Abstract
The thermophile Cupriavidus sp. strain S-6 accumulated polyhydroxybutyrate (PHB) from glucose at 50°C. A 9.0-kbp EcoRI fragment cloned from the genomic DNA of Cupriavidus sp. S-6 enabled Escherichia coli XL1-Blue to synthesize PHB at 45°C. Nucleotide sequence analysis showed a pha locus in the clone. The thermophilic polyhydroxyalkanoate (PHA) synthase (PhaCCsp) shared 81% identity with mesophilic PhaC of Cupriavidus necator H16. The diversity between these two strains was found dominantly on their N and C termini, while the middle regions were highly homologous (92% identity). We constructed four chimeras of mesophilic and thermophilic phaC genes to explore the mutations related to its thermostability. Among the chimeras, only PhaCH16β, which was PhaCH16 bearing 30 point mutations derived from the middle region of PhaCCsp, accumulated a high content of PHB (65% [dry weight]) at 45°C. The chimera phaCH16β and two parental PHA synthase genes were overexpressed in E. coli BLR(DE3) cells and purified. At 30°C, the specific activity of the chimera PhaCH16β (172 ± 17.8 U/mg) was 3.45-fold higher than that of the parental enzyme PhaCH16 (50 ± 5.2 U/mg). At 45°C, the half-life of the chimera PhaCH16β (11.2 h) was 127-fold longer than that of PhaCH16 (5.3 min). Furthermore, the chimera PhaCH16β accumulated 1.55-fold (59% [dry weight]) more PHA content than the parental enzyme PhaCH16 (38% [dry weight]) at 37°C. This study reveals a limited number of point mutations which enhance not only thermostability but also PhaCH16 activity. The highly thermostable and active PHA synthase will provide advantages for its promising applications to in vitro PHA synthesis and recombinant E. coli PHA fermentation.
INTRODUCTION
Polyhydroxyalkanoates (PHAs) are a type of biopolyester. Numerous bacteria accumulate PHAs intracellularly as a carbon source and reducing power sink. PHAs have been intensively studied since Lemoigne discovered poly(β-hydroxybutyrate) (PHB) in the bacterium Bacillus megaterium in 1926 (2, 19, 21, 22). Bacterial fermentation is a method commonly used for the mass production of PHA (49, 50). Numerous bacteria, such as Cupriavidus necator H16 (formerly Ralstonia eutropha H16) (46), Alcaligenes latus, Methylobacterium organophilum, and recombinant Escherichia coli, have been studied for the production of PHA to a high concentration with a high level of productivity (6). However, the cost of production of PHA with bacterial fermentation is higher than that of petroleum-based plastics (6). Transgenic plants may provide a cost-effective solution for PHA production in the future (45), although the feasibility of this approach has been questioned (10). Moreover, in vitro polymerization has been demonstrated for PHA production (11). By use of this approach, it is possible to control the properties of PHAs, such as the molecular weight of polymers and the monomer composition of PHAs.
Nowadays, the high cost of production of PHA predominantly hampers its applications (6). To resolve this issue, improvements made in the performance of PHA synthase is a promising direction (25). PHA synthase is the key enzyme of PHA biosynthesis (22). Its function correlates with the monomer composition of PHA, the molecular weight of the synthesized polymer, and the PHA contents of bacterial fermentations (1, 25, 39, 40). There have been many efforts dedicated to improving the enzyme activity of PHA synthase. Because the crystal structure of PHA synthase has not been determined, a rational design method is not applicable for accomplishing this goal. Nonrational design methods, i.e., in vitro and in vivo evolution approaches, are commonly applied to many PHA synthases, including those of C. necator H16 (41), Aeromonas caviae (1, 18), Pseudomonas sp. strain 61-3 (35, 36), and Pseudomonas putida GPo1 (33). The above-mentioned studies succeeded in obtaining point mutations which improve the performance of PHA synthase by changing the substrate specificity or enhancing the enzyme activity.
Once beneficial mutations are found, subsequent saturation mutagenesis of these spots optimizes the beneficial effect (44). For example, in the PHA synthase of C. necator H16 (PhaCH16), a G4D mutation increases the protein expression level of PHA synthase (26). An F420S mutation enhances the specific activity of PHA synthase (41). A G4D or F420S mutation results in more PHB accumulation in recombinant E. coli (26, 41). Another beneficial mutation, A510D(E), leads PhaCH16 to synthesize a higher-molecular-weight polymer (43). The thermostability of PHA synthase has never been the aim of protein engineering. A highly thermostable and active PHA synthase will lead to promising applications for in vitro PHB polymerization and recombinant E. coli fermentation (25). So far, over 59 PHA synthase genes from 45 bacterial strains have been cloned and characterized (29), and most of them are from mesophiles. Few reports have mentioned thermophilic PHA synthases (14). In these limited reports, thermophiles exhibited a more efficient ability for PHA accumulation than mesophiles (16, 17, 31). The results of those studies also support the assumption that thermophilic PHA synthase has a higher level of enzyme activity.
Cupriavidus sp. strain S-6 is a thermophile, isolated from a hot spring in Southern Taiwan. It accumulates PHB from glucose at 50°C. Its 16S rRNA gene is 99% identical to that of the mesophile C. necator H16, a well-studied PHA synthesis strain formerly known as Ralstonia eutropha H16 (28, 46). A phaC gene fragment of Cupriavidus sp. S-6 amplified by colony PCR (34) showed a high level of identity with the phaC gene of the mesophile C. necator H16. The PHA synthase of the thermophile Cupriavidus sp. S-6 is a proper model to explore mutations related to the thermostability of PHA synthase.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are described below. Escherichia coli XL1-Blue and plasmid pBlusescript II KS were used for library construction. The thermophile Cupriavidus sp. S-6 and the mesophile C. necator H16 were grown in Luria-Bertani (LB) medium at 45°C and 30°C, respectively. PCR primers CspCNdeIF and CspCHindIIIR (Table 1) amplified the PHA synthase gene of Cupriavidus sp. S-6, which was cloned into the NdeI and HindIII sites of pET-23a (Novagen). The thermophilic PHA synthase overexpression vector was designated pECspC. The chimeric PHA synthase gene phaCH16β was amplified with primers EcoRI23SDF and 23BamHIR and was cloned into the EcoRI and BamHI sites of pET-23a. The PhaCH16β overexpression vector was designated pEH16β.
Table 1.
Primers used in this study
| Primer | Sequencea |
|---|---|
| EcoRI23SDF | 5′-CGGAATTCGAAGGAGATATACATATG-3′ |
| 23BamHIR | 5′-AGCGGATCCTGGTGCTCGAGTGC-3′ |
| CspCNdeIF | 5′-AGAGACACGACCATATGGCGACCGGC-3′ |
| CspCHindIIIR | 5′-CCGGAAGCTTAAGCCTTGGCCTTGA-3′ |
| CspAB–BamHIF | 5′-CAAGGATCCCAACCGTTACCGAAGGC-3′ |
| CspAB–NotIR | 5′-ATAGTTTAGCGGCCGCTCGGTCAGCCCATGTG-3′ |
| H16AB–BamHIF | 5′-TCCCTCCGGATCCCATTGAAAGGACT-3′ |
| H16AB–NotIR | 5′-TGAACCAGGGCGGCCGCTCAGCCCATAT-3′ |
| CspC505F | 5′-GATGCGATGTCGCCGGCCAATTTCCT-3′ |
| CspC530R | 5′-AGGAAATTGGCCGGCGACATCGCATC-3′ |
| CspC790F | 5′-AACAAGTACTACATCCTCGACCTGCAG-3′ |
| CspC816R | 5′-CTGCAGGTCGAGGATGTAGTACTTGTT-3′ |
| CspC1253F | 5′-TGGTCTGGAACTACGTGGTCGACAACT-3′ |
| CspC1279R | 5′-AGTTGTCGACCACGTAGTTCCAGACCA-3′ |
| CspC1582F | 5′-GTCATCAATCCGCCGGCGAAGAACAA-3′ |
| CspC1607R | 5′-TTGTTCTTCGCCGGCGGATTGATGAC-3′ |
| PhaCF1 | 5′-ATCAACAAGTWCTACRTCYTSGACCT-3′ |
| PhaCR2 | 5′-GTSTTCRTSRTSWSCTGGCGCAACCC-3′ |
| G286AF | 5′-ACGCCAGCATGGCCGCGAGCACCTGGGACG-3′ |
| G286AR | 5′-CGTCCCAGGTGCTCGCGGCCATGCTGGCGT-3′ |
| A341QF | 5′-GCGAGCACCCGGCCCAGAGCGTCACGCTGC-3′ |
| A341QR | 5′-GCAGCGTGACGCTCTGGGCCGGGTGCTCGC-3′ |
The sequence recognized by the restriction enzyme is underlined; the ribosome binding site sequence is double underlined. The sequences of CspC530R, CspC816R, CspC1279R, and Csp1607R are complementary to those of CspC505F, CspC790F, CspC1253F, and CspC1582F, respectively. Gray shading indicates the mutation site.
