNA hybrids were removed from the solution using derivatized magnetic microbeads. These RNA preparations are referred to below as mRNA. RNA immunoprecipitations were performed using mRNA prepared from JSE1880 and the RNase III D70A protein, a mutant of S. coelicolor RNase III that binds to RNase III substrates but does not cleave them (16). Binding mixtures contained mRNA equivalent to 10 μg of starting total RNA, 500 ng of RNase III D70A, and other components as described previously (16). Binding mixtures were incubated at 37°C for 10 min and were then treated with Dynabeads protein A (Invitrogen) which had been coated with polyclonal, affinity-purified antibody to RNase III D70A, according to the manufacturer's instructions. Following a 20-min incubation at room temperature with shaking, the Dynabeads-antibody-RNA-RNase III D70A complexes (referred to here as BARD) were isolated by biomagnetic separation, and the complexes were washed three times with 200 μl of phosphate-buffered saline–0.1% Tween 20 containing 20 units/ml of RNasin (Promega). Following the third wash, 100 μl of 10 mM Tris-HCl (pH 8.0)-1 mM Na2EDTA was added to the BARD complexes, and the suspensions were extracted with an equal volume of phenol-chloroform-isoamyl alcohol. RNAs were then collected by ethanol precipitation. cDNAs were synthesized from mRNA or from the BARD RNAs using random primers as described previously (9). The RNA preparations used for the M145 versus 1880 RNA-Seq comparison and those used for RNA immunoprecipitation were prepared from two separate mycelial samples, both grown as described above.
RNA-Seq analysis.
cDNAs prepared from M145 and JSE1880 mRNA and BARD RNAs were sheared to an average fragment size of 300 bases using acoustic focusing. The SPRI-Works platform was used to create libraries, which were sequenced on an Illumina GAII× with a read length of 50 bases. The resulting sequences were extracted and demultiplexed using Illumina's SCS2.7 software. The data were then analyzed using the RNA-Seq component of the CLC Genomics Workbench package. The imported reads were trimmed to remove low-quality sequences as well as any reads that were shorter than 36 bases. The reads were mapped to the published S. coelicolor genome sequence, using the prokaryote settings and RPKM (reads per kilobase of exon model per million mapped reads) (32) as the method of determining the expression level of the gene regions. Using S. coelicolor M145 as the control, the percent change in the JSE1880 reads was determined. The results were double-checked using normal reference mapping, to ensure proper alignments. To reduce the error rate, a Bonferroni correction was used with a confidence value of 0.01. The expression values were then exported to Excel files.
For the BARD analysis, the Chip-seq component of the CLC Genomics Workbench was used to compare the BARD and JSE1880 transcripts. The parameters were optimized to find known restriction sites. The JSE1880 sample was used as the control, and the optimal peak size was determined to be 100. Since the BARD contained transcripts precipitated via the RNA immunoprecipitation procedure, the coverage peaks represented RNase III binding sites and possible cleavage sites. To provide some confirmation of the RNA-Seq analysis, selected genes were examined to ensure that RNase III binding peaks were present for those transcripts known to contain RNase III cleavage sites.
Excel files containing the data described in this report may be obtained from the corresponding author upon request.
RT-PCR and RNase III cleavage assays.
Reverse transcription-PCRs (RT-PCRs) were performed using random primers for reverse transcription and gene-specific primers for PCR as described previously (8, 16). The results shown in Fig. 1 were obtained at 20 PCR cycles when the band intensity was linearly proportional to the number of cycles. Primer sequences are shown in Table S1 in the supplemental material. Primers rps1 and +16R are complementary to the readthrough transcript from the rpsO-pnp operon of S. coelicolor, while primers ramR F1 and R1 identified the ramR transcript. SCO5737 (encoding polynucleotide phosphorylase) and SCO3982 to SCO3988 were amplified using primer pairs SCPNPF3 and pnpB1 and SCO3982F1 and R1, respectively (see Table S1), and products were cloned into pCR2.1TOPO as described previously (10). Nonradioactive runoff transcripts from these constructs were prepared using T7 RNA polymerase, as described previously (16). The organization of SCO3982 to SCO3988 suggested that all seven genes are transcribed to produce a polycistronic mRNA. Therefore, the transcript used in the RNase cleavage assays was transcribed from a PCR product representing all seven contiguous genes. RNase III digestions of transcripts and primer extensions of digestion products were performed as described previously (10). Sequences of the primers used for primer extension, SCINTLF1, 3987R2, and 3988 R1, are shown in Table S1 in the supplemental material.
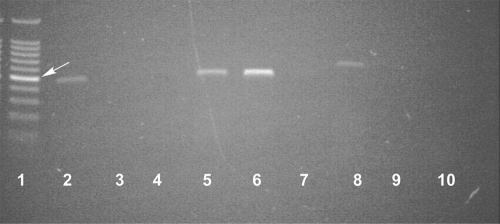
Fig 1.
RT-PCR analysis of transcripts from S. coelicolor M145 (parental strain) and JSE1880 (rnc-null mutant). RT-PCR was performed as described previously (8, 16), and products were analyzed after 20 PCR cycles to ensure linearity between band intensity and the number of cycles. Lane 1, size standards (the arrow indicates the 500-bp standard); lane 2, mRNA from M145 as the template for reverse transcription; lane 3, M145 BARD RNA; lane 4, M145 mock BARD RNA; lanes 5 and 8, JSE1880 mRNA; lanes 6 and 9, JSE1880 BARD RNA; lanes 7 and 10, JSE1880, mock BARD RNA. The primers used for the PCRs for lanes 2 to 7 were specific for the readthrough transcript from the S. coelicolor rpsO-pnp operon and produced a 460-bp fragment, while the primers used for lanes 8 to 10 were specific for ramR, producing a 583-bp fragment.
RESULTS AND DISCUSSION
RNA-Seq analysis of the S. coelicolor transcriptome.
The RNA substrates used in these studies were derived from total S. coelicolor RNA prepared from mycelium grown on solid medium to mid-log phase. Neither of the pigmented antibiotics produced by S. coelicolor was visible at 20 h postinoculation when mycelium was harvested. As described in Materials and Methods, total RNAs, isolated ca. 20 h postinoculation from S. coelicolor M145 and from JSE1880, the rnc-null mutant, were treated by the MICROBExpress procedure to reduce the levels of rRNA in these preparations. These RNA preparations are referred to below as S. coelicolor mRNAs. RNA-Seq analysis of these preparations indicated that mRNA reads represented 64% of the total for M145 and 76% of the total for JSE1880. As ribosomal RNAs normally account for 85 to 90% of the total RNA in bacteria, the MICROBExpress procedure produced a considerable enrichment of mRNAs in our preparations.
