Abstract
Alp7A is a bacterial actin from Bacillus subtilis plasmid pLS20 that functions in plasmid segregation. Alp7A's function requires that it assemble into filaments that treadmill and exhibit dynamic instability. These dynamic properties require the two other components of the alp7A operon, the downstream alp7R gene and the upstream alp7C sequence, as does the ability of Alp7A to form filaments at its physiological concentration in the cell. Here, we show that these two other components of the operon also determine the amount of Alp7A that is produced in the cell. The deletion of alp7R leads to overproduction of Alp7A, which assembles into large, amorphous, static filaments that disrupt chromosome segregation and cell division. The product of the alp7R gene is a DNA-binding protein that represses transcription of the alp7A operon. Purified Alp7R protein binds specifically to alp7C, which contains two σA promoters embedded within a series of near-repeats of a 10-mer. Alp7R also shows the typical nonspecific binding activity of a DNA-binding protein: Alp7R-GFP (green fluorescent protein) associates with the chromosomes of cells that lack alp7C. When Alp7A-GFP is produced in B. subtilis along with untagged Alp7R, Alp7A-GFP also colocalizes with the chromosome, indicating that Alp7R associates with Alp7A. Hence Alp7R, determines both the activity and the cellular concentration of Alp7A, and it can associate with Alp7A even if it is not bound to alp7C.
INTRODUCTION
Bacteria have very many actin-like proteins (Alps) whose amino acid sequences indicate that they are only distantly related to the very highly conserved eukaryotic actin (4). The Alps are themselves evolutionarily diverse and so, in order to lay the groundwork for understanding this diversity, we have organized them into families whose members show at least 30% amino acid identity (4, 47). Although only a small number of Alp families have been studied to date, it has become apparent that the Alps share the basic properties of eukaryotic actin. Their crystal structures approximate that of actin and they polymerize into filaments (35). The Alps participate in a variety of central processes relating to cell architecture and growth or to the movement and positioning of large structures within the cell.
There are fundamental differences between the few families of Alps that are encoded on bacterial chromosomes and the many families that are encoded on mobile genetic elements. The chromosomal Alps are highly conserved. Members of the MreB family, for example, are required for the proper cell morphology of nonspherical bacteria and are present in most rod-shaped bacteria (8, 9, 22, 24, 43, 49). FtsA, the other prominent chromosomal Alp, is a critical component of the cell division machinery and is well represented in eubacteria (7, 14, 21, 48). There are, in contrast, at least 30 distinct families of Alps that are encoded on a variety of mobile genetic elements, and many of these Alps perform the same function. For example, Alp7A, AlfA, and ParM represent three distantly related families of Alps; they are encoded on three different plasmids, yet they all segregate plasmids (1, 4, 27, 32, 47). Their identical function notwithstanding, these Alps have been shown to differ in their cell biological and in their biophysical properties (1, 4, 10, 11, 26, 32). Studying these proteins provides an opportunity for us to comprehend the scope of actin-based solutions to the challenge of plasmid segregation and at the same time to gain insights into the plasticity of actin that the study of eukaryotic actin or the few chromosomal Alps of bacteria cannot provide.
We have focused on Alp7A, a plasmid segregation protein from the 55-kb Bacillus subtilis plasmid pLS20 (4, 25). Alp7A shares only 13% primary sequence identity with eukaryotic actin but contains its five signature nucleotide binding motifs. Alp7A-GFP (green fluorescent protein), which retains the full function of Alp7A in plasmid segregation, polymerizes into filaments in B. subtilis that display the characteristic dynamic properties of both eukaryotic actin and tubulin; Alp7A filaments treadmill and exhibit dynamic stability. Mutations of amino acids that would be expected, in the manner of eukaryotic actin, to be involved in the binding or hydrolysis of nucleotide either eliminate these dynamic properties or prevent Alp7A from assembling into filaments (4).
The alp7A gene is the first of two genes that comprise a small operon adjacent to the pLS20 origin of replication (4, 25). A mini-pLS20 plasmid built from the operon and adjacent origin is, like pLS20 itself, completely stable over more than 30 generations of exponential growth. Deletion of the operon or of the alp7A gene from the operon leads to the rapid loss of mini-pLS20. Alp7A must be able to assemble into dynamic actin-like filaments in order to function in plasmid segregation; the mutations that prevent Alp7A filament formation or deprive Alp7A filaments of their dynamic properties also destabilize the plasmid. The dynamic properties of Alp7A also require that the other components of the operon be present in the cell either in cis or in trans. In the absence of the promoter region and the smaller downstream gene alp7R, only static filaments are formed, and these lack the uniform appearance of their dynamic counterparts. The other components of the operon also determine the critical concentration for Alp7A filament formation in the cell. When these components are present, dynamic filaments are observed if Alp7A is present at the level ordinarily found in a cell containing pLS20. When they are absent, filaments are observed only if Alp7A is present at more than 5-fold higher levels, and these filaments are static (4).
In the case of alp7A then, the activity and therefore the function of a bacterial actin is regulated by the other components of the operon. There is regulation of actin function and activity in other actin-based plasmid segregation systems, but it can be of a very different sort. In the parMR operon of the Escherichia coli plasmid R1, for example, the parM gene codes for another bacterial actin that is distantly related to eukaryotic actin, to the alp7 family, and to the other bacterial actin families (4, 12, 13, 45, 47). ParM, like Alp7A, is a plasmid segregation determinant, and filaments of both this prototypical ParM and the ParM of plasmid pB171 exhibit dynamic instability (10, 11, 34, 37, 41). Biophysical experiments with the ParM proteins have demonstrated, though, that this dynamic instability requires no extrinsic factors and that the critical concentration for filament formation is low enough that filaments would be expected to form spontaneously within the cell (10, 41). Experiments with an in vitro reconstitution of the entire system indicate that the product of the downstream parR gene, bound to the parC site that is situated upstream of parM, squelches the dynamic instability of ParM filaments and prevents their catastrophic disassembly (11).
The parMR promoter is embedded in the parC site such that by binding at parC, the ParR protein also represses transcription of the operon. Transcriptional repression has been demonstrated in the parM operon from plasmid R1 and also in the parM operon of plasmid pB171 (20, 26, 40). Complete repression of the R1 operon requires all 10 of the short repeat elements that comprise parC (2).
Comparatively little is known about alp7R and alp7C, the corresponding components of the alp7A operon, although they are a key to understanding the fundamental differences in behavior between the ParM and Alp7A systems. As more of these systems are studied closely, it has become apparent that even though all are actin-based plasmid segregation machineries, they do not function identically. Among the systems that have been studied—ParM, AlfA, the ParM of plasmid pSK41 (actually distantly related to the prototypical ParM and a member of another Alp family), and Alp7A—differences have been observed in nucleotide binding and hydrolysis parameters, in the kinetics of filament formation, and in filament structure and filament behavior (4, 10, 11, 26, 32–37, 41). These differences no doubt reflect the great evolutionary distances between these Alp families. As a necessary step toward cataloging and explaining the differences between the Alp7A system and these other actin-based plasmid segregations systems, we turned our attention to the regulatory components of the alp7A operon.
MATERIALS AND METHODS
Molecular biology.
The standard techniques of molecular biology were used. Genomic DNA was purified from Bacillus with a modification of a protocol developed for Gram-negative bacteria (29). Plasmids and DNA fragments, including products of PCR amplifications, were purified with kits from Qiagen, Invitrogen, or Fermentas. DNA modification enzymes, polymerases, and other molecular biology reagents were obtained from New England BioLabs unless noted otherwise. Shrimp alkaline phosphatase was obtained from Roche Diagnostics, and RNase was obtained from Qiagen. Deoxynucleoside triphosphates (dNTPs) were obtained from a variety of sources, including Roche Diagnostics and Fermentas. Other biochemicals and chemicals were obtained from Fisher, VWR, or Sigma. Agarose for routine electrophoresis was obtained from Denville Scientific. Oligonucleotide primers (see Table S1 in the supplemental material) were synthesized by Allele Biotechnology and Pharmaceuticals or by Integrated DNA Technologies. DNA sequencing was performed by Eton Bioscience or by Genewiz.