Construction and screening of a genomic library.
Genomic DNA of Cupriavidus sp. S-6 was extracted with an Illustra bacterial genomicPrep Minispin kit (GE Healthcare) and was digested with EcoRI (Fermentas) overnight. One microgram of EcoRI-digested genomic DNA was ligated with 50 ng of EcoRI and alkaline phosphatase (New England BioLabs)-treated pBluescript II KS by T4 DNA ligase (Fermentas) at 8°C. The ligation product was transformed into E. coli XL1-Blue cells by electroporation as described previously (33). Transformants were spread onto an LB agar plate containing 1.5% glucose and 100 μg/ml ampicillin and incubated at 37°C for 24 to 48 h. Colonies with PHA accumulation were screened based on an opaque colony appearance.
Construction of chimeric PHA synthase genes.
The construction of a chimeric phaC gene included two parts, the fragmentation of the phaC gene and the assembly of the chimeric gene. The PHA synthase genes phaCH16 and phaCCsp were fragmented into three fragments with corresponding degenerate primer pairs (Table 1 and see Fig. 2). PCR primers EcoRI23SDF and CspC530R amplified the a and α fragments. Primers CspC505F and CspC1607R amplified the b and β fragments. Primers CspC1582F and 23BamHIR amplified the c and γ fragments. The PCR mixture contained 1× PCR amplification buffer (Finnzymes), 2.2 mM MgCl2, 180 μM (each) deoxynucleoside triphosphate (dNTP), 1 μM (each) primers, 2% dimethyl sulfoxide (DMSO) (Sigma), 0.6 U DyNAzyme II DNA polymerase (Finnzymes), 0.5 U Pfu DNA polymerase (Fermentas), and plasmid pEReC (pET-23a containing the phaCH16 gene) (32) or pECspC (pET-23a containing the phaCCsp gene) as the template in a 50-μl reaction mixture volume. The thermal cycle program consisted of 94°C for 5 min, 47°C for 30 s, 72°C for 1 min 40 s, and 35 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 1 min 40 s; the reaction was then followed by incubation at 72°C for 1 min and stopped at 15°C, all attained on a 2720 thermal cycler (Applied Biosystems). The amplified gene fragments were gel purified to remove template DNA and primers (QIAquick gel extraction kit; Qiagen).
Fig 2.
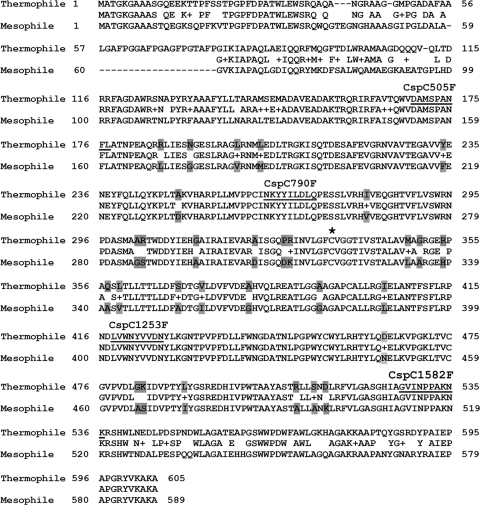
Alignment of PHA synthases from C. necator H16 and Cupriavidus sp. S-6. Thermophile, PHA synthase of Cupriavidus sp. S-6; Mesophile, PHA synthase of C. necator H16. The range of the a fragment is from the 1st amino acid to the 152nd amino acid of the mesophile PhaCH16. The b fragment is from the 153rd to the 510th amino acid of PhaCH16. The c fragment is from the 511th to the 589th amino acid of PhaCH16. The βN fragment is from the 248th to the 410th amino acid of PhaCH16. Cysteine (C), marked by an asterisk, is the catalytic site of PHA synthase. The gray box indicates the difference between the middle regions of mesophile and thermophile PHA synthases. The underlined sequences are the primer recognition sites for chimera construction.
Selected DNA fragments of phaCH16 (a, b, and c) and phaCCsp (α, β, and γ) were mixed in an equal molar ratio. Approximately 100 ng of the DNA mixture was added to a 20 μl of PCR mixture containing 1× PCR amplification buffer (Finnzymes), 2.2 mM MgCl2, 180 μM (each) dNTP, 2% DMSO, 0.3 U of DyNAzyme II DNA polymerase (Finnzymes), and 0.3 U Pfu DNA polymerase (Fermentas) to perform primerless assembly PCR. The PCR program consisted of 94°C for 5 min and then 32 cycles of 94°C for 30 s, 55°C for 60 s, and 72°C for 40 s (+2 s/cycle); the reaction was then followed by incubation at 72°C for 2 min and was stopped at 15°C. Finally, for chimeric gene reamplification, the 50-μl PCR mixture contained 1 μl of the assembly PCR product, 0.8 μM primers (EcoRI23SDF and 23BamHIR), and 2% DMSO; PCR was performed for 20 cycles (94°C for 30 s, 52°C for 30 s, and 72°C for 2 min). The amplified chimeric phaC genes were cloned into the pGEM-T Easy vector (Promega) for DNA sequence analysis.
Site-directed mutagenesis.
The site-directed mutagenesis of the PHA synthase gene was performed directly on plasmid pBCspAB-H16 by using a QuikChange site-directed mutagenesis kit (Stratagene). The oligonucleotide purification cartridge (OPC)-purified mutagenic primers and cloning primers were purchased from MB Biotech (Taipei, Taiwan) (Table 1).
Construction of an artificial pha operon.