RT-PCR was used to analyze the quality of these RNA preparations prior to the RNA-Seq study. Typical results are presented in Fig. 1. Lanes 2 and 5 show the products obtained using M145 and JSE1880 mRNAs, respectively, as templates for reverse transcription, with primers complementary to the readthrough transcript from the rpsO-pnp operon of S. coelicolor employed for PCR. The readthrough transcript produced by the rpsO-pnp operon is known to be cleaved by RNase III in S. coelicolor (10). Thus, the rspO-pnp transcript was expected to be present in both M145 and JSE1880, as is shown in Fig. 1, with higher levels of this transcript present in the mutant strain lacking RNase III (15). Quantification of the band intensities indicated that the JSE1880 mRNA preparation (lane 5) contained ca. 2.5 times as much of the readthrough transcript as the M145 preparation (lane 2), a value consistent with the levels observed in Northern blots of RNAs from M145 and JSE1880 (15). Figure 1 also shows that the JSE1880 mRNA preparation contained transcripts of the ramR gene (lane 8), which encodes a response regulator (35). Other RT-PCR experiments revealed the presence in the mRNAs of transcripts for relA, encoding the (p)ppGpp synthetase; hrdB, encoding the major vegetative sigma factor in S. coelicolor; and cca, encoding the S. coelicolor tRNA nucleotidyltransferase (data not shown). These results demonstrated that transcripts corresponding to a diverse array of genes were present in the mRNA preparations.
cDNAs were then prepared from these mRNA preparations via reverse transcription with random primers, and the cDNAs were subjected to high-throughput sequencing using the Illumina platform. Sequences obtained from the amplified cDNAs were analyzed using the RNA-Seq component of the CLC Genomics Workbench package. A total of ca. 9 million reads were obtained for M145 and 8 million for JSE1880, equivalent to 36× coverage of the S. coelicolor genome. The annotated S. coelicolor genome database (http://strepdb.Streptomyces.org.uk) lists 7,846 known or putative protein-coding genes and 86 genes encoding structural RNAs and tRNAs. The RNA-Seq analysis identified 7,800 putative protein-coding transcripts in S. coelicolor M145 and JSE1880, 65 tRNA genes, and 18 genes encoding ribosomal RNAs. Notably and as expected, the rnc (RNase III) transcript was absent from JSE1880.
It was surprising to detect as many as 7,800 transcripts in the S. coelicolor mRNA pool during the exponential phase of growth. We suspect that this number reflects the high sensitivity of the RNA-Seq procedure. The use of amplified cDNA libraries as templates for sequencing allows the detection of even low-abundance transcripts. To estimate the number of transcripts that might fall into this low-abundance category, we examined the levels of expression of six genes that are transcribed maximally during the stationary phase of growth of S. coelicolor rather than in exponential phase. Five of these genes (SCO5315, SCO5316, SCO5317, SCO5318, SCO5321) are components of the whiE locus, responsible for the synthesis of the polyketide spore pigment in S. coelicolor, and the sixth (SCO4035) encodes the RNA polymerase sigma factor, SigF (28). We considered the RPKM values for these six genes and obtained an average of 59.74 (range, 39.63 to 76.94). We then determined the number of genes with RPKM values above this average. That number was 4,702. Thus, we suggest that the expression of at least 4,700 genes is required for the exponential growth of S. coelicolor under our conditions. The remaining ca. 3,100 genes may be transcribed at low levels during exponential growth, perhaps from leaky promoters. Antisense transcripts, which have been identified in S. coelicolor and other bacteria (39, 41, 42), may also contribute to the low-abundance category.
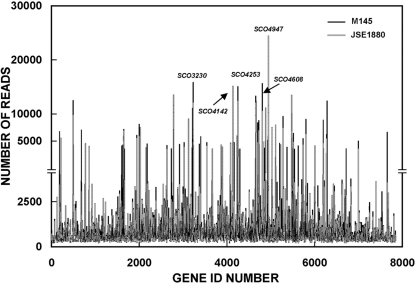
The transcriptomes of S. coelicolor M145 and JSE1880 are shown with single-gene resolution in Fig. 2. Several of the transcripts that yielded the largest number of sequencing reads for each mRNA pool are highlighted in the figure. The largest number of reads from the M145 sequencing data was obtained for SCO3230, which encodes peptide synthetase I for the calcium-dependent antibiotic (CDA) synthesized by S. coelicolor. SCO3231 and SCO3232 are contained in a cluster of genes that are involved in CDA synthesis (20) and were present at levels well above the average level of expression of genes in M145. SCO4142 encodes a putative phosphate binding protein precursor, while SCO4253 encodes a homolog of a phage tail sheath protein. The function of the latter protein in S. coelicolor, if there is one, and the reasons for its high level of expression are unknown at this point. SCO4808 encodes succinyl coenzyme A (CoA) synthetase β chain. Succinyl CoA has recently been shown to be involved in the synthesis of a number of natural products, including some antibiotics (48). The largest number of reads from either S. coelicolor M145 or JSE1880 was obtained for SCO4947, which encodes nitrate reductase α chain, NarG3. The level of this transcript was ca. 4-fold higher in JSE1880 than in M145. S. coelicolor contains operons for three nitrate reductases (14), and transcript levels for other subunits of reductase 3 and for several subunits of reductase 2 also increased in JSE1880, by 2.0- to 3.4-fold (see Table S2 in the supplemental material).
Fig 2.
Single-gene resolution plot of the transcriptomes of S. coelicolor M145 and JSE1880. The RNA-Seq analysis was performed as described in Materials and Methods. The transcripts that are noted specifically in the plot are those for which the largest numbers of sequencing reads were observed with either M145 or JSE1880 mRNAs as templates. It should be noted that the S. coelicolor chromosome is linear (4), thus the gene ID numbers indicate the order of genes beginning at the left end of the chromosome.
