Abstract
The type IV pili are helical filaments found on many Gram-negative pathogenic bacteria, with multiple diverse roles in pathogenesis, including microcolony formation, adhesion, and twitching motility. Many pathogenic enterotoxigenic Escherichia coli (ETEC) isolates express one of two type IV pili belonging to the type IVb subclass: CFA/III or Longus. Here we show a direct correlation between CFA/III expression and ETEC aggregation, suggesting that these pili, like the Vibrio cholerae toxin-coregulated pili (TCP), mediate microcolony formation. We report a 1.26-Å resolution crystal structure of CofA, the major pilin subunit from CFA/III. CofA is very similar in structure to V. cholerae TcpA but possesses a 10-amino-acid insertion that replaces part of the α2-helix with an irregular loop containing a 310-helix. Homology modeling suggests a very similar structure for the Longus LngA pilin. A model for the CFA/III pilus filament was generated using the TCP electron microscopy reconstruction as a template. The unique 310-helix insert fits perfectly within the gap between CofA globular domains. This insert, together with differences in surface-exposed residues, produces a filament that is smoother and more negatively charged than TCP. To explore the specificity of the type IV pilus assembly apparatus, CofA was expressed heterologously in V. cholerae by replacing the tcpA gene with that of cofA within the tcp operon. Although CofA was synthesized and processed by V. cholerae, no CFA/III filaments were detected, suggesting that the components of the type IVb pilus assembly system are highly specific to their pilin substrates.
INTRODUCTION
A number of Gram-negative bacterial pathogens utilize a class of pili, the type IVb pili, to colonize the human intestinal epithelium and cause gastrointestinal diseases. Vibrio cholerae relies on the toxin-coregulated pili (TCP) to self-associate in microcolonies in the small intestine (53), where it secretes cholera toxin, an ADP-ribosylating enterotoxin responsible for the severe diarrhea that is characteristic of cholera disease. The type IVb pili of Salmonella enterica serovar Typhi, the causative agent of typhoid fever, are implicated in self-association and adhesion, as they bind to the cystic fibrosis transmembrane receptor on intestinal epithelial cells (43, 54). The bundle-forming pili (BFP) of enteropathogenic Escherichia coli (EPEC) are required for microcolony formation and possibly adherence to host cells in the small intestine (1, 18, 25). EPEC is a common cause of childhood diarrhea, particularly in developing countries. Enterotoxigenic E. coli (ETEC), which also causes childhood diarrhea as well as traveler's diarrhea in developing countries, displays several pilus types, including one of two type IVb pili, CFA/III or Longus, depending on the strain (19, 23, 52). The CFA/III and Longus pili have not been as well characterized as other type IVb pilins, in part because no animal models have been identified to study their role in ETEC pathogenesis. However, both pilus types have been implicated in bacterial aggregation, adhesion to intestinal epithelial cells, and mouse colonization. CFA/III expression is associated with ETEC colonization of infant mice and rabbits (23) and with adhesion to human colon carcinoma cells (51). Similarly, Longus is required by ETEC strain E9034A for self-aggregation, protection from antimicrobial agents (7), twitching motility, and adherence to cultured intestinal epithelial cells (34). Anti-Longus antibodies are capable of inhibiting ETEC cell adhesion (34) and have been detected in diarrheal patients (44), suggesting that these pili play a clinical role in ETEC infection. Thus, the ETEC type IVb pili, like those of V. cholerae, S. Typhi, and EPEC, appear to be critical virulence factors.
The ETEC type IVb pili are closely related to V. cholerae TCP. For each type IVb pilus system, the genes encoding the pilin subunit and pilus assembly proteins are clustered in an operon. The cof operon, which encodes CFA/III pili, is located on a 55-kb virulence plasmid and the lng operon is carried on a 70-kb virulence plasmid in ETEC, whereas the tcp operon is located on chromosome I in V. cholerae. The synteny for genes encoding the pilin subunits and the pilus biogenesis proteins is identical in the three operons. These similarities and the protein sequence conservation among the pilus components suggest a common evolutionary origin for these pili. The pilin subunit encoded by the ETEC cofA gene (CofA) shares 76% amino acid identity with LngA and 37% identity with the TCP subunit TcpA. Importantly, the three proteins share close to 100% sequence identity in the first 30 residues. This N-terminal segment acts as the inner membrane anchor for the pilin subunits prior to pilus assembly and as a polymerization domain for the assembled pili. Sequence conservation in this critical dual-function segment suggests a common assembly mechanism and filament architecture.
The type IVb pili are a subgroup of the type IV pilus family. Type IV pilins possess an N-methylated N-terminal residue, a conserved hydrophobic ∼25-residue N terminus with an almost-invariant glutamate at residue 5, a cysteine pair in the C terminus, and common components for pilus biogenesis. The type IVa and IVb pilins are distinguished based on the length and sequence of their signal peptide, the identity of the N-terminal residue, and the length of the mature protein (9, 17). Interestingly, type IVb pili are found almost exclusively on enteric bacterial pathogens, whereas type IVa pili are found on human pathogens with diverse tissue and organ specificities, including Pseudomonas aeruginosa, Haemophilus influenzae, and pathogenic Neisseria spp. Structural studies on three full-length type IV pilins (10, 11, 21, 40) and several recombinant pilins lacking the N-terminal polymerization domain (3, 10, 22, 31, 37, 45, 58) have revealed a common architecture: an extended 53-residue α-helix (α1), the C-terminal half of which is embedded in a globular domain with a central antiparallel β-sheet. Yet, the type IVa and IVb pilins have distinct protein folds. In the type IVa pilins, the β-sheet has continuous nearest-neighbor connectivity and is followed by a C-terminal loop that lies at the periphery of the globular domain, whereas the C terminus of the type IVb pilins is buried in the globular domain as the central strand of the β-sheet. The significance of the different protein folds is not known and may be a consequence of evolution within a common niche or of adaptation to a particular environment.
Pseudo-atomic resolution structures of two pilus filaments, the type IVa GC pilus from Neisseria gonorrhoeae and the type IVb TCP from V. cholerae, have been determined by transmission electron microscopy (EM) and image reconstruction followed by fitting of pilin crystal structures into the EM density envelope (11, 29). In both GC pili and TCP, pilin subunits are held together primarily by hydrophobic interactions among the N-terminal α-helices, which form a helical array in the core of the filament. The pilin globular domains are loosely packed on the filament surface, anchored by the N-terminal α-helices. Thus, the most conserved region of the protein, the N-terminal α-helix, mediates filament formation, whereas the more-variable globular domain is exposed and defines the diverse interactions and functions of the pili. The helical symmetries of the filaments are similar for GC pili and TCP, suggesting that the assembly mechanism is conserved between the two type IV pilus subtypes.
The type IV pilus biogenesis process is not well understood. The type IVa pilus assembly requires as many as 40 gene products encoded in different parts of the genome, whereas the type IVb system comprises less than a dozen proteins, all encoded within the pilus operon (50). Both systems employ a dedicated prepilin peptidase that removes the signal sequence from the pilin subunit, an assembly ATPase, a conserved polytopic inner membrane assembly protein, additional nonconserved bitopic inner membrane proteins, and an outer membrane secretin, through which the pilus grows for display on the bacterial surface (4, 8, 41). The type IVa pilus machinery is relatively promiscuous in terms of the filaments it can assemble. P. aeruginosa is capable of assembling type IVa pili from Dichelobacter nodosus and Moraxella bovis (5, 33), and N. gonorrhoeae can assemble type IVa pili from P. aeruginosa (57), Francisella tularensis (47), and even from the Gram-positive Clostridium perfringens (46). In addition, P. aeruginosa from one strain can assemble distinct type IV pili from related strains (2, 56). In contrast, the type IVb pilus machinery may be more selective, as EPEC was not able to assemble the V. cholerae TCP (35). In that study, the entire bundlin gene encoding the signal peptide and the mature pilin was replaced with the entire tcpA gene, resulting in incomplete processing of TcpA by the prepilin peptidase BfpP. While the presence of unprocessed TcpA may have impaired TCP assembly in EPEC, a significant amount of processed TcpA was detected yet no pili were observed, suggesting further incompatibilities between the BFP assembly apparatus and the V. cholerae pilin. These heterologous expression systems can contribute to understanding the pilus assembly mechanism and may prove useful for generating pilus-based vaccines.