Plasmids were introduced into E. coli DH5α or BL21(DE3) by electroporation with a Gene Pulser Xcell (Bio-Rad) or by transformation of chemically competent cells (18).
Plasmids and plasmid constructions.
The plasmids used in this study are listed in Table 1. The construction of plasmids pAID3129 (mini-pLS20), pAID3195 [mini-pLS20 (alp7A-gfp), where gfp encodes green fluorescent protein], pAID3169 [mini-pLS20 alp7A(D212A)], and pAID3171 (mini-pLS20 Δalp7AR) was described in our previous study (4).
Table 1.
Plasmids and strains used in this study
| Plasmid or strain | Description or genotype | Reference and/or source |
|---|---|---|
| Plasmids | ||
| pAID3129 | Mini-pLS20 | 4 |
| pAID3169 | Mini-pLS20 alp7A(D212A) | 4 |
| pAID3171 | Mini-pLS20 Δalp7AR | 4 |
| pAID3195 | Mini-pLS20 alp7A-gfp | 4 |
| pAID3204 | pPxylalp7R | This study |
| pAID3219 | pPxylalp7R-gfp | This study |
| pAID3226 | Mini-pLS20 alp7A(D212A) Δalp7R | This study |
| pAID3232 | Mini-pLS20 alp7A(D212A)-gfp Δalp7R | This study |
| pAID3244 | Mini-pLS20 Δalp7R | This study |
| pAID3261 | Mini-pLS20 alp7A-gfp Δalp7R | This study |
| pAID3277 | pPT7His6-alp7R | This study |
| pME7 | pParaalp7R | This study |
| Strains | ||
| B. subtilis | ||
| PY79 | Prototroph, 168 lineage | 53 |
| JP3133 | pAID3129 [mini-pLS20]/PY79 | 4 |
| JP3161 | PY79 thrC::(xylR+ Pxylalp7A-gfperm) | 4 |
| JP3169 | pAID3169/PY79 | 4 |
| JP3171 | pAID3171/PY79 | 4 |
| JP3196 | pAID3195/PY79 | 4 |
| JP3210 | PY79 thrC::(xylR+ Pxylalp7R erm) | This study |
| JP3223 | pAID3219/PY79 | This study |
| JP3233 | pAID3232/JP3210 | This study |
| JP3245 | pAID3261/JP3210 | This study |
| JP3247 | pAID3226/JP3210 | This study |
| JP3248 | pAID3226/PY79 | This study |
| JP3300 | PY79 amyE::(alp7C cat) | This study |
| JP3302 | pAID3244/JP3210 | This study |
| JP3309 | PY79 amyE::(alp7C cat) thrC::(xylR+ Pxylalp7A-gfp erm) | This study |
| JP3311 | pAID3204/JP3309 | This study |
| JP3315 | pAID3204/JP3161 | This study |
| JP3322 | pAID3219/JP3300 | This study |
| E. coli | ||
| JP313 | F−ΔaraBAD-714 [araD139]B/r? Δ(argF-lac)169 λ− e14−flhD5301 Δ(fruK-yeiR)725(fruA25) relA1rpsL150(Strr) rbsR22 Δ(fimB-fimE)632(::IS1) deoC1 | 6 |
| JP3227 | pME7/JP313 | This study |
| JP3277 | pAID3277/BL21(DE3) | This study |
(i) Expression of alp7R and production of Alp7R.
Plasmid pME7 (pParaalp7R) was constructed by PCR amplification with oligonucleotide primers P7 and P8 of the section of plasmid pAID3147 (mini-pLS20 Δalp7A [4]) corresponding to 45 bp upstream of the alp7A initiation codon, the alp7A in-frame deletion, the 10-bp intergenic region, the alp7R gene, and the transcription terminator, followed by restriction of the amplicon with PstI and SphI and ligation of the product to plasmid pBAD33 (16) restricted with PstI and SphI. Plasmid pAID3204 (pPxylalp7R) was constructed by PCR amplification with oligonucleotide primers P1 and P2 of a section of plasmid pAID3147 corresponding to 45 bp upstream of the alp7A initiation codon, the alp7A in-frame deletion, the 10-bp intergenic region, the alp7R gene, and the transcription terminator, followed by restriction of the amplicon with BamHI and SphI and ligation of the product to plasmid pWH1520 (42) restricted with BamHI and SphI. Plasmid pAID3219 (pPxylalp7R-gfp) was constructed from plasmid pAID3211, which contains a fusion of gfp to alp7R. pAID3211 was constructed by PCR amplification with oligonucleotide primers P3 and P4 of a section of plasmid pAID3147 corresponding to 45 bp upstream of the alp7A initiation codon, the alp7A in-frame deletion, the 10-bp intergenic region, and the alp7R gene lacking its termination codon, followed by restriction of the amplicon with EagI and ligation of the product to pMUTIN-GFP+ (23) restricted with EagI. The alp7R-gfp gene codes for the 4-amino-acid linker ASID. pAID3211 DNA was amplified with oligonucleotide primers P5 and P6, and the amplicon was restricted with XmaI and SphI and ligated to pWH1520 restricted with XmaI and SphI. pAID3219 includes the trpA transcription terminator that follows gfp in pMUTIN-GFP+. Plasmid pAID3277 (pPT7His6-alp7R) was constructed from a preliminary pCR2.1-TOPO clone (Invitrogen). The alp7R gene lacking its initiation codon was amplified from pAID3147 by PCR with oligonucleotide primers P11 and P12, the amplicon was cloned into the pCR2.1-TOPO vector, and candidates were screened by DNA sequencing. The TOPO clone was restricted with NheI, and the 411-bp fragment containing the alp7R gene was ligated to pET28a (Novagen) restricted with NheI. The insert's orientation was determined by DNA sequencing.
(ii) Deletion of alp7R.
Plasmid pAID3226 [mini-pLS20 alp7A(D212A) Δalp7R] was constructed via a modification of the standard PCR-based site-directed mutagenesis protocol with mutagenic oligonucleotide primers P9 and P10 (52). The template for this mutagenesis was a derivative of plasmid pAID3169 that was generated by monomolecular ligation of its largest (6,847 bp) AatII restriction fragment (4). The 1,645-bp fragment resulting from restriction of pAID3169 with BsrGI and NheI was then replaced with the corresponding fragment from the mutagenized plasmid. Plasmid pAID3244 (mini-pLS20 Δalp7R) was constructed by replacing the 1,035-bp BsrGI-BbvCI fragment of pAID3226 with the corresponding fragment from pAID3129. Plasmid pAID3232 [mini-pLS20 alp7A(D212A)-gfp Δalp7R] was constructed by replacing the 2,840-bp AvrII-SphI fragment from pAID3195 (4) with a fragment that was generated by amplification of pAID3226 with oligonucleotide primers P11 and P12 and then restriction with AvrII and SphI. Plasmid pAID3261 [mini-pLS20 (alp7A-gfp) Δalp7R] was constructed by replacing the 695-bp AgeI fragment of pAID3244 with the corresponding fragment from pAID3195. All plasmids were recovered as transformants of strain JP3227. Transformants were isolated in the presence of 0.2% l-arabinose and were maintained and stored in the presence of 0.2% arabinose.
Bacterial strains, strain construction, and growth of bacteria.