It is relevant at this point to investigate the extent to which the number of sequencing reads shown for each transcript represented in Fig. 2 reflects the abundance of that transcript in the mRNA pools from S. coelicolor M145 and JSE1880. To examine this issue, we calculated the relative amounts of two transcripts from the rpsO-pnp operon from the RNA-Seq results shown in Fig. 2 and compared those results with previous data obtained for these two transcripts by Northern blotting. Our Northern blotting studies indicated a 1.6-fold increase in the rpsO transcript level, encoding ribosomal protein S15, in the mRNA pools from JSE1880 compared to that of M145 during exponential growth (15). The corresponding ratio obtained from the RNA-Seq data was ca. 1.5. Similarly, Northern blotting results indicated a 2.7-fold increase in the level of the readthrough transcript in JSE1880 compared with that in M145 during exponential growth (15). To measure the level of the readthrough transcript from the RNA-Seq data, we determined the number of sequencing reads for the intergenic region between rpsO and pnp, converted the number of reads to RPKM (32), and compared the RPKM values for M145 and JSE1880. The ratio obtained was ca. 2.5. Thus, our Northern blotting results compare favorably with the data obtained from the RNA-Seq analysis, and at least for these two transcripts, our results suggest that the number of sequencing reads does reflect transcript abundance in the mRNA pools. We note, however, that we do not have Northern blotting data for other transcripts at this point.
To facilitate analysis of the RNA-Seq data, we elected to examine those transcripts whose levels changed by at least 2-fold in JSE1880 compared with levels in M145. The RNA-Seq analysis identified 143 such transcripts, of which 105 gave RPKM values above the low-abundance cutoff value indicated above. We organized those 143 transcripts into the 16 functional categories listed in Table 1. A complete listing of the genes corresponding to these transcripts can be found in Table S2 in the supplemental material. These transcripts were derived from genes with potential regulatory functions (e.g., bldK [34]), whose transcript levels increased by ca. 2.5-fold in JSE1880 compared with those in M145 and cdaR (38), whose transcript levels decreased by 2.1-fold. The transcript categories also include metabolic genes and genes of unknown function. Interestingly, most of the transcripts whose levels changed were increased; less than 20% of the transcripts whose levels changed by a factor of ≥2 were present at decreased levels. tRNA transcripts are included in Table 1 even though not all of those transcripts changed by 2-fold. Specifically, we observed that eight tRNAs increased by ≥2-fold in JSE1880, while two decreased by that level.
Table 1.
Functional categories of transcripts whose levels changed by at least 2-fold in S. coelicolor JSE1880 as compared with those of M145
| Functional category | No. of genes whose transcript levelsa: |
|
|---|---|---|
| Decreased | Increased | |
| Putative regulatory genes | 2 (0) | 15 (10) |
| ABC transporters | 2 (0) | 7 (5) |
| N and P metabolism | 0 (0) | 13 (12) |
| Carbohydrate metabolism | 0 (0) | 10 (8) |
| ATP-GTP binding proteins | 2 (0) | 0 (0) |
| Cytochromes | 0 (0) | 3 (3) |
| Membrane ATPases | 1 (0) | 0 (0) |
| Other membrane proteins | 3 (0) | 11 (10) |
| Transposases and other mobile elements | 2 (0) | 3 (1) |
| Glycerol utilization and metabolism | 2 (0) | 0 (0) |
| Lipid metabolism | 1 (0) | 1 (1) |
| Miscellaneous proteins with homologs in other species | 8 (1) | 12 (7) |
| Hypothetical proteins | 6 (2) | 37 (16) |
| Ribosomal proteins | 0 (0) | 2 (0) |
| tRNAs | 2 (0) | 8 (1) |
The numbers represent the numbers of genes in a particular category whose transcript levels decreased or increased by at least 2-fold in JSE1880 as compared with those of M145. The numbers in parentheses indicate the numbers of those transcripts that were present in the BARD.
RNA immunoprecipitation.
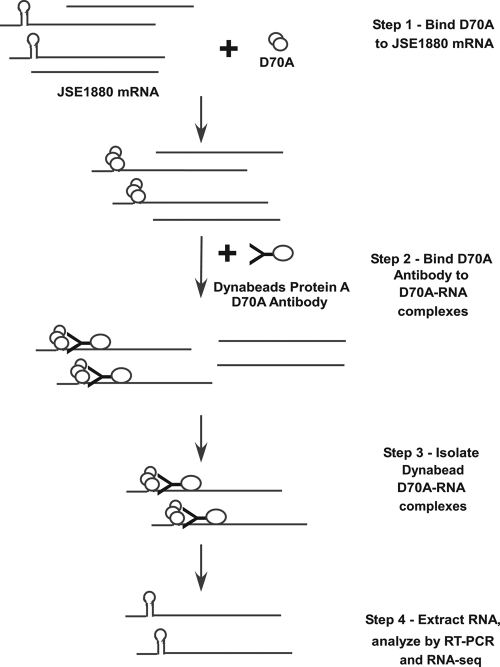
The RNA-Seq data provided a revealing look at the differences between the transcriptomes of S. coelicolor M145 and JSE1880, but those data include changes that are both direct and indirect results of the effects of RNase III on gene expression. To obtain a more direct look at those effects, we coupled RNA immunoprecipitation (RIP) with the RNA-Seq procedure. An outline of the RIP procedure is shown in Fig. 3. Briefly, JSE1880 mRNAs were allowed to bind to RNase III D70A, a mutant form of the enzyme that binds substrates but will not cleave them (16). We used JSE1880 mRNA for these experiments, because that mRNA preparation would contain transcripts that had not been cleaved by RNase III in vivo. The RNase III D70A-mRNA complexes were then treated with antibody to RNase III D70A coupled to Dynabeads protein A conjugates. After biomagnetic separation and washing, RNA was extracted from the Dynabeads complexes. These RNAs are referred to as the BARD (bead-antibody-RNA- D70A).
Fig 3.
Outline of the RNA immunoprecipitation (RIP) procedure. As described in detail in Materials and Methods, the procedure involved binding the RNase III mutant protein, RNase III D70A, to S. coelicolor mRNA, binding anti-RNase III D70A antibody to the RNA-RNase III D70A complexes, and isolation of the antibody-RNA-RNase III D70A complexes using magnetic beads.