To further characterize the ETEC type IVb pili, we examined the role of CFA/III in ETEC aggregation. We further determined a 1.26-Å crystal structure of N-terminally truncated CofA and derived a computational model for the CFA/III pilus by using the V. cholerae TCP structure as a template. Our analysis revealed strong functional and structural similarities between CFA/III and TCP, yet V. cholerae is unable to synthesize CFA/III filaments when CofA is expressed in place of TcpA. The implications for these findings on pilus assembly and functions are discussed.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains, plasmids, and primers are listed in Table 1. V. cholerae strains O395 and SJK70 and ETEC strains 31-10 and 31-10p were provided by Ronald Taylor (Dartmouth Medical School). E. coli strains were grown in Luria-Bertani (LB) broth at 37°C. V. cholerae strains were grown under TCP-expressing conditions in LB broth (starting pH, 6.5) for 16 h at 30°C (26). ETEC strains were grown overnight in colonization factor antigen (CFA) agar (1% Casamino Acids [Difco], 0.15% yeast extract, 0.005% MgSO4, and 0.0005% MnCl2 with 2% agar added [16]) at 37°C. Antibiotics were used at final concentrations of 100 μg/ml streptomycin, 45 μg/ml kanamycin, 30 μg/ml gentamicin, and 100 μg/ml ampicillin.
Table 1.
Bacterial strains, plasmids, and primers
| Reagent | Description or sequence | Source or reference |
|---|---|---|
| Bacterial strains | ||
| V. cholerae O395 | O1 classical Ogawa, wild type, Smr | 53 |
| E. coli S17 λpir | thi pro recAhsdR17(rK− mK+)[RP4-2-Tc::Mu::Kmr Tn7] λpir; Tpr Smr | 13 |
| V. cholerae SJK70 | O395; ΔtcpA; Smr | R. K. Taylor |
| V. cholerae SI003 | SJK70, pMin1 | This study |
| V. cholerae JR003 | O395, ΔtcpAcofA | This study |
| ETEC 31-10 | Wild-type ETEC, carries 55-kb virulence plasmid with cof operon encoding CFA/III | 24 |
| ETEC 31-10p | ETEC 31-10, spontaneous CFA/III-deficient strain lacking 55-kb virulence plasmid | 51 |
| E. coli Origami(DE3) | Δ(ara-leu)7697 ΔlacX74 ΔphoAPvuII phoR araD139 ahpC galE galK rpsL F′[lac+lacIqpro] (DE3) gor522::Tn10trxB; Kmr Smr Tcr | Novagen |
| Plasmids | ||
| pTK1 | pKAS32::tcpACl-rplL; Apr | 26 |
| pTK1-ΔBbsI | pTK1, BbsI site deleted | This study |
| pJR2 | pTK1::cofA within tcpACl genomic flanking sequences | This study |
| pMin1 | pACYC184; Gmr Cmr | 38 |
| pMT5 | toxT; Apr | 14 |
| Primers | ||
| Ec-cofA-fpcr | GGAATTCCATATGGACTCACGTACTGTTTCTGAATTGGTC | This study |
| Ec-cofA-rpcr | CGGGATCCTTAACGGCTCGCCAAAGTAATAGAGTTA | This study |
| oOMP-fB2 | CATGGAAGACCGTTCAAATAAGTTTGTTTAACTTAATCTTAACG | This study |
| oOMP-rB1 | CATGGAAGACCGACCCTCTTGACCGGTTTTC | This study |
| oOMP-fA1 | CATGGAAGACCGGGGTATGAGCCTTCTGGAAGTCATCATC | This study |
| oOMP-rA2 | CATGGAAGACCGTGAATTAACGGCTCGCCAAAGTAATAG | This study |
| Vc-tcpA-f1.36 | GCAATGATCATCATGACAGATCTGC | This study |
| Vc-tcpA-flank-F2 | GTAAAATGGTGGAGTTACATAAATATGC | This study |
| Vc-tcpA-flank-R2 | GATACCACATTGAATTTAATAAGTTGC | This study |
Electron microscopy.
ETEC strains 31-10 and 31-10p were grown overnight on CFA agar plates. Cells were applied to glow-discharged carbon-coated copper grids (EM Sciences) by gently touching a single colony with the grid. Cells were stained with 1% phosphotungstic acid and imaged on a Hitachi 8000 STEM microscope at 5,000 to 6,000× magnification.
Autoaggregation assay.
ETEC strains 31-10 and 31-10p were grown overnight on CFA agar plates for induction of CFA/III pili. Cells were scraped from the plate and gently suspended in phosphate-buffered saline (PBS, pH 6.0 and 7.0) or Tris-buffered saline (TBS, pH 8.5) with various concentrations of NaCl (10, 100, or 200 mM) and left to settle for 30 to 60 min. Ten-microliter aliquots were placed on glass slides, covered with a glass coverslip, and sealed with nail enamel. Cell aggregation was screened using light microscopy, and phase-contrast microscopy images were collected using a Leica DMI 4000B microscope at 100× magnification.
Expression and purification of N-terminally truncated CofA.
The gene fragment encoding CofA residues 29 to 208 was PCR amplified from ETEC 31-10 genomic DNA using primers Ec-cofA-fpcr and Ec-cofA-rpcr and cloned into pET15b (Novagen) at the BamHI and NdeI sites to provide an N-terminal hexahistidine tag (His tag) for purification by metal affinity chromatography. N-terminally truncated CofA (ΔN-CofA) was expressed in Escherichia coli Origami(DE3) cells. Cells were grown to an optical density (at 600 nm) of 0.4 in LB broth containing ampicillin at 37°C. ΔN-CofA expression was induced with 0.4 mM isopropyl-β-d-1-thiogalactopyranoside, and cells were grown for a further 18 h at 19°C. Cells were harvested by centrifugation at 5,000 × g for 30 min. Cell pellets were flash-frozen in liquid nitrogen and stored at −80°C until required. Frozen cell pellets from 6-liter cultures were thawed and suspended in lysis buffer containing 50 mM NaH2PO4/Na2HPO4 (pH 7.4), 500 mM NaCl, and EDTA-free protease inhibitor cocktail (Roche). The suspension was incubated at room temperature with lysozyme for 1 h, and then cells were lysed by sonication. Cell debris was removed by centrifugation at 40,000 × g. The supernatant was loaded onto a HisTrap column (GE Healthcare) preequilibrated with buffer A (50 mM NaH2PO4/Na2HPO4 [pH 7.4], 50 mM imidazole, 500 mM NaCl). The column was washed with buffer A, and protein was eluted with buffer B (50 mM NaH2PO4/Na2HPO4 [pH 7.4], 500 mM NaCl, 50 to 300 mM imidazole in a linear gradient). Fractions containing the protein were pooled and dialyzed against buffer C (20 mM Tris-HCl [pH 7.4], 50 mM NaCl, 1 mM EDTA). The His tag was removed by thrombin cleavage, and ΔN-CofA was further purified on a Sephacryl S-100 HR size exclusion column (GE Healthcare) preequilibrated with buffer C. Fractions were analyzed by SDS-PAGE, and peak fractions were combined and concentrated to 15 mg/ml by using an Amicon stirred cell concentrator with a 3,500-Da molecular mass cutoff filter (Millipore). Protein was flash-frozen in liquid nitrogen and stored at −80°C.