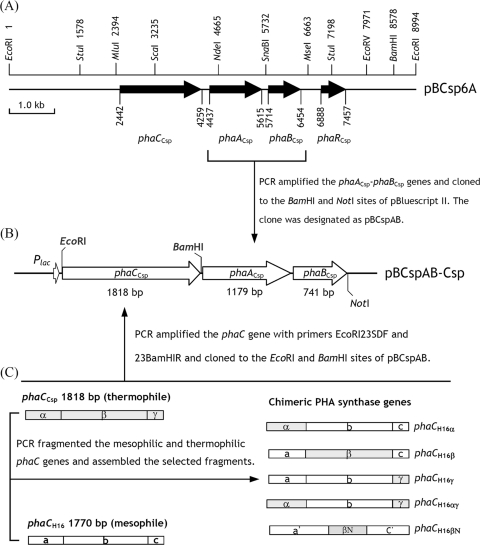
An artificial pha operon, consisting of phaC-phaACsp-phaBCsp, was constructed downstream from the lac promoter of the pBluescript II plasmid (Fig. 1B). First, the phaACsp-phaBCsp gene fragment of Cupriavidus sp. S-6 was amplified with primers CspAB-BamHIF and CspAB-NotIR (Table 1 and Fig. 1B) and was cloned into the BamHI and NotI sites of pBluescript II. The 50-μl PCR mixture contained 1 μl genomic DNA (30 ng), 0.8 μM primers, and 1 M betaine as a PCR additive. After heating for 5 min at 94°C, PCR was performed for 2 cycles pre-PCR (94°C for 20 s, 52°C for 30 s, and 72°C for 3 min), followed by 26 cycles of PCR (94°C for 20 s, 63°C for 30 s, and 72°C for 3 min). The gel-purified PCR product was cloned to the BamHI and NotI sites of pBluescript II KS. The plasmid carrying the phaACsp and phaBCsp genes was designated pBCspAB. Plasmid pBH16AB was constructed by use of the same method, but the phaAH16-phaBH16 gene fragment of C. necator H16 was amplified with primers H16AB-BamHI and H16AB-NotIR (Table 1). Subsequently, the wild-type or chimeric phaC gene was PCR amplified with primers EcoRI23SDF and 23BamHIR and cloned into the EcoRI and BamHI sites of plasmid pBCspAB or pBH16AB.
Fig 1.
Organization of the pha locus in Cupriavidus sp. S-6 and construction of the artificial pha operon and chimeric phaC genes. (A) Organization of the phaCCsp, phaACsp, and phaBCsp genes in the pha operon from Cupriavidus sp. S-6 and restriction map of the 9-kb EcoRI-cloned DNA fragment. (B) Construction of the artificial pha operon. (C) Construction of chimeric phaC genes. Details of the construction of the artificial pha operon and chimeric phaC are described in Materials and Methods.
PHA accumulation and analysis.
Recombinant E. coli cells carrying the artificial pha operon was precultured in LB medium containing 100 μg/ml ampicillin at 30°C overnight. The PHA accumulation experiment was carried out by inoculating 2% of a culture grown overnight into a 250-ml Erlenmeyer flask containing 30 ml LB broth, 100 μg/ml ampicillin, 1.5% sodium gluconate, and 50 μM isopropyl-β-d-1-thiogalactopyranoside (IPTG). After 48 h of cultivation at 30°C, 37°C, 42°C, and 45°C, respectively, in an orbital shaker at 200 rpm, the bacterial cells were harvested, washed twice with saline, and lyophilized. The lyophilized cells were subjected to methylation. The PHA content was determined by gas chromatography as described in a previous report (33).
Overexpression and purification of PHA synthases.
E. coli BLR(DE3) cells (Novagen) harboring overexpression plasmids were precultured overnight in 2 ml 2× YT medium (32) containing 100 μg/ml ampicillin at 30°C. One milliliter of the culture grown overnight was seeded into a 500-ml Erlenmeyer flask containing 100 ml 2× YT medium and 200 μg/ml of ampicillin and incubated at 30°C. Once the optical density at 600 nm (OD600) reached approximately 0.6, a final concentration of 0.3 mM IPTG was added to the medium, and the culture was cultivated at 20°C for an additional 40 h to induce the overexpression of PHA synthase. The crude lysate preparation, protein purification with ammonium sulfate precipitation, and methyl hydrophobic interaction chromatography were performed as described previously (32). After a final concentration of 20% glycerol was added, the purified enzyme was concentrated with an Amicon Ultra instrument (30-kDa-molecular-mass cutoff) to approximately 1 mg/ml and stored at −70°C. The purified enzymes were separated by SDS-PAGE. Subsequently, Coomassie brilliant blue R-250 staining revealed the electrophoretic homogeneity of the purified recombinant PHA synthases (see Fig. S1A in the supplemental material). Activity staining (Fig. S1B) and Western blotting (Fig. S1C) showed the activities of polymerization and their identities. Anti-PhaCH16 antiserum was the first antibody used for Western blotting (42). Western blotting and activity staining were performed as described previously (32, 42).
PHA synthase activity assay.
The substrate of PHA synthase, β-hydroxybutyryl-coenzyme A (CoA) (3-HBCoA), was prepared as reported previously (32, 37). PHA synthase activity was assayed by a discontinuous method by monitoring the CoA released from the substrate 3-ΗΒCoA (12). A 0.3-ml reaction mixture (100 mM Tricine [pH 8.0], 1 mM 3-ΗΒCoA, 0.2% glycerol, and 0.05% Hecameg) in an Eppendorf tube was preincubated at 30°C or an assigned temperature for 5 min. The addition of 12 to 50 nM PHA synthase to the mixture initiated the reaction. Aliquots (15 μl) were removed at intervals (20 s) and quenched by immediate mixing with 40 μl of 0.5% trichloroacetic acid. The quenched mixture (55 μl) was added to 50 μl of 2 mM 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) in 150 mM Tris (pH 8.0) and incubated at room temperature for 2 min. After 300 μl deionized water was added, the optical absorbance of the yellow mixture was measured at 412 nm. The concentration of CoA released was calculated with Beer's law (ε = 13.6 mM−1 cm−1). The linear range of the increase in the OD412 was taken to calculate the reaction rate. The release of CoA in the blank control, a reaction mixture without PHA synthase, was also monitored to check the stability of 3-HBCoA at temperatures of 40°C to 60°C.
Optimal temperature and thermostability measurements.
The optimal temperature for the enzyme activity assay was determined by using the standard assay at temperatures ranging from 15°C to 60°C at 5°C intervals. To determine thermostability, purified PHA synthase (0.14 mg/ml) in 50 mM potassium phosphate buffer (pH 7.5) containing 20% glycerol (Invitrogen) and 0.005% Hecameg (Calbiochem) was incubated at different temperatures (between 4°C and 60°C) for 30 min. After the samples were cooled on ice for 5 min, the residual activity was determined by using the standard method at their optimal temperature. The pH of the reaction buffer (Tricine, pH 8.0) was adjusted at room temperature.
Half-lives of thermal inactivation.
Purified PHA synthase (0.14 mg/ml) in 50 mM potassium phosphate buffer (pH 7.5) containing 20% glycerol (Invitrogen) and 0.005% Hecameg was incubated at 45°C. Incubation was carried out with a precisely temperature-controlled water bath (model B402H; Firstek, Taiwan). Aliquots removed at various time intervals were incubated on ice for 5 min prior to enzyme activity measurements. The residual activity was assayed at optimal temperatures (PhaCH16 at 37°C, PhaCH16β at 45°C, and PhaCCsp at 50°C). The heat inactivation experiment was monitored until >80% of the activity was lost. The plot of the percent log residual activity versus time was linear. First-order rate constants of thermal inactivation were obtained by linear regression with semilogarithmic coordinates. The inactivation rate constant (kinact) was obtained from the slope, and the half-life of PHA synthase was estimated.
Nucleotide sequence accession numbers.
The complete nucleotide sequence of the 8,999-bp Cupriavidus sp. S-6 DNA fragment appears in the EMBL, GenBank, and DDBJ nucleotide sequence database under accession no. HE610111. The accession no. of the 16S rRNA gene sequence of Cupriavidus sp. strain S-6 in the EMBL/GenBank/DBJ database is HE660045.
RESULTS
Cupriavidus sp. S-6.