This analysis identified 1,401 transcripts to which RNase III D70A and presumably wild-type RNase III can bind. To examine the specificity of this binding, we performed RT-PCR analysis on several BARD preparations. Figure 1 shows RT-PCR results obtained for the rpsO-pnp readthrough transcript in the BARD from S. coelicolor M145 (lane 3) and JSE1880 (lane 6). Quantification of these bands indicated that the JSE1880 BARD contained ca. 2.5 times the amount of the readthrough transcript as did the JSE1880 mRNA preparation. Thus, RNA immunoprecipitation did enrich the BARD in transcripts that are recognized by RNase III. No PCR product corresponding to the readthrough transcript was observed in the M145 BARD, consistent with our observation that the half-life of the readthrough transcript is significantly shorter in vivo in M145 than in JSE1880 (15). We also examined the BARD for another transcript, ramR, which has been shown not to be a substrate for RNase III (46). It is apparent from Fig. 1 that although this transcript is present in the JSE1880 mRNA preparation (lane 8), it is absent from the JSE1880 BARD (lane 9). Thus, the ramR transcript was not precipitated in the RIP procedure. Mock BARDs were prepared from reaction mixtures lacking RNase III D70A. RT-PCR analyses of those preparations (lanes 4, 7, and 10) showed that little nonspecific precipitation of the rpsO-pnp and ramR transcripts occurred during the RNA immunoprecipitation.
Table 1 (see also the supplemental material) shows the distribution of transcripts in the BARD among the various functional categories. Of the 143 transcripts whose levels changed by 2-fold, 79 were present in the JSE1880 BARD. Notably, SCO4947, the transcript of the nitrate reductase α chain, was present in the BARD, as were a number of transcripts for ABC transporters (see Table S2 in the supplemental material and see below). The genes for the latter group of transcripts are likely targets for RNase III regulation of antibiotic production, as ABC transporters are known to be involved in antibiotic production in Streptomyces (19, 36). It is also interesting to note that one tRNA transcript, SCOt26, encoding tRNAala (CGC), increased by 3.6-fold in JSE1880 and was present in the BARD.
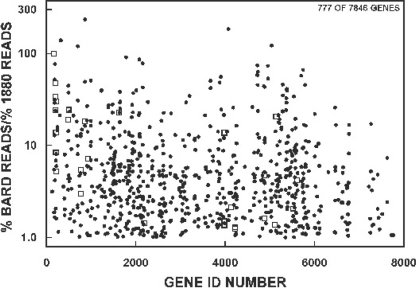
To further refine our RIP results, we calculated an enrichment ratio for each of the 1,401 transcripts in the BARD, defined as the percentage of the total reads for each transcript in the BARD divided by the percentage of the total reads for the same transcript in the starting material for the RIP procedure, JSE1880 mRNA. This calculation revealed levels of enrichment in the BARD ranging from 0.01 to over 200. An enrichment level of less than 1.0 indicates that the percentage of reads for a particular transcript in the BARD was actually lower than the corresponding percentage of reads in the mRNA pool. We reasoned that these transcripts represented low-affinity substrates for RNase III and/or products precipitated nonspecifically by RNase III D70A or the antibody. These transcripts were thus eliminated from further analysis. With the elimination of those transcripts, as shown in Fig. 4, 777 transcripts in the BARD were found to be present at a level equal to or greater than the level of their representation in the JSE1880 mRNA pool. Twenty-eight of these transcripts also increased by ≥2-fold in JSE1880 compared with levels in M145 (open squares in Fig. 4). These transcripts are listed in Table S3 in the supplemental material.
Fig 4.
Scatter plot of the enrichment ratios for BARD transcripts. Enrichment ratios, defined as the percentage of the total reads for each transcript in the BARD divided by the percentage of the total reads for the same transcript in the starting JSE1880 mRNA pool, were calculated and are plotted against gene ID number. The data are plotted using a semilog scale to spread the points over the area of the graph. Open squares represent the enrichment ratios for the 28 genes whose transcript levels increased by ≥2-fold in the JSE1880 mRNA pool compared with those of M145.
Table 2 lists the 10 genes whose transcripts were enriched to the greatest extent in the BARD. It is noteworthy that two of these genes encode putative regulatory proteins and a third encodes an ECF sigma factor. ECF sigma factors have been shown to be involved in the regulation of antibiotic production in Streptomyces antibioticus and Bacillus subtilis (26, 31). It is also noteworthy that the two most abundant transcripts in JSE1880, SCO4142 and SCO4947, were not enriched in the BARD (enrichment ratios = 0.04 and 0.13, respectively). Thus, the increased levels of these transcripts observed in JSE1880 may result from an indirect effect of RNase III rather than from cleavage of the transcripts themselves.
Table 2.
The 10 genes whose transcripts were enriched to the greatest extents in the BARD
| Gene ID no. | Function or protein the gene encodes | Fold increase in JSE1880 | Fold enrichment in BARD |
|---|---|---|---|
| SCO0168 | Putative transcriptional regulator | 2.41 | 100 |
| SCO0323 | Unknown | 1.62 | 138 |
| SCO0703 | Putative regulator, similar to AbaA, orfD (13) | 1.36 | 119 |
| SCO0864 | Probable ECF sigma factor | 1.70 | 234 |
| SCO1783 | Unknown | 1.23 | 91 |
| SCO2081 | Unknown | 1.23 | 86 |
| SCO2154 | Putative integral membrane protein | 1.38 | 77 |
| SCO4077 | Unknown | −1.01 | 184 |
| SCO5040 | Unknown, contains TTA codon | 1.22 | 122 |
| SCO5123 | Putative small membrane protein | 1.24 | 247 |
We argue that the 777 genes whose transcripts were identified via the analysis shown in Fig. 4 represent specific targets for RNase III for the following reasons. First, only 1,401 of ca. 7,800 transcripts were initially identified in the BARD. Some transcripts present at reasonable levels of abundance in JSE1880 were not precipitated by the RIP procedure. Thus, the RIP procedure did not simply precipitate transcripts indiscriminately from the mRNA pool. Second, the level of a given transcript in the BARD was not a simple function of its level in the mRNA pool. That is, the BARD does not contain only the most abundant transcripts in the mRNA pool. Many transcripts that were of relatively lower abundance in the mRNA pool were significantly enriched in the BARD. Finally, as indicated above, we eliminated those transcripts whose enrichment level in the BARD was <1.0 compared with the mRNA pool. Given these considerations, we suspect that the 777 genes whose transcripts are represented in Fig. 4 approximate the size of the RNase III regulon in S. coelicolor.
Comparison of RNA-Seq and microarray analyses of S. coelicolor.
The results of three microarray studies can be productively compared with our RNA-Seq results (Table 3). In the first, Huang et al. performed a global microarray analysis of gene expression in S. coelicolor M145 (22). They looked at several time points during growth in liquid cultures using modified R5 medium (29). Our data were performed using RNAs obtained from mycelium grown on solid media (see Materials and Methods), and we examined only one time point on the growth curve. We estimate that our data were taken at a time of growth corresponding to a time between points T4 and T5 (around 20 h postinoculation) on the growth curve obtained by Huang et al. Noteworthy in this regard is their observation of an increase in the levels of the CDA peptide synthetase I (SCO3230) between time points T3 and T4 in their studies. We also observed significant levels of this transcript in our studies (Fig. 2), and we suspect that we would have detected even higher levels at later points along the growth curve.