Crystallization and X-ray data collection.
Crystals of ΔN-CofA were grown in hanging drops by vapor diffusion in 100 mM Tris-HCl (pH 8.5), 2.5 M ammonium sulfate, and 1.04 mM N,N-dimethyldodecylamine N-oxide, conditions that were optimized from those identified using the Hampton crystal screen (HR2-110) with Hampton detergent screen I as an additive. Crystals were flash-frozen in liquid nitrogen using 20% (vol/vol) glycerol in mother liquor as cryoprotectant. Diffraction data were collected on beamline X26C of the National Synchrotron Light Source (NSLS), Brookhaven National Laboratory, and processed using HKL2000 (28). The data collection and refinement are summarized in Table 2.
Table 2.
Crystallographic data collection and analysis
| Data collected | ΔN-CofA value(s) |
|---|---|
| Beamline | NSLS X26C |
| Space group | P1 |
| Cell a, b, c (Å) | 29.50, 34.15, 64.23 |
| Cell α, β, γ | 95.7, 97.4, 94.0 |
| Resolution range (Å) | 50–1.26 |
| Wavelength (Å) | 0.82 |
| Completeness (%)a | 96.01/87.56 |
| Wilson B-value (Å2) | 10.9 |
| No. of observed reflections | 263,810 |
| No. of unique reflections | 66,018 |
| Rsym (%)a,b | 6.7/50.1 |
| I/σa | 33.2/3.7 |
| Redundancy | 4.2/3.5 |
| Mosaicity | 0.3 |
| Refinement statistics | |
| Resolution limits (Å) | 25–1.26 |
| Molecules/unit cell | 2 |
| Rcryst (%)c | 16.0 |
| Rfree (%)d | 19.4 |
| Avg B-factor (Å2) | 14.4 |
| No. of protein atoms | 2625 |
| No. of water molecules | 510 |
| RMSD bond angles | 0.009 |
| RMSD bond lengths | 1.336 |
| Ramachandran plot | |
| Most favored (%) | 90.6 |
| Allowed (%) | 9.4 |
Overall/last shell (resolution range for last shell is 1.31–1.26).
Rsym is the unweighted R value on I between symmetry mates.
Rcryst = Σhkl||Fobs(hkl)| − |Fcalc(hkl)||/Σhkl|Fobs(hkl)|.
Rfree is the cross validation R factor for 5% of reflections against which the model was not refined.
Structure determination and refinement.
The ΔN-CofA structure was solved by molecular replacement using the N-terminally truncated V. cholerae TcpA crystal structure (PDB ID 1OQV) as the search model. Only the core β-sheet and α-helices of TcpA were used for the model. All nonidentical residues between the CofA and TcpA globular domains were changed to Ser, Ala, or Gly using COOT (15). PHASER (49) yielded two distinct solutions with relatively high Z-scores (8.61 and 7.10). Several cycles of simulated annealing refinement (Cartesian annealing) were performed using CNS (6), which provided an interpretable electron density map with two molecules in the unit cell. Further refinement using REFMAC5 (36) significantly improved the quality of the map, revealing electron density for most of the missing side chains and also the main chain of missing loops. The map was further improved by iterative rounds of fitting and refinement using COOT and REFMAC5 (36). The ARP/wARP program (42) was used to locate water oxygen atoms, which were confirmed by visual inspection of the electron density map and of a composite annealed omit map, calculated using CNS. Final cycles of restrained individual anisotropic refinement brought the Rwork and Rfree values to 0.160 and 0.194, respectively. The model was validated with PROCHECK, SFCHECK, and Molprobity (12, 27, 55) (Table 2).
Construction of the CFA/III filament model.
The CFA/III pilus filament model was generated using the V. cholerae TCP structure, obtained by fitting of the TcpA crystal structure into our EM reconstruction volume (29). First, the N-terminal 28 residues that are missing in the CofA crystal structure were modeled using the coordinates for the corresponding residues in the full-length PAK pilin crystal structure, as was done for TcpA. The full-length CofA structure was then superimposed onto a TcpA subunit in the TCP model using the model-building program in COOT (15). The symmetry operators determined for the TCP reconstruction (8.4-Å axial rise, 96.8° azimuthal rotation) were applied to the subunit to generate a CFA/III filament model using COOT.
Generation of V. cholerae cofA strain.
Seamless mutagenesis with the type IIS restriction enzyme BbsI (32) was used to replace the V. cholerae O395 tcpA gene fragment encoding the mature TcpA protein with that encoding mature CofA in the suicide vector pTK1, a derivative of pKAS32, which carries the tcpA gene and its genomic flanking regions (48). First, pTK1 was mutagenized to delete a single BbsI site using QuikChange (Stratagene). Next, the resultant vector, pTK1-ΔBbsI, was linearized by inverse PCR amplification using primers oOMP-fB2 and oOMP-rB1 (Table 1). This step added BbsI digestion sites to each end of the PCR product (PCR fragment A) and eliminated the sequence encoding the mature TcpA protein, leaving the tcpA gene-flanking regions, including the segment encoding the TcpA signal peptide. The 626-bp cofA gene encoding the mature pilin protein was PCR amplified from ETEC 31-10 genomic DNA using primers oOMP-fA1 and oOMP-rA2, both of which carry BbsI sites (PCR fragment B). Fragments A and B were digested with BbsI and ligated together to produce pJR2, which was transformed into E. coli S17λpir. Following plasmid amplification and purification using a Qiagen spin miniprep kit, the cofA gene sequence was verified by DNA sequencing using primer Vc.tcpA.f1.36 (University of British Columbia NAPS Unit). The E. coli S17-pJR2 strain was then mated with V. cholerae ΔtcpA strain SJK70 by allelic exchange as described previously (48) to replace the tcpA gene with cofA. The correct location and sequence of the cofA gene in the new strain, V. cholerae JR003, were verified by DNA sequencing in both directions using primers TcpA-flank-F2 and TcpA-flank-R2.
Analysis of pilin and pilus expression in the V. cholerae cofA strain.
Expression of CofA was assessed by growing V. cholerae JR003 under pilus-inducing conditions (LB, pH 6.5, 30°C, 16 h on a Ferris wheel rotator), analyzing 20 μl of cell culture by SDS-PAGE, and immunoblotting with rabbit anti-CofA antibodies. For comparison, ETEC 31-10 cells expressing CofA and ETEC-31-10p, which lack the cof operon, were grown in CFA broth at 37°C shaking for 16 h. Pilus assembly was assessed by physically shearing the pili from the cells by using an Ultra-Turrax T8.01 disperser, filtering the cell homogenate, and analyzing it by SDS-PAGE and immunoblotting. DnaK was included as a loading control and blotted with anti-DnaK antibodies (Enzo Life Sciences). Pilus assembly was also assessed by EM imaging of a negatively stained V. cholerae JR003 cell culture. Five-microliter aliquots of overnight culture were applied to carbon-coated copper grids (Cedarlane Labs), excess fluid was wicked away after 2 min, and the grids were stained with 1% phosphotungstic acid, pH 7.0. Cells were imaged using an FEI F20 Supertwin electron microscope.
Production of anti-CofA antibodies.
The ΔN-CofA protein used for crystallization was used as an antigen for polyclonal rabbit anti-CofA antibody production (Pacific Immunology).
Figure preparation.
Molecular structure figures were prepared using MacPyMol (http://www.pymol.org).
Protein structure accession number.
The atomic coordinates for ΔN-CofA have been deposited in the Protein Data Bank under accession number 3S0T.
RESULTS
ETEC autoaggregation correlates with CFA/III expression.