Cupriavidus sp. S-6, a Gram-negative bacterium, was isolated from a hot spring in Southern Taiwan. Its 16S rRNA gene, amplified with primers 27F and 1492R (38), is 99% identical to that of the mesophile Cupriavidus necator H16. Phylogenetically, Cupriavidus sp. S-6 was very closely related to the mesophile C. necator H16. However, the optimal growth temperature of Cupriavidus sp. S-6 was markedly different from that of C. necator H16. Cupriavidus sp. S-6 was a thermophile. Its optimal growth temperature was around 50°C. Cupriavidus sp. S-6 accumulated 49% (dry weight) of PHB from gluconate as the carbon source at 50°C. Furthermore, it was also capable of accumulating 18% (dry weight) poly(3-hydroxybutyrate-co-3-hydroxyvalerate) copolymer with 78 mol% 3-hydroxyvalerate monomer from sodium valerate as a sole carbon source at 45°C. The ability for the biosynthesis of PHB at 50°C supported that the PHA synthase of Cupriavidus sp. S-6 was thermostable. In contrast, C. necator H16, a mesophile, was commonly cultivated at 30°C for bacterial growth and PHA accumulation (27, 30). It did not grow at temperatures over 37°C (data not shown). Colony PCR was used to clone a phaC gene fragment (806 bp) of Cupriavidus sp. S-6 (phaCCsp) by use of primers PhaCF1 and PhaCR2 (34). The partial gene of phaCCsp encoded 268 amino acids, which exhibited 91% identity and 94% similarity to the PHA synthase of the mesophile C. necator H16. The results of PHA accumulation experiments and PCR cloning supported that the protein sequences of the PHA synthases of the thermophile Cupriavidus sp. S-6 and the mesophile C. necator H16 were highly similar; however, their thermostabilities were different.
Cloning of the pha operon of Cupriavidus sp. S-6.
A genomic DNA library (∼10,000 colonies) of Cupriavidus sp. S-6 was constructed with E. coli XL1-Blue and cultivated on an LB agar plate containing 1.5% glucose and ampicillin. After 2 days of cultivation at 30°C, an opaque colony was isolated. The positive candidate was further purified by streak culturing and was subjected to PHA accumulation experiments. It accumulated 40% (dry weight) and 12% (dry weight) PHB contents from glucose as a carbon source at 30°C and 45°C, respectively. The plasmid of the candidate clone was designated pBCsp6A. EcoRI digestion and agarose gel electrophoresis analysis showed an approximately 9-kbp inserted DNA fragment in plasmid pBCsp6A. Colony PCR detected the existence of the PHA synthase gene in pBCsp6A. The DNA sequence was the same as that amplified from the genomic DNA of the thermophile Cupriavidus sp. S-6 (data not shown). PHA accumulation and PCR results revealed that plasmid pBCsp6A possessed a thermostable PHA synthase gene, which was derived from the thermophile Cupriavidus sp. S-6.
Nucleotide sequences of the PHA biosynthesis genes.
The nucleotide sequence of pBCsp6A in both strands was determined. Sequence analysis showed a pha operon identified by BLAST (Fig. 1A). The pha locus of Cupriavidus sp. S-6 consisted of a phaCAB operon, in which the organization was the same as that of C. necator H16 (27, 30). The PHA synthase (phaCCsp), β-ketothiolase (phaACsp), and acetoacetyl-CoA reductase (phaBCsp) genes of the thermophile Cupriavidus sp. S-6 showed 81%, 92%, and 96% identity, respectively, to the corresponding genes of the mesophile C. necator H16 (27, 30). Obviously, the β-ketothiolase and acetoacetyl-CoA reductase genes of Cupriavidus sp. S-6 and C. necator H16 were highly homologous; however, the PHA synthase genes were not (81% identity). According to the alignment results (Fig. 2), the diversity of the two PHA synthases was dominant on the N-terminal (corresponding to amino acids 1 to 152 of PhaCH16) and C-terminal (corresponding to amino acids 522 to 589 of PhaCH16) regions (Fig. 2). The middle regions (corresponding to amino acids 153 to 521 of PhaCH16) of both PHA synthases were highly homologous (92% identity). The middle region involved the whole α/β-hydrolase fold region of PHA synthase analyzed by Pfam (7). In summary, the unique diversity of the thermophilic and mesophilic PHA synthases, which were highly diverse in the N and C termini but highly homologous in the middle region, provides an opportunity to explore the correlation between mutations and thermostability.
Establishment of an in vivo system for evaluation of the thermostability of PHA synthases.
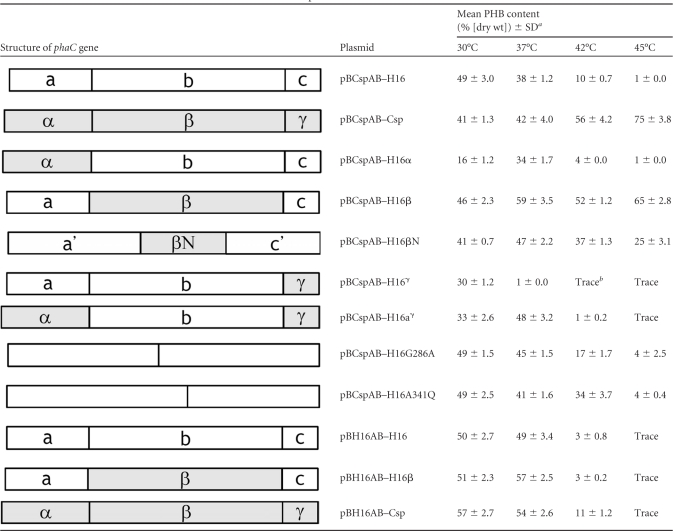
In order to evaluate the thermostability of PHA synthases, an artificial pha operon, phaC-phaACsp-phaBCsp, was constructed downstream of the lac promoter of pBluescript II (pBCspAB) (Fig. 1B). The addition of IPTG triggered the expression of the pha operon. In the artificial operon, the β-ketothiolase and acetoacetyl-CoA reductase genes of the thermophile Cupriavidus sp. S-6 were employed. The candidate phaC gene was cloned into the EcoRI and BamHI sites of the plasmid. The phaAH16 and phaBH16 genes of the mesophile C. necator H16 were also evaluated as the component genes of the artificial pha operon for PHA accumulation at high temperatures. However, the mesophilic phaAH16 and phaBH16 genes did not support the accumulation of PHB in recombinant E. coli cells at 45°C, even though the thermophilic PHA synthase gene phaCCsp was in the artificial pha operon (pBH16AB-Csp) (Table 2).
Table 2.
PHA accumulation in recombinant E. coli at different temperaturesa
The data shown are the means of data from three individual experiments. Plasmid pBCspAB–H16 carried the mesophilic phaCH16 gene from C. nector H16; pBCspAB–Csp carried the thermophilic phaCCsp gene from Cupriavidus sp. S-6. Others carried the chimera of the phaCH16 and phaCH16 genes or the mutants. The pBCspAB serial plasmids carried the thermophilic phaA and phaB genes of the thermophile Cupriavidus sp. S-6 to build the metabolic pathway for the provision of R-3-hydroxybutyryl-CoA; the pBH16AB serial plasmids carried the mesophilic phaA and phaB genes of C. necator H16.
Trace indicates a PHB content of <0.2%.
Recombinant E. coli strain XL1-Blue/pBCspAB-Csp, harboring the wild-type PHA synthase gene of the thermophile Cupriavidus sp. S-6, accumulated 41% (dry weight) PHB at 30°C (Table 2). With the elevation of the cultivation temperature, the PHB content increased and reached its highest level at 45°C (75% [dry weight]) (pBCspAB-Csp) (Table 2). In contrast, E. coli XL1-Blue/pBCspAB-H16, harboring the PHA synthase of the mesophile C. necator H16, accumulated the highest content of PHB (49% [dry weight]) at 30°C, and the content then drastically decreased with the increase of the cultivation temperature (pBCspAB-H16) (Table 2). At 45°C, only 1% (dry weight) PHB content was detected (Table 2). In this assay system, the thermostabilities of thermophilic (PhaCCsp) and mesophilic (PhaCH16) PHA synthases were differentiated distinctly based on the highest level of PHB accumulation at different temperatures. The results demonstrated the feasibility of using this system to differentiate the thermostabilities of PHA synthases.