Table 3.
Comparison of RNA-Seq and microarray analyses of parental and mutant strains of Streptomyces coelicolor
| Transcript | Fold increase or decreasea | Protein encoded by the gene | Total no. | Present in BARD |
|---|---|---|---|---|
| Transcripts whose levels changed by ≥2-fold in the RNA-seq analysis and by ≥6-fold in the microarray analysis | ||||
| SCO3142 | 2.23 | Conserved hypothetical protein | No | |
| SCO3152 | 2.73 | Conserved hypothetical protein | Yes | |
| SCO3216 | −4.81 | Membrane ATPase | No | |
| SCO3217 | −2.18 | CdaR | No | |
| SCO3223 | −13.76 | Putative ABC transporter | No | |
| SCO3224 | −9.71 | Putative ABC transporter ATP-binding protein | No | |
| SCO3236 | −3.02 | Possible antibiotic oxygenase | No | |
| SCO3244 | −4.19 | Putative secreted protein | No | |
| SCO3984 | 2.23 | Conserved hypothetical protein | Yes | |
| SCO4843 | 2.20 | Putative integral membrane protein | No | |
| SCO4880 | 3.91 | Possible transferase | Yes | |
| SCO4947 | 3.23 | Nitrate reductase α chain, NarG3 | Yes | |
| SCO4948 | 3.77 | Nitrate reductase β chain, NarH3 | Yes | |
| SCO5107 | 3.16 | Putative succinate dehydrogenase membrane subunit | Yes | |
| SCO6276 | −4.89 | Epoxide hydrolase | No | |
| SCO6277 | −4.95 | Putative secreted protein | Yes | |
| SCO6278 | −4.06 | Integral membrane transport protein | No | |
| SCO6280 | −2.00 | Similar to Streptomyces nogalater nogalamycin biosynthesis activator, SnoA | No | |
| SCO6282 | −15.73 | Probable 3-oxoacyl-[acyl-carrier protein] reductase | Yes | |
| SCO6283 | −2.56 | Conserved hypothetical protein | No | |
| SCO6682 | −3.80 | Conserved hypothetical protein | 21 | No |
| Transcripts whose levels changed by ≥2-fold in the RNA-Seq analysis and by <6-fold in the microarray analysis | 51 | |||
| Transcripts whose levels changed by ≥2-fold in the RNA-Seq analysis but which were not listed in the microarray analysis | 72 | |||
| Transcripts whose levels changed by ≥6-fold in the microarray analysis but by <2-fold in the RNA-Seq analysis | 165 |
Fold increase or decrease in the level of a given transcript in JSE1880 as compared with that of M145.
In a second study, Huang et al. compared the levels of gene expression in an S. coelicolor parental strain and an RNase III mutant (23). In these studies, the authors isolated RNA from mycelium grown on solid R5 medium, which is similar to the R2YE medium used in the present study (29). Nevertheless, several caveats apply to the comparison between the data of Huang et al. and our studies. First, Huang et al. used a different parental strain, J1501, a strain that lacks the phage growth limitation (Pgl) system and which possesses nearly 1,000 kb of additional DNA, present as long terminal inverted repeats, compared with M145 (44). Second, Huang et al. did not use an RNase III-null mutant; rather, they used a point mutant, C120, whose RNase III has been shown to retain some catalytic activity (16). RT-PCR analyses and our RNA-Seq results show that JSE1880 is a true RNase III-null mutant. As noted above, there were also slight differences in the growth medium used in their study (R5) and the one employed in the studies reported here (R2YE). Finally, their strains contained plasmids bearing the tipA promoter, and these plasmids were used to overexpress selected genes in their experiments. Despite these differences, a comparison between the results of Huang et al. and those presented here is illuminating. At 28 h postinoculation of their medium, Huang et al. observed ca. 200 genes whose transcript levels changed by at least 6-fold between J1501 and C120, a number similar to the number of transcripts whose levels changed by ≥2-fold between M145 and JSE1880. A more detailed comparison of the microarray and RNA-Seq results is presented in Table 3. We tallied 21 transcripts whose levels changed by ≥2-fold in our RNA-Seq analyses and by ≥6-fold in the microarray analyses. Of these 21, 8 were also present in the BARD. Fifty-one transcripts changed by ≥2-fold in the RNA-Seq analysis but by <6-fold in the microarray study, while 72 transcripts changed by ≥2-fold in the RNA-Seq analysis but were not listed among the transcripts identified by Huang et al. Finally, 165 transcripts changed by ≥6-fold in the microarray analysis but by <2-fold in our RNA-Seq study. Some of the differences between our results and those of Huang et al. are almost certainly due to the differences between the experimental conditions used in the two studies, but some probably reflect intrinsic differences in the microarray and RNA-Seq approaches.
In a third study, Karoonuthaisiri et al. examined the regional organization of gene expression in S. coelicolor using microarrays (27). These studies were based on the prediction from the genome sequence that a ca. 4.9-Mb central region of the linear S. coelicolor chromosome would be more active transcriptionally than the regions flanking this central core (4). Karoonuthaisiri et al. observed an average 4-fold difference in the levels of gene expression between the central core and the flanking arms. We examined the same question by totaling the sequencing reads obtained for the 1.5-Mb left arm, the 2.3-Mb right arm, and the 4.9-Mb central region of the S. coelicolor chromosome. Our results were less dramatic than those of Karoonuthaisiri et al. but were consistent with the prediction. We observed an average of ca. 500 reads per gene for the left and right arms of the chromosome and ca. 800 reads per gene for the central core. We note that Vockenhuber et al. (42) reported that half of the annotated transcripts they identified were transcribed from genes in the central core, and 28% and 22% were derived from the left and right arms of the chromosome, values similar to those obtained in the analysis reported here.
RNA-Seq analysis of S. coelicolor small noncoding RNA.
Two recent studies have identified small, noncoding RNAs in S. coelicolor. Using bioinformatic approaches, Swiercz et al. identified 114 intergenic regions that contained putative sRNA genes and verified the presence of nine sRNAs via Northern blotting and cDNA cloning (39). Vockenhuber et al. recently reported an RNA-Seq analysis of the S. coelicolor transcriptome and identified 63 possible sRNAs (42). They verified the presence of 11 of these RNAs in vivo by Northern blotting.