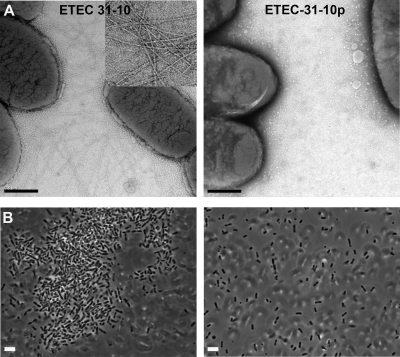
To establish a role for CFA/III in ETEC autoaggregation, ETEC strains were grown overnight on CFA agar, which induces CFA/III expression (16). ETEC strain 31-10 cells, which carry the 55-kb virulence plasmid containing the cof operon, are highly piliated under these conditions, as shown by transmission electron microscopy (TEM) (Fig. 1A, left). In contrast, ETEC strain 31-10p cells, which lack this plasmid, displayed no pili on their surfaces when grown in the same medium (Fig. 1A, right). ETEC 31-10 cells grown under various other conditions, including CFA broth, LB agar, and LB broth, displayed very few pili (data not shown). Cells from CFA plates were suspended in PBS (pH 6.0 or 7.0) or TBS (pH 8.5) with various concentrations of NaCl (10 mM, 100 mM, or 200 mM) and allowed to settle, and then aggregation was observed using light microscopy or phase-contrast microscopy. ETEC 31-10 cells aggregated into large clusters under all buffer conditions tested (Fig. 1B, left). In contrast, no aggregation was observed for ETEC 31-10p grown on CFA plates (Fig. 1B, right) or for strain 31-10 grown in liquid culture. Thus, ETEC autoaggregation correlates well with CFA/III display.
Fig 1.
ETEC pilus expression correlates with bacterial aggregation. (A) TEM images of ETEC strains 31-10 and 31-10p after overnight growth on CFA agar plates. Inset: close-up of pili on 31-10 cells. Scale bar, 0.5 μm. (B) Phase-contrast images of ETEC 31-10 and 31-10p grown on CFA agar, suspended in PBS (pH 7.0, 100 mM NaCl), and allowed to settle for 60 min.
Crystal structure of ETEC CofA.
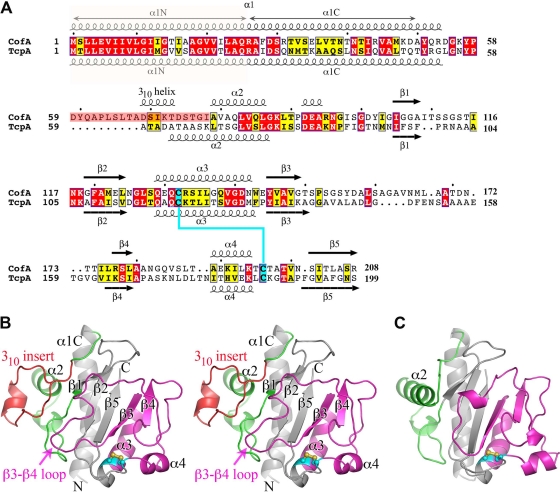
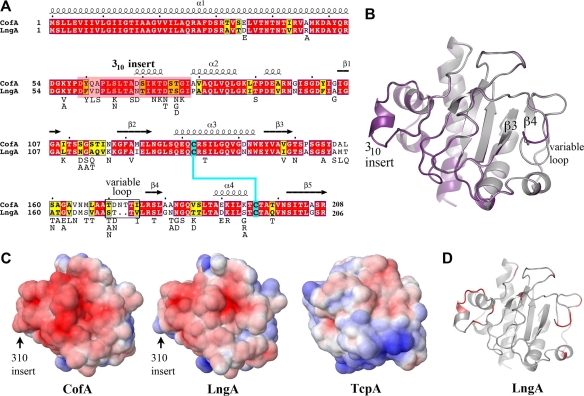
To characterize the CFA/III pili, we determined the crystal structure of N-terminally truncated CofA (ΔN-CofA, residues 29 to 208) to 1.26-Å resolution by molecular replacement with V. cholerae ΔN-TcpA O1, classical biotype (PDB ID 1OQV) (Table 2). Although CofA and TcpA share 37% amino acid sequence identity overall, they are only 30% identical for the globular domains that were crystallized (Fig. 2A). Thus, a number of modified ΔN-TcpA models were generated and used as search models to obtain a molecular replacement solution for ΔN-CofA. ΔN-CofA crystallized in the P1 space group with two molecules in the unit cell. All residues were readily fit into the electron density map for molecule A, whereas residues 113 to 115 are disordered in molecule B. As the two molecules are highly similar (root mean square deviation [RMSD], 0.39 Å for main chain atoms), only molecule A is shown. CofA has the characteristic fold of the type IVb pilins with an N-terminal α-helix, α1C (residues 29 to 53), and a twisted 5-stranded antiparallel β-sheet with non-nearest-neighbor connectivity, where the C-terminal 8-residue segment forms the central strand (β5) (Fig. 2B). Between α1C and the β-sheet is the αβ-loop (residues 54 to 104), which begins with a broad loop (residues 54 to 68), followed by a 1.5-turn 310-helix (residues 69 to 73), a 2-turn α-helix (α2, residues 79 to 86), an extended irregular loop (87 to 105) containing a single-turn 310-helix (residues 92 to 94), and then a short β-hairpin-like loop with a type II turn. The αβ-loop is followed by the first two strands of the central β-sheet, β1 and β2, between which lies a large loop (residues 109 to 119). Following β2 the polypeptide chain exits the β-sheet to form a 4-turn α-helix (α3, residues 129 to 142) that is distorted at its C-terminal end by two glycines, Gly137 and Gly140. α3 runs almost parallel to α1C and interacts with α1C and the β-sheet via hydrophobic side chains. After α3 the chain reverses direction to form β3. The 27-residue-long loop that follows β3 (the β3-β4 loop) winds around the “front” surface of the globular domain and buries much of the β-sheet. This loop feeds into the edge strand of the β-sheet (β4, residues 175 to 177), after which the polypeptide chain reverses direction via a type II turn, followed by an extended stretch along the edge of the globular domain. The chain turns 90° to form a 1.5-turn α-helix, α4 (residues 189 to 194). Finally, an irregular loop connects α4 with β5, the central strand of the β-sheet (residues 201 to 208). The C-terminal residue, Arg208, forms a salt bridge with Asp144 at the N terminus of β3. Conserved cysteines at positions 132 and 196 form a disulfide bond that links the N terminus of α3 with the loop that follows α4. The D-region, defined as the 64-residue section between the two cysteines, and the αβ-loop flank the β-sheet and together comprise a large portion of the globular domain. These two regions display considerable sequence and structural variability among the type IV pilins and house residues critical to pilus functions (8).
Fig 2.
Comparison of amino acid sequences and atomic structures of ETEC CofA and V. cholerae TcpA. (A) Amino acid sequence alignment of ETEC CofA (strain 31-10; matches GenBank accession number BAA07174) and V. cholerae TcpA (classical strain O395; GenBank accession number ABQ19609), based on alignment of the two crystal structures. Only the mature regions of the pilin proteins are shown. The secondary structure is indicated above and below the sequences. The α1N α-helix was predicted based on amino acid sequence homology with the corresponding regions in the full-length pilin crystal structures (PDB ID 1OQW, 2HI2). Identical residues are shown in white text with red shading; homologous residues are shown in black text with yellow shading. The 310 insert is shaded pink in CofA. Disulfide-bonded cysteines are shaded cyan. (B) Crystal structure of ΔN-CofA (residues 28 to 208) at 1.26-Å resolution, shown in stereo view with the αβ-loop in green and the D-region in magenta. The 310 insert within the αβ-loop is shown in red. Side chains for cysteines Cys132 and Cys196 are shown in a ball-and-stick representation. (C) ΔN-TcpA crystal structure (PDB ID 1OQV), shown for comparison with ΔN-CofA, colored as described for panel B. Side chains for cysteines Cys120 and Cys186 are shown in a ball-and-stick representation.