The strains used in this study are listed in Table 1. E. coli strain JP3227 was constructed by transformation of strain JP313 (6) with plasmid pME7. All physiology and microscopy experiments were carried out at 30°C in B. subtilis strain PY79 (53) or in derivatives of PY79. The construction of strain JP3161 (PY79 thrC::xylR+ Pxylalp7A-gfp erm) was described in our previous study (4). Strain JP3210 [PY79 thrC::(xylR+ Pxylalp7R erm)] was constructed by integration into the PY79 chromosome of a segment of plasmid pAID3204 containing the xylR gene and Pxylalp7R. The derivative of chromosomal integration vector pDG1664 (15) containing Pxylalp7A that had been constructed for making strain JP3206 [PY79 thrC::(xylR+ Pxylalp7A erm)] (4) was restricted with SpeI and NruI, and the 7,703-bp fragment was ligated to the 823-bp fragment generated by restriction of pAID3204 with SpeI and NruI. The segment containing xylR and Pxylalp7R was then integrated into the PY79 chromosome at thrC by a double recombination event. Strain JP3300 (PY79 amyE::alp7C) was constructed by integration into the PY79 chromosome of a 531-bp segment corresponding to the 477 bp directly upstream of alp7A, the first 15 codons of alp7A, and the sequence ACCTAATGA. The segment was generated by PCR amplification of pAID3129 with oligonucleotide primers P15 and P16 and restriction of the amplicon with BamHI and HindIII. The product was ligated to B. subtilis chromosomal integration vector pDG1662 (15) restricted with BamHI and HindIII. The cloned segment was then integrated into the PY79 chromosome at amyE by a double recombination event. The chromosomal insert was verified by DNA sequencing. Strain JP3309 (PY79 amyE::alp7C thrC::xylR+ Pxylalp7A-gfp) was constructed by transformation of JP3300 with genomic DNA from strain JP3161 (4). Strain JP3311 (pPxylalp7R/PY79 amyE::alp7C thrC::xylR+ Pxylalp7A-gfp) was constructed by transformation of strain JP3309 with plasmid pAID3204. All other B. subtilis strains were constructed by standard transformation of PY79 or of derivatives of PY79 with the plasmids described above (5).
Medium components were manufactured by Becton, Dickinson, and Co. Our LB formulation is 1% Bacto-tryptone, 0.5% yeast extract, 0.5% NaCl per liter, with no pH adjustment. Most antibiotics were obtained from Sigma. Media for strains containing derivatives of pWH1520, including mini-pLS20 and its derivatives, were supplemented with 100 μg ml−1 ampicillin or carbenicillin for E. coli or with 10 μg ml−1 tetracycline for Bacillus strains. Chloramphenicol was used at 5 μg ml−1 and erythromycin at 2 μg ml−1 for Bacillus strains, kanamycin at 50 μg ml−1 for E. coli, and spectinomycin at 100 μg ml−1 for either Bacillus strains or E. coli.
Plasmid stability assays.
Plasmid stability assays were carried out as described previously (4), except that the starter cultures contained 10 μg ml−1 tetracycline and 0.5% d-xylose, the shake flask cultures requiring xylose were supplemented hourly with xylose to 0.5%, and the nonselective medium for the plating of samples contained 0.5% xylose. For assays of plasmid retention, colonies were patched onto LB agar plates containing 0.5% xylose and onto LB agar plates containing 10 μg ml−1 tetracycline and 0.5% xylose.
Microscopy.
Agarose pads contained 1.2% agarose (Invitrogen UltraPure 15510-027), 25% LB medium, and 0.2 μg ml−1 FM4-64 [N-(3-triethylammoniumpropyl)-4-{6-[4-(diethylamino)phenyl]hexatrienyl}pyridinium dibromide; Molecular Probes/Invitrogen]. DAPI (4′,6-diamidino-2-phenylindole, dihydrochloride; Invitrogen), when included, was present at 0.01 μg ml−1. Xylose was present at the concentrations indicated below. Pads were inoculated with colonies from an LB agar plate containing selective antibiotic and, when required for viability, 0.5% xylose, that had been streaked the previous day from frozen glycerol stocks and incubated overnight at 30°C. The pads were incubated at 30°C prior to and during imaging in the Weather Station temperature-controlled chamber outfitted to the microscope (Precision Control LLC.).
For slides prepared from liquid cultures, FM4-64 was added to 1 ml of culture to a concentration of 2 μg ml−1 and DAPI to a concentration of 2 μg ml−1, the cells were pelleted for 0.5 min at 3,300 × g in a microcentrifuge and resuspended in approximately 5% of the supernatant, and 5.0 μl of the resuspension was applied to a polylysine-coated coverslip for imaging.
Microscopy was carried out as described previously (4). Unless otherwise noted, micrographs of GFP and DNA (DAPI) were prepared from undeconvolved images, and micrographs of membranes (FM4-64) were prepared from deconvolved images.
Immunoblotting.
Cultures for immunoblotting were started with colonies from an LB agar plate containing 10 μg ml−1 tetracycline that had been streaked the previous day from frozen glycerol stocks and incubated overnight at 30°C. Several colonies were suspended in a small volume of LB medium, typically 0.5 ml, and equal volumes of the suspension were used to inoculate a set of 6-ml LB cultures containing 10 μg ml−1 tetracycline and the appropriate amount of xylose. The cultures were rolled at 30°C and supplemented hourly with xylose at the designated concentration.
Lysates were prepared from cells in exponential phase, at an optical density at 600 nm (OD600) of approximately 0.5. Trichloroacetic acid was added to a final concentration of 5% to a volume of cells corresponding to 1 OD600, and the cells were incubated on ice for 30 min, pelleted, and washed as described previously (31). Cells were lysed with 1 mg ml−1 chicken egg white lysozyme for 30 min at 37°C in 60 μl of a buffer consisting of 35% sucrose, 900 μM EDTA, 30 mM Tris-HCl, pH 8.0, that was supplemented with 160 μg ml−1 phenylmethylsulfonyl fluoride and 0.6 μl of a protease inhibitor cocktail (Sigma P2714). Two microliters of Triton X-100 was added at the end of the 30-min incubation. After the addition of 140 μl 2× SDS-PAGE sample preparation buffer, the samples were boiled for 5 min, vortexed, boiled once more for 3 min, and then pelleted at ambient temperature in a microcentrifuge at maximum speed. Fifteen microliters (0.075 OD600) of the supernatant was loaded onto a polyacrylamide gel for SDS-PAGE. The preparation of the hexahistidine-tagged Alp7A, which was used as a molecular-weight standard, has been described previously (4).
Proteins were transferred to polyvinylidene fluoride (PDVF) membranes in Towbin buffer containing 20% (vol/vol) methanol (51). Incubation of the membrane with the primary antibody, a polyclonal antibody raised against Alp7A (4), used at 1:10,000, and with the secondary antibody, a horseradish peroxidase-linked anti-rabbit IgG (GE Healthcare), used at 1:3,000 or 1:5,000, was carried out in TBST, pH 8.0 (150 mM NaCl, 10 mM Tris-HCl, pH 8.0), to which was added 0.06% Tween 20 (vol/vol) and 5% (wt/vol) instant nonfat dry milk (Safeway). Washes were performed in TBST. The ECL Plus Western blotting detection system (GE Healthcare) was used for detection. Relative band intensities were computed from multiple exposures of a blot with the gel analysis feature of ImageJ 1.45m (38).
Purification of Alp7R.
Shake flask cultures of strain JP3277 were grown at 30°C in LB medium supplemented with 50 μg ml−1 kanamycin, induced at an OD600 of 0.4 with 750 μM isopropyl-β-d-thiogalactopyranoside (IPTG), and harvested 6 h later. The cells were lysed in CelLytic B cell lysis reagent (product no. B7435; Sigma) supplemented with 200 μg ml−1 chicken egg white lysozyme, 1 μg ml−1 RNase A (Qiagen), a protease inhibitor cocktail (product no. P2714; Sigma), and 150 μg ml−1 PMSF. The lysate was then treated with 40 units of DNase I (product code M0303; New England BioLabs) in the presence of 10 mM MgCl2 and then centrifuged after supplementation with NaCl to 300 mM, imidazole to 12.5 mM, and HEPES to 50 mM, pH 8.0. His6-Alp7R was purified from the lysate by nickel affinity chromatography. The lysate was loaded onto a column of His-Select nickel affinity gel (product no. P6611; Sigma), the column was washed with 300 mM NaCl, 12.5 mM imidazole, 0.1% Triton X-100, 50 mM HEPES, pH 8.0, and the protein was eluted in 300 mM NaCl, 250 mM imidazole, 50 mM HEPES, pH 8.0.