Mutations enhance the thermostability of mesophilic PhaCH16.
In this study, α, β, and γ denote the N-terminal (corresponding to amino acids 1 to 152 of PhaCH16), middle (corresponding to amino acids 153 to 521 of PhaCH16), and C-terminal (corresponding to amino acids 522 to 589 of PhaCH16) regions of the thermophilic PHA synthase PhaCCsp, respectively (Fig. 1C). In addition, a, b, and c represent the N-terminal, middle, and C-terminal regions of the mesophilic PHA synthase PhaCH16, respectively (Fig. 1C). In order to explore mutations of PhaCCsp and their relationships to thermostability, a region-selected approach was used to construct four chimeric PHA synthase genes (phaCH16α, phaCH16β, phaCH16γ, and phaCH16αγ) of phaCH16 and phaCCsp (Fig. 1C), in which the N-terminal, middle, and C-terminal regions of the thermophilic PHA synthase gene (phaCCsp) replaced the corresponding regions of the mesophilic PHA synthase gene (phaCH16). Figure 1C shows the gene structures of the chimeras. The PCR-constructed chimeric gene was cloned in to the phaC gene position of the artificial pha operon and transformed into E. coli XL-1 Blue for PHA accumulation experiments.
The in vivo thermostabilities of chimeric PHA synthases were evaluated based on the PHB content accumulated at 30°C, 37°C, 42°C, and 45°C. Four chimeras were capable of accumulating PHB at 30°C; the PHB content ranged from 16 to 46% (dry weight) (Table 2). All chimeras were enzymatically active. With the increase of the cultivation temperature, chimeras accumulated different amounts of PHB in the cell. The temperature at which the cells accumulated the highest contents of PHB was the index to differentiate the thermostabilities of chimeras. In the four chimeras, at 45°C, only PhaCH16β accumulated a high content of PHB (65% [dry weight]) (pBCspAB-H16β) (Table 2); phaCΗ16α, phaCΗ16γ, and phaCΗ16αγ accumulated less than 1% (dry weight) PHB at 45°C, which was similar to the amount accumulated with the mesophilic parental enzyme PhaCH16 (pBCspAB-H16α, pBCspAB-H16γ, and pBCspAB-H16αγ) (Table 2). The results showed that only chimeric PhaCH16β was thermostable. Moreover, only mutations from the β region of thermophilic PhaCCsp enhanced the thermostability of the mesophilic PHA synthase PhaCH16; mutations from α, γ, or the combined α and γ regions of PhaCCsp did not improve the thermostability of PhaCH16. Although the chimera PhaCH16β accumulated a high content of PHB at 45°C (65% [dry weight]), the PHB content was still lower than that with PhaCCsp (75% [dry weight]). Hence, the thermostabilities of the chimera PhaCH16β and the parental enzyme PhaCCsp might be close but not the same. The α region, γ region, or both regions of PhaCCsp were still possessing mutations that contributed to the thermostability of PhaC, even though the contribution was minor.
In order to explore the scope of the β region that actually contributed to the enhancement of the thermostability of PhaCH16, another chimeric gene, phaCH16βN, was constructed. The chimeric phaCH16βN gene was the phaCH16 gene with a partial β region sequence (βN) of phaCCsp. Figure 2 shows the βN region of PhaCCsp (CspC790F to CspC1253F). With the βN region, 17 point mutations surrounding the catalytic site (GXCXGG) of PHA synthase were introduced into PhaCH16 (Fig. 2). The chimera PhaCH16βN accumulated 41% (dry weight) PHB at 30°C and reached a peak at 37°C (47% [dry weight]). Subsequently, the PHB content decreased with the elevation of the cultivation temperature (pBCspAB-H16βN) (Table 2). Nonetheless, chimeric PhaCH16βN still accumulated 25% (dry weight) PHB at 45°C (Table 2). Obviously, the point mutations derived from the partial β region (βN) improved the thermostability of PhaCH16. However, the thermostability of PhaCH16βN was not equal to that of chimeric PhaCH16β. The results suggest that the essential mutations related to the thermostability of PhaCCsp are mainly scattered on the whole β region rather than on a partial area. Preliminary site-directed mutagenesis experiments showed that the A341Q mutant of PhaCH16 accumulated 34% (dry weight) PHB at 42°C and accumulated a PHB content comparable to that of the wild type at 30°C and 37°C (pBCspAB-H16A341Q) (Table 2). The results suggested that the A341Q substitution enhanced the thermostability of PhaCH16. The G286A mutant of PhaCH16 accumulated a higher content of PHB (45% [dry weight]) than the wild type (38% [dry weight]) at 37°C and accumulated 17% (dry weight) PHB at 42°C (pBCspAB-H16G286A) (Table 2). The G286A mutation enhanced the enzyme activity and slightly enhanced the thermostability.
Effects of temperature on the activity of PHA synthases.
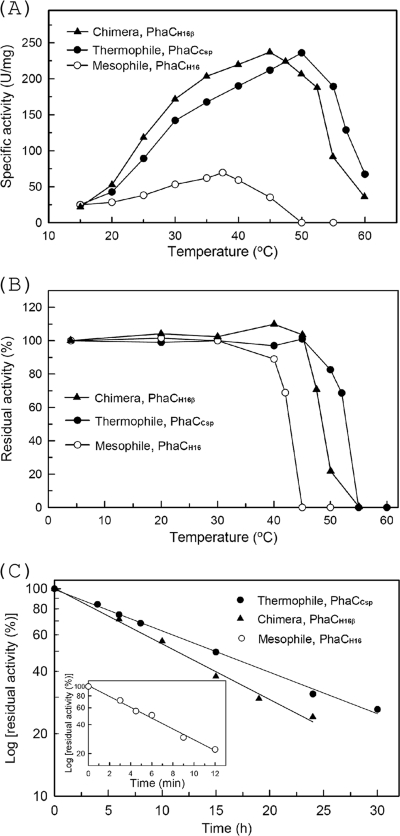
The parental and chimeric PHA synthase genes phaCH16, phaCCsp, and phaCH16β were overexpressed in E. coli BLR(DE3) cells and purified. Figure 3A shows the activity-temperature profiles of chimeric and parental PHA synthases. The parental enzymes PhaCH16 and PhaCCsp had temperature optima of 37°C and 50°C, respectively; the optimal temperature of the chimera PhaCH16β was 45°C. At their optimal temperatures, PhaCH16, PhaCCsp, and the chimera PhaCH16β had specific activities of 70 ± 8.8 U/mg, 236 ± 19.4 U/mg, and 237 ± 11.8 U/mg, respectively. Although the optimal temperature of chimeric PhaCH16β was 5°C lower than that of its parental enzyme PhaCCsp, its specific activity was the same as that of PhaCCsp at their optimal temperatures.
Fig 3.
Optimal temperature, thermal stability, and half-lives of thermal inactivation of PHA synthases. (A) Activity-temperature profiles of wild-type and chimeric PHA synthases. (B) Thermal stability of PHA synthases. (C) Kinetics of thermal inactivation of PHA synthases at 45°C. To estimate the thermal stabilities of PHA synthases, the enzyme was preincubated at the indicated temperatures for 30 min and then incubated on ice for 5 min. The residual activities were then determined by using a discontinuous method at 37°C (mesophile, PhaCH16), 45°C (chimera, PhaCH16β), or 50°C (thermophile, PhaCCsp). PhaCCsp and PhaCH16 are PHA synthases from the thermophile Cupriavidus sp. S-6 and the mesophile C. necator H16, respectively. PhaCH16β is a chimera of PhaCH16 and PhaCCsp. The data are the means of data from three individual experiments.