By mapping our RNA-Seq reads to the unannotated S. coelicolor genome sequence, we were able to measure the number of reads in the intergenic regions corresponding to the 20 sRNAs verified by Northern blotting in the studies of Swiercz et al. All of these sRNAs except one (scr3974) were present in both the M145 and JSE1880 mRNA pools. The numbers of reads observed for 17 of the remaining sRNAs were essentially the same in the two mRNA preparations. Two sRNAs, scr6925 and scr2101, were present at higher levels in JSE1880 than in M145. Thus, the level of scr6925 was 2-fold higher in JSE1880 and scr2101 was 7-fold higher in JSE1880 than in M145. Reads corresponding to both scr6925 and scr2101 were observed in the BARD, but the enrichment ratio for scr6925 was only 0.10 and that for scr2101 was 0.57.
SCO6925 encodes a putative membrane protein. SCO6925 transcripts were not significantly increased in JSE1880 compared with M145 and were not present in the BARD. SCO2101 encodes a putative carotenoid dehydrogenase, and its transcript levels were increased by 1.6 in JSE1880 compared with those of M145. SCO2101 transcripts were also present in the BARD and their levels were enriched by a factor of 3.6 compared with those of the JSE1880 mRNA pool. It is possible that scr2101 is regulated indirectly by RNase III rather than serving as a substrate for the enzyme and that the loss of this regulation in JSE1880 leads to increased levels of the SCO2101 transcript in the rnc mutant.
RNA immunoprecipitation identifies substrates for RNase cleavage and targets for RNase III binding.
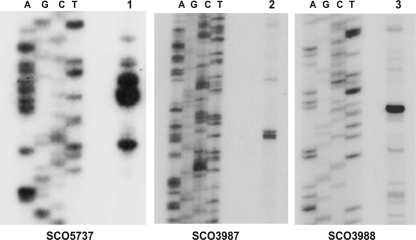
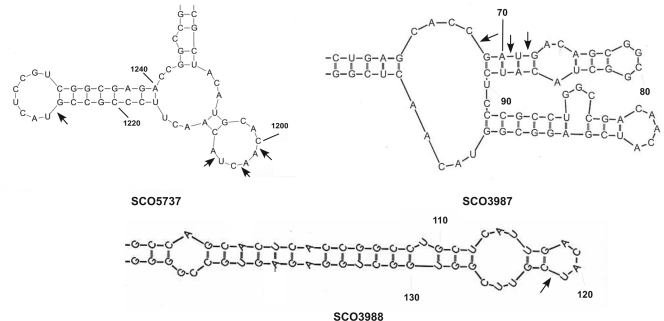
The binding of RNase III D70A to specific transcripts in the BARD suggested the likely possibility that at least some of those transcripts would be substrates for RNase III cleavage. We chose two transcripts for which the BARD data suggested multiple RNase III binding sites and cloned the genes corresponding to those transcripts in pCR2.1 TOPO. The genes selected for this analysis were SCO5737, encoding polynucleotide phosphorylase, and SCO3982 to SCO3988, encoding several putative transcriptional regulators, including a putative member of the GntR family, SCO3986. SCO3982 and SCO3983 encode hypothetical proteins but both contain TTA codons, suggesting that they may be targets for bldA regulation. The SCO5737 transcript level was enriched by 3.6-fold in the BARD compared to that in the JSE1880 mRNA pool, while transcript levels corresponding to the SCO3982 to SCO3988 region were enriched by 1.4- to 14-fold. Radioactive and nonradioactive transcripts of SCO5737 and SCO3982 to SCO3988 were prepared as described previously (10, 16). Digestion of radioactive transcripts of these genes showed that RNase III did cleave those transcripts (data not shown). Nonradioactive runoff transcripts were then digested with RNase III, and the digestion products were subjected to primer extension using primers complementary to regions downstream of the suspected cleavage sites (see Table S1 in the supplemental material). Those suspected sites were identified based on the sequence regions identified by sequencing the BARD cDNAs. Primer extension products were fractionated on a sequencing gel along with a sequencing ladder obtained using the same primer employed for primer extension. Results of these experiments are shown in Fig. 5. It is apparent that there are RNase III cleavage sites within the SCO5737 coding region (Fig. 5, left). In addition, RNase III cleavage sites were observed in the SCO3982 to SCO3988 transcript in regions corresponding to the SCO3987 and SCO3988 open reading frames (Fig. 5, center and right). Mfold modeling (49) of the regions containing the cleavage sites suggested that all of the observed sites occurred in stem-loop structures (Fig. 6), as has been observed for other substrates of S. coelicolor RNase III (10, 46). These results show clearly that the RNA immunoprecipitation procedure does identify substrates for RNase III. The RNA-Seq approach will allow us to identify RNase III cleavage sites globally in S. coelicolor using the 5′-end-sequencing strategy described by Wurtzel et al. (45).
Fig 5.
Primer extension of products from RNase III cleavage assays. RNase III digestions of nonradioactive transcripts of SCO5737 and SCO3982 to SCO3988 were performed as described previously (10). Primer extension of digestion products was performed as described previously (10), and the same primer used for extension was also used to generate the sequencing ladders shown in the figure. Lanes 1, 2, and 3 show the extension products obtained representing RNase III cleavages in SCO5837, SCO3987, and SCO3988. The results shown in the left panel were taken from a previously published figure (15) and are reproduced here with permission from the Journal of Bacteriology.
Fig 6.
Mfold (49) models of the regions of SCO5737, SCO3987, and SCO3988 that contain RNase III cleavage sites. Bases are numbered from the first base in the translation start codon for each gene, and arrows indicate the major RNase III cleavage sites determined by primer extension.
Although the results of Fig. 5 indicate that the BARD does contain transcripts that are cleaved by RNase III, we think it unlikely that all of the transcripts represented in Fig. 4 are RNase III substrates. Indeed, examination of the RNA immunoprecipitation results identified one transcript, sti1 (SCO0762), encoding a protease inhibitor precursor, which is present in the BARD (although at an enrichment ratio of 0.80) even though the sti1 transcript has been shown not to be cleaved by RNase III (46). It has been proposed that in E. coli, RNase III regulates the expression of some genes by binding to their transcripts without cleaving them (12, 25, 33). Rather than using a brute-force approach to identify such transcripts in S. coelicolor, it should be possible to test this hypothesis directly by performing RNA-Seq analysis on a strain lacking RNase III but expressing the RNase III D70A mutant protein that binds transcripts but does not cleave them. Transcripts whose levels are changed in that strain compared with M145 will be candidates for regulation by RNase III binding without subsequent cleavage.