The N-terminal 28 residues of CofA (α1N) are not present in the ΔN-CofA structure but are predicted based on sequence homology to form the first half of an extended α-helix, α1, as seen in the full-length P. aeruginosa PAK pilin and N. gonorrhoeae GC pilin structures (10, 11). In the PAK and GC pilins, α1 is a continuous 53-residue α-helix with a curved S-shape. The C-terminal half of α1, α1C, is embedded in the globular domain, and the N-terminal half, α1N, protrudes like a stalk. The curvature of α1 in PAK and GC pilins is induced by three helix-disrupting residues, Gly14, Pro22, and Pro/Gly42. CofA also has a glycine at position 14, but it has isoleucine and threonine at positions 22 and 42, respectively. However, CofA has 3 glycine residues within α1N (Gly11, Gly14, and Gly18), and α1C has a distinct curve despite the absence of a glycine at position 42. Thus, the CofA α1 is also expected to be curved. The helix curvature and flexibility in α1N due to the preponderance of glycines likely contribute to packing of this helix in the pilus filament.
Although the sequence similarities between CofA and TcpA are limited within the globular domain, the two proteins are very similar in structure (RMSD, 1.40 Å for all Cα) (Fig. 2A to C). The most notable difference between CofA and TcpA is a 10-amino-acid insertion in the αβ-loop of CofA that diverts the polypeptide chain between residues 59 and 78 relative to the TcpA structure. In TcpA, the loop emerging from α1 folds into the 4-turn α-helix, α2, which lies at a right angle to α1. In CofA, the polypeptide follows the same path as TcpA for residues 54 to 58, but this loop continues downward and outward relative to α1 and then loops back to form a 310-helix before entering a short α2 corresponding to the C-terminal half of α2 in TcpA. We refer to the region within the αβ-loop of CofA that diverges from TcpA as the 310 insert (residues 59 to 78). The two pilin structures also differ somewhat in the large irregular β3-β4 loop on the surface of the globular domain, which interacts with the 310 insert in CofA, and between β4 and the disulfide-bonded C196 (Fig. 2B and C). These regions, together with the 310 insert, are the least-well-conserved in sequence between the two proteins.
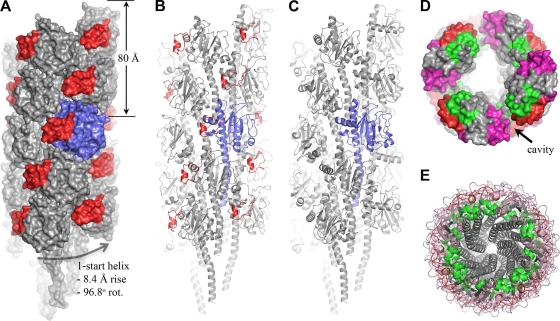
CFA/III filament model.
Because of the structural similarities between CofA and TcpA, we used the TCP EM reconstruction to derive a model for the CFA/III pilus. In the TCP structure, TcpA subunits are arranged along a right-handed one-start helix with an axial rise of 8.4 Å and an azimuthal rotation of 96.8° per subunit (29). A full-length CofA subunit was first generated by appending the N-terminal 28 residues of the full-length PAK pilin structure (PDB ID 1OQW; 39% sequence identity, 75% sequence similarity over these residues) onto ΔN-CofA and changing individual residues in silico to the corresponding CofA residues. Full-length CofA was then superimposed onto a single TcpA subunit within the TCP filament, and the CFA/III filament was generated by imposing the helical symmetry operators on this subunit (Fig. 3A). The CofA subunits pack well in the CFA/III model, with no major steric clashes, as expected due to their structural similarity to TcpA. Importantly, the 310 insert fits into the cavity that lies between the globular domains, narrowing this gap considerably (Fig. 3A to D). The CofA αβ-loops are oriented inward and contact the N-terminal α-helix of their neighboring subunits, whereas the D-regions lie on the filament surface (Fig. 3D). The D-regions do not protrude from the filament surface to the extent that they do in TCP, and this feature, together with the 310 insert that partially fills the gaps between subunits, produces a smoother filament surface for CFA/III than for TCP. Because the N-terminal α-helix is not present in the CofA and TcpA crystal structures and this region is not well-resolved in the TCP EM reconstructions (29), its position in CFA/III is not known with precision. Nonetheless, this segment, as modeled using the corresponding PAK pilin coordinates, packs exceptionally well into the filament core, almost filling it completely without introducing major steric clashes (Fig. 3E).
Fig 3.
CFA/III filament model and comparison with TCP structure. (A and B) CFA/III filament model shown in space-filling (A) and cartoon (B) representations. A single subunit is colored blue, and the 310 insert is shown in red for all subunits. The α1N segment of each subunit was modeled using the X-ray coordinates for the corresponding region in PAK pilin. (C) Pseudo-atomic resolution TCP structure derived from fitting of the TcpA subunit into the EM density map (29), shown for comparison with the CFA/III filament model. (D) End view of CFA/III in space-filling representation to demonstrate subunit packing and positioning of the 310 insert in the depression between subunits. An 80-Å slab is shown, as described for panel A, to better illustrate the cavity depth. The core appears hollow because the entire α1 is not shown for all subunits. (E) End view of the entire 18-subunit CFA/III model, showing the putative arrangement of N-terminal α-helices in the core of the filament.
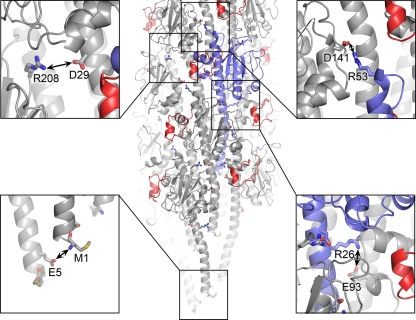
The largest subunit-subunit interface occurs among the N-terminal α-helices in the hydrophobic core of the filament, but there are also a number of complementary charged residues on the globular domains that are in close proximity in neighboring subunits and are likely to form salt bridges. We had previously identified TcpA residues Arg26 in α1N and Glu83 in the αβ-loop as being important for filament assembly (30). These residues are located within salt bridging distance in the TCP model in neighboring subunits in the left-handed three-start helix. In the CFA/III model, Arg26 NH1 is within 5.2 Å of Glu93 Oε1, which corresponds to Glu83 in TcpA (Fig. 4). Small adjustments in dihedral angles of the side chains of these residues would allow formation of a stabilizing salt bridge in CFA/III. Salt bridges may also be present between charged residues within the CofA globular domains: Asp29 and Arg208 in the right-handed 4-start helix and Arg53 and Asp141 in neighboring subunits in the three-start helix (Fig. 4). In addition, the invariant Glu5 is located at the same axial position and within salt-bridging distance of the positively charged N-terminal amine in the adjacent subunit in the 1-start helix. These complementary charges occur at the same level in the filament due to the helical symmetry, with each subunit staggered along the 1-start helix by 8.4 Å. A salt bridge between these two residues would neutralize their charges in the otherwise-hydrophobic environment of the filament core (Fig. 4) and may provide a driving force for filament assembly (11).
Fig 4.
Complementary electrostatic interactions between subunits in the CFA/III model. The filament is colored as described for Fig. 3A. Oxygen atoms are shown in red and nitrogens are blue in the insets.
Computational model of LngA.
