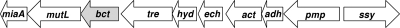Abstract
Geobacter metallireducens is a Fe(III)-respiring deltaproteobacterium and serves as a model organism for aromatic compound-degrading, obligately anaerobic bacteria. In this study, a genetic system was established for G. metallireducens using nitrate as an alternative electron acceptor. Surprisingly, disruption of the benzoate-induced bamY gene, encoding a benzoate coenzyme A (CoA) ligase, reproducibly showed an increased biomass yield in comparison to the wild type during growth with benzoate but not during growth with acetate. Complementation of bamY in trans converted the biomass yield back to the wild-type level. Growth of the bamY mutant with benzoate can be rationalized by the identification of a previously unknown succinyl-CoA:benzoate CoA transferase activity; it represents an additional, energetically less demanding mode of benzoate activation. The activity was highly enriched from extracts of cells grown on benzoate, yielding a 50-kDa protein band; mass spectrometric analysis identified the corresponding benzoate-induced gene annotated as a CoA transferase. It was heterologously expressed in Escherichia coli and characterized as a specific succinyl-CoA:benzoate CoA transferase. The newly identified enzyme in conjunction with a benzoate-induced succinyl-CoA synthetase links the tricarboxylic acid cycle to the upper benzoyl-CoA degradation pathway during growth on aromatic growth substrates.
INTRODUCTION
Aromatic compounds are widely distributed in nature and comprise many components that are harmful to human health and/or the environment. They can only be fully degraded to CO2 by aerobic and anaerobic microorganisms. The strategies for the catabolism of aromatic growth substrates differ fundamentally in aerobic and anaerobic bacteria. Aerobic bacteria make use of oxygenases that are involved in aromatic ring hydroxylation/cleavage. In contrast, a completely different set of degradation pathways and enzymes is used in aromatic compound-degrading anaerobes. The anaerobic degradation pathways of monocyclic aromatic compounds have initially been studied in facultative anaerobes such as Thauera sp., Azoarcus sp., Magnetospirillum sp., Rhodopseudomonas palustris, and “Aromatoleum aromaticum” (for recent reviews, see references 11, 19, 20, and 37). In contrast, most information about the anaerobic aromatic degradation pathways in obligate anaerobes derive from studies with the Fe(III)-respiring model organism Geobacter metallireducens. This organism was isolated and described by Lovley et al. and uses, e.g., phenol, toluene, p-cresol, or benzoate as carbon sources and metal oxides or nitrate as electron acceptors in anoxic respiratory chains (33, 34).
In anaerobic bacteria the majority of monocyclic aromatic growth substrates are converted to the central intermediate benzoyl-coenzyme A (BCoA) by enzymes of the peripheral aromatic catabolism. The anaerobic degradation of benzenes, phenols, cresols, benzoates, aromatic amino acids, and many other monocyclic aromatic compounds involves the activation of aryl carboxylate substrates/intermediates to the corresponding CoA esters. In all cases investigated thus far this activation is catalyzed by AMP-forming aryl carboxylate CoA ligases. Benzoate CoA ligases have been isolated and characterized in both facultatively and obligately anaerobic bacteria (4, 6, 17, 21, 23, 29, 31, 40, 46, 50). For example, G. metallireducens contains a single gene, bamY, encoding a benzoate CoA ligase. The expression of bamY, as well as the synthesis and activity of its product, was induced during growth on benzoate and other aromatic compounds (39, 50).
The next step in the so-called benzoyl-CoA degradation pathway involves the energetically and mechanistically difficult reduction of benzoyl-CoA to cyclohexa-1,5-diene-1-carboxyl-CoA (1,5-dienoyl-CoA) (7, 8). The benzoyl-CoA/1,5-dienoyl-CoA redox pair is the most negative one of a substrate/product couple in biology, and the highly endergonic electron transfer from a physiological donor to the aromatic ring has to be coupled to an exergonic reaction (27). There are two completely different classes of benzoyl-CoA reductases (BCRs), which both yield 1,5-dienoyl-CoA. Facultative anaerobes use an ATP-dependent class I BCR (9, 49), whereas obligate anaerobes are supposed to contain an ATP-independent, high-molecular-weight subunit complex harboring W, Zn, FAD, FeS clusters, and selenocysteine as cofactors (28, 32). The subsequent steps involved in 1,5-dienoyl-CoA degradation comprise modified β-oxidation reactions that are catalyzed by similar enzymes in facultatively and obligately anaerobic bacteria (30, 41). In bacteria with an anaerobic respiratory chain, the product acetyl-CoA is finally oxidized to CO2 in the tricarboxylic acid (TCA) cycle.
To study the unprecedented enzymes involved in the anaerobic degradation of aromatic compounds, genetic systems have successfully been developed in a number of facultatively anaerobic bacteria (1, 14, 18, 22, 42, 51). Very recently, a first genetic system was described for the obligately anaerobic G. metallireducens that included growth on solid medium with Fe(III)-citrate as an electron acceptor, markerless gene mutation, and gene complementation (48). The limitations of this system included poor growth on solid medium with Fe(III)-citrate as an electron acceptor, the requirement for harvesting cells in early-log phase, and the dependency on supplements such as yeast extract.
In the present study, we establish an alternative genetic system for G. metallireducens using a pure mineral medium with nitrate as a highly soluble electron acceptor allowing high growth yields. Using this system, the gene coding for a benzoate CoA ligase, previously considered essential for benzoate utilization, was found to be dispensable. Instead, a previously unknown succinyl-CoA:benzoate CoA transferase activity was identified and characterized as an additional enzyme involved in benzoate activation.
MATERIALS AND METHODS
Bacterial strains and plasmids.
G. metallireducens was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ 7210). Chemically competent Escherichia coli JM109 was purchased from Promega (Madison, WI). The Champion pET101 directional TOPO expression kit and chemically competent E. coli OneShotTOP10 and BL21Star(DE3)OneShot were purchased from Life Technologies (Grand Island, NY).
Growth media, culturing conditions, and determination of antibiotic resistance.
E. coli strains were cultivated as described previously (43). G. metallireducens was cultivated at 30°C under strictly anoxic conditions in a mineral salt medium (35), in which nitrate (15 mM) was used as the electron acceptor and either acetate (30 mM) or benzoate (5 mM) was used as the electron donor. Plating on solid medium containing 1.5% agar was performed in an anaerobic glove box (Coy Laboratory Products, Inc., Grass Lake, MI) under an H2/N2 atmosphere (5:95 [vol/vol]). Cultivation on solid medium was performed in anaerobic containers that were flushed with N2/CO2 (80:20 [vol/vol]) at a 0.5-bar overpressure; the containers were transferred to the anaerobic glove box during cultivation. The sensitivity of G. metallireducens to antibiotics was tested in liquid mineral medium. All antibiotic stock solutions were sterile and anoxic.