Obviously, PhaCH16β inherited high enzyme activity from its parental enzyme PhaCCsp despite having a lower optimal temperature. At 30°C, the specific activities of PhaCH16, PhaCCsp, and chimeric PhaCH16β were 50 ± 5.2, 142 ± 11.7, and 172 ± 17.8 U/mg, respectively. The specific activity of chimeric PhaCH16β was 1.21-fold higher than that of PhaCCsp (142 ± 11.7 U/mg) and 3.45-fold higher than that of PhaCH16 (50 ± 5.2 U/mg). At 15°C, PhaCH16, PhaCCsp, and chimeric PhaCH16β showed nearly the same specific activities (Fig. 3A). Obviously, chimeric PhaCH16β showed better activity than those of its parental enzymes PhaCH16 and PhaCCsp at 30°C (Fig. 3A). At a temperature of 30°C, high-cell-density fermentation for the mass production of PHB was reported (49, 50). At this temperature, chimeric PhaCH16β has a higher specific activity than those of its parental enzymes PhaCCsp and PhaCH16. This finding suggests that chimeric PhaCH16β will accumulate higher levels of PHB than PhaCCsp and PhaCH16 in recombinant E. coli fermentations.
At 37°C, PhaCH16β accumulated 1.55-fold more PHB than did PhaCH16 (Table 2). This is in good agreement with the correlation between the specific activity of PHA synthase and the PHB content (40). However, at 30°C, PhaCH16 accumulated a higher level of PHB than those of the higher-activity enzymes PhaCH16β and PhaCCsp (Table 2). This finding conflicts with the above-described results. We postulate that the conflict may be due to the use of the thermophilic genes phaACsp and phaBCsp in the artificial operon. Previous reports suggested that a thermophilic enzyme shows a more rigid structure but less flexibility than a mesophilic enzyme (47, 48). This means that thermophilic enzymes will present lower levels of activity at lower temperatures than mesophilic enzymes. If the inference was correlative in our case, then thermophilic PhaACsp and PhaBCsp provided lower concentrations of PHA synthase substrate than did mesophilic PhaAH16 and PhaBH16.
To further investigate this observation, another artificial pha operon was constructed, in which the β-ketothiolase and acetoacetyl-CoA reductase genes were derived from the mesophile C. necator H16. In pBH16AB-X-based plasmids (where X is H16, Csp, or H16β), the thermostable PHA synthases PhaCCsp and chimeric PhaCH16β accumulated higher levels of PHB than did PhaCH16 at 30°C and 37°C (Table 2). In the pBH16AB serial plasmids, the PHB content and specific activity of PHA synthase revealed a positive correlation at 30°C and 37°C. Furthermore, the PHB content accumulated by E. coli/pBH16AB-X serial strains was higher than that accumulated by E. coli/pBCspAB-X serial strains (Table 2). The results suggest that not only PHA synthase but also β-ketothiolase and acetoacetyl-CoA reductase are critical for efficient PHA accumulation.
Thermostability of PHA synthases.
With regard to heat stability, mesophilic parental enzyme PhaCH16 was stable at 30°C for 30 min (Fig. 3B). It retained about 70% of its maximum activity at 42°C and became inactive at 45°C. The thermophilic parental enzyme PhaCCsp and chimera PhaCH16β were stable at up to 45°C and retained about 70% of their maximum activity at 47°C and 52°C, respectively; both became inactive at 55°C (Fig. 3B). The kinetics of heat inactivation, reported as the rate of thermal inactivation (half-life of heat inactivation [t1/2]), was also measured at 45°C. As shown in Fig. 3C, the parental enzymes PhaCH16 and PhaCCsp and chimera PhaCH16β exhibited first-order inactivation kinetics, with t1/2 values of 5.3 min, 15.06 h, and 11.20 h, respectively. Substantially, the half-life of PhaCH16 (5.3 min) was enhanced 127-fold (11.20 h for PhaCH16β) when the β region of PhaCCsp was introduced. Nonetheless, the half-life of the chimera PhaCH16β (11.20 h) was a little shorter than that of thermophilic parental enzyme PhaCCsp (15.06 h). This suggests that the amino acid residues contributing to the thermostability of PhaCCsp were not all in the β region and that some were located in the α or γ region. This result is in good agreement with the results of the PHA accumulation experiments (pBCspAB-Csp and pBCspAB-H16β) (Table 2).
DISCUSSION
This study presents a thermostable PHA synthase (PhaCCsp) from the thermophile Cupriavidus sp. S-6 that has a high level of activity and a high level of thermostability. The most important characteristic of PhaCCsp is its primary structure, which is highly similar to that of the PHA synthase of the mesophile C. necator H16. By use of the region-selected method, 30 point mutations derived from the thermophilic PHA synthase were verified to have an association with the enhancement of thermostability and the activity of the mesophilic PHA synthase PhaCH16. This study establishes an in vivo system in E. coli which is applicable for differentiating the thermostability of PHA synthases. The system differentiated the thermostability of PHA synthases based on the accumulation of PHA at various temperatures. It did not require tedious work with protein purification and biochemical analyses. Thus, it was suitable for the rapid screening of thermostable PhaC candidates.
This study employed E. coli XL1-Blue, a strain commonly used for PHA fermentation (4, 49, 50), as the host cell line for the PHA accumulation experiment. Fotadar et al. and Hoffman et al. reported previously that E. coli DH5α is capable of growth at temperatures of up to 49°C, although growth is prohibitive beyond 40°C (8, 15). The effects of temperature on the growth of E. coli XL1-Blue were similar; however, E. coli XL1-Blue had no growth when the cultivation temperature went above 45°C (data not shown). At 42°C and 45°C, the growth and PHA accumulation of recombinant E. coli XL1-Blue were monitored for 4 days. The turbidity and PHA content of recombinant E. coli reached a plateau on the second day and slightly decreased with time at 42°C and 45°C (data not shown). In this study, recombinant E. coli XL1-Blue strains accumulated PHA at temperatures of 30°C to 45°C. Theoretically, a more thermostable PHA synthase should cause a peak in the PHB content at higher temperatures. Accordingly, the thermostable PHA synthase can be discriminated.
The gene structure of the chimera phaCH16β is that of the phaCH16 gene possessing the β region sequence of phaCCsp. In the β region, 30 point mutations were introduced into mesophilic PhaCH16. The 30 point mutations derived from PhaCCsp were naturally evolved and not artificially generated. Figure 2 exhibits the point mutations in PhaCH16β. Taguchi et al. previously reported a point mutation, S80P, of PhaCH16 that enhances the thermostability of PhaCH16 but results in 27% enzyme activity (41). In this study, the S80P mutation was not included in PhaCH16β; position S80 is not in the middle range of PhaCH16. Hence, an effect of S80P on the thermostability of PhaCH16β was ruled out. Kumar et al. indicated previously that the appearance of arginine (Arg) and tyrosine (Tyr) is significantly more frequent in thermophilic proteins based on a comparison of sequences of homologous thermophile-mesophile protein pairs (20). In the original sequence of 30 point mutations (PhaCH16), there are no Arg and Tyr residues. However, five Arg mutations (L170R, S287R, K312R, H338R, and A493R) and one Tyr mutation (F218Y) were observed for PhaCH16β (Fig. 2). Accordingly, the Arg and Tyr mutations should be highly promising in relation to the thermostability of PhaCH16β. Watanabe and Suzuki previously proposed the proline rule for protein thermostabilization (51). Those authors suggested that replacements with proline residues reduce the conformational freedom of the polypeptide chain and thus increase protein thermostabilization (51). In PhaCH16β, one amino acid substitution, D311P, was generated. Based on the proline rule, the D311P mutation might be related to the thermostability of PhaCH16β.
