Abstract
N-linked glycosylation of protein is a posttranslational modification found in all three domains of life. The flagellin proteins of the archaeon Methanococcus maripaludis are known to be modified with an N-linked tetrasaccharide consisting of N-acetylgalactosamine (GalNAc), a diacetylated glucuronic acid (GlcNAc3NAc), an acetylated and acetamidino-modified mannuronic acid with a substituted threonine group (ManNAc3NAmA6Thr), and a novel terminal sugar residue [(5S)-2-acetamido-2,4-dideoxy-5-O-methyl-α-l-erythro-hexos-5-ulo-1,5-pyranose]. To identify genes involved in biosynthesis of the component sugars of this glycan, three genes, mmp1081, mmp1082, and mmp1083, were targeted for in-frame deletion, based on their annotation and proximity to glycosyltransferase genes known to be involved in assembly of the glycan. Mutants carrying a deletion in any of these three genes remained flagellated and motile. A strain with a deletion of mmp1081 had lower-molecular-mass flagellins in Western blots. Mass spectrometry of purified flagella revealed a truncated glycan with the terminal sugar absent and the threonine residue and the acetamidino group missing from the third sugar. No glycan modification was seen in either the Δmmp1082 or Δmmp1083 mutant grown in complex Balch III medium. However, a glycan identical to the Δmmp1081 glycan was observed when the Δmmp1082 or Δmmp1083 mutant was grown under ammonia-limited conditions. We hypothesize that MMP1082 generates ammonia and tunnels it through MMP1083 to MMP1081, which acts as the amidotransferase, modifying the third sugar residue of the M. maripaludis glycan with the acetamidino group.
INTRODUCTION
N-linked glycosylation of proteins is a posttranslational modification found in all three domains of life (12, 23, 43). While its distribution is apparently rather limited throughout the domain Bacteria, N-linked glycosylation is widespread in the Archaea. A gene for the oligosaccharyltransferase required for the final step in the process, i.e., transfer of the assembled glycan from its lipid carrier to specific sites on the target protein, has been found in all but two sequenced archaeal genomes (33).
The N-linked glycosylation process in archaea is thought to be similar to that reported for bacteria (3, 22, 43), where the glycan is first assembled on a lipid carrier on the cytoplasmic side of the cytoplasmic membrane. However, archaea use dolichol mono- or pyrophosphate as the lipid carrier (like eukaryotes), while bacteria use undecaprenol pyrophosphate (18, 19, 31). A series of glycosyltransferases assemble the glycan from nucleotide-activated sugars, and once complete, the glycan is flipped across the membrane by an as yet unidentified flippase in archaea. It is then transferred by the oligosaccharyltransferase (AglB) to the target protein at selected asparagine residues located within an N-X-S/T motif (X cannot be proline) (1, 48). AglB is homologous to the bacterial oligosaccharyltransferase (PglB) and to the catalytic subunit (STT3) of the oligosaccharyltransferase complex of eukaryotes (12, 13, 21, 35).
Within the domain Archaea, N-linked glycans have been reported on a number of surface-exposed proteins, such as S layers (2, 12, 17, 37), flagellins (13, 25, 45, 47), and, most recently, pilins (42). However, both genetic and structural studies on N-linked glycans have been reported for only a very limited number of archaeal species. These organisms are the mesophilic methanogens Methanococcus voltae and Methanococcus maripaludis, the extreme halophile Haloferax volcanii (12, 23), and the thermoacidophile Sulfolobus acidocaldarius (38, 44). In these organisms, genes involved mainly in the assembly of the glycan, i.e., glycosyltransferases and the oligosaccharyltransferase, have been identified (2, 4, 5, 13, 24, 45, 50), while only a limited number of genes involved in the actual biosynthesis of the sugars comprising the glycan have been characterized (34, 38, 46, 49, 50).