In order to estimate the plating efficiency of G. metallireducens the cell number of a pre-stationary-phase liquid culture was determined by using the cell counting chamber. The number of CFU in these cultures was determined by plating 10-fold serial dilutions. For the assessment of plasmid stability G. metallireducens was electrotransformed with broad-host-range IncQ plasmids RSF1010 (45) and pCD342 (16), respectively. The resulting streptomycin- and kanamycin-resistant liquid cultures were transferred over at least 10 passages (>100 generations) into nonselective liquid medium. Tenfold serial dilutions of each passage were plated onto selective and nonselective solid medium, respectively. The resulting CFU ml−1 on selective and nonselective media were compared.
Preparation of electrocompetent G. metallireducens and electroporation procedure.
Preparation of electrocompetent G. metallireducens cells was performed in a glove box in an H2/N2 atmosphere (5:95 [vol/vol]), and this was followed a modified protocol described previously (13). All steps were performed anaerobically. Portions (100 ml) of mid-log-phase cultures in mineral medium with acetate (optical density at 600 nm [OD600] of 0.4 to 0.5 corresponding to 4 × 108 to 5 × 108 cells ml−1) were harvested by centrifugation at 4°C for 10 min at 7,000 × g. The cells were washed twice in 100 ml of electroporation buffer (13) and suspended in 1 ml of the same buffer containing 10% dimethyl sulfoxide (DMSO). The resulting electrocompetent cells were stored in liquid nitrogen. Electroporation was performed in 1-mm-gap electroporation cuvettes using an Eporator (Eppendorf, Germany). Then, 100-μl portions electrocompetent G. metallireducens were pulsed at 1.7 kV cm−1 for 4 ms. After electroporation, the cells were immediately transferred into 1 ml of room temperature mineral medium with acetate as a carbon source and allowed to recover for around 5 h, prior to plating on selective solid medium or cultivation in liquid growth medium.
General DNA techniques.
The genomic DNA of G. metallireducens was isolated as described previously by Cheng and Jiang (12). PCR products were purified from agarose gels using the Wizard SV Gel and PCR Clean-Up system (Promega). Plasmid DNA was purified by using the Wizard Plus Minipreps DNA purification system (Promega). Primers were provided by Metabion (Martinsried, Germany). For cloning reactions using the Champion pET101 directional TOPO expression kit, PCR amplification was performed using Pfu DNA polymerase (Thermo Scientific). All other PCR amplifications were performed using the GoTaq Green Master Mix (Promega) according to the manufacturer's instructions. The sequences of all of the primers used are listed in Table S1 in the supplemental material.
Homologous expression and purification of bamY.
For homologous expression of bamY (gi78194605), plasmid pCD342 was used (16). The bamY gene was amplified from the G. metallireducens genome using the primers bamY_Bam+SDGm_for and bamY_Hind+cHis_rev (for oligonucleotide primer sequences see Table S1 in the supplemental material). The forward primer included restriction sites for BamHI and the Shine-Dalgarno sequence of bamY from G. metallireducens (26) in addition to the annealing nucleotides. The reverse primer included the restriction site for HindIII and a C-terminal 6-fold His tag sequence in addition to the annealing nucleotides. The resulting PCR product was digested with BamHI and HindIII and inserted into the corresponding sites of pCD342 to form the expression plasmid pCDbamY. After transformation, the resulting kanamycin-resistant cultures were grown at 30°C in 2 liters of mineral medium containing 30 mM acetate as electron donor. Stationary-phase cultures were harvested by centrifugation at 7,000 × g for 10 min at 4°C. The cells were suspended in 50 mM Tris-HCl (pH 6.8), MgCl2, 10% glycerol (vol/vol), DNase I (10 μg/g of cells), and lysozyme (50 μg/g of cells) and were subsequently disrupted by sonication (7 W, 5 to 7 pulses of 30 s). After centrifugation at 100,000 × g, His-tagged BamY was purified from the supernatant at 4°C by Ni-Sepharose high-performance affinity column (HisTrap HP; GE Healthcare). After equilibration of the 1-ml column with 20 mM Tris-HCl (pH 7.4), 500 mM NaCl, 20 mM imidazole, and 10% (vol/vol) glycerol, the extract was applied to the column at a flow rate of 0.75 ml min−1 and washed with 10 column volumes, followed by a linear gradient over 30 column volumes from 20 to 500 mM imidazole in buffer.
Enrichment of succinyl-CoA:benzoate CoA transferase and heterologous expression of its gene.
The succinyl-CoA:benzoate CoA transferase activity was partially purified from benzoate-grown cells of wild-type G. metallireducens in four chromatographic steps. Since the enzyme was not sensitive to oxygen, all purification steps were carried out under oxic conditions. Cells (10 g [wet mass]) were suspended in 15 ml of 50 mM Tris-HCl (pH 6.8) containing 4 mM MgCl2, 5 mM KH2PO4, 1 mM dithioerythritol (DTE), and 10% (vol/vol) glycerol. After the addition of DNase I and lysozyme, the cells were disrupted by sonication as described above. The extract was centrifuged at 100,000 × g, and the supernatant was dialyzed overnight in suspension buffer.