Approximately 2 mg of His6-Alp7R from the most abundant fraction were treated with thrombin agarose (product no. T7151; Sigma) in 10 mM CaCl2, 300 mM NaCl, 50 mM HEPES, pH 8.0, for 1 h or 2 h at ambient temperature. SDS-PAGE indicated that this treatment resulted in complete removal of the hexahistidine tag. The Alp7R protein was then dialyzed against 300 mM NaCl, 1 mM EDTA, 20 mM HEPES, pH 8.0. Protein concentration was determined on the basis of the calculated extinction coefficient of 7,450 M−1 cm−1 (30).
Electrophoretic mobility shift assay (EMSA).
DNA substrates were generated by amplification of pAID3129 with oligonucleotide primers P17 and P18 (alp7C), P19 and P20 (alp7R), and P21 and P22 (ori rep). The alp7C amplicon was restricted with SspI. The sequences of the three amplicons are as follows: alp7C (182 bp), 5′-CGTAAAGCCCCGGGCCTGAAATCACTTTTCTCTACTGATTTCACTGATTTCATTTTTATTATATAATCCTCAAATAGCCTGTATTCACTGATTTTAAATGTGATTTCATTTTATTGACTTTAGTGATATAAGATGCTAGTATTGAGGAAAGTGAAATCAAAGGAGAGAATAAAAATATGAAT-3′; alp7R (189 bp), 5′-GAGCAGATTAGTACATTCAGTAAAGGTAAGAGTAAGGGTACCTTTAGAGAGTATGCCTTTCAGCTCATAGAAAGGGACATGCAACAACAGAAAGAGGAACAGCAGAATAGAGAAAAAGATCGTCATGTTCATGATGAATTAATTGCCATGAGAGAAGAAATGAAGAAAGAATTTCGTGATTTGAGGAAG-3′; and ori rep (191 bp), 5′-GACAGACATAGGCAATCGATCAGGATTTGAAACTAGCGTCATAGAGACGTCTGAGGTTTCCAGCTCTGCCTTGCTATCGCCAGGCTTTCGCCTGCCATGACCTTTTTACATACAATGCTTGTCCTGTATGCAACTTCTATGGGGTTTGTCTCGTGTTCTCTCACACGGTCACACTCAATTGTGTGCCGCTG-3′.
The DNA substrate was mixed with Alp7R protein in a 20-μl reaction mixture consisting of 300 mM NaCl, 5% polyethylene glycol (PEG) 8000, 1 mM EDTA, 25 μg ml−1 poly(dI-dC) (Sigma), 500 μg ml−1 bovine serum albumin (BSA; New England BioLabs), 20 mM HEPES, pH 7.5, and the mixture was incubated at ambient temperature for approximately 30 min. Typically, 100 ng of the alp7C DNA substrate and molar equivalents of the other two substrates were used. Two microliters of a dye mixture (2% xylene cyanol, 2% bromphenol blue, 300 mM NaCl, 1 mM EDTA, 20 mM HEPES, pH 7.5) was added to each reaction mixture, and the mixtures were fractionated on a 5% polyacrylamide gel (acrylamide/bis-acrylamide, 29:1 [Bio-Rad], 1× Tris-borate-EDTA [TBE], 1% PEG 8000, 10% glycerol) in 1× TBE after a 30-min prerun. The gel was stained and destained briefly with ethidium bromide and then either photographed or visualized with a Typhoon 9400 variable mode imager (GE Healthcare). Band intensities were quantified with ImageJ 1.45m (38).
RESULTS
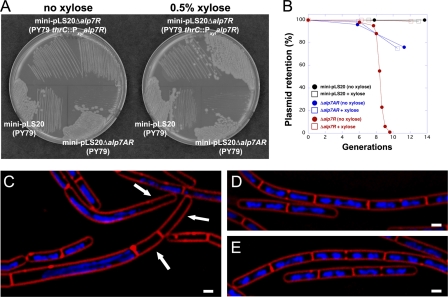
The alp7A operon of B. subtilis plasmid pLS20 is a plasmid stability determinant. A mini-pLS20 plasmid containing the alp7A operon and the adjacent pLS20 origin of replication is completely stable; deletion of alp7A or of the entire operon leads to loss of the plasmid (4). The alp7A gene is followed by a smaller gene, which we have designated alp7R (Fig. 1). We deleted alp7R from mini-pLS20 and assessed the stability of the resulting plasmid. The introduction of mini-pLS20 Δalp7R into B. subtilis required that alp7R be present elsewhere in the genome (Fig. 2A), so the gene was placed under the control of the inducible Pxyl promoter and installed on the chromosome. This requirement for alp7R suggested that mini-pLS20 Δalp7R would be unstable, and moreover, that the plasmid most likely could not be tolerated by the cell. Plasmid stability assays confirmed this. A culture was grown for several generations in the presence of xylose and selective antibiotic and then assayed for antibiotic resistance at intervals following subculture into unsupplemented medium. Plasmid retention fell off precipitously starting at between 6 and 10 generations after the withdrawal of xylose, such that by about 10 generations, the plasmid was lost completely (Fig. 2B, red circles). During this period, the growth rate slowed greatly. Under the microscope, we observed aberrations in cell morphology, chromosome segregation, and cell division (Fig. 2C). Many cells appeared to be anucleate (Fig. 2C, arrows). If the culture was instead maintained in the presence of xylose (Fig. 2B, red squares), the plasmid was stable, the cells grew normally, and they were indistinguishable from cells of the strain containing intact mini-pLS20 (Fig. 2D and E).
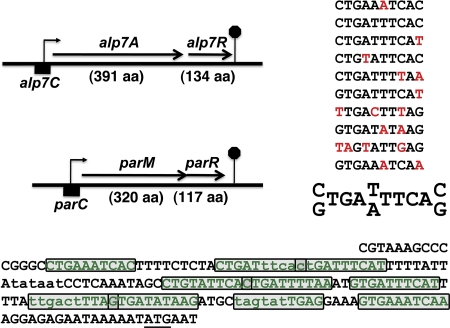
Fig 1.
The alp7AR and parMR operons have similar structures. The alp7AR and parMR operons are diagrammed to scale, with the lengths of their gene products in amino acids (aa) shown. The alp7C sequence through the second codon of alp7A is presented below, with the alp7A initiation codon underlined. Two potential σA promoters are denoted in lowercase (17). The sequence contained in a set of near-repeat elements is indicated as green text; the individual repeats are boxed. As construed here, six of the repeats overlap an adjoining repeat by one base. An inferred consensus is presented in the upper right. The presence of a base or a pair of bases in the consensus requires at least six or more appearances at that position; deviations are indicated in red. Some deviations, as in repeats 7 and 9, appear to reflect promoter element constraints.
Fig 2.
Deletion of alp7R destabilizes mini-pLS20 and is lethal to cells carrying the plasmid. (A) LB agar plates containing 10 μg ml−1 tetracycline with or without 0.5% d-xylose were incubated at 30°C for 24 h. Lower left, JP3133 (PY79 carrying mini-pLS20 [pLS20 origin of replication and alp7AR operon]); lower right, JP3171 (PY79 carrying mini-pLS20 Δalp7AR [pLS20 origin of replication only]); top, JP3302 (PY79 with an integrated xylose-inducible copy of alp7R, carrying mini-pLS20 Δalp7R [pLS20 origin of replication and alp7AR operon lacking alp7R]). (B) Exponential cultures grown in the absence of selection were sampled at regular intervals for plasmid retention as described in Materials and Methods. Black, JP3133; blue, JP3171; red, JP3302. Open squares, hourly supplementation with xylose to 0.5%; filled circles, no xylose supplementation. (C to E) Fluorescence microscopy images of strains supplemented or not supplemented with xylose. Exponential cultures grown in the absence of selection as in the plasmid stability assay were sampled for microscopy. Membranes were stained with FM4-64 (red), and DNA was stained with DAPI (blue). Scale bar equals 1 μm; all panels are at the same scale. (C) JP3302, with no xylose supplementation, at 4 h. Arrows indicate anucleate cells. (D) JP3302, supplemented hourly with 0.5% xylose, at 4 h. (E) JP3133, supplemented hourly with xylose to 0.5%, at 3 h.