In M. maripaludis, the flagella are composed of three major and highly conserved proteins, called flagellins FlaB1, FlaB2, and FlaB3. Both FlaB1 and FlaB2 are major components of the filament, while FlaB3 is responsible for the curved portion or hook region (14). All three flagellins are modified with a tetrasaccharide linked to asparagine residues at multiple positions within each flagellin via N-acetylgalactosamine (25). There are 3 sites of glycan attachment in FlaB1, 4 sites in FlaB2, and 2 sites in FlaB3. The second sugar of the glycan is a diacetylated glucuronic acid, while the third sugar is a mannuronic acid derivative with a threonine substitution at position 6 and an acetamidino group at position 3. The final sugar is a unique diglycoside of an aldulose, representing the first of its kind found naturally.
Genetic studies have shown that gene deletions resulting in a truncated glycan have significant effects on flagellum assembly and/or function in M. maripaludis (45, 46). Flagellins with a glycan that lacks the third and fourth sugar residues can still be assembled into a working flagellum, although swimming function is impaired compared to that of wild-type cells that have flagellins still bearing the tetrasaccharide. However, in mutants where the flagellins have only a single sugar attached or are completely nonglycosylated, flagella are not formed (45). Similar findings were reported initially for M. voltae (13).
In vitro analyses have revealed the roles of several gene products in the biosynthesis of sugars that act as precursors to those found in the glycan structure (41), although genetic studies that identify genes critical for biosynthesis/modification of the sugar components of the glycan are lacking for M. maripaludis and scant for archaea in general. Here we report on the genetic and structural analysis of a three-gene operon located adjacent to genes encoding glycosyltransferases previously identified as involved in the assembly of the glycan (45). We present evidence that these three genes, mmp1081 to -1083, act as an archaeal version of the wbuX-wbuY-wbuZ system responsible for the acetamidation of l-FucNAc to l-FucNAm in lipopolysaccharide (LPS) biosynthesis in Escherichia coli O145 (16). Our model indicates that MMP1081 to -1083 are responsible for the addition of the acetamidino group present on the third sugar, likely using ammonia generated by the glutaminase-like ability of MMP1082 and tunneled through MMP1083 to MMP1081, which acts as the actual amidotransferase.
MATERIALS AND METHODS
Strains and growth conditions.
M. maripaludis strain Mm900 (39) was grown anaerobically under a headspace gas of CO2-H2 (20:80) at 30°C in Balch III medium (8), with McCas medium (39) used during transformation events. Neomycin (1 mg/ml), puromycin (2.5 μg/ml), and 8-azahypoxanthine (240 μg/ml) were used for selection as necessary. For ammonia-free and complementation experiments, anaerobic nitrogen-free minimal medium was used (11) and was supplemented with either NH4Cl (10 mM) or l-alanine (10 mM) from a sterile anaerobic stock solution as the nitrogen source. Escherichia coli strain DH5α, utilized for plasmid cloning, was grown at 37°C in Luria-Bertani medium, with ampicillin (100 μg/ml) added for selection when necessary.
RT-PCR.
To determine if the mmp1081 to -1083 genes formed a cotranscribed unit, reverse transcriptase PCR (RT-PCR) was performed using primers (listed in Table S1 in the supplemental material) designed to amplify sequences across the intergenic regions of neighboring genes. An RNA template was extracted from wild-type cells by use of an RNeasy miniprep kit (Qiagen Inc., Mississauga, Ontario, Canada) with optional DNase digestion (Qiagen Inc.) per the manufacturer's protocol. cDNA was amplified using a One-Step RT-PCR kit (Qiagen Inc.) in accordance with the supplied protocol. In addition, two standard PCR amplifications were performed with each reaction, using the same primer combination but a different template, with one amplifying genomic DNA and the other using the purified RNA as a control to rule out genomic DNA contamination of the RNA sample.
Plasmid construction.
Plasmids used for generation of in-frame deletions of mmp1081, mmp1082, and mmp1083 were constructed as previously described (39, 46) and are listed in Table 1. Briefly, primers were designed (see Table S1 in the supplemental material) to amplify ∼1,000 bp of the flanking upstream and downstream regions of the respective gene, resulting in the deletion of the majority of the gene while leaving the remaining 3′ and 5′ ends in frame. Internal primers had AscI restriction sites incorporated to permit ligation of flanking segments, which would allow for in-frame deletion of the majority of the gene of interest. BamHI sites were included in external primers to allow for ligation into pCRPrtNeo (39).