The dialyzed protein solution was applied to a DEAE-Sepharose column at a flow rate of 0.75 ml min−1 (Fast Flow [GE Healthcare]; diameter, 16 mm; volume, 10 ml), which had been equilibrated with 10 mM Tris-HCl (pH 7.8) containing 2 mM MgCl2, 5 mM KH2PO4, 1 mM dithioerythritol, and 10% (vol/vol) glycerol (buffer A). The column was washed with 30 ml of buffer A at a flow rate of 1 ml min−1 and afterward with 70 ml of 50 mM KCl in buffer A. The succinyl-CoA:benzoate CoA transferase activity eluted in a linear gradient of 50 to 120 mM KCl in buffer A (100 ml). Pooled fractions with succinyl-CoA:benzoate CoA transferase activity were applied to a hydroxyapatite column (ceramic hydroxyapatite type I, 40 μm [MacroPrep; Bio-Rad]; diameter, 16 mm; volume, 7 ml) that had been equilibrated with buffer A. The transferase activity eluted at 50 mM KH2PO4 in buffer A at a flow rate of 1 ml min−1. Next, 15 ml of the active hydroxyapatite fraction was concentrated (Vivaspin 6 [Sartorius], 10,000 molecular weight cutoff) to a volume of 5 ml and subsequently applied to a MonoQ column (GE Healthcare; volume, 1 ml), which had been equilibrated with buffer A. The column was washed with 10 ml of buffer A, followed by 100 mM KCl in buffer A at a flow rate of 0.5 ml min−1. The transferase activity eluted in a linear gradient of 100 to 200 mM KCl in buffer A. The active fractions were concentrated, and 0.5 ml were applied to a Reactive Green Agarose affinity chromatography column (GE Healthcare; diameter, 5 mm; volume, 1 ml) which had been equilibrated with buffer A. The column was washed with 10 ml of buffer A and 10 ml of 50 mM KCl in buffer A at a flow rate of 0.5 ml min−1. Activity eluted in a step gradient of 100 mM KCl in buffer A. Purity control was carried out by SDS-PAGE analysis.
For the heterologous expression of the gene putatively coding for succinyl-CoA:benzoate CoA transferase (gi78194516), the Champion pET101 Directional TOPO Expression Kit was used (Life Technologies). The primers Gm_act-pET/D_for and Gm_act-pET/D_rev were designed according to the manufacturer's instructions. Ligation, cloning, and expression were performed according to the manufacturer's protocol. Cells were suspended and disrupted, after which the His-tagged succinyl-CoA:benzoate transferase was purified by Ni-affinity chromatography as described above for BamY.
Construction of a bamY::kan disruption mutant and complementation in trans.
For the construction of a bamY disruption mutant, the plasmid pK18mob was selected (44). The plasmid contains a kanamycin resistance cassette and a pMB1 origin of replication which prevents the replication in G. metallireducens.
A central 672-bp-comprising fragment of bamY (referred to as bamY′) was amplified from the G. metallireducens chromosome using primers bamYDelHind_for and bamYDelXba_rev (see Fig. S3 in the supplemental material). In addition to annealing nucleotides, the primer sequences contained restriction sites for subsequent cloning of the PCR product. The resulting PCR product was digested with HindIII and XbaI and inserted into the corresponding sites of pK18mob to form plasmid pKbamY′. After transformation with 125 ng of pKbamY′, positive clones that had incorporated the plasmid into the chromosome were identified by growth with kanamycin-containing medium. For the verification of the bamY disruption, several PCRs using the primers bamY_Bam+SDGm_for (P1), R24 (P2), and F24 (P3) and bamY_Hind+cHis_rev (P4) were performed using GoTaq Green Master Mix (Promega) (see Table S1 and Fig. S3 in the supplemental material). In addition, PCR control reactions were performed using Phusion High-Fidelity DNA polymerase (Finnzymes, Finland) for large PCR products according to the manufacturer's instructions.
In order to complement the bamY::kan mutation, bamY was expressed in trans using the plasmid pCDbamYSm. This plasmid was obtained from pCDbamY by replacing the kanamycin resistance cassette by a streptomycin resistance cassette. It was amplified from plasmid RSF1010 (45) using the primers Sm_for_BglII and Sm_rev_BglII, which included the restriction site for BglII. The resulting PCR product was digested with BglII and inserted into the corresponding sites of pCDbamY. For the complementation of the bamY::kan mutation, 100 ng of plasmid pCDbamYSm was introduced into the mutant strain by electroporation.
Enzyme assays.
Benzoate CoA ligase activity was assayed in a coupled spectrophometric assay as described previously (46). Succinyl-CoA:benzoate CoA transferase activity was determined in a discontinuous high-pressure liquid chromatography (HPLC) assay (C18 reversed-phase HPLC; Eurosphere-100 10 μm, Waters Alliance 2695 [Waters, Eschborn, Germany]) by monitoring the substrate consumption and product formation at 30°C by diode array detection. The assay solution contained 100 mM Tris-HCl (pH 7.8), 5 mM MgCl2, and 0.2 mM succinyl-CoA or benzoyl-CoA. After the addition of 1 to 3 mg of cell extract or 10 to 50 μg of enriched protein to 200 μl of assay solution, the reaction was started by the addition of 0.15 mM carboxylic acid. Samples (50 μl) were taken at different time points, and the reaction was stopped by the addition of 5 μl of 10% (vol/vol) formic acid.
Determination of kinetic parameters and molecular mass.
Km values for benzoate and succinate were estimated by varying the concentration of the respective carboxylic acid from 0.025 to 0.5 mM. The data obtained were analyzed by fitting them to the Michaelis-Menten equation using the Prism software package (GraphPad, CA). The conversion of succinate, acetate, malate, fumarate, crotonate, glutarate, cyclohexanecarboxylate, cylohex-1-ene-1-carboxylate, and p-hydroxybenzoate as CoA acceptors was tested at 0.125 mM concentrations. The native molecular mass of the purified His-tagged succinyl-CoA:benzoate CoA transferase was determined by using an FPLC Superdex 200 gel filtration column (GE Healthcare; diameter, 10 mm; volume, 25 ml). After equilibration with 50 mM Tris-HCl (pH 6.8) containing 4 mM MgCl2, 5 mM KH2PO4, 1 mM DTE 10% (vol/vol) glycerol, and 150 mM KCl, 200-μl portions of the protein solution (∼2.5 mg) were applied at a flow rate of 0.3 ml min−1.
Mass spectrometry analysis.
Protein identification was performed by in-gel tryptic digestion, followed by UPLC-LTQ Orbitrap MS/MS analysis as described previously (29). Mascot searches of the raw data were done against the genome entry of Geobacter metallireducens GS-15.
RESULTS
Gene expression system in G. metallireducens with nitrate as an electron acceptor.
G. metallireducens is known to grow with nitrate as an alternative electron acceptor in liquid cultures, which is reduced to ammonia (35). Under these conditions, an increased requirement for Fe(III) in the culture medium was described previously (47). However, no growth on solid medium with nitrate as electron acceptor has yet been described, which could be explained by an increased sensitivity to oxidative stress caused by reactive nitrogen oxide intermediates. In particular during growth on aromatic compounds, enzymes involved in oxidative stress defense are known to be induced in G. metallireducens (25).