Margarit et al. found previously that G147A and G189A mutations of a neutral protease from Bacillus subtilis were more stable against irreversible thermal inactivation (23). Ganter and Plückthun showed previously that one Gly-to-Ala substitution (G316A) in glyceraldehyde-3-phosphate dehydrogenase strongly stabilized a mutant under conditions of irreversible heat denaturation (9). Menéndez-Arias and Argos compared the amino acid sequences of thermophilic and mesophilic molecules from six different protein families. They showed that the Gly→Ala substitution is the top residue substitution for helices and strands from mesophiles to thermophiles (24). Those studies strongly suggest that a position-specific Gly→Ala substitution in an enzyme is highly related to the enhancement of thermostability. In this study, three Gly→Ala substitutions, G286A, G365A, and G377A, were found in PhaCH16β (Fig. 2). The Gly→Ala substitutions in PhaCH16β should be correlative with its thermostability. In contrast, three Ala→Gly substitutions, A296G, A334G, and A466G, were also observed for PhaCH16β (Fig. 2). The Ala→Gly substitution in the helix is helix destabilizing (5). This means that the Ala→Gly substitution is promising to decrease the thermostability of proteins. Thus, the role of the Ala→Gly substitutions A296G, A334G, and A466G in PhaCH16β needs to be further investigated.
Previous research described an F420S mutation in PhaCH16 which enhanced specific activity 2.4-fold. However, the thermostability of the F420S mutant is lower than that of the wild type (41). In the sequence of PhaCH16β, F420 is conservative. The high level of activity of PhaCH16β does not relate to the F420S mutation. PhaCH16β should bear beneficial mutations related to activity enhancement that have not yet been reported. Recently, Bhubalan et al. reported a highly active PHA synthase from Chromobacterium sp. strain USM2, PhaCCs, which revealed a homology of 46% with PhaCH16 and exhibited a specific activity of 238 ± 98 U/mg at 30°C (3). This is the highest specific activity reported for a purified PHA synthase (3). In this study, the chimera PhaCH16β has a specific activity of 172 ± 17.8 U/mg at 30°C. Despite a lower specific activity than that of Chromobacterium sp. USM2 at 30°C, the chimera PhaCH16β exhibited comparable activity at its optimal temperature of 45°C (237 ± 11.8 U/mg). Furthermore, the chimera PhaCH16β is thermostable and only 30 amino acids different from PhaCH16. Thus, the chimera PhaCH16β is a proper model to explore the activity- and thermostability-enhancing substitutions in PHA synthase.
The in vitro synthesis approach is an ideal method to produce a tailor-made PHA polymer; however, the availability of a stable and catalysis-efficient PHA synthase and the substrate 3-hydroxyacyl-CoA limits its application (11). The thermostable and highly active PHA synthase will give a partial answer to questions regarding in vitro PHA synthesis (13). The chimera PhaCH16β showed a long half-life at 45°C (11.2 h) and possessed high specific activity at 30°C and 45°C. Thus, it is a good candidate for in vitro PHA polymerization. In addition, PhaCH16β possesses a higher level of activity than the mesophilic PhaCH16 and thermophilic PhaCCsp parental enzymes at 20°C to 40°C (Fig. 3A). The chimeric phaCH16β gene is more appropriate for E. coli PHA fermentation than the mesophilic phaCH16 gene.
Regarding the optimal temperature, enzyme activity, thermostability, or kinetics of heat inactivation, the chimera PhaCH16β shows better characteristics than the parental enzyme PhaCH16. The high levels of thermostability and enzyme activity of PhaCH16β are the effect of the introduction of 30 point mutations derived from the thermophilic enzyme PhaCCsp. The introduced point mutations should include activity-improving and thermostability-enhancing residues. Due to the limited numbers of mutations generated in PhaCH16β, it is possible to explore the role of each mutation by site-directed mutagenesis in future studies.
Supplementary Material
ACKNOWLEDGMENTS
This work is supported by grant NSC 96-2311-B-022-002-MY2 from the National Science Council (Taipei, Taiwan).
We are grateful to Anthony J. Sinskey and Christopher J. Biogham (Massachusetts Institute of Technology) for their generous donation of the specific antiserum against the PHA synthase of C. necator H16.
Footnotes
Published ahead of print 9 March 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Amara AA, Steinbüchel A, Rehm BH. 2002. In vivo evolution of the Aeromonas punctata polyhydroxyalkanoate (PHA) synthase: isolation and characterization of modified PHA synthases with enhanced activity. Appl. Microbiol. Biotechnol. 59:477–482 [DOI] [PubMed] [Google Scholar]
- 2. Anderson AJ, Dawes EA. 1990. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54:450–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bhubalan K, et al. 2011. Characterization of the highly active polyhydroxyalkanoate synthase of Chromobacterium sp. strain USM2. Appl. Environ. Microbiol. 77:2926–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bullock WO, Fernandez JM, Short JM. 1987. XL1-Blue—a high-efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. Biotechniques 5:376 [Google Scholar]
- 5. Chakrabartty A, Schellman JA, Baldwin RL. 1991. Large differences in the helix propensities of alanine and glycine. Science 351:586–588 [DOI] [PubMed] [Google Scholar]
- 6. Choi J, Lee SY. 1999. Factors affecting the economics of polyhydroxyalkanoate production by bacterial fermentation. Appl. Microbiol. Biotechnol. 51:13–21 [Google Scholar]
- 7. Finn RD, et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38(Suppl 1):D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fotadar U, Zaveloff P, Terracio L. 2005. Growth of Escherichia coli at elevated temperatures. J. Basic Microbiol. 45:403–404 [DOI] [PubMed] [Google Scholar]
- 9. Ganter C, Plückthun A. 1990. Glycine to alanine substitutions in helices of glyceraldehyde-3-phosphate dehydrogenase: effects on stability. Biochemistry 29:9395–9402 [DOI] [PubMed] [Google Scholar]
- 10. Gerngross TU. 1999. Can biotechnology move us toward a sustainable society? Nat. Biotechnol. 17:541–544 [DOI] [PubMed] [Google Scholar]
- 11. Gerngross TU, Martin DP. 1995. Enzyme-catalyzed synthesis of poly[(R)-(−)-3-hydroxybutyrate]: formation of macroscopic granules in vitro. Proc. Natl. Acad. Sci. U. S. A. 92:6279–6283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gerngross TU, et al. 1994. Overexpression and purification of the soluble polyhydroxyalkanoate synthase from Alcaligenes eutrophus: evidence for a required posttranslational modification for catalytic activity. Biochemistry 33:9311–9320 [DOI] [PubMed] [Google Scholar]
- 13. Gross RA, Ganesh M, Lu W-H. 2010. Enzyme-catalysis breathes new life into polyester condensation polymerizations. Trends Biotechnol. 28:435–443 [DOI] [PubMed] [Google Scholar]
- 14. Hezayen FF, Steinbüchel A, Rehm BH. 2002. Biochemical and enzymological properties of the polyhydroxybutyrate synthase from the extremely halophilic archaeon strain 56. Arch. Biochem. Biophys. 403:284–291 [DOI] [PubMed] [Google Scholar]
