Abstract
Human listeriosis outbreaks in Canada have been predominantly caused by serotype 1/2a isolates with highly similar pulsed-field gel electrophoresis (PFGE) patterns. Multilocus sequence typing (MLST) and multi-virulence-locus sequence typing (MVLST) each identified a diverse population of Listeria monocytogenes isolates, and within that, both methods had congruent subtypes that substantiated a predominant clone (clonal complex 8; virulence type 59; proposed epidemic clone 5 [ECV]) that has been causing human illness across Canada for more than 2 decades.
TEXT
Genotyping of the food-borne pathogen Listeria monocytogenes for public health surveillance and outbreak detection under the PulseNet system is accomplished using pulsed-field gel electrophoresis (PFGE) (12). DNA sequence-based subtyping methods, such as multilocus sequence typing (MLST) (3, 8, 10) and multi-virulence-locus sequence typing (MVLST) (2, 14), have also been used for phylogenetic studies and to categorize isolates into higher-level groups, such as evolutionary lineages, clonal complexes, and epidemic clones. The term epidemic clone has been defined as a group of genetically related isolates implicated in geographically and temporally unrelated outbreaks that are presumably of a common ancestor (1, 2, 4, 7, 14).
Within Canada, laboratory surveillance indicated that L. monocytogenes serotype 1/2a isolates with very similar PFGE patterns caused a substantial proportion of the sporadic cases, clusters, and outbreaks (represented by PulseNet Canada AscI/ApaI PFGE macrorestriction profile LMACI.0001/LMAAI.0001) (see Fig. S1 in the supplemental material) (5, 6). This predominant PFGE clone included a large nationwide outbreak of listeriosis in 2008 that was caused by consumption of contaminated ready-to-eat (RTE) deli meats (6). In this study, multiple sequence-based subtyping methods were used to investigate the genetic diversity within Canadian human clinical L. monocytogenes isolates and to determine the place of the predominant PFGE clone within these phylogenies.
A 71-isolate panel was selected from isolates submitted to the Canadian National Microbiology Laboratory (NML) by provincial microbiology laboratories or to the Bureau of Microbial Hazards for identification and serotyping services as part of the national Listeriosis Reference Service (9). From these, a diverse selection of mostly clinical isolates was chosen to represent (i) the 10 most frequently observed PFGE types (and similar patterns) in the PulseNet Canada database, (ii) the five L. monocytogenes serotypes representing lineages I and II, and (iii) clinical cases from eight provinces spanning 30 years (see Table S1 in the supplemental material). The panel included 41 isolates of the predominant PFGE pattern (31 with AscI PFGE pattern LMACI.0001 and 10 isolates with the highly similar PFGE pattern LMACI.0040) (see Fig. S1 in the supplemental material) (6).
In addition to serotyping (11) and PFGE (5), sequence-based subtyping of housekeeping genes (10), virulence genes (2), and comK prophage junction fragments (13) was performed. Comparative genomics of publicly available L. monocytogenes genomes, including those of the 2008 Canadian listeriosis outbreak (6), was used to identify unique genetic markers on the chromosome backbone (LM5578_2229) and a 50-kb Listeria genomic island (LGI1; marker locus LM5578_1886). The panel was screened for these markers using PCR (see Table S2 in the supplemental material).
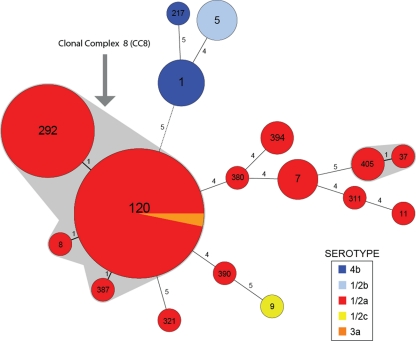
MLST detected a total of 17 different sequence types (STs) in the 71-isolate panel. All 41 isolates representing the predominant PFGE pattern were members of L. monocytogenes clonal complex 8 (CC8), which comprised four highly related STs (Fig. 1; see also Table S1 in the supplemental material). These CC8 STs were all single-locus variants of ST120 and individually differed from ST120 by no more than one single nucleotide polymorphism (SNP). The total number of CC8 isolates in the panel was 49, and all were serotype 1/2a with the exception of a single serotype 3a isolate. All other observed STs were not members of CC8, with these isolates having 4 or more alleles that were different from those of ST120 (Fig. 1). None of these more distantly related STs included isolates with PFGE patterns similar to the predominant pattern. Hence, the possibility of a predominant serotype 1/2a clone, first suggested through PFGE, was further supported by the membership of these same strains in CC8.