In order to establish a genetic system, including growth on solid medium with nitrate (15 mM) as electron acceptor, the preparation of agar plates and the inoculation of G. metallireducens was carried out in an anaerobic glove box under an N2/H2 atmosphere (95:5 [vol/vol]). Incubation of plates was performed in custom-made gas-tight containers in an N2/CO2 atmosphere with 0.5-bar overpressure using acetate (30 mM) or benzoate (5 mM) as electron donors. Under these conditions, the formation of red colonies was observed after 4 to 6 days at 25 to 30°C that were 1 to 2 mm in diameter (see Fig. S1 in the supplemental material). The plating efficiency was ca. 60%. No growth of G. metallireducens was observed in the presence of the following antibiotics at the concentrations as indicated: kanamycin (100 μg ml−1), streptomycin (100 μg ml−1), ampicillin (100 μg ml−1), tetracycline (15 μg ml−1), chloramphenicol (30 μg ml−1), and gentamicin (50 μg ml−1).
For establishing a protocol for homologous expression of a gene in G. metallireducens during growth on nitrate, bamY, encoding benzoate-CoA ligase, was introduced into vector pCD342 (16) with a 6-fold His tag. This gene was previously heterologously expressed in E. coli (50) and served as a proof of principle. The resulting pCDbamY was introduced into G. metallireducens by electroporation with transformation efficiencies of ∼104 CFU μg of DNA−1. To assess the plasmid stability, the colonies formed on kanamycin were transferred over at least 10 passages. Comparison of the CFU ml−1 on selective and nonselective media indicated that >70% of the cells still contained the plasmid displaying a high stability of pCD342 in G. metallireducens.
The homologously expressed, His-tagged BamY was purified using a Ni-chelating affinity chromatography column. A protein band of ∼60 kDa was highly enriched that contained a benzoate CoA ligase activity of 13 μmol min−1 mg−1, which is comparable to the previously described BamY heterologously produced in E. coli (50) (see Fig. S2 in the supplemental material).
Characterization of a bamY disruption mutant.
A central 672-bp fragment of bamY was inserted into plasmid pK18mob carrying a kanamycin resistance cassette; it is not replicated in G. metallireducens due to the pMB1 origin of replication (see Fig. S3 in the supplemental material). The resulting plasmid was introduced into G. metallireducens by electroporation. G. metallireducens bamY::kan strains that had incorporated the knockout plasmid into the chromosome by homologous recombination were identified by growth on mineral medium containing kanamycin. The insertion of the plasmid into the bamY gene was verified by sequencing PCR amplicons obtained using appropriate oligonucleotide primer pairs (see Fig. S3 in the supplemental material).
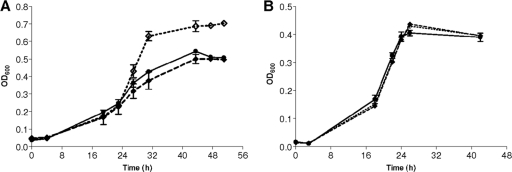
Extracts from cells of the G. metallireducens bamY::kan strain fully lost benzoate CoA ligase activity (<0.01 nmol min−1 mg−1; activity in the wild type, 0.5 to 1 nmol min−1 mg−1). However, the G. metallireducens bamY::kan strain was still able to grow with benzoate and nitrate as the sole sources of cell carbon and energy. Surprisingly, the maximal OD600 reached (5 mM benzoate) was reproducibly 1.35- to 1.5-fold higher than in the wild-type strain (Fig. 1A). This unexpected finding suggested that the expression of bamY was dispensable or even disadvantageous for growth on benzoate under the conditions used. In contrast, growth on acetate was highly similar in the wild type and in the bamY::kan strain (Fig. 1B). In summary, these findings suggest that, next to BamY, an additional, thus-far-unknown enzyme had to be involved in the initial step of benzoate degradation.
Fig 1.
Growth of G. metallireducens strains on A. benzoate and B. acetate. Wild type (●), bamY::kan mutant (♢), and complemented mutant (♦). Error bars indicate the standard deviations of biological triplicates.
The bamY gene was complemented in trans in the G. metallireducens bamY::kan mutant using the plasmid pCDbamYSm. This plasmid was constructed from the expression plasmid pCDbamY by replacing the kanamycin resistance with a streptomycin resistance cassette for selection of the complemented mutant from the kanamycin-resistant bamY::kan mutant. In the corresponding complemented bamY::kan mutant strain that expressed an intact bamY gene in trans, the observed phenotype of an increased biomass yield was reverted back to the wild-type level (Fig. 1A). This result confirms that disruption of bamY had a positive effect for growth on benzoate. Again, no effect was observed during growth on acetate (Fig. 1B).
In vitro carboxylic acid activation activities.
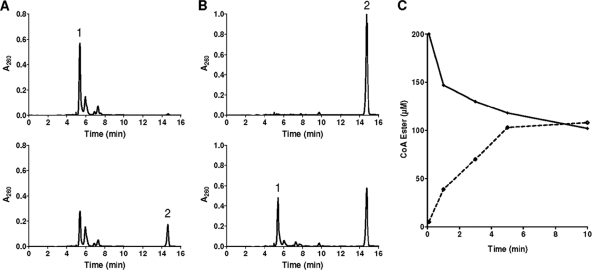
The potential alternative formation of benzoyl-CoA by a CoA transferase was tested by in vitro HPLC assays using extracts of the wild type and the bamY::kan mutant; acetyl-CoA, crotonyl-CoA, and succinyl-CoA served as potential CoA donors, and benzoate served as the CoA acceptor. Only with succinyl-CoA was time- and protein-dependent formation of benzoyl-CoA observed during HPLC analysis of CoA ester substrates consumed and products formed. The specific activity was 13 nmol min−1 mg−1 (for the results of representative HPLC analyses with enriched enzyme, see Fig. 2A). The activity was nearly identical in the wild type and in the bamY::kan mutant; virtually no activity was determined in extracts from wild-type cells grown on acetate (<0.01 nmol min−1 mg−1). The reaction could also be monitored in the reverse reaction at a similar rate (Fig. 2B). It leveled off when succinyl-CoA and benzoyl-CoA were present in an almost 1:1 ratio, indicating that the reaction proceeded close to thermodynamic equilibrium (ΔG = 0, Fig. 2C). In conclusion, the presence of the benzoate-induced succinyl-CoA:benzoate CoA transferase activity enabled growth on benzoate, even when the benzoate CoA ligase activity was knocked out.
Fig 2.