Table 1.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pCRPrtNeo | hmv promoter-hpt fusion plus Neor cassette in pCR2.1Topo; Ampr | 39 |
| pKJ735 | pCRPrtNeo with in-frame deletion of mmp1081 | This study |
| pKJ751 | pCRPrtNeo with in-frame deletion of mmp1082 | This study |
| pKJ757 | pCRPrtNeo with in-frame deletion of mmp1083 | This study |
| pHW40 | nif promoter-lacZ fusion plus Purr cassette; Ampr | John Leigh |
| pKJ903 | pCRII-TOPO vector with mmp1082 | This study |
| pKJ752 | pHW40 with mmp1081 complement | This study |
| pKJ952 | pHW40 with mmp1082 complement | This study |
| pKJ951 | pHW40 with mmp1083 complement | This study |
Plasmids used for complementation experiments (Table 1) were constructed as previously reported (14, 30). For ease of cloning, one internal NsiI site in mmp1082 was removed by use of a silent T-to-C mutation inserted by site-directed mutagenesis using the primers MMP1082_For_SDM and MMP1082_Rev_SDM, with plasmid pKJ903 carrying the wild-type version of the gene as the template. All plasmid inserts were sequenced to confirm the integrity of the construct and the in-frame deletion of the targeted gene.
M. maripaludis mutant generation.
Markerless in-frame deletions of mmp1081, mmp1082, and mmp1083 were created as previously described (39). Mutants were identified from among transformants by PCRs using whole cells as a template and primers that would amplify across the targeted gene. After agarose gel electrophoresis, the size of the PCR product obtained was compared to the size predicted by the gene deletion and to the size of the product obtained using wild-type cells as the template. DNA sequencing of the PCR products was performed to confirm that the deletion was in frame. Southern blot analysis was performed to validate deletions, in accordance with a previously described protocol (14).
Western blotting.
Western blots of whole-cell lysates were run and developed as previously described, but using an antiserum specific to flagellin FlaB2 (46). This antiserum was raised in chickens by injecting the purified N-terminally His-tagged variable region of FlaB2. This variable region extends from amino acids 85 to 139 of the mature protein and is unique to FlaB2 among the three flagellins. Antibodies were purified from egg yolks and shown to react only with FlaB2.
Semisolid swarm motility plate assay.
Five-milliliter samples of M. maripaludis strains grown overnight were pelleted anaerobically and then resuspended gently in 200 μl of fresh medium. Ten-microliter samples were used to inoculate the center of 0.25% (wt/vol) agar Balch III plates, and the plates were incubated for 7 days at 30°C (45).
Flagellum isolation.
M. maripaludis flagella were isolated from the mmp1081, mmp1082, and mmp1083 deletion strains by using a previously described protocol (9). Strains were grown in either Balch III medium (4.7 mM NH4Cl) or nitrogen-free medium supplemented with l-alanine.
Mass spectrometry.
Purified flagella were subjected to mass spectrometry as previously reported (25). Briefly, flagellin isolates (50 μg) were digested overnight with trypsin (Promega, Madison, WI) at a ratio of 30:1 (protein to enzyme [vol/vol]) in 50 mM ammonium bicarbonate at 37°C. The flagellin digests were analyzed by nano-liquid chromatography–tandem mass spectrometry (nano-LC–MS/MS) using a QTOF Ultima hybrid-quadrupole time-of-flight mass spectrometer coupled to a NanoAquity UPLC system (Waters, Milford, MA). The digests were injected onto a 5-mm by 300-μm-internal-diameter Acclaim PepMap100 C18 μ-precolumn (Dionex/Thermo Scientific, Sunnyvale, CA) and separated on a 100-μm by 100-mm-internal-diameter by 1.7-μm BEH130 C18 column (Waters) under the following gradient conditions: 5% to 45% acetonitrile (ACN) in 0.2% formic acid over 35 min and 45% to 95% ACN over 5 min, at 450 nl/min. MS/MS spectra were acquired on doubly, triply, and quadruply charged ions and searched against the NCBInr database by using the Mascot search engine (Matrix Science Ltd., London, United Kingdom). Glycopeptide MS/MS spectra were interpreted by hand.