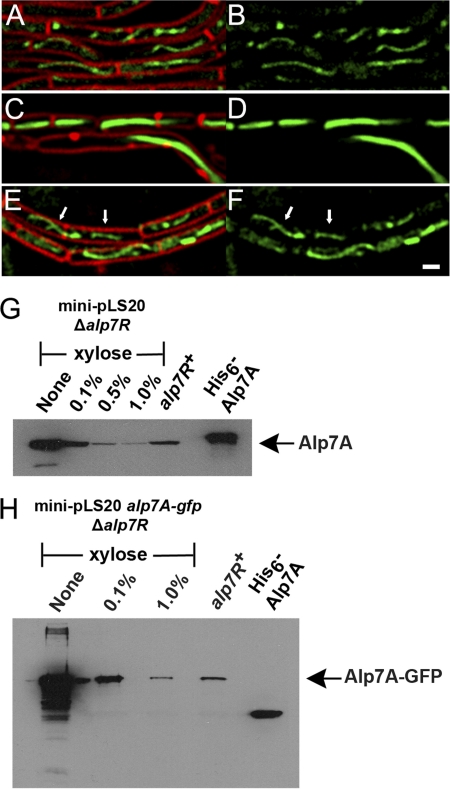
We had seen similar disruptive effects on cell growth and viability in connection with the abnormal filament formation that follows from overproduction of Alp7A. Observations from a xylose titration were indeed consistent with a scenario in which limiting the amount of Alp7R results in Alp7A-GFP overproduction. When little or no xylose was provided to a culture, so that little or no Alp7R was produced, many cells contained thick ribbon-like filaments that seemed to fill them completely and in many cases distorted them to the point of inhibiting their normal growth and division (Fig. 3C and D). All of these filaments were static (see Fig. S1 in the supplemental material). At a high xylose concentration, many cells contained filaments that appeared normal and exhibited dynamic instability (Fig. 3E and F), a property of the fully functional protein (Fig. 3A and B; see also Fig. S2 and S3 in the supplemental material).
Fig 3.
Depletion of Alp7R leads to overproduction of Alp7A. (A to F) Fluorescence microscopy images of strains grown in the presence or absence of d-xylose (agarose pads; scale bar equals 1 μm; all panels are at the same scale). (A and B) JP3196 (PY79 carrying mini-pLS20 alp7A-gfp) in the absence of xylose. (C to F) JP3245 (PY79 with an integrated xylose-inducible copy of alp7R, carrying mini-pLS20 alp7A-gfp Δalp7R) in the absence of xylose (C and D) or in the presence of 0.5% xylose (E and F). Arrows denote filaments whose dynamic behavior can be tracked in the lower left section of the corresponding movie (see Fig. S2 in the supplemental material). (A, C, and E) Cell membranes (FM4-64) and GFP. (B, D, and F) GFP only. See also Fig. S1, S2, and S3 in the supplemental material. (G and H) Immunoblots prepared from lysates of JP3302 (PY79 with an integrated xylose-inducible copy of alp7R, carrying mini-pLS20 Δalp7R) (G) or JP3245 (PY79 with an integrated xylose-inducible copy of alp7R, carrying mini-pLS20 alp7A-gfp Δalp7R) (H) grown in the presence of various concentrations of xylose. The filter was probed with a polyclonal anti-Alp7A serum. Lanes are labeled with the xylose concentrations used. The lane labeled “alp7R+” shows the steady-state level of Alp7A that is present in JP3133 (PY79 carrying mini-pLS20) (G) or of Alp7A-GFP that is present in JP3196 (PY79 carrying mini-pLS20 alp7A-gfp) (H). The rightmost lane contains purified His6-Alp7A (G and H).
It appeared then that Alp7R was functioning as a negative regulator of the alp7A gene. It was nevertheless possible that Alp7R had no effect on the amount of Alp7A that was produced in the cell and that its function was simply to enable the Alp7A that was present to be assembled into normal dynamic filaments. In order to distinguish these two possibilities, we measured the steady-state levels of Alp7A in the presence and absence of Alp7R. When the chromosomal alp7R gene was left uninduced in strains containing Δalp7R plasmids, the cells contained five to 10 times the Alp7A of a strain containing mini-pLS20 (Fig. 3G) and 50 times the Alp7A-GFP of a strain containing mini-pLS20 alp7A-gfp (Fig. 3H). Induction of the alp7R gene depressed Alp7A and Alp7A-GFP levels to that of mini-pLS20 or below (Fig. 3G and H).
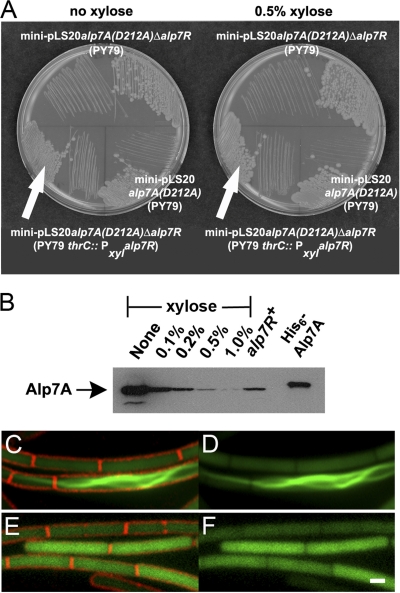
We substituted alp7A(D212A) for the wild-type alp7A in mini-pLS20 Δalp7R because this point mutation prevents filament formation when present in mini-pLS20 (4). We hoped by doing this to avoid the potential complications of studying regulation under lethal conditions. The resulting Δalp7R strain did not require xylose for viability (Fig. 4A), but the steady-state level of Alp7A(D212A) was again elevated, to 20 times that found in the cell when the alp7R gene was intact (Fig. 4B). Although we had not seen Alp7A(D212A)-GFP polymerize into filaments before, this level of overproduction was sufficient to drive much of the protein into filaments, albeit filaments lacking dynamic properties (Fig. 4C and D; see also Fig. S4 in the supplemental material). As was the case for wild-type Alp7A, induction of the chromosomal alp7R gene led to a reduction in the steady-state level of Alp7A(D212A) or of Alp7A(D212)-GFP: the greater the induction, the lower the steady-state level (Fig. 4B to F). At the highest induction level, no filaments were observed, and soluble Alp7A(D212A)-GFP was distributed uniformly throughout the cell, as it was in a strain containing mini-pLS20 alp7A(D212A) with an intact alp7R gene (Fig. 4E and F).
Fig 4.
Deletion of alp7R causes Alp7A(D212A) to assemble into filaments. (A) LB agar plates containing 10 μg ml−1 tetracycline with or without 0.5% d-xylose were incubated overnight at 30°C. Lower left, JP3247 [PY79 with an integrated xylose-inducible copy of alp7R, carrying mini-pLS20 alp7A(D212A) Δalp7R]; lower right, JP3169 [PY79 carrying mini-pLS20 alp7A(D212A)]; top, JP3248 [PY79 carrying mini-pLS20 alp7A(D212A) Δalp7R]. (B) Immunoblot prepared from lysates of JP3247 grown in the presence of various concentrations of xylose. The filter was probed with a polyclonal anti-Alp7A serum. Lanes are labeled with the xylose concentrations used. The lane labeled “alp7R+” shows the steady-state level of Alp7A(D212A) that is present in JP3169. The rightmost lane contains purified His6-Alp7A. (C to F) Fluorescence microscopy images of strain overproducing Alp7A(D212A) (agarose pads; scale bar equals 1 μm, all panels are at the same scale). JP3233 [PY79 with an integrated xylose-inducible copy of alp7R, carrying mini-pLS20 alp7A(D212A)-gfp Δalp7R] in the absence of xylose (C and D) or in the presence of 0.5% xylose (E and F). (C and E) Cell membranes (FM4-64) and GFP. (D and F) GFP only. See also Fig. S4 in the supplemental material.