HPLC analysis of succinyl-CoA:benzoate CoA transferase reaction. (A) Time-dependent consumption of 0.2 mM succinyl-CoA (peak 1) and formation of benzoyl-CoA (peak 2) in the presence of 0.5 mM benzoate. Upper panel, after 0.1 min of incubation; lower panel, after 10 min of incubation. (B) Time-dependent consumption of 0.2 mM benzoyl-CoA (peak 2) and formation of succinyl-CoA (peak 1) in the presence of 0.5 mM succinate. Upper panel, after 0.1 min of incubation; lower panel, after 10 min of incubation. (C) Time course of succinyl-CoA:benzoate CoA transferase reaction starting with succinyl-CoA. Solid line, succinyl-CoA; dashed line, benzoyl-CoA.
In addition to this succinyl-CoA-dependent transferase, a benzoate induced, ATP-dependent succinyl-CoA synthetase activity was determined (16.5 nmol min−1 mg−1), which was missing in extracts from cells grown on acetate (<0.1 nmol min−1 mg−1). The induction of the putative genes coding for this TCA enzyme during growth on benzoate were previously described (10, 50). The combined induction of genes coding for a succinyl-CoA synthetase/CoA transferase during growth on aromatic compounds directly links benzoate activation to the TCA cycle.
G. metallireducens was predicted to contain three different enzymes involved in acetyl-CoA formation during growth on acetate, including the essentially irreversible AMP-forming acetyl-CoA synthetase and the fully reversible succinyl-CoA:acetate CoA transferase and acetate kinase/phosphotransacetylase (2, 10). Determination of in vitro activities of the putative acetate activating enzymes in extracts from cells grown on acetate confirmed the presence of a highly active succinyl-CoA:acetate CoA transferase (550 nmol min−1 mg−1) and an acetate CoA ligase (60 nmol min−1 mg−1), whereas no acetate kinase activity was determined (<0.1 nmol min−1 mg−1). In summary, G. metallireducens appears to use two different modes for acetate and benzoate activation via specific carboxylic acid CoA synthetases and CoA transferases.
Enrichment of succinyl-CoA:benzoate CoA transferase from the wild type and identification of its gene.
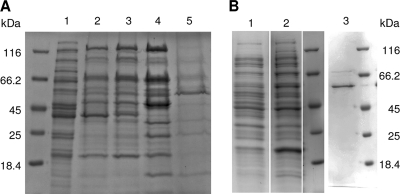
The succinyl-CoA:benzoate CoA transferase activity was 53-fold enriched from extracts of wild-type cells grown on benzoate using four chromatographic steps (Table 1 and Fig. 3A). After affinity chromatography on Reactive Green agarose, the pooled protein fractions containing succinyl-CoA:benzoate CoA transferase activity showed a highly enriched protein band on SDS polyacrylamide gels at ∼50 kDa. The protein was excised from the gel and digested with trypsin, and the peptides obtained were subsequently analyzed by mass spectrometry. Using the MASCOT platform, the best matches were obtained with peptides from a deduced gene product in the genome of G. metallireducens, which was annotated as a CoA transferase (gi78194516; theoretical mass, 48.1 kDa). The protein identification was based on four peptides (sequence coverage, 13%; protein summary score, 208.5). The gene coding for succinyl-CoA:benzoate CoA transferase was previously identified as a gene at the downstream edge of the benzoate-induced cluster IB that includes genes encoding enzymes involved in β-oxidation reactions (50). The genomic context is shown in Fig. 4.
Table 1.
Enrichment of succinyl-CoA:benzoate CoA transferasea
| Purification step | Protein (mg) | Sp act (mU mg−1) | Total activity (mU) | Recovery (%) | Purification (fold) |
|---|---|---|---|---|---|
| Cell extract | 630 | 13 | 7,900 | 100 | 1 |
| DEAE | 118 | 74 | 8,750 | 110 | 6 |
| Hydroxyapatite | 44 | 98 | 4,330 | 55 | 8 |
| MonoQ | 1.2 | 261 | 315 | 4 | 21 |
| Reactive Green | 0.06 | 673 | 37 | 0.5 | 53 |
The starting material was 10 g (wet mass) of wild-type G. metallireducens grown on benzoate. “1 mU” corresponds to the formation of 1 nmol of benzoyl-CoA min−1.
Fig 3.
SDS-PAGE analysis of succinyl-CoA:benzoate CoA transferase enrichment from the wild type (A) and after heterologous expression of the gene in E. coli (B). (A) Lanes: 1, cell extracts of G. metallireducens; 2, after DEAE chromatography; 3, ceramic hydroxyapatite; 4, MonoQ; 5, Reactive Green agarose. (B) Lanes: 1 and 2, cell extract of E. coli before (lane 1) and after (lane 2) IPTG induction; 3, purified His-tagged protein after Ni-chelating chromatography.
Fig 4.
Genomic context of the gene coding for succinyl-CoA:benzoate CoA transferase. The gene referred to as bct is indicated by the gray-shaded arrow (gi78194516). Abbreviations for annotated predicted gene products: miaA, DNA mismatch repair protein; mutL, tRNS isopentenyl transferase; tre, transcriptional regulator with PAS sensor; hyd, hydrolase; ech, enoyl-CoA hydratase/isomerase; act, acetyl-CoA transferase; adh, short-chain dehydrogenase; pmp, fusaric acid resistance protein; ssy, sodium symporter.
Using BLAST, highly homologous genes with deduced amino acid sequence identities greater than 75% were only found in aromatic compound-degrading members of the Geobacteraceae, including G. bemidjensis, G. daltonii, and Geobacter species M21. Notably, in each of the genomes two copies of such highly similar transferase genes are present. Similarities to experimentally characterized CoA transferases from other organisms clearly indicate that the enzyme from G. metallireducens belongs to the Ia family of CoA transferases. Members of this family contain a conserved active-site glutamate (E248 in the transferase of G. metallireducens), which forms a covalent mixed anhydride intermediate during catalysis (24, 36).
Heterologous production and characterization of succinyl-CoA:benzoate CoA transferase.
The putative gene coding for succinyl-CoA:benzoate CoA transferase identified by mass spectrometry was heterologously expressed in E. coli with a 6-fold C-terminal His tag (Fig. 3B). After purification by Ni-chelating affinity chromatography, a highly enriched protein band that migrated at a slightly higher mass than wild-type succinyl-CoA:benzoate CoA transferase was obtained which can be explained by the additional 6-fold His tag. The specific activity of the heterologously expressed CoA transferase was 265 nmol min−1 mg−1 and ca. 40% of the highly enriched wild-type enzyme.