Electron microscopy.
Cells were washed using 50 mM MgSO4 and negatively stained with 2% phosphotungstic acid (pH 7.0). Samples were examined on Formvar-coated gold grids and imaged on a Hitachi H-700 transmission electron microscope at 75 kV.
RESULTS
Previously, two glycosyltransferase genes (mmp1079 and mmp1080) involved in flagellin glycan assembly were identified (45). Bioinformatics analysis revealed that immediately downstream of mmp1080 was a cluster of three genes, mmp1081, mmp1082, and mmp1083, which showed high sequence similarity (see Fig. S1 in the supplemental material) to three E. coli genes (wbuX, wbuY, and wbuZ) implicated in the biosynthesis of 2,6-dideoxy-2-acetamidino-l-galactose (l-FucNAm) of the LPS O antigen (16). Therefore, these three genes were potential candidates responsible for the acetamidino group attached to the third sugar of the M. maripaludis flagellin glycan. The protein encoded by mmp1081 shows homology to the bacterial N-acetyl sugar amidotransferase WbuX (41% identity, 60% similarity), while mmp1082 and mmp1083 encode proteins that show homology to the WbuX accessory proteins, i.e., WbuY (42% identity, 62% similarity) and WbuZ (47% identity, 68% similarity), identified in the study by Feng et al. (16).
RT-PCR analysis of the mmp1081 to -1083 genes.
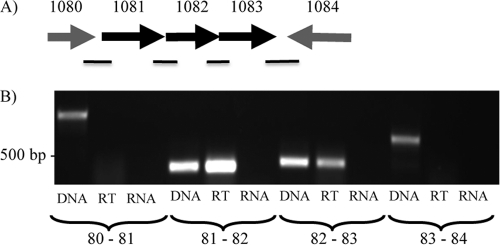
Based on their orientation and short intergenic regions, it was thought that mmp1081 to -1083 may be cotranscribed as a single operon (Fig. 1A). To test this hypothesis, primers were designed to amplify sequences across the intergenic regions of genes located between mmp1080 and mmp1084 (Fig. 1B). RT-PCR and standard PCR were performed on isolated RNA from M. maripaludis cells, and a standard PCR using the same primer pair was also performed using genomic DNA as the template to confirm the amplicon size and primer specificity (Fig. 1B). Amplification occurred for each of the primer pairs in a standard PCR amplification with genomic DNA as the template. No such amplification was seen in the PCR using isolated RNA, confirming that the RNA sample was free of genomic DNA contamination. The RT-PCR results show that mmp1081 to -1083 are cotranscribed, while mmp1080, though transcribed in the same orientation as mmp1081, is not part of this operon. There is a large intergenic region of slightly over 700 bp between mmp1080 and mmp1081. Additionally, mmp1081, mmp1082, and mmp1083 are cotranscribed independently of mmp1084, as expected, since mmp1084 is transcribed from the other strand.
Fig 1.
The mmp1081-mmp1082-mmp1083 operon. (A) Representation of the gene region surrounding mmp1081, mmp1082, and mmp1083, which was targeted for RT-PCR to examine possible cotranscription. The black lines below the genes represent regions between genes that were targeted for amplification. (B) RT-PCR experiment indicating cotranscription of mmp1081, mmp1082, and mmp1083. Each primer set was run using a standard PCR and genomic Mm900 DNA (DNA) for amplicon size confirmation. An RT-PCR (RT) was run using the same primers and purified Mm900 mRNA to establish cotranscription of genes. DNA bands in the RT lanes are indicative of genes that are cotranscribed. Additionally, a standard PCR was run using the purified mRNA as a template as a control against genomic DNA contamination of the RNA (RNA).
Generation of in-frame deletions of mmp1081, mmp1082, and mmp1083.