We note finally with regard to the transcripts identified in our RNA-Seq analysis that, with the exception of the rpsO-pnp transcript and possibly the transcript of the SCO3982 to SCO3988 region discussed above, our analyses did not identify polycistronic or readthrough transcripts. We will analyze our data to examine such transcripts in a subsequent study.
The role of RNase III in the regulation of antibiotic production in S. coelicolor.
The analyses reported above identified a number of genes with three significant characteristics: (i) their transcripts were present at higher levels in the RNase III mutant strain than in S. coelicolor M145, the parental strain; (ii) their transcripts were present in the BARD at enrichment ratios of greater than 1; and (iii) the products of these genes have potential regulatory activity. To cite just three of a number of genes that meet these criteria, the transcript for SCO5608, encoding a putative transcriptional regulator, was increased in the rnc-null mutant by a factor of 2.74 and was enriched in the BARD by a factor of 4.22. The transcript for SCO0168, encoding a putative cyclic nucleotide binding transcriptional regulator, was increased in the rnc-null mutant by a factor of 2.41 and was enriched in the BARD by a factor of 100, while the transcript for SCO0864, encoding a probable ECF sigma factor, was increased in the rnc-null mutant by a factor of 1.7 and enriched in the BARD by a factor of 234. All of these genes might be involved in the regulation of antibiotic production in S. coelicolor.
This possibility raises the question of whether there is a single target for RNase III regulation of antibiotic production or whether antibiotic production is affected by the sheer number of genes whose transcripts are cleaved by RNase III. There is a precedent for the first mechanism represented by AdpA, a member of the AraC/XylS family of transcriptional regulators, which is known to regulate morphological differentiation in S. coelicolor in an RNase III-dependent fashion. Thus, AdpA is required for sporulation, and AdpA and RNase III participate in a feedback loop that regulates the levels of both proteins in S. coelicolor (47). It is clear from these studies, however, that AdpA is not a global regulator of antibiotic production in S. coelicolor and, therefore, that RNase III does not regulate antibiotic production globally via AdpA (47). We note here that the adpA transcript level decreased by ca. 10% in the S. coelicolor JSE1880 mRNA pool compared with that of M145 but was present in the BARD at an enrichment ratio of 22.
Given the number of genes identified in the present study whose transcripts are likely to be substrates for RNase III or whose transcript levels are likely to be regulated indirectly by RNase III, we favor the hypothesis that there is not a single global regulator of antibiotic production whose levels are controlled by RNase III but rather that RNase III exerts a pleiotropic effect, affecting at least one gene which is required for the production of each of the antibiotics synthesized by S. coelicolor. We will examine this hypothesis experimentally by disrupting genes identified by our RNA immunoprecipitation analyses and assessing the effect of gene disruption on antibiotic production.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation (grant no. MCB-0817177 to G.H.J.).
We thank the Emory Genome Center for the sequencing, as well as Chad Haase for his input on sample preparation for the project.
Footnotes
Published ahead of print 2 March 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Aceti DJ, Champness W. 1998. Transcriptional regulation of Streptomyces coelicolor pathway-specific antibiotic regulators by the absA and absB loci. J. Bacteriol. 180:3100–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adamis T, Champness W. 1992. Genetic analysis of absB, a Streptomyces coelicolor locus involved in global antibiotic regulation. J. Bacteriol. 174:4622–4628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bardwell JC, et al. 1989. Autoregulation of RNase III operon by mRNA processing. EMBO J. 8:3401–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bentley SD, et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147 [DOI] [PubMed] [Google Scholar]
- 5. Berdy J. 1980. Recent advances in and prospects of antibiotic research. Process. Biochem. 15:28–35 [Google Scholar]
- 6. Bibb MJ. 1996. The regulation of antibiotic production in Streptomyces coelicolor. Microbiology 142:1335–1344 [DOI] [PubMed] [Google Scholar]
- 7. Bibb MJ. 2005. Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8:208–215 [DOI] [PubMed] [Google Scholar]
- 8. Bralley P, Jones GH. 2004. Organization and expression of the polynucleotide phosphorylase gene (pnp) of Streptomyces: processing of pnp transcripts in Streptomyces antibioticus. J. Bacteriol. 186:3160–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bralley P, Jones GH. 2003. Overexpression of the polynucleotide phosphorylase gene (pnp) of Streptomyces antibioticus affects mRNA stability and poly(A) tail length but not ppGpp levels. Microbiology 149:2173–2182 [DOI] [PubMed] [Google Scholar]
- 10. Chang SA, Bralley P, Jones GH. 2005. The absB gene encodes a double-strand specific endoribonuclease that cleaves the readthrough transcript of the rspO-pnp operon in Streptomyces coelicolor. J. Biol. Chem. 280:33213–33219 [DOI] [PubMed] [Google Scholar]
- 11. Chater KF. 1998. Taking a genetic scalpel to the Streptomyces colony. Microbiology 144:1465–1478 [DOI] [PubMed] [Google Scholar]
- 12. Drider D, Condon C. 2004. The continuing story of endoribonuclease III. J. Mol. Microbiol. Biotechnol. 8:195–200 [DOI] [PubMed] [Google Scholar]
- 13. Fernandez-Moreno MA, et al. 1992. abaA, a new pleiotropic regulatory locus for antibiotic production in Streptomyces coelicolor. J. Bacteriol. 174:2958–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fischer M, Alderson J, van Keulen G, White J, Sawers RG. 2010. The obligate aerobe Streptomyces coelicolor A3(2) synthesizes three active respiratory nitrate reductases. Microbiology 156:3166–3179 [DOI] [PubMed] [Google Scholar]
- 15. Gatewood ML, Bralley P, Jones GH. 2011. RNase III-dependent expression of the rpsO-pnp operon of Streptomyces coelicolor. J. Bacteriol. 193:4371–4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gravenbeek ML, Jones GH. 2008. The endonuclease activity of RNase III is required for the regulation of antibiotic production by Streptomyces coelicolor. Microbiology 154:3547–3555 [DOI] [PubMed] [Google Scholar]