The native molecular mass as determined by gel filtration was ∼90 kDa, suggesting an α2-composition. The Km value for benzoate was 55 ± 5 μM (mean value ± the standard deviation). Next to benzoate, only cyclohex-1-ene-1-carboxylate was used as a CoA acceptor at a comparable rate, whereas other nonaromatic carboxylic acids tested were virtually not converted (Table 2). Together, these findings suggest a high preference for benzoate or closely related analogues as CoA acceptor. In the reverse reaction with benzoyl-CoA as CoA donor no nonaromatic carboxylic acid other than succinate was converted (Table 2).
Table 2.
Substrate preference of heterologously expressed succinyl-CoA:benzoate CoA transferase from G. metallireducens
| Substrate | Activity (%)a |
|
|---|---|---|
| CoA donor: benzoyl-CoA | CoA donor: succinyl-CoA | |
| Succinate | 100 | ND |
| Acetate | <0.1 | <0.1 |
| Malate | <0.1 | ND |
| Fumarate | <0.1 | ND |
| Crotonate | ND | <0.1 |
| Glutarate | ND | <0.1 |
| Benzoate | ND | 100 |
| Cyclohex-1-ene-carboxylate | ND | 126 |
| Cylohexanecarboxylate | ND | <0.1 |
| p-Hydroxybenzoate | ND | 11 |
Carboxylic acids (0.125 mM) were tested as CoA acceptors in the presence of benzoyl-CoA or succinyl-CoA as CoA donors. ND, not determined.
DISCUSSION
Genetic system for G. metallireducens grown with nitrate as an electron acceptor.
Very recently, two different genetic systems became available for an obligately anaerobic, aromatic compound-degrading bacterium. The systems described by Tremblay et al. (48) and in the present study have different strengths and weaknesses and may be suitable for different individual purposes. The system described by Tremblay et al. enables the introduction of markerless mutations, which is recommended for the insertion of multiple markerless mutations and the corresponding multiple complementations. For example, the formation of a bamY bct double mutant cannot be achieved in the system established here due to its dependency on the introduction of antibiotic resistance cassettes. On the other side, the advantage of the system established here enabled a much higher growth yield due to the use of nitrate as an electron acceptor and, in contrast to the system by Tremblay et al., the cells can be harvested in the mid-exponential-growth phase. Moreover, it does not depend on supplements such as yeast extract, which may have a negative effect on the expression of catabolic enzymes.
The system established here will in particular be useful for studying the physiology of deltaproteobacteria during growth on aromatic growth substrates, including Fe(III)- and sulfate-reducing as well as fermenting bacteria. For example, the study of the highly complex class II BCRs, which are composed of eight subunits with W, FeS, and selenocysteine cofactors requiring complex cofactor assembly machineries will benefit from the system established (28, 50). Although G. metallireducens does not use polycyclic aromatic hydrocarbons (PAHs), the system established may be helpful for expressing enzymes from related deltaproteobacteria that are involved in the degradation of PAHs, e.g., established anaerobic pure/enrichment cultures that are capable of using naphthalene or phenanthrene as the sole carbon source belong to the sulfate-reducing deltaproteobacteria (5, 15, 38).
Benzoyl-CoA formation in aromatic compound-degrading anaerobes.
Thus far, the activation of benzoate or hydroxylated, aminated, methylated, or halogenated analogues was always found to be catalyzed by AMP-forming aryl carboxylate CoA ligases. This finding was in particular surprising as obligate anaerobes such as sulfate-reducing or fermenting bacteria have a very poor energy yield during growth on aromatic compounds. With the succinyl-CoA:benzoate CoA transferase, an alternative, energetically less demanding enzyme has been identified to be involved in aryl carboxyl-CoA formation. It appears to be present in all aromatic compound degrading members of the Fe(III)-respiring Geobacteraceae but might also be present in sulfate-reducing or fermenting bacteria.
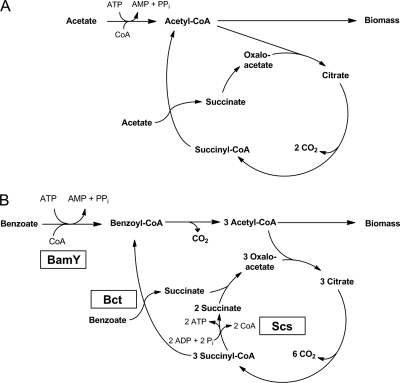
Since three acetyl-CoAs are formed per benzoate oxidized, the succinyl-CoA:benzoate CoA transferase as the only catabolic enzyme involved in succinate formation from succinyl-CoA would not be sufficient for maintaining the TCA cycle (Fig. 5B). For this reason, the additional induction of a succinate regenerating succinyl-CoA synthetase is essential for growth on benzoate. In contrast, during growth on acetate no succinyl-CoA synthetase is required as per acetyl-CoA oxidized one succinate is formed by succinyl-CoA:acetate CoA transferase (Fig. 5A). In conclusion, no succinyl-CoA synthetase activity was determined during growth on acetate.
Fig 5.
Activation of carboxylic acid growth substrates and TCA cycle in G. metallireducens. (A) Acetate as carbon source; (B) benzoate as carbon source. During growth on benzoate a succinyl-CoA synthetase is required as only maximally one of the three succinyl-CoA formed per benzoyl-CoA oxidized is regenerated by succinyl-CoA:benzoate CoA transferase. BamY, benzoate-CoA ligase; Bct, succinyl-CoA:benzoate CoA transferase (benzoate CoA transferase); Scs, succinyl-CoA synthetase.
Disruption of the bamY gene in G. metallireducens resulted in the unexpected phenotype of the observed increased biomass yield (i.e., the OD600 reached per mol of benzoate consumed). This, on first view, curious finding suggests that the loss of BamY activity was apparently rather advantageous to the cell, since benzoate activation by a CoA transferase enables a higher ATP yield during growth on benzoate. The bamY gene is integrated in the benzoate induced bam gene cluster II, which is supposed to be acquired by G. metallireducens as a whole by lateral transfer. This assumption is substantiated by the finding that in G. metallireducens the genes coding for the degradation of aromatic compounds are organized in a genomic island (2, 10). The gene coding for succinyl-CoA:benzoate CoA transferase in G. metallireducens is part of the benzoate induced cluster IB, and its acquisition could have made bamY dispensable under certain conditions. This hypothesis is supported by the finding that the closely related, benzoate-degrading G. bemidjiensis contains only a nonfunctional bamY, in which a mutation caused an internal TGA stop codon (3). Functional bamY genes contain a codon for glutamate at this position. Indeed, cells of G. bemidjiensis grown on benzoate do not contain a benzoate CoA ligase activity but a succinyl-CoA:benzoate CoA transferase activity (our unpublished results). In conclusion, the artificial inactivation of the bamY gene carried out here during the establishment of a genetic system obviously occurred in nature in a related Geobacter species.