We used the ΔN-CofA crystal structure to derive a homology model for ΔN-LngA, the major pilin subunit of the second ETEC type IVb pilus, Longus (Fig. 5). Due to their high sequence identity (76%) LngA is predicted to be very close in structure to CofA and to possess the 310 insert within the αβ-loop. LngA has two fewer amino acids than CofA, which results in a shorter loop immediately preceding β4 (Fig. 5A and B, variable loop). Based on the amino acid similarities between CofA and LngA, the Longus filament is also expected to closely resemble that of CFA/III. Residues predicted to be involved in subunit-subunit interactions in CFA/III are well-conserved between the two pilins. Amino acid differences occur mainly on the solvent-exposed surface within the D-region. Nonetheless, both the shape and the chemistry of this exposed face are similar for CofA and LngA, and both have an overall negative charge (Fig. 5C). Amino acid differences between the ETEC pilins and V. cholerae TcpA also occur mainly on the solvent-exposed surface. These differences, including those in the αβ-loop that form the 310 insert in the ETEC pilins, result in different surface topographies and chemistries for TcpA and the ETEC pilins (Fig. 5C and D) and their corresponding pilus surfaces.
Fig 5.
Comparison of the CofA and LngA amino acid sequences and structures. (A) Amino acid sequence alignment of CofA and LngA (ETEC strain E9034A; GenBank accession number AAC33154). CofA and LngA share 76% amino acid sequence identity and 87% similarity. Sequences were aligned manually based on superposition of the LngA homology model with CofA. The color scheme is as described for Fig. 1A. The secondary structure of CofA is indicated above the sequence. LngA sequences were compared further, and variant residues are listed below the LngA sequence. (B) Superposition of the ΔN-LngA homology model (purple) on the ΔN-CofA crystal structure (gray), showing the divergent “variable loop” that precedes strand β4. (C) Electrostatic surfaces of CofA, LngA, and TcpA globular domains, showing the pilin face predicted to be exposed on the pilus filament. The orientation of each structure is similar to that shown in panel B. Proteins are colored according to electrostatic potential, calculated using DELPHI (39), with red representing negative charge, blue is positive, and white is uncharged (scale, −7 kT to +7 kT). (D) LngA homology model, showing that variable residues (red) are localized to the exposed LngA face.
Heterologous expression of CofA in V. cholerae.
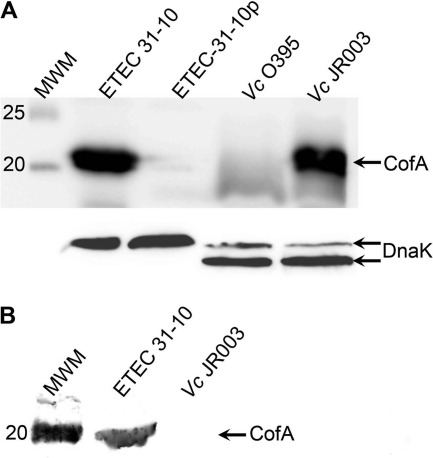
Because of the structural similarity between CofA and TcpA and the sequence conservation in the interfaces between pilin subunits in the pilus filament models, we hypothesized that the V. cholerae TCP biogenesis apparatus would assemble CFA/III pili if provided with the CofA subunit. The V. cholerae tcpA gene within the tcp operon was replaced with the cofA gene by allelic exchange, leaving the segment encoding the TcpA signal peptide in place to allow for processing by the V. cholerae prepilin peptidase TcpJ. CofA expression in this V. cholerae strain, JR003, was analyzed by SDS-PAGE and immunoblotting, and CFA/III assembly was assessed by EM and by shearing the pili off the cell surfaces. CofA is expressed in V. cholerae JR003 and processed to its mature form when grown under pilus-inducing conditions (Fig. 6A). However, the CofA band in sheared cell supernatants was almost undetectable, whereas a strong band was observed for ETEC 31-10 (Fig. 6B). No pilus filaments were observed by EM for JR003, whether grown under TCP-inducing conditions (LB, pH 6.5, 30°C) or CFA/III-inducing conditions (CFA agar, 37°C) (data not shown). These results demonstrate that CofA is synthesized and processed by V. cholerae, but the TCP assembly apparatus cannot efficiently incorporate CofA into pilus filaments.
Fig 6.
Heterologous expression of CofA in Vibrio cholerae. (A) Immunoblot showing CofA in whole-cell culture of wild-type ETEC 31-10 and V. cholerae (Vc) JR003, in which the gene segment encoding the mature TcpA was replaced with the corresponding cofA gene fragment within the tcp operon. CofA from both strains ran at the expected molecular mass of the processed pilin (calculated molecular mass, 21.6 kDa) rather than the unprocessed prepilin (calculated molecular mass, 25.3 kDa). Negative controls: ETEC 31-10p, which lacks the cof operon; V. cholerae O395, the parental strain for JR003. An immunoblot for DnaK (shown below) served as a loading control. (B) Immunoblot analysis of whole-cell supernatants, indicating the presence of intact CFA/III filaments in ETEC 31-10 but not V. cholerae JR003.
DISCUSSION
We have provided here evidence that CFA/III pili mediate ETEC aggregation. ETEC cells aggregate when the 55-kb virulence plasmid housing the cof operon is present and CFA/III pili are expressed, whereas no aggregation is observed in an ETEC strain lacking the virulence plasmid or when cells are grown under non-pilus-expressing conditions. The correlation between pilus expression and ETEC aggregation suggests that it is CFA/III self-association and not the work of another protein whose gene is carried on the virulence plasmid that is responsible for aggregation, although we cannot yet rule out the latter possibility. These observations parallel those of TCP, which mediate V. cholerae aggregation, referred to as autoagglutination, as it can be visualized by eye after overnight growth in liquid culture. Our results suggest that CFA/III, like TCP, may mediate microcolony formation to facilitate colonization of the human gut.
The ETEC ΔN-CofA structure is very similar to ΔN-TcpA from V. cholerae. CofA has the characteristic type IVb pilin fold first identified in TcpA (10) but differs from TcpA in the αβ-loop, where the first half of α2 is replaced with an irregular loop containing a 310-helix. LngA is predicted to share the CofA structure, including the 310 insert, based on their sequence similarities. CofA and TcpA are more similar to each other in amino acid sequence and structure than to other type IVb pilins of known structure: the EPEC bundlin (45) and S. Typhi PilS (58), which share the type IVb pilin fold but have markedly different αβ-loop and D-regions.
As expected from its structural similarity to TcpA, CofA fits well into the TCP pseudo-atomic resolution structure, with its 310 insert partially filling the gap between the pilin globular domains on the filament surface. Residues involved in subunit-subunit interactions are well-conserved among CofA, LngA, and TcpA, whereas amino acid differences between the ETEC type IV pilins and V. cholerae TcpA are predicted to give these filaments distinctly different surfaces with respect to their chemistries and topographies. The presence of the 310 insert in CofA, together with its less-pronounced D-region, produces a smoother and less undulating filament surface for CFA/III than observed for TCP. We proposed previously that TCP self-association occurs via intercalation of protruding TcpA D-regions into cavities between subunits on adjacent TCP proteins to mediate V. cholerae autoagglutination (31). Such intercalation appears unlikely for CFA/III and Longus, but surface complementarity between adjacent filaments may be sufficient to drive ETEC aggregation. Although the CFA/III surface is predicted to have a net negative charge, we observed ETEC aggregation at and near pH 7.