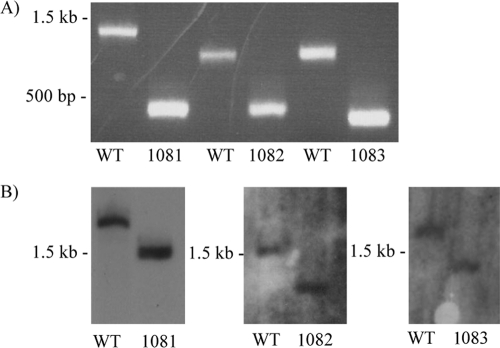
In an effort to determine the role, if any, that the mmp1081, mmp1082, and mmp1083 genes play in the biosynthesis of the sugars of the flagellin N-linked glycan, in-frame deletions of the genes were generated. Deletion strains were identified by screening individual transformants by PCR, using primers that would amplify across each targeted gene (Fig. 2A); in each case, the PCR product obtained from the deletion strain was smaller than the product obtained with the same primers but with wild-type cells as the template, by the theoretically predicted amount. For each deletion, PCR products were sequenced to confirm that the deletion was in frame (data not shown). Additionally, each mutant was subjected to Southern blot analysis (Fig. 2B), and again, the size of the band hybridized to the digoxigenin (DIG)-labeled probe for each mutant strain was smaller than the corresponding band for the wild-type DNA, by the predicted amount.
Fig 2.
Confirmation of deletion of mmp1081, mmp1082, and mmp1083. (A) PCRs used whole cells of the wild type (WT; Mm900) or the deletion strains as templates for gene-specific primers. In all cases, the predicted sizes of the amplicons were obtained. (B) Southern blot confirmation of the deletion of mmp1081, mmp1082, or mmp1083. The decrease in the size of the band hybridized to the labeled probe in each mutant compared to strain Mm900 (WT) corresponded to the predicted loss for each target gene. For mmp1081, genomic DNAs from wild-type and Δmmp1081 cells were digested with NdeI; for mmp1082, genomic DNAs from wild-type and Δmmp1082 cells were digested with SspI; and for mmp1083, genomic DNAs from wild-type and Δmmp1083 cells were digested with EcoRI.
Western blot analysis of flagellins from the mutant strains.
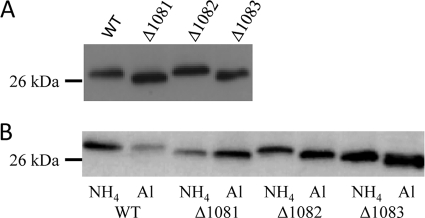
A reduction in the molecular mass of the flagellin subunits is known to occur in mutants of M. maripaludis harboring deletions of genes involved in the N-linked glycosylation pathway (45). This reduction can be detected in Western blots developed with anti-flagellin antiserum. When Western blots were run using whole-cell lysates of the mmp1081, mmp1082, and mmp1083 deletion strains, a decrease in flagellin molecular mass was observed in the mmp1081 mutant, with a smaller shift observed in the mmp1083 mutant. No difference in flagellin molecular mass could be seen in the mmp1082 mutant grown under these conditions (Fig. 3A). These data suggested a role for this operon in flagellin glycosylation.
Fig 3.
Western blots of total cell lysates of Mm900 (WT) and the mmp1081, mmp1082, and mmp1083 deletion strains probed with anti-FlaB2 antibodies. A decrease in flagellin size in mutants is typically the initial evidence of a modified glycan. (A) Cells were grown in Balch III medium. (B) Cells were grown in nitrogen-free medium supplemented with either NH4Cl (NH4) or alanine (Al).
Mass spectrometry analysis of flagellin proteins.