Alp7R is therefore a negative regulator of alp7A gene expression and presumably of the alp7A operon. The structure of the alp7A operon closely resembles that of the parM operon (Fig. 1). The actin gene, alp7A or parM, is followed by a smaller gene, alp7R or parR, that codes for a protein with a high percentage of charged amino acid residues, many of which show up in short runs of two or more (4). ParR is a DNA-binding protein that binds at parC, a sequence directly upstream of the parM gene that contains 10 imperfect iterations of a 10-mer sequence which straddle the promoter (2, 3). The sequence directly upstream of alp7A contains a series of 10 imperfect iterations of a 10-mer sequence in which two putative σA promoters are embedded (Fig. 1). When expressed in B. subtilis PY79, an Alp7R-GFP fusion protein delineated the chromosome in the manner of DAPI staining, indicating that it was bound to DNA nonspecifically (Fig. 5A, left). When the fusion protein was expressed in a variant of PY79 in which the alp7C sequence had been installed on the chromosome, discrete foci could be observed (Fig. 5A, right). In cells that contained these foci, the number ranged from one to four, as would be expected under these growth conditions if a single focus were formed at each alp7C site. These findings suggested that Alp7R was a DNA-binding protein like ParR and that it bound specifically at alp7C.
Fig 5.
Alp7R is a DNA-binding protein that binds specifically at alp7C. (A) Fluorescence microscopy of strains expressing alp7R-gfp (agarose pads; scale bar equals 1 μm, all panels are at the same scale). Left, JP3223 (PY79 carrying pPxylalp7R-gfp) in the presence of 0.5% xylose. Right, JP3322 (PY79 with an integrated copy of alp7C, carrying pPxylalp7R-gfp) in the presence of 0.1% xylose. Top, cell membranes (FM4-64) and DNA (DAPI). Bottom, cell membranes and GFP. (B and C) Electrophoretic mobility shift assays (EMSA) of alp7C DNA by the Alp7R protein. See Materials and Methods. The number above each lane indicates the molar ratio of Alp7R protein to the DNA amplicon. (B) Competitor poly(dI-dC) was omitted from the binding reaction mixture. The high-molecular-weight material consists entirely of complex. (C) Competitor poly(dI-dC) was present in the binding reaction mixture at 25 μg ml−1. At the higher Alp7R-to-alp7C ratios, the high-molecular-weight mate- rial contains both competitor and complex. (D) The percentage of unshifted DNA in each lane of panels B and C is plotted against the molar ratio of Alp7R to DNA. Black filled symbols, alp7C amplicon with competitor, from the experiment represented in panel C; black open symbols, alp7C amplicon without competitor, from the experiment represented in panel B; blue, alp7R amplicon with competitor, from the experiment represented in panel C; green, origin of replication amplicon with competitor, from the experiment represented in panel C.
EMSAs were therefore carried out to assess the nature and specificity of the binding. As the molar ratio of purified Alp7R protein to alp7C DNA was increased from 1 to 25, we observed a shift of the DNA from its normal migration position, owing to its incorporation into high-molecular-weight complexes (Fig. 5B and D). There was also shifting, but considerably diminished, of either of two alternative DNA fragments of similar size, one corresponding to part of the alp7R structural gene and the other to part of the pLS20 origin of replication. This difference was particularly pronounced when competitor was included in the binding reaction mixtures (Fig. 5C). Even at a 25 molar excess of Alp7R, where more than 80% of the alp7C DNA shifted, nearly 80% of the other two DNA fragments failed to shift (Fig. 5C and D). These data confirmed our in vivo findings that Alp7R, although capable of binding to DNA nonspecifically, binds specifically to alp7C.
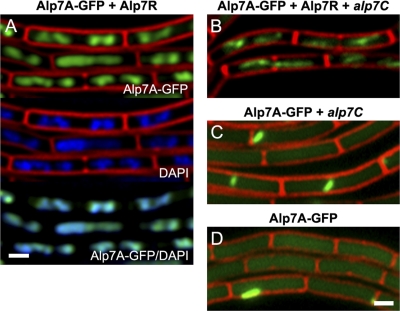
Having demonstrated binding of Alp7R at alp7C, we attempted to determine how Alp7A interacts with these other components. In order to do this, we took advantage of the observation that in the absence of alp7C, Alp7R binds nonspecifically to DNA (Fig. 5A, left). In place of alp7R-gfp, we expressed alp7R coordinately with alp7A-gfp. Interaction of Alp7A-GFP and Alp7R would be expected to give rise to a fluorescence pattern that resembled that of Alp7R-GFP. This turned out to be the case; the chromosome was again delineated in the manner of DAPI staining (Fig. 6A). Hence, Alp7A interacts with Alp7R and it can do so even if there is no alp7C present in the cell for Alp7R to bind to.
Fig 6.
Alp7R and Alp7A interact; Alp7R is indispensable for the formation of dynamic Alp7A filaments. (A to D) Fluorescence microscopy images of strains expressing Alp7A-GFP by itself or in the presence of other components of the system (agarose pads containing 0.5% xylose; scale bar equals 1 μm, all panels are at the same scale). (A) JP3315 (PY79 with an integrated xylose-inducible copy of alp7A-gfp, carrying pPxylalp7R). Top, cell membranes (FM4-64) and GFP (Alp7A-GFP); middle, cell membranes (FM4-64) and DNA (DAPI); bottom, GFP (Alp7A-GFP) and DNA (DAPI). GFP panels were prepared from deconvolved images. (B) JP3311 (PY79 with an integrated xylose-inducible copy of alp7A-gfp and an integrated copy of alp7C, carrying pPxylalp7R). (C) JP3309 (PY79 with an integrated xylose-inducible copy of alp7A-gfp and an integrated copy of alp7C). (D) JP3161 (PY79 carrying an integrated xylose-inducible copy of alp7A-gfp). (B to D) Cell membranes and GFP. See also Fig. S5 to S7 in the supplemental material.
When we introduced alp7C, we reconstituted the complete system, albeit with its components dispersed: alp7C was on the chromosome, alp7A-gfp was in single copy elsewhere on the chromosome, and alp7R was on a multicopy plasmid. It was therefore not surprising that coordinate induction of alp7A-gfp and alp7R eventually gave rise to dynamic filaments in these cells (Fig. 6B; see also Fig. S5 in the supplemental material). Alp7R was required for this. In its absence, in a strain that contained only the chromosomal copies of alp7C and alp7A-gfp but no alp7R plasmid, only small static filaments were observed (Fig. 6C; see also Fig. S6 in the supplemental material), and this profile was indistinguishable from that of a strain containing only the chromosomal copy of alp7A-gfp (Fig. 6D; see also Fig. S7 in the supplemental material). So alp7C in the absence of alp7R has no effect on Alp7A activity, and Alp7R is therefore indispensable for the formation of dynamic filaments. There was also no fluorescent delineation of the chromosome in these cells (Fig. 6C and D), indicating that Alp7A-GFP is unable to interact directly with DNA and confirming that the fluorescence pattern in Fig. 6A could be generated only through an interaction between Alp7A-GFP and Alp7R.
DISCUSSION
In our previous study, we found that alp7R and alp7C regulate the activity of Alp7A, that alp7R and alp7C are required for Alp7A to assemble into dynamic filaments at its physiological concentration (4). In the present study, we have found that Alp7R and alp7C also regulate the production of Alp7A. When alp7R was deleted from mini-pLS20, the steady-state levels of Alp7A were increased up to 50-fold, and the cells were filled with large filaments of irregular shape that lacked dynamic properties. These rigid filaments that filled much of the cells' interior space were likely to interfere with chromosome segregation or with cell division and presumably account for the lethality that we observed.