Fig 1.
Minimum spanning tree analysis of Canadian human clinical L. monocytogenes isolates obtained between 1988 and 2010 based on the MLST method of Ragon et al. (10). The data set is based on isolates presented in Table S1 in the supplemental material. Circles correspond to sequence types (STs), while numbers on branches are the numbers of allele differences between connected STs. Clonal complexes (STs with single allele differences) are shaded gray, and clonal complex 8 (CC8) is indicated. The size of each circle is proportional to the number of isolates in each ST. The alignment and minimum spanning tree were created using BioNumerics v5.10.
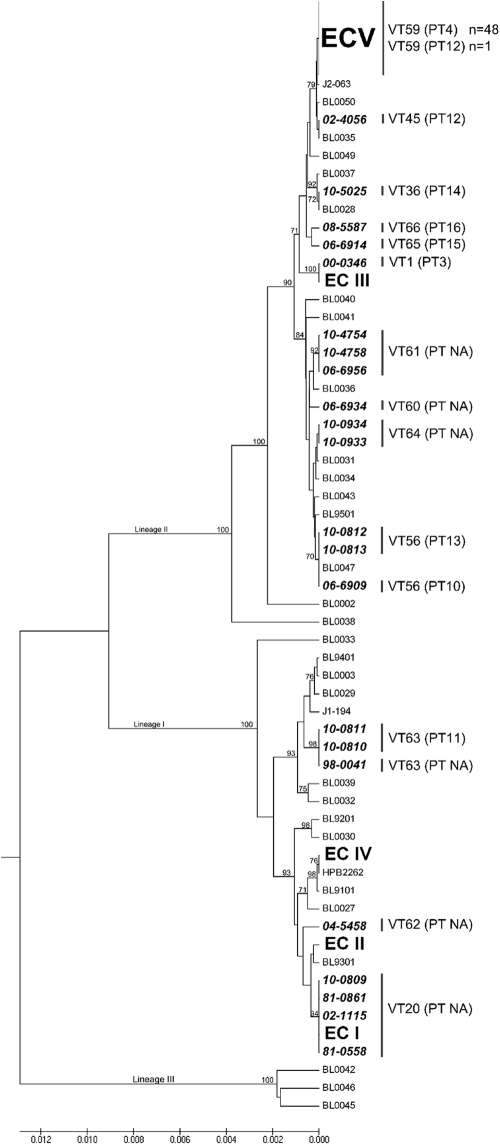
Thirteen total MVLST profiles were observed in the 71-isolate panel, and all 49 of the CC8 isolates yielded identical sequences for the six virulence genes in the MVLST scheme (Fig. 2; see also Table S1 in the supplemental material). This combination of virulence alleles, designated virulence type 59 (VT59), while predominant among Canadian cases, had never been reported previously among global isolates tested (Fig. 2).
Fig 2.
Unrooted unweighted-pair group method using average linkages (UPGMA) tree based on MVLST with 6 virulence gene fragments that shows the placement of the proposed epidemic clone V (ECV) isolates among other ECs and non-ECs of L. monocytogenes. Non-ECV strains that were part of the 71 isolates analyzed in the present study are listed in bold with the NML of Canada isolate identification number. BL, isolates from Knabel's MVLST database that were not part of the 71 isolates analyzed in the present study but that were added for comparison purposes. Lineage information is shown as I, II, and III.