Longus pili are more prevalent than CFA/III in ETEC patient isolates. A PCR-based study of 56 ETEC isolates that employed cofA- and lngA-specific primers identified 38 lngA-positive strains and only two cofA-positive strains (20). We compared published nonredundant LngA amino acid sequences and mapped the nonconserved residues to the LngA homology model (Fig. 5A and D; see also Fig. S1 in the supplemental material). Amino acid differences among LngA variants, like those of LngA, CofA, and TcpA, are localized to the exposed surface of the pilin (Fig. 5D). This may simply reflect the need to conserve residues involved in protein structure and pilus assembly, with the exposed residues being nonessential. However, since the exposed residues define the pilus filament surface, variation of these residues may profoundly alter pilus interactions with each other and with molecules in the environment, such as host receptors and molecules of the immune system. We showed previously that single amino acid substitutions on the V. cholerae TcpA surface can completely disrupt TCP-mediated autoagglutination without any apparent effects on pilin structure or pilus assembly (31). Amino acid differences among LngA variants may allow different ETEC strains to recognize their own kind through pilus-pilus interactions. It is notable that variation among LngA sequences is comparable to that between LngA and CofA (Fig. 5A). For example, ETEC E9034A LngA is 81% identical to ECOR27 LngA and is 76% identical to strain 31-10 CofA (see Fig. S1). The other proteins encoded on these pilus operons, most of which are involved in pilus assembly, are more highly conserved, both within Longus-expressing strains and between Longus and CFA/III systems (20). Thus, the cof operon can be considered a variant of the lng operon that is transmitted on its own plasmid, an idea supported by phylogenetic analyses of available cofA and lngA sequences (20).
In spite of the structural similarities between CofA and TcpA, as well as the amino acid conservation among residues involved in subunit-subunit interactions within the intact pilus, V. cholerae is not able to assemble CFA/III pili. This result suggests that one or more components of the TCP biogenesis apparatus are unable to recognize CofA. This outcome is somewhat surprising given that a number of type IVa pili have been expressed and assembled in heterologous type IVa systems (2, 5, 33, 46, 47, 56, 57), although in some cases a retraction-deficient background was required. In all of these type IVa pilus expression systems, the heterologous pilins are much less similar to the host pilins than CofA is to TcpA, and the type IVa pilus assembly apparatus is more complex than that of type IVb (4). The inability of V. cholerae to express CFA/III pili is, however, consistent with earlier work showing that EPEC is unable to assemble V. cholerae TCP when the bfpA gene is replaced with the tcpA gene in the bfp operon (35). In this earlier study, TCP assembly likely failed in part because the TcpA signal peptide was not recognized by the bundlin prepilin peptidase. We avoided this pitfall here by inserting only the region of the cofA gene encoding the mature pilin, allowing the prepilin peptidase TcpJ to recognize and remove the TcpA signal peptide. Furthermore, TcpA is more closely related to CofA than to BfpA in terms of sequence and structure. Nonetheless, we were unable to detect any pili in our CofA-expressing V. cholerae strain grown under either TCP- or CFA/III-inducing conditions. CofA and TcpA are highly conserved in their N-terminal α1N segment (Fig. 1A), which serves as a membrane anchor for the pilin subunit prior to its incorporation into pilus filaments. This suggests that the TCP assembly apparatus failed to recognize the less-conserved CofA globular domain. The inner membrane proteins required for TCP assembly, TcpE, TcpR, and TcpD (53), are not highly similar to the corresponding proteins, CofI, CofE, and CofF (37%, 0%, and 23%, respectively), which may explain their inability to recognize CofA. Certainly the 310 insert of CofA is a distinguishing feature of CofA, and it may not allow interaction with TCP assembly components, but even single amino acid differences in α1 or elsewhere could potentially disrupt this specificity. The type IVb pilus assembly apparatus appears to be more specific than that of the type IVa pili, despite it being a simpler system comprising fewer components. These heterologous expression systems may provide valuable tools to dissect the pairwise interactions of the pilus assembly apparatus.
In summary, our results reveal close structural similarities between the ETEC and V. cholerae type IVb pili with respect to their subunit folds and pilus architectures and also reveal unique features that may impart distinct functions to the ETEC pili. The architectures of the pilin subunits and the pilus filament maximize the variability among the pili to provide diverse functions within a conserved filament structure. Our results reinforce the previously observed specificity of the type IVb pilus biogenesis apparatus, which contrasts with the somewhat promiscuous type IVa pilus machinery. Our demonstration of CFA/III-mediated ETEC aggregation together with our new structure and models provides a system and a rational basis for investigating ETEC pilus interactions and type IV pilus assembly.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dixon Ng for electron microscopy images, Dennis Wong, Sumaiya Islam, and Juliana Li for technical assistance, and the beamline staff at the Brookhaven National Synchrotron Light Source for their assistance in data collection.
This work was supported by operating grants from the National Institutes of Health (grant AI061051) and the Canadian Institutes of Health Research (CIHR) and by salary awards to L.C. from CIHR and the Michael Smith Foundation for Health Research.
Footnotes
Published ahead of print 23 March 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Anantha RP, Stone KD, Donnenberg MS. 1998. Role of BfpF, a member of the PilT family of putative nucleotide-binding proteins, in type IV pilus biogenesis and in interactions between enteropathogenic Escherichia coli and host cells. Infect. Immun. 66:122–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Asikyan ML, Kus JV, Burrows LL. 2008. Novel proteins that modulate type IV pilus retraction dynamics in Pseudomonas aeruginosa. J. Bacteriol. 190:7022–7034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Audette GF, Irvin RT, Hazes B. 2004. Crystallographic analysis of the Pseudomonas aeruginosa strain K122-4 monomeric pilin reveals a conserved receptor-binding architecture. Biochemistry 43:11427–11435 [DOI] [PubMed] [Google Scholar]
- 4. Ayers M, Howell PL, Burrows LL. 2010. Architecture of the type II secretion and type IV pilus machineries. Future Microbiol. 5:1203–1218 [DOI] [PubMed] [Google Scholar]
- 5. Beard MK, et al. 1990. Morphogenetic expression of Moraxella bovis fimbriae (pili) in Pseudomonas aeruginosa. J. Bacteriol. 172:2601–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brunger AT, et al. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905–921 [DOI] [PubMed] [Google Scholar]
- 7. Clavijo AP, Bai J, Gomez-Duarte OG. 2010. The Longus type IV pilus of enterotoxigenic Escherichia coli (ETEC) mediates bacterial self-aggregation and protection from antimicrobial agents. Microb. Pathog. 48:230–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Craig L, Li J. 2008. Type IV pili: paradoxes in form and function. Curr. Opin. Struct. Biol. 18:267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Craig L, Pique ME, Tainer JA. 2004. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2:363–378 [DOI] [PubMed] [Google Scholar]
- 10. Craig L, et al. 2003. Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol. Cell 11:1139–1150 [DOI] [PubMed] [Google Scholar]
- 11. Craig L, et al. 2006. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol. Cell 23:651–662 [DOI] [PubMed] [Google Scholar]
- 12. Davis IW, et al. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35:W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. De Lorenzo V, Timmis K. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386–405 [DOI] [PubMed] [Google Scholar]