- 15. Hoffman H, Valdina J, Frank ME. 1966. Effects of high incubation temperature upon the cell wall of Escherichia coli. J. Bacteriol. 91:1635–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ibrahim MHA, Steinbüchel A. 2009. Poly(3-hydroxybutyrate) production from glycerol by Zobellella denitrificans MW1 via high-cell-density fed-batch fermentation and simplified solvent extract. Appl. Environ. Microbiol. 75:6222–6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ibrahim MHA, Steinbüchel A. 2010. High-cell-density cyclic fed-batch fermentation of a poly(3-hydroxybutyrate)-accumulating thermophile, Chelatococcus sp. strain MW10. Appl. Environ. Microbiol. 76:7890–7895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kichise T, Taguchi S, Yoshiharu D. 2002. Enhanced accumulation and changed monomer composition in polyhydroxyalkanoate (PHA) copolyester by in vitro evolution of Aeromonas caviae PHA synthase. Appl. Environ. Microbiol. 68:2411–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim YB, Lenz RW. 2001. Polyesters from microorganisms. Adv. Biochem. Eng. Biotechnol. 71:51–79 [DOI] [PubMed] [Google Scholar]
- 20. Kumar S, Tsai C-J, Nussinov R. 2000. Factors enhancing protein thermostability. Protein Eng. 13:179–191 [DOI] [PubMed] [Google Scholar]
- 21. Lemoigne M. 1926. Products of dehydration and of polymerization of β-hydroxybutyric acid. Bull. Soc. Chem. Biol. 8:770–782 [Google Scholar]
- 22. Madison LL, Huisman GW. 1999. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol. Mol. Biol. Rev. 63:21–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Margarit I, et al. 1992. Cumulative stabilizing effects of glycine to alanine substitutions in Bacillus subtilis neutral protease. Protein Eng. 5:543–550 [DOI] [PubMed] [Google Scholar]
- 24. Menéndez-Arias L, Argos P. 1989. Engineering protein thermal stability: sequence statistics point to residue substitutions in alpha-helices. J. Mol. Biol. 206:397–406 [DOI] [PubMed] [Google Scholar]
- 25. Nomura CT, Taguchi S. 2007. PHA synthase engineering toward superbiocatalysts for custom-made biopolymers. Appl. Microbiol. Biotechnol. 73:969–979 [DOI] [PubMed] [Google Scholar]
- 26. Normi YM, et al. 2005. Characterization and properties of G4X mutants of Ralstonia eutropha PHA synthase for poly(3-hydroxybutyrate) biosynthesis in Escherichia coli. Macromol. Biosci. 5:197–206 [DOI] [PubMed] [Google Scholar]
- 27. Peoples OP, Sinskey AJ. 1989. Poly-β-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16. Identification and characterization of the PHB polymerase gene (phbC). J. Biol. Chem. 264:15298–15303 [PubMed] [Google Scholar]
- 28. Pohlmann A, et al. 2006. Genome sequence of the bioplastic-producing “Knallgas” bacterium Ralstonia eutropha H16. Nat. Biotechnol. 24:1257–1262 [DOI] [PubMed] [Google Scholar]
- 29. Rehm BH. 2003. Polyester synthases: natural catalysts for plastics. Biochem. J. 376:15–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schubert P, Steinbüchel A, Schlegel HG. 1988. Cloning of the Alcaligenes eutrophus genes for synthesis of poly-β-hydroxybutyrate (PHB) and synthesis of PHB in Escherichia coli. J. Bacteriol. 170:5837–5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sheu D-S, Chen W-M, Yang J-Y, Chang R-C. 2009. Thermophilic bacterium Caldimonas taiwanensis produces poly(3-hydroxybutyrate-co-3-hydroxyvalerate) from starch and valerate as carbon sources. Enzyme Microb. Technol. 44:289–294 [Google Scholar]
- 32. Sheu D-S, Lai Y-W, Chang R-C, Chen W-M. 2009. Detection of polyhydroxyalkanoate synthase activity on a polyacrylamide gel. Anal. Biochem. 393:62–66 [DOI] [PubMed] [Google Scholar]
- 33. Sheu D-S, Lee C-Y. 2004. Altering the substrate specificity of olyhydroxyalkanoate synthase 1 derived from Pseudomonas putida GPo1 by localized semirandom mutagenesis. J. Bacteriol. 186:4177–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sheu D-S, Wang Y-T, Lee C-Y. 2000. Rapid detection of polyhydroxyalkanoate-accumulated bacteria isolated from the environment by colony PCR. Microbiology 146:2019–2025 [DOI] [PubMed] [Google Scholar]
- 35. Shozui F, Matsumoto K, Sasaki T, Taguchi S. 2009. Engineering of polyhydroxyallanoate synthase by Ser477X/Gln481X saturation mutagenesis for efficient production of 3-hydroxybutyrate-based copolyesters. Appl. Microbiol. Biotechnol. 84:1117–1124 [DOI] [PubMed] [Google Scholar]
- 36. Shozui F, et al. 2010. A new beneficial mutation in Pseudomonas sp. 61-3 polyhydroxyalkanoate (PHA) synthase for enhanced cellular content of 3-hydroxybutyrate-based PHA explored using its enzyme homolog as a mutation template. Biosci. Biotechnol. Biochem. 74:1710–1712 [DOI] [PubMed] [Google Scholar]
- 37. Song J, Zhang S, Lenz RW, Goodwin S. 2000. In vitro polymerization and copolymerization of 3-hydroxypropionyl-CoA with the PHB synthase from Ralstonia eutropha. Biomacromolecules 1:433–439 [DOI] [PubMed] [Google Scholar]
- 38. Suzuki MT, Giovannoni SJ. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Taguchi S, Doi Y. 2004. Evolution of polyhydroxyalkanoate (PHA) production system by “enzyme evolution”: successful case studies of directed evolution. Macromol. Biosci. 4:146–156 [DOI] [PubMed] [Google Scholar]
- 40. Taguchi S, et al. 2001. Analysis of mutational effects of polyhydroxybutyrate (PHB) polymerase on bacterial PHB accumulation using an in vivo assay system. FEMS Microbiol. Lett. 198:65–71 [DOI] [PubMed] [Google Scholar]
- 41. Taguchi S, Nakamura H, Hiraishi T, Yamato I, Doi Y. 2002. In vitro evolution of a polyhydroxybutyrate synthase by intragenic suppression-type mutagenesis. J. Biochem. 131:801–806 [DOI] [PubMed] [Google Scholar]
- 42. Tian J, et al. 2005. Analysis of transient polyhydroxybutyrate production in Wautersia eutropha H16 by quantitative Western analysis and transmission electron microscopy, J. Bacteriol. 187:3825–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tsuge T, et al. 2004. Mutation effects of a conserved alanine (A510) in type I polyhydroxyalkanoate synthase from Ralstonia eutropha on polyester biosynthesis. Macromol. Biosci. 4:963–970 [DOI] [PubMed] [Google Scholar]
- 44. Tsuge T, et al. 2007. Variation in copolymer composition and molecular weight of polyhydroxyalkanoate generated by saturation mutagenesis of Aeromonas caviae PHA synthase. Macromol. Biosci. 7:846–854 [DOI] [PubMed] [Google Scholar]
- 45. Valentin HE, et al. 1999. PHA production, from bacteria to plants. Int. J. Biol. Macromol. 25:303–306 [DOI] [PubMed] [Google Scholar]
- 46. Vandamme P, Coenye T. 2004. Taxonomy of the genus Cupriavidus: a tale of lost and found. Int. J. Syst. Evol. Microbiol. 54:2285–2289 [DOI] [PubMed] [Google Scholar]
- 47. Vieille C, Zeikus GJ. 2001. Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65:1–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vihinen M. 1987. Relationship of protein flexibility to thermostability. Protein Eng. 1:477–480 [DOI] [PubMed] [Google Scholar]
- 49. Wang FL, Lee SY. 1997. Production of poly(3-hydroxybutyrate) by fed-batch culture of filamentation-suppressed recombinant Escherichia coli. Appl. Environ. Microbiol. 63:4765–4769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang FL, Lee SY. 1998. High cell density culture of metabolically engineered Escherichia coli for the production of poly(3-hydroxybutyrate) in a defined medium. Biotechnol. Bioeng. 58:325–328 [PubMed] [Google Scholar]
- 51. Watanabe K, Suzuki Y. 1998. Protein thermostabilization by proline substitutions. J. Mol. Catal. B Enzym. 4:167–180 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.