- 17. Hesketh A, Chen WJ, Ryding J, Chang S, Bibb M. 2007. The global role of ppGpp synthesis in morphological differentiation and antibiotic production in Streptomyces coelicolor A3(2). Genome Biol. 8:R161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hesketh A, et al. 2011. Genome-wide dynamics of a bacterial response to antibiotics that target the cell envelope. BMC Genomics 12:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hillerich B, Westpheling J. 2006. A new GntR family transcriptional regulator in Streptomyces coelicolor is required for morphogenesis and antibiotic production and controls transcription of an ABC transporter in response to carbon source. J. Bacteriol. 188:7477–7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hojati Z, et al. 2002. Structure, biosynthetic origin, and engineered biosynthesis of calcium-dependent antibiotics from Streptomyces coelicolor. Chem. Biol. 9:1175–1187 [DOI] [PubMed] [Google Scholar]
- 21. Hsieh C-J, Jones GH. 1995. Nucleotide sequence, transcriptional analysis and glucose regulation of the phenoxazinone synthase gene from Streptomyces antibioticus. J. Bacteriol. 177:5740–5747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang J, Lih C- J, Pan K-H, Cohen SN. 2001. Global analysis of growth phase responsive gene expression and regulation of antibiotic biosynthetic pathways in Streptomyces coelicolor using DNA microarrays. Genes Dev. 15:3183–3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang J, et al. 2005. Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol. Microbiol. 58:1276–1287 [DOI] [PubMed] [Google Scholar]
- 24. Jayapal KP, Lian W, Glod F, Sherman DH, Hu WS. 2007. Comparative genomic hybridizations reveal absence of large Streptomyces coelicolor genomic islands in Streptomyces lividans. BMC Genomics 8:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ji X. 2006. Structural basis for non-catalytic and catalytic activities of ribonuclease III. Acta Crystallog. D Biol. Crystallogr. 62:933–940 [DOI] [PubMed] [Google Scholar]
- 26. Jones GH, Paget MSB, Chamberlin L, Buttner MJ. 1997. Sigma-E is required for the production of the antibiotic actinomycin in Streptomyces antibioticus. Mol. Microbiol. 23:169–178 [DOI] [PubMed] [Google Scholar]
- 27. Karoonuthaisiri N, Weaver D, Huang J, Cohen SN, Kao CM. 2005. Regional organization of gene expression in Streptomyces coelicolor. Gene 353:53–66 [DOI] [PubMed] [Google Scholar]
- 28. Kelemen GH, et al. 1998. Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3(2). J. Bacteriol. 180:2515–2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, England [Google Scholar]
- 30. Kirby KS, Fox-Carter E, Guest M. 1967. Isolation of deoxyribonucleic acid and ribosomal ribonucleic acid from bacteria. Biochem. J. 104:258–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Luo Y, Helmann JD. 2009. Extracytoplasmic function sigma factors with overlapping promoter specificity regulate sublancin production in Bacillus subtilis. J. Bacteriol. 191:4951–4958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5:621–628 [DOI] [PubMed] [Google Scholar]
- 33. Nicholson AW. 1999. Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol. Rev. 23:371–390 [DOI] [PubMed] [Google Scholar]
- 34. Nodwell JR, McGovern K, Losick R. 1996. An oligopeptide permease responsible for the import of an extracellular signal governing aerial mycelium formation in Streptomyces coelicolor. Mol. Microbiol. 22:881–893 [DOI] [PubMed] [Google Scholar]
- 35. O'Connor TJ, Nodwell JR. 2005. Pivotal roles for the receiver domain in the mechanism of action of the response regulator RamR of Streptomyces coelicolor. J. Mol. Biol. 351:1030–1047 [DOI] [PubMed] [Google Scholar]
- 36. Ostash B, et al. 2003. Targeted disruption of Streptomyces globisporus lndF and lndL cyclase genes involved in landomycin E biosynthesis. Folia Microbiol. 48:484–488 [DOI] [PubMed] [Google Scholar]
- 37. Rodriguez-Garcia A, Barreiro C, Santos-Beneit F, Sola-Landa A, Martin JF. 2007. Genome-wide transcriptomic and proteomic analysis of the primary response to phosphate limitation in Streptomyces coelicolor M145 and in a ΔphoP mutant. Proteomics 7:2410–2429 [DOI] [PubMed] [Google Scholar]
- 38. Ryding NJ, Anderson TB, Champness WC. 2002. Regulation of the Streptomyces coelicolor calcium-dependent antibiotic by absA, encoding a cluster-linked two-component system. J. Bacteriol. 184:794–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Swiercz JP, Hindra, et al. 2008. Small non-coding RNAs in Streptomyces coelicolor. Nucleic Acids Res. 36:7240–7251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Takata R, Mukai T, Hori K. 1987. RNA processing by RNase III is involved in the synthesis of Escherichia coli polynucleotide phosphorylase. Mol. Gen. Genet. 209:28–32 [DOI] [PubMed] [Google Scholar]
- 41. Thomason MK, Storz G. 2010. Bacterial antisense RNAs: how many are there, and what are they doing? Annu. Rev. Genet. 44:167–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vockenhuber MP, et al. 2011. Deep sequencing-based identification of small non-coding RNAs in Streptomyces coelicolor. RNA Biol. 8:468–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vohradsky J, Branny P, Thompson CJ. 2007. Comparative analysis of gene expression on mRNA and protein level during development of Streptomyces cultures by using singular value decomposition. Proteomics 7:3853–3866 [DOI] [PubMed] [Google Scholar]
- 44. Weaver D, et al. 2004. Genome plasticity in Streptomyces: identification of 1 Mb TIRs in the S. coelicolor A3(2) chromosome. Mol. Microbiol. 51:1535–1550 [DOI] [PubMed] [Google Scholar]
- 45. Wurtzel O, et al. 2010. A single-base resolution map of an archaeal transcriptome. Genome Res. 20:133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xu W, Huang J, Cohen SN. 2008. Autoregulation of AbsB (RNase III) expression in Streptomyces coelicolor by endoribonucleolytic cleavage of absB operon transcripts. J. Bacteriol. 190:5526–5530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xu W, Huang J, Lin R, Shi J, Cohen SN. 2010. Regulation of morphological differentiation in S. coelicolor by RNase III (AbsB) cleavage of mRNA encoding the AdpA transcription factor. Mol. Microbiol. 75:781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang W, Bolla ML, Kahne D, Walsh CT. 2010. A three enzyme pathway for 2-amino-3-hydroxycyclopent-2-enone formation and incorporation in natural product biosynthesis. J. Am. Chem. Soc. 132:6402–6411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zuker M. 2003. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.