Additional genetic markers, identified through comparative genomics (6), were also congruent with the CC8/VT59 clone. The chromosomal locus LM5578_2229 was detected in all CC8/VT59 isolates tested with both primer pairs in Table S2 in the supplemental material and detected in only a single isolate unrelated to CC8/VT59 (06-6914; ST390/VT65) (see Table S1 in the supplemental material). However, while all CC8/VT59 isolates showed 100% sequence identity at this locus, the amplicon sequence from 06-6914 had 6 SNPs and 1 gap in LM5578_2229 along with a 34-bp insertion in the 3′ flanking intergenic region compared to the sequences of the CC8/VT59 isolates. Notably, isolates J2-063, BL0050, 02-4056, and BL0035 were closely related to VT59 (Fig. 2), but none of these strains showed amplification of chromosomal locus LM5578_2229.
PCR screening for LGI1 (using locus LM5578_1886) confirmed that this island was present in 43 of 49 CC8/VT59 isolates and absent in all 22 of the non-CC8/non-VT59 isolates (see Table S1 in the supplemental material); therefore, LGI1 is not universally present in all CC8/VT59 isolates. Lastly, 48 of 49 (98%) CC8/VT59 isolates shared 100% nucleotide identity in their comK prophage junction fragment sequences, and this shared genotype was designated prophage type 4 (PT4) (Fig. 2; see also Table S1 in the supplemental material). A single CC8/VT59 isolate (isolate 10-0819) was PT12 rather than PT4, and the downstream junction fragment sequences of PT4 and PT12 shared a nucleotide sequence identity of 84.1%.
The presence of the CC8/VT59 clone in clinical isolates from two Canadian provinces in 1988 demonstrated that this clone has been causing disease in Canada for at least the past 2 decades (see Table S1 in the supplemental material), and nationwide distribution occurred by the mid-1990s, when CC8/VT59 isolates were associated with invasive listeriosis across Canada. To further estimate the historical prevalence in Canada of the predominant PFGE/CC8/VT59 clone, laboratory surveillance data from 1,061 isolates submitted to the NML between 1995 and 2010 for serotyping and PFGE testing were reviewed. During this period, the majority of isolates were serotype 1/2a (54.8%) and 237 (22.3%) were typical of the predominant PFGE patterns associated with CC8/VT59 (see Fig. S1 in the supplemental material). Furthermore, of the 41 listeriosis case clusters detected during the same period, 31 (75.6%) were caused by serotype 1/2a isolates and 16 (39%) were caused by isolates with the predominant PFGE pattern or similar PFGE patterns.
Reasons for the relatively high prevalence of the CC8/VT59 clone in Canada are unknown; however, the existence of a predominant clone within an otherwise genetically diverse L. monocytogenes total population suggests that determinants of virulence and survival encoded by CC8/VT59 isolates have been successful in both human and environmental niches, respectively. Regarding virulence, the available epidemiologic information indicated that the majority of CC8/VT59 cases occurred in elderly or immunocompromised individuals and that no maternofetal cases were reported among clusters/outbreaks caused by this clone. The rates of human disease may also be affected by the frequency and degree of exposure to CC8/VT59. A recent study (13) suggested that CC8/VT59 strains possess a strong capacity for biofilm formation, which may support persistence within food production environments and subsequent contamination of foods.
The congruence of multiple genetic subtyping methods substantiated the presence of a distinct clone of L. monocytogenes capable of causing human disease, and although the origins of this clone are not known, we believe that these Canadian isolates share an ancestor. This clone belonged to lineage II, most commonly serotype 1/2a, and belonged to MLST CC8 with MVLST profile VT59. All isolates also encoded the LM5578_2229 marker locus, and the majority were PT4 and also encoded the large LGI1 region. Following the epidemic clone nomenclature for L. monocytogenes (defined using MVLST and association with multiple outbreaks), we propose that VT59 represents a newly identified epidemic clone (epidemic clone five [ECV]). Presumptive PCR detection of ECV can be accomplished using locus LM5578_2229, but results should be confirmed using MVLST. For public health and food safety investigations, the observation of highly prevalent molecular subtypes also highlights the need for improved molecular epidemiologic tools (including whole-genome sequencing) that can identify additional markers beyond ST membership to properly distinguish individual clusters of disease and trace outbreaks back to contaminated food products.