The availability of genes coding for both a benzoate CoA ligase and a succinyl-CoA:benzoate CoA transferase could be advantageous under certain conditions in nature. The essentially irreversible benzoate CoA ligase should facilitate growth on aromatic compounds at very low concentrations compared to the fully reversible CoA transferase. Low concentrations of aromatic compounds probably prevail more often at natural habitats than the high concentrations that are generally applied in pure/enrichment laboratory cultures. In conclusion, the apparent advantageous inactivation of bamY observed under experimental conditions might be rather disadvantageous at low substrate concentrations at environmental sites.
Supplementary Material
ACKNOWLEDGMENTS
We thank Javier F. Juárez, Madrid, Spain, for providing plasmids and for many helpful suggestions during the establishment of the genetic system.
This study was funded by the Deutsche Forschungsgemeinschaft (BO 1565/10-2).
Footnotes
Published ahead of print 9 March 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Achong GR, Rodriguez AM, Spormann AM. 2001. Benzylsuccinate synthase of Azoarcus sp. strain T: cloning, sequencing, transcriptional organization, and its role in anaerobic toluene and m-xylene mineralization. J. Bacteriol. 183:6763–6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aklujkar M, et al. 2009. The genome sequence of Geobacter metallireducens: features of metabolism, physiology and regulation common and dissimilar to Geobacter sulfurreducens. BMC Microbiol. 9:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aklujkar M, et al. 2010. The genome of Geobacter bemidjiensis, exemplar for the subsurface clade of Geobacter species that predominate in Fe(III)-reducing subsurface environments. BMC Genomics 11:490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Auburger G, Winter J. 1992. Purification and characterization of benzoyl-CoA ligase from a syntrophic, benzoate-degrading, anaerobic mixed culture. Appl. Microbiol. Biotechnol. 37:789–795 [DOI] [PubMed] [Google Scholar]
- 5. Bergmann FD, Selesi D, Meckenstock RU. 2011. Identification of new enzymes potentially involved in anaerobic naphthalene degradation by the sulfate-reducing enrichment culture N47. Arch. Microbiol. 193:241–250 [DOI] [PubMed] [Google Scholar]
- 6. Biegert T, Altenschmidt U, Eckerskorn C, Fuchs G. 1993. Enzymes of anaerobic metabolism of phenolic compounds: 4-hydroxybenzoate-CoA ligase from a denitrifying Pseudomonas species. Eur. J. Biochem. 213:555–561 [DOI] [PubMed] [Google Scholar]
- 7. Boll M. 2005. Dearomatizing benzene ring reductases. J. Mol. Microbiol. Biotechnol. 10:132–142 [DOI] [PubMed] [Google Scholar]
- 8. Boll M. 2005. Key enzymes in the anaerobic aromatic metabolism catalysing Birch-like reductions. Biochim. Biophys. Acta 1707:34–50 [DOI] [PubMed] [Google Scholar]
- 9. Boll M, Fuchs G. 1995. Benzoyl-coenzyme A reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. ATP dependence of the reaction, purification and some properties of the enzyme from Thauera aromatica strain K172. Eur. J. Biochem. 234:921–933 [DOI] [PubMed] [Google Scholar]
- 10. Butler JE, et al. 2007. Genomic and microarray analysis of aromatics degradation in Geobacter metallireducens and comparison to a Geobacter isolate from a contaminated field site. BMC Genomics 8:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carmona M, et al. 2009. Anaerobic catabolism of aromatic compounds: a genetic and genomic view. Microbiol. Mol. Biol. Rev. 73:71–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng HR, Jiang N. 2006. Extremely rapid extraction of DNA from bacteria and yeasts. Biotechnol. Lett. 28:55–59 [DOI] [PubMed] [Google Scholar]
- 13. Coppi MV, Leang C, Sandler SJ, Lovley DR. 2001. Development of a genetic system for Geobacter sulfurreducens. Appl. Environ. Microbiol. 67:3180–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coschigano PW. 2002. Construction and characterization of insertion/deletion mutations of the tutF, tutD, and tutG genes of Thauera aromatica strain T1. FEMS Microbiol. Lett. 217:37–42 [DOI] [PubMed] [Google Scholar]
- 15. Davidova IA, Gieg LM, Duncan KE, Suflita JM. 2007. Anaerobic phenanthrene mineralization by a carboxylating sulfate-reducing bacterial enrichment. ISME J. 1:436–442 [DOI] [PubMed] [Google Scholar]
- 16. Dehio M, Knorre A, Lanz C, Dehio C. 1998. Construction of versatile high-level expression vectors for Bartonella henselae and the use of green fluorescent protein as a new expression marker. Gene 215:223–229 [DOI] [PubMed] [Google Scholar]
- 17. Egland PG, Gibson J, Harwood CS. 1995. Benzoate-coenzyme A ligase, encoded by badA, is one of three ligases able to catalyze benzoyl-coenzyme A formation during anaerobic growth of Rhodopseudomonas palustris on benzoate. J. Bacteriol. 177:6545–6551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Egland PG, Pelletier DA, Dispensa M, Gibson J, Harwood CS. 1997. A cluster of bacterial genes for anaerobic benzene ring biodegradation. Proc. Natl. Acad. Sci. U. S. A. 94:6484–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fuchs G. 2008. Anaerobic metabolism of aromatic compounds. Ann. N. Y. Acad. Sci. 1125:82–99 [DOI] [PubMed] [Google Scholar]
- 20. Fuchs G, Boll M, Heider J. 2011. Microbial degradation of aromatic compounds: from one strategy to four. Nat. Rev. Microbiol. 9:803–816 [DOI] [PubMed] [Google Scholar]
- 21. Geissler JF, Harwood CS, Gibson J. 1988. Purification and properties of benzoate-coenzyme A ligase, a Rhodopseudomonas palustris enzyme involved in the anaerobic degradation of benzoate. J. Bacteriol. 170:1709–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gescher J, Zaar A, Mohamed M, Schagger H, Fuchs G. 2002. Genes coding for a new pathway of aerobic benzoate metabolism in Azoarcus evansii. J. Bacteriol. 184:6301–6315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gibson J, Dispensa M, Fogg GC, Evans DT, Harwood CS. 1994. 4-Hydroxybenzoate-coenzyme A ligase from Rhodopseudomonas palustris: purification, gene sequence, and role in anaerobic degradation. J. Bacteriol. 176:634–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heider J. 2001. A new family of CoA-transferases. FEBS Lett. 509:345–349 [DOI] [PubMed] [Google Scholar]