Although Alp7R may contribute in more than one way to setting Alp7A levels in the cell, its principal contribution is at the level of transcription. Alp7R is a negative regulator of alp7A gene expression. The region immediately upstream of alp7A, which we call alp7C in analogy with parC, the corresponding region in the parMR operon, contains two potential σA promoters. The EMSA and cell biology data indicate that Alp7R binds preferentially at alp7C. Although we cannot rule out more sophisticated mechanisms, Alp7R seems to function as a simple transcriptional repressor, impeding transcription by promoter occlusion. In this respect, alp7AR would resemble every actin-based bacterial plasmid segregation system whose regulation has been studied to date, including the parMR systems from plasmids R1 and pB171 (3, 20, 40) and two distantly related systems, the parMR system from plasmid pSK41 (46) and the alfAB system from plasmid pLS32 (1, 50). In each case, the product of the downstream gene is a DNA-binding protein that binds upstream of the actin gene and represses transcription. Alp7R presumably represses transcription of the entire operon, including the alp7R gene. The negative feedback loop that is established necessarily ensures that the appropriate amount of Alp7R is present in the cell to promote the formation of normal dynamic filaments. Disruption of this feedback loop leads to aberrant filament formation, perturbation of plasmid segregation, and cell death.
The sequence of Alp7R resembles that of ParR in its length and charge distribution (4). The crystal structures of ParR from pB171 and of the N-terminal portion of the DNA-binding protein from pSK41 in complex with a 20-mer derived from its binding site revealed that both proteins contain N-terminal ribbon-helix-helix (RHH) domains, which places them in the MetJ/Arc family of gene regulators (28, 39, 46). The RHH domain, or more properly the surface that is formed from a pair of RHH domains upon protein dimerization, constitutes the DNA-binding moiety. The sequence diversity among MetJ/Arc family members and the small size of the DNA-binding domain make it difficult to determine, in the absence of structural information, whether Alp7R is a member of this family as well (44). That said, it is not hard to spot stretches of similarity between Alp7R and some RHH family members. For example, the sequence near the N terminus of Alp7R (PLFNVR) resembles that of the bacteriophage P22 repressor proteins Arc (PQFNLR) and Mnt (PHFNFR), two founding members of the family; in the RHH dimer, this short sequence gives rise to the β-sheet, or “ribbon,” that contacts the DNA (44).
RHH protein dimers tend to interact cooperatively and give rise to higher-order structures (28, 44). This has turned out to be true in the case of the DNA-binding proteins associated with actin-driven plasmid segregation systems. The structure of the ParR-DNA complex was inferred from the pB171 ParR crystal structure and electron micrographs of the ParR protein of plasmid R1 with parC DNA. ParR dimers assembled into a ringlike helical formation, with 12 dimers comprising a full 360° turn of the ring (28). The cocrystal of the pSK41 RHH domain with its 20-mer binding site yielded a similar structure, a finding which was supported by electron microscopy. Each 20-mer was bound by two dimers, and as in the case of ParR, a total of 12 dimers were required for a full turn of the ring (46). In both structures, the bound DNA was wrapped around the outer circumference of the ring and the actin filament was proposed to interact with the interior of the ring (28, 46).
It is likely that alp7C is also bound by many copies of Alp7R. Alp7R-GFP forms bright foci on bacterial chromosomes that contain alp7C, and a single protein or dimer could not be expected to produce so bright a focus. The high-molecular-weight material generated in the EMSA experiments migrates as a diffuse band and may represent a collection of species, yet the apparent size of this collection does not change above a certain Alp7R-to-alp7C ratio (Fig. 5B). So the complexes are probably discrete species, with a fixed number of Alp7R monomers or dimers bound at alp7C. If one assumes that each complex contains a single copy of alp7C, the median molecular weight of the collection is consistent with a complex containing from 35 to 40 copies of Alp7R. It would follow that each of the 10 repeats is bound by about four copies of Alp7R, or two Alp7R dimers. This is a rough reckoning, but the numbers are of the same order as those in the ParR and pSK41 complexes, so it would not be surprising if the Alp7R/alp7C complex is of a similar configuration. And binding is likely to be cooperative as well, because Hill plots of the EMSA data yield Hill coefficients (nH values) of 3.0 or greater (not shown).
Alp7R binds specifically at alp7C, but it also binds nonspecifically to unrelated DNA sequences, as is typical of DNA-binding proteins. When expressed in strains that lack alp7C, Alp7R-GFP delineated the chromosome. We found that replacement of Alp7R-GFP with Alp7R and Alp7A-GFP gave rise to a similar pattern of fluorescence, indicating that Alp7A interacts with Alp7R under these circumstances. We had inferred an interaction between Alp7A and Alp7R from our earlier observation that Alp7R/alp7C is required for Alp7A to assemble into dynamic filaments at its physiological concentration (4), but this is the first demonstration of the interaction. The interaction is likely to be direct, although we cannot rule out the possibility that it is mediated by other cellular proteins. It is noteworthy, though, that this interaction requires neither the Alp7R-alp7C centrosome complex nor that Alp7A be filamentous. We had expected the interaction between Alp7A and Alp7R to require binding of Alp7R at alp7C. Instead, Alp7R, bound nonspecifically to DNA in its search for alp7C, may already be associated with Alp7A.
We undertook the present study to extend our understanding of the Alp7A system and as part of an ongoing effort in the field to sort out which features are conserved and which vary among the great many actin-based plasmid segregation systems. Our findings add to the evidence that the DNA-binding protein in these systems, in addition to its function in the mechanics of plasmid segregation, invariably functions to regulate transcription of the operon. Evolution has allowed for differences in polymer structure, in the kinetics of polymerization, in the role that the DNA-binding protein has in regulating these parameters. But the DNA-binding protein as regulator of transcription would seem to be a constant.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Sharon Torigoe for many helpful discussions, to Vera Alverdi for performing matrix-assisted laser desorption ionization (MALDI)-mass spectrometry on the purified Alp7R protein, to Joseph Lucas for supplying reagents, to Andrew Doedens for assistance with imaging equipment, and to Megana Roopreddy and Jason Royal for assistance with strain construction.
This material is based on work supported under grants to J.P. from the NIH (grants 2R01-GM073898 and R-GM084334Z).
Footnotes
Published ahead of print 16 March 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Becker E, et al. 2006. DNA segregation by the bacterial actin AlfA during Bacillus subtilis growth and development. EMBO J. 25:5919–5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Breuner A, Jensen RB, Dam M, Pedersen S, Gerdes K. 1996. The centromere-like parC locus of plasmid R1. Mol. Microbiol. 20:581–592 [DOI] [PubMed] [Google Scholar]
- 3. Dam M, Gerdes K. 1994. Partitioning of plasmid R1. Ten direct repeats flanking the parA promoter constitute a centromere-like partition site parC, that expresses incompatibility. J. Mol. Biol. 236:1289–1298 [DOI] [PubMed] [Google Scholar]
- 4. Derman AI, et al. 2009. Phylogenetic analysis identifies many uncharacterized actin-like proteins (Alps) in bacteria: regulated polymerization, dynamic instability and treadmilling in Alp7A. Mol. Microbiol. 73:534–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dubnau D, Davidoff-Abelson R. 1971. Fate of transforming DNA after uptake by competent Bacillus subtilis. J. Mol. Biol. 56:209–221 [DOI] [PubMed] [Google Scholar]
- 6. Economou A, Pogliano JA, Beckwith J, Oliver DB, Wickner W. 1995. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell 83:1171–1181 [DOI] [PubMed] [Google Scholar]
- 7. Feucht A, Lucet I, Yudkin MD, Errington J. 2001. Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol. Microbiol. 40:115–125 [DOI] [PubMed] [Google Scholar]
- 8. Figge RM, Divakaruni AV, Gober JW. 2004. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 51:1321–1332 [DOI] [PubMed] [Google Scholar]
- 9. Formstone A, Errington J. 2005. A magnesium-dependent mreB null mutant: implications for the role of mreB in Bacillus subtilis. Mol. Microbiol. 55:1646–1657 [DOI] [PubMed] [Google Scholar]
- 10. Garner EC, Campbell CS, Mullins RD. 2004. Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science 306:1021–1025 [DOI] [PubMed] [Google Scholar]
- 11. Garner EC, Campbell CS, Weibel DB, Mullins RD. 2007. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science 315:1270–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gerdes K, Moller-Jensen J, Ebersbach G, Kruse T, Nordstrom K. 2004. Bacterial mitotic machineries. Cell 116:359–366 [DOI] [PubMed] [Google Scholar]
- 13. Gerdes K, Moller-Jensen J, Jensen RB. 2000. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 37:455–466 [DOI] [PubMed] [Google Scholar]
- 14. Goley ED, et al. 2011. Assembly of the Caulobacter cell division machine. Mol. Microbiol. 80:1680–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guerout-Fleury AM, Frandsen N, Stragier P. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57–61 [DOI] [PubMed] [Google Scholar]
- 16. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haldenwang WG. 1995. The sigma factors of Bacillus subtilis. Microbiol. Rev. 59:1–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hanahan D. 1985. Techniques for transformation of E. coli. In Glover DM. (ed), DNA cloning: a practical approach. RL Press, Oxford, England [Google Scholar]
- 19. Reference deleted.