Nucleotide sequence accession numbers.
DNA sequences of the virulence gene amplicons were deposited in GenBank under accession numbers JQ436380 through JQ436451 (see Table S3 in the supplemental material).
Supplementary Material
ACKNOWLEDGMENTS
We thank P. Hemmons, M. Jerome, H. Tabor, C.-L. Cross, C. Singh, C. Clark, K. Bernard, E. Ballegeer, A. Desrochers, R. Weinberg, the Institute Pasteur, S. Brisse, the Canadian Food Inspection Agency, the NML Genomics Core, and the Genomics Core Facility at Penn State University.
Research conducted in Stephen J. Knabel's laboratory was supported by a USDA Special Grant on Milk Safety.
The CPHLN consists of the British Columbia Centre for Disease Control, Alberta ProvLab, Saskatchewan Disease Control Laboratory, Cadham Provincial Laboratory (Manitoba), Ontario Agency for Health Protection and Promotion, Laboratoire de Santé Publique du Québec, Queen Elizabeth II Health Sciences Centre (Nova Scotia), and Newfoundland Public Health Laboratory.
Footnotes
Published ahead of print 15 February 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Chen Y, Knabel S. 2008. Strain typing. In Liu D. (ed), Handbook of Listeria monocytogenes. CRC Press, New York, NY [Google Scholar]
- 2. Chen Y, Zhang W, Knabel SJ. 2007. Multi-virulence-locus sequence typing identifies single nucleotide polymorphisms which differentiate epidemic clones and outbreak strains of Listeria monocytogenes. J. Clin. Microbiol. 45:835–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chenal-Francisque V, et al. 2011. Worldwide distribution of major clones of Listeria monocytogenes. Emerg. Infect. Dis. 17:1110–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cheng Y, Siletzky RM, Kathariou S. 2008. Genomic divisions/lineages, epidemic clones, and population structure. In Liu D. (ed), Handbook of Listeria monocytogenes. CRC Press, New York, NY [Google Scholar]
- 5. Clark CG, et al. 2010. Surveillance for Listeria monocytogenes and listeriosis, 1995-2004. Epidemiol. Infect. 138:559–572 [DOI] [PubMed] [Google Scholar]
- 6. Gilmour MW, et al. 2010. High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genomics 11:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kathariou S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65:1811–1829 [DOI] [PubMed] [Google Scholar]
- 8. Maiden MC, et al. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95:3140–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pagotto F, Ng LK, Clark C, Farber J, Canadian Public Health Laboratory Network 2006. Canadian listeriosis reference service. Foodborne Pathog. Dis. 3:132–137 [DOI] [PubMed] [Google Scholar]
- 10. Ragon M, et al. 2008. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 4:e1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Seeliger HPR, Hohne K. 1979. Serotyping of Listeria monocytogenes and related species. In Bergan T, Norris JR. (ed), Methods in microbiology, vol 13 Academic Press, New York, NY [Google Scholar]
- 12. Swaminathan B, Barrett TJ, Hunter SB, Tauxe RV, CDC PulseNet Task Force 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Verghese B, et al. 2011. comK prophage junction fragments as markers for Listeria monocytogenes genotypes unique to individual meat and poultry processing plants and a model for rapid niche-specific adaptation, biofilm formation, and persistence. Appl. Environ. Microbiol. 77:3279–3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang W, Jayarao BM, Knabel SJ. 2004. Multi-virulence-locus sequence typing of Listeria monocytogenes. Appl. Environ. Microbiol. 70:913–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.