- 25. Heintz D, et al. 2009. Differential membrane proteome analysis reveals novel proteins involved in the degradation of aromatic compounds in Geobacter metallireducens. Mol. Cell. Proteomics 8:2159–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Juarez JF, et al. 2011. Identification of the Geobacter metallireducens bamVW two-component system, involved in transcriptional regulation of aromatic degradation. Appl. Environ. Microbiol. 76:383–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kung JW, et al. 2010. Reversible biological birch reduction at an extremely low redox potential. J. Am. Chem. Soc. 132:9850–9856 [DOI] [PubMed] [Google Scholar]
- 28. Kung JW, et al. 2009. Identification and characterization of the tungsten-containing class of benzoyl-coenzyme A reductases. Proc. Natl. Acad. Sci. U. S. A. 106:17687–17692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuntze K, et al. 2011. Enzymes involved in the anaerobic degradation of meta-substituted halobenzoates. Mol. Microbiol. 82:758–769 [DOI] [PubMed] [Google Scholar]
- 30. Kuntze K, et al. 2008. 6-Oxocyclohex-1-ene-1-carbonyl-coenzyme A hydrolases from obligately anaerobic bacteria: characterization and identification of its gene as a functional marker for aromatic compounds degrading anaerobes. Environ. Microbiol. 10:1547–1556 [DOI] [PubMed] [Google Scholar]
- 31. Lochmeyer C, Koch J, Fuchs G. 1992. Anaerobic degradation of 2-aminobenzoic acid (anthranilic acid) via benzoyl-coenzyme A (CoA) and cyclohex-1-enecarboxyl-CoA in a denitrifying bacterium. J. Bacteriol. 174:3621–3628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Löffler C, et al. 2011. Occurrence, genes and expression of the W/Se-containing class II benzoyl-coenzyme A reductases in anaerobic bacteria. Environ. Microbiol. 13:696–709 [DOI] [PubMed] [Google Scholar]
- 33. Lovley DR, et al. 1993. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch. Microbiol. 159:336–344 [DOI] [PubMed] [Google Scholar]
- 34. Lovley DR, Lonergan DJ. 1990. Anaerobic oxidation of toluene, phenol, and p-cresol by the dissimilatory iron-reducing organism, GS-15. Appl. Environ. Microbiol. 56:1858–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lovley DR, Phillips EJ. 1988. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl. Environ. Microbiol. 54:1472–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Macieira S, Zhang J, Velarde M, Buckel W, Messerschmidt A. 2009. Crystal structure of 4-hydroxybutyrate CoA-transferase from Clostridium aminobutyricum. Biol. Chem. 390:1251–1263 [DOI] [PubMed] [Google Scholar]
- 37. Meckenstock RU, Mouttaki H. 2011. Anaerobic degradation of non-substituted aromatic hydrocarbons. Curr. Opin. Biotechnol. 22:406–414 [DOI] [PubMed] [Google Scholar]
- 38. Musat F, et al. 2009. Anaerobic degradation of naphthalene and 2-methylnaphthalene by strains of marine sulfate-reducing bacteria. Environ. Microbiol. 11:209–219 [DOI] [PubMed] [Google Scholar]
- 39. Peters F, Heintz D, Johannes J, van Dorsselaer A, Boll M. 2007. Genes, enzymes, and regulation of para-cresol metabolism in Geobacter metallireducens. J. Bacteriol. 189:4729–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Peters F, Rother M, Boll M. 2004. Selenocysteine-containing proteins in anaerobic benzoate metabolism of Desulfococcus multivorans. J. Bacteriol. 186:2156–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Peters F, Shinoda Y, McInerney MJ, Boll M. 2007. Cyclohexa-1,5-diene-1-carbonyl-coenzyme A (CoA) hydratases of Geobacter metallireducens and Syntrophus aciditrophicus: evidence for a common benzoyl-CoA degradation pathway in facultative and strict anaerobes. J. Bacteriol. 189:1055–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rost R, Haas S, Hammer E, Herrmann H, Burchhardt G. 2002. Molecular analysis of aerobic phenylacetate degradation in Azoarcus evansii. Mol. Genet. Genomics 267:656–663 [DOI] [PubMed] [Google Scholar]
- 43. Sambrook J, Fritsch E, TManiatis (ed). 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 44. Schäfer A, Tauch A, WJäger Kalinowski J, Thierbach G, Puhler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73 [DOI] [PubMed] [Google Scholar]
- 45. Scholz P, et al. 1989. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene 75:271–288 [DOI] [PubMed] [Google Scholar]
- 46. Schühle K, et al. 2003. Benzoate-coenzyme A ligase from Thauera aromatica: an enzyme acting in anaerobic and aerobic pathways. J. Bacteriol. 185:4920–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Senko JM, Stolz JF. 2001. Evidence for iron-dependent nitrate respiration in the dissimilatory iron-reducing bacterium Geobacter metallireducens. Appl. Environ. Microbiol. 67:3750–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tremblay P-L, Aklujkar M, Leang C, Nevin K, Lovley DR. 2012. A genetic system for Geobacter metallireducens: role of the flagellin and pilin in the reduction of Fe(III) oxide. Environ. Microbiol. Rep. 4:82–88 [DOI] [PubMed] [Google Scholar]
- 49. Unciuleac M, Boll M. 2001. Mechanism of ATP-driven electron transfer catalyzed by the benzene ring-reducing enzyme benzoyl-CoA reductase. Proc. Natl. Acad. Sci. U. S. A. 98:13619–13624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wischgoll S, et al. 2005. Gene clusters involved in anaerobic benzoate degradation of Geobacter metallireducens. Mol. Microbiol. 58:1238–1252 [DOI] [PubMed] [Google Scholar]
- 51. Wöhlbrand L, Rabus R. 2009. Development of a genetic system for the denitrifying bacterium “Aromatoleum aromaticum” strain EbN1. J. Mol. Microbiol. Biotechnol. 17:41–52 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.