- 20. Jensen RB, Dam M, Gerdes K. 1994. Partitioning of plasmid R1. The parA operon is autoregulated by ParR and its transcription is highly stimulated by a downstream activating element. J. Mol. Biol. 236:1299–1309 [DOI] [PubMed] [Google Scholar]
- 21. Jensen SO, Thompson LS, Harry EJ. 2005. Cell division in Bacillus subtilis: FtsZ and FtsA association is Z-ring independent, and FtsA is required for efficient midcell Z-ring assembly. J. Bacteriol. 187:6536–6544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jones L, Carballido-Lopez R, Errington J. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913–922 [DOI] [PubMed] [Google Scholar]
- 23. Kaltwasser M, Wiegert T, Schumann W. 2002. Construction and application of epitope- and green fluorescent protein-tagging integration vectors for Bacillus subtilis. Appl. Environ. Microbiol. 68:2624–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kruse T, Bork-Jensen J, Gerdes K. 2005. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol. Microbiol. 55:78–89 [DOI] [PubMed] [Google Scholar]
- 25. Meijer WJ, de Boer AJ, van Tongeren S, Venema G, Bron S. 1995. Characterization of the replication region of the Bacillus subtilis plasmid pLS20: a novel type of replicon. Nucleic Acids Res. 23:3214–3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moller-Jensen J, et al. 2003. Bacterial mitosis: ParM of plasmid R1 moves plasmid DNA by an actin-like insertional polymerization mechanism. Mol. Cell 12:1477–1487 [DOI] [PubMed] [Google Scholar]
- 27. Moller-Jensen J, Jensen R, Lowe J, Gerdes K. 2002. Prokaryotic DNA segregation by an actin-like filament. EMBO J. 21:3119–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moller-Jensen J, Ringgaard S, Mercogliano CP, Gerdes K, Lowe J. 2007. Structural analysis of the ParR/parC plasmid partition complex. EMBO J. 26:4413–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Neumann B, Pospiech A, Schairer HU. 1992. Rapid isolation of genomic DNA from Gram-negative bacteria. Trends Genet. 8:332–333 [DOI] [PubMed] [Google Scholar]
- 30. Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. 1995. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4:2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pogliano K, Hofmeister AE, Losick R. 1997. Disappearance of the sigma E transcription factor from the forespore and the SpoIIE phosphatase from the mother cell contributes to establishment of cell-specific gene expression during sporulation in Bacillus subtilis. J. Bacteriol. 179:3331–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Polka JK, Kollman JM, Agard DA, Mullins RD. 2009. The structure and assembly dynamics of plasmid actin AlfA imply a novel mechanism of DNA segregation. J. Bacteriol. 191:6219–6230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Popp D, et al. 2010. Polymeric structures and dynamic properties of the bacterial actin AlfA. J. Mol. Biol. 397:1031–1041 [DOI] [PubMed] [Google Scholar]
- 34. Popp D, et al. 2008. Molecular structure of the ParM polymer and the mechanism leading to its nucleotide-driven dynamic instability. EMBO J. 27:570–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Popp D, Robinson RC. 2011. Many ways to build an actin filament. Mol. Microbiol. 80:300–308 [DOI] [PubMed] [Google Scholar]
- 36. Popp D, et al. 2010. Structure and filament dynamics of the pSK41 actin-like ParM protein: implications for plasmid DNA segregation. J. Biol. Chem. 285:10130–10140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Popp D, et al. 2007. Concerning the dynamic instability of actin homolog ParM. Biochem. Biophys. Res. Commun. 353:109–114 [DOI] [PubMed] [Google Scholar]
- 38. Rasband WS. 2011. ImageJ. National Institutes of Health, Bethesda, MD: http://imagej.nih.gov/ij/ [Google Scholar]
- 39. Raumann BE, Brown BM, Sauer RT. 1994. Major groove DNA recognition by β-sheets: the ribbon-helix-helix family of gene regulatory proteins. Curr. Opin. Struct. Biol. 4:36–43 [Google Scholar]
- 40. Ringgaard S, Ebersbach G, Borch J, Gerdes K. 2007. Regulatory cross-talk in the double par locus of plasmid pB171. J. Biol. Chem. 282:3134–3145 [DOI] [PubMed] [Google Scholar]
- 41. Rivera CR, Kollman JM, Polka JK, Agard DA, Mullins RD. 2011. Architecture and assembly of a divergent member of the ParM family of bacterial actin-like proteins. J. Biol. Chem. 286:14282–14290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rygus T, Hillen W. 1991. Inducible high-level expression of heterologous genes in Bacillus megaterium using the regulatory elements of the xylose-utilization operon. Appl. Microbiol. Biotechnol. 35:594–599 [DOI] [PubMed] [Google Scholar]
- 43. Schirner K, Errington J. 2009. Influence of heterologous MreB proteins on cell morphology of Bacillus subtilis. Microbiology 155:3611–3621 [DOI] [PubMed] [Google Scholar]
- 44. Schreiter ER, Drennan CL. 2007. Ribbon-helix-helix transcription factors: variations on a theme. Nat. Rev. Microbiol. 5:710–720 [DOI] [PubMed] [Google Scholar]
- 45. Schumacher M. 2008. Structural biology of plasmid partition: uncovering the molecular mechanisms of DNA segregation. Biochem. J. 412:1–18 [DOI] [PubMed] [Google Scholar]
- 46. Schumacher MA, et al. 2007. Segrosome structure revealed by a complex of ParR with centromere DNA. Nature 450:1268–1271 [DOI] [PubMed] [Google Scholar]
- 47. Shaevitz JW, Gitai Z. 2010. The structure and function of bacterial actin homologs. Cold Spring Harb. Perspect. Biol. 2:a000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shiomi D, Margolin W. 2007. Dimerization or oligomerization of the actin-like FtsA protein enhances the integrity of the cytokinetic Z ring. Mol. Microbiol. 66:1396–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Soufo HJ, Graumann PL. 2010. Bacillus subtilis MreB paralogues have different filament architectures and lead to shape remodelling of a heterologous cell system. Mol. Microbiol. 78:1145–1158 [DOI] [PubMed] [Google Scholar]
- 50. Tanaka T. 2010. Functional analysis of the stability determinant AlfB of pBET131, a miniplasmid derivative of Bacillus subtilis (natto) plasmid pLS32. J. Bacteriol. 192:1221–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Towbin H, Staehelin T, Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U. S. A. 76:4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang W, Malcolm BA. 1999. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange site-directed mutagenesis. Biotechniques 26:680–682 [DOI] [PubMed] [Google Scholar]
- 53. Youngman P, Perkins JB, Sandman K. (ed). 1984. New genetic methods, molecular cloning strategies, and gene fusion techniques for Bacillus subtilis which take advantage of Tn917 insertional mutagenesis. Academic Press, New York, NY [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.