- 14. DiRita VJ, Neely M, Taylor RK, Bruss PM. 1996. Differential expression of the ToxR regulon in classical and E1 Tor biotypes of Vibrio cholerae is due to biotype-specific control over toxT expression. Proc. Natl. Acad. Sci. U. S. A. 93:7991–7995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Emsley P, Cowtan K. 2004. COOT: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 16. Evans DG, Evans DJ, Jr, Tjoa W. 1977. Hemagglutination of human group A erythrocytes by enterotoxigenic Escherichia coli isolated from adults with diarrhea: correlation with colonization factor. Infect. Immun. 18:330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Giron JA, Gomez-Duarte OG, Jarvis KG, Kaper JB. 1997. Longus pilus of enterotoxigenic Escherichia coli and its relatedness to other type-4 pili: a minireview. Gene 192:39–43 [DOI] [PubMed] [Google Scholar]
- 18. Giron JA, Ho AS, Schoolnik GK. 1991. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science 254:710–713 [DOI] [PubMed] [Google Scholar]
- 19. Giron JA, Levine MM, Kaper JB. 1994. Longus: a long pilus ultrastructure produced by human enterotoxigenic Escherichia coli. Mol. Microbiol. 12:71–82 [DOI] [PubMed] [Google Scholar]
- 20. Gomez-Duarte OG, et al. 2007. Genetic diversity of the gene cluster encoding Longus, a type IV pilus of enterotoxigenic Escherichia coli. J. Bacteriol. 189:9145–9149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hartung S, et al. 2011. Ultra-high resolution and full-length pilin structures with insights for filament assembly, pathogenic functions and vaccine potential. J. Biol. Chem. 286:44254–44265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hazes B, Sastry PA, Hayakawa K, Read RJ, Irvin RT. 2000. Crystal structure of Pseudomonas aeruginosa PAK pilin suggests a main-chain-dominated mode of receptor binding. J. Mol. Biol. 299:1005–1017 [DOI] [PubMed] [Google Scholar]
- 23. Honda T, Arita M, Miwatani T. 1984. Characterization of new hydrophobic pili of human enterotoxigenic Escherichia coli: a possible new colonization factor. Infect. Immun. 43:959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Honda T, Wetprasit N, Arita M, Miwatani T. 1989. Production and characterization of monoclonal antibodies to a pilus colonization factor (colonization factor antigen III) of human enterotoxigenic Escherichia coli. Infect. Immun. 57:3452–3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hyland RM, et al. 2008. The bundlin pilin protein of enteropathogenic Escherichia coli is an N-acetyllactosamine-specific lectin. Cell. Microbiol. 10:177–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kirn TJ, Lafferty MJ, Sandoe CM, Taylor RK. 2000. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol. Microbiol. 35:896–910 [DOI] [PubMed] [Google Scholar]
- 27. Laskowski RA, McArthur MW, Moss DS, Thornton JM. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291 [Google Scholar]
- 28. Leslie AGW, Brick P, Wonacott A. 1986. Daresbury Laboratory information quarterly for protein crystallography, vol 18, p 33–39 Daresbury Laboratory, Warrinton, United Kingdom [Google Scholar]
- 29. Li J, Egelman E, Craig L. 2012. Structure of the Vibrio cholerae type IVb pilus and stability comparison with the Neisseria gonorrhoeae type IVa pilus. J. Mol. Biol. 418:47–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li J, et al. 2008. Vibrio cholerae toxin-coregulated pilus structure analyzed by hydrogen/deuterium exchange mass spectrometry. Structure 16:137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lim MS, et al. 2010. Vibrio cholerae El Tor TcpA crystal structure and mechanism for pilus-mediated microcolony formation. Mol. Microbiol. 77:755–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lu Q. 2005. Seamless cloning and gene fusion. Trends Biotechnol. 23:199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mattick JS, et al. 1987. Morphogenetic expression of Bacteroides nodosus fimbriae in Pseudomonas aeruginosa. J. Bacteriol. 169:33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mazariego-Espinosa K, Cruz A, Ledesma MA, Ochoa SA, Xicohtencatl-Cortes J. 2010. Longus, a type IV pilus of enterotoxigenic Escherichia coli, is involved in adherence to intestinal epithelial cells. J. Bacteriol. 192:2791–2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McNamara BP, Donnenberg MS. 2000. Evidence for specificity in type 4 pilus biogenesis by enteropathogenic Escherichia coli. Microbiology 146:719–729 [DOI] [PubMed] [Google Scholar]
- 36. Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255 [DOI] [PubMed] [Google Scholar]
- 37. Nguyen Y, Jackson SG, Aidoo F, Junop M, Burrows LL. 2010. Structural characterization of novel Pseudomonas aeruginosa type IV pilins. J. Mol. Biol. 395:491–503 [DOI] [PubMed] [Google Scholar]
- 38. Nye MB, Pfau JD, Skorupski K, Taylor RK. 2000. Vibrio cholerae H-NS silences virulence gene expression at multiple steps in the ToxR regulatory cascade. J. Bacteriol. 182:4295–4303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oron A, Wolfson H, Gunasekaran K, Nussinov R. 2003. Using DelPhi to compute electrostatic potentials and assess their contribution to interactions. Curr. Protoc. Bioinformatics 8:Unit 8.4 [DOI] [PubMed] [Google Scholar]
- 40. Parge HE, et al. 1995. Structure of the fibre-forming protein pilin at 2.6 Å resolution. Nature 378:32–38 [DOI] [PubMed] [Google Scholar]
- 41. Pelicic V. 2008. Type IV pili: e pluribus unum? Mol. Microbiol. 68:827–837 [DOI] [PubMed] [Google Scholar]
- 42. Perrakis A, Morris R, Lamzin VS. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6:458–463 [DOI] [PubMed] [Google Scholar]
- 43. Pier GB, et al. 1998. Salmonella typhi uses CFTR to enter intestinal epithelial cells. Nature 393:79–82 [DOI] [PubMed] [Google Scholar]
- 44. Qadri F, et al. 2000. Human antibody response to Longus type IV pilus and study of its prevalence among enterotoxigenic Escherichia coli in Bangladesh by using monoclonal antibodies. J. Infect. Dis. 181:2071–2074 [DOI] [PubMed] [Google Scholar]
- 45. Ramboarina S, et al. 2005. Structure of the bundle-forming pilus from enteropathogenic Escherichia coli. J. Biol. Chem. 48:40252–40260 [DOI] [PubMed] [Google Scholar]
- 46. Rodgers K, Arvidson CG, Melville S. 2011. Expression of a Clostridium perfringens type IV pilin by Neisseria gonorrhoeae mediates adherence to muscle cells. Infect. Immun. 79:3096–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Salomonsson E, et al. 2009. Functional analyses of pilin-like proteins from Francisella tularensis: complementation of type IV pilus phenotypes in Neisseria gonorrhoeae. Microbiology 155:2546–2559 [DOI] [PubMed] [Google Scholar]
- 48. Skorupski K, Taylor RK. 1996. Positive selection vectors for allelic exchange. Gene 169:47–52 [DOI] [PubMed] [Google Scholar]
- 49. Storoni LC, McCoy AJ, Read RJ. 2004. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 60:432–438 [DOI] [PubMed] [Google Scholar]
- 50. Strom MS, Lory S. 1993. Structure-function and biogenesis of the type IV pili. Annu. Rev. Microbiol. 47:565–596 [DOI] [PubMed] [Google Scholar]
- 51. Taniguchi T, et al. 2001. Gene cluster for assembly of pilus colonization factor antigen III of enterotoxigenic Escherichia coli. Infect. Immun. 69:5864–5873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Taniguchi T, Fujino Y, Yamamoto K, Miwatani T, Honda T. 1995. Sequencing of the gene encoding the major pilin of pilus colonization factor antigen III (CFA/III) of human enterotoxigenic Escherichia coli and evidence that CFA/III is related to type IV pili. Infect. Immun. 63:724–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. U. S. A. 84:2833–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tsui IS, Yip CM, Hackett J, Morris C. 2003. The type IVB pili of Salmonella enterica serovar Typhi bind to the cystic fibrosis transmembrane conductance regulator. Infect. Immun. 71:6049–6050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vaguine AA, Richelle J, Wodak SJ. 1999. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr. D Biol. Crystallogr. 55:191–205 [DOI] [PubMed] [Google Scholar]
- 56. Watson AA, Mattick JS, Alm RA. 1996. Functional expression of heterologous type 4 fimbriae in Pseudomonas aeruginosa. Gene 175:143–150 [DOI] [PubMed] [Google Scholar]
- 57. Winther-Larsen HC, et al. 2007. Pseudomonas aeruginosa type IV pilus expression in Neisseria gonorrhoeae: effects of pilin subunit composition on function and organelle dynamics. J. Bacteriol. 189:6676–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xu XF, et al. 2004. NMR structure of a type IVb pilin from Salmonella typhi and its assembly into pilus. J. Biol. Chem. 279:31599–31605 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.