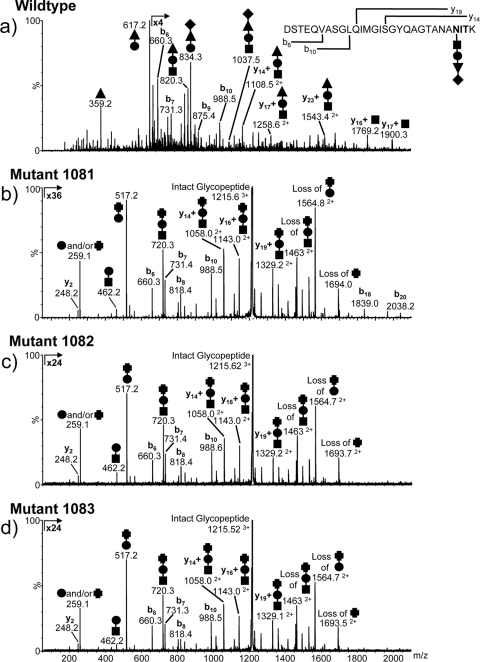
To ascertain if the deletions resulted in a defective glycan structure, flagella were purified from each mutant strain grown in Balch III medium. LC-MS/MS analysis of the flagellar tryptic peptides from the mmp1081 mutant indicated that the glycan structure was truncated and lacked the terminal sugar and the threonine modification of the third sugar (Fig. 4b). In addition, the third sugar had lost the acetamidino group and was likely di-N-acetyl mannuronic acid. Similar analyses of flagellar tryptic peptides from the Δmmp1082 and Δmmp1083 mutants demonstrated that the glycans present were predominantly wild type, although a small amount of glycan truncated in a manner identical to that observed for the Δmmp1081 mutant was observed (data not shown). Furthermore, the Δmmp1083 mutant contained a single Ser-to-Ala mutation that eliminated one of the N-glycosylation consensus sequons on FlaB1. The sequence of the wild-type T158–178 tryptic glycopeptide is GDAVALIVDVNASFAGEIPER, while the modified sequence is GDAVALIVDVNAAFAGEIPER. This sequence change was not observed in the wild type and the other mutant strains. However, this mutation does not explain why the Δmmp1083 mutant flagellin appeared to migrate faster in Western blots than the wild-type or Δmmp1082 mutant flagellin, since the Western blots were developed with antiserum specific to FlaB2. Subsequently, the flaB2 gene from the Δmmp1083 mutant was sequenced, and a single amino acid change was also found. This change, from Asn to Asp (due to a single A-to-G mutation at position 364 of the gene), results in a loss of the N-glycosylation site at this position. This explains why the flagellin in the Δmmp1083 mutant appears smaller than the wild-type or Δmmp1082 flagellin in Western blots.
Fig 4.
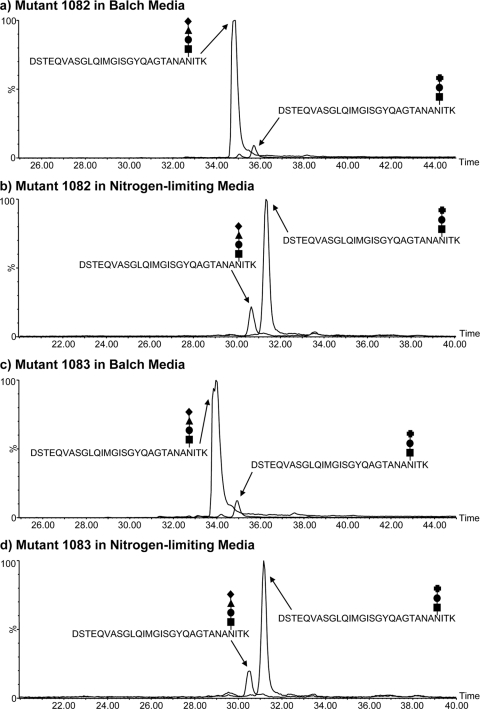
Nano-LC–MS/MS analysis of the FlaB2 tryptic glycopeptide T53–81 from the wild-type (a) and mmp1081 (b), mmp1082 (c), and mmp1083 (d) mutant strains of M. maripaludis. The major carbohydrate oxonium ions are identified in the MS/MS spectra by using symbols to indicate the sugar residues present [■, GalNAc; ●, GlcNAc3NAcA; ▲, ManNAc3NAmA6Thr; ♦, (5S)-2-acetamido-2,4-dideoxy-5-O-methyl-α-l-erythro-hexos-5-ulo-1,5-pyranose; and ✚, ManNAc3NAcA]. The b and y ions arising from fragmentation of the peptide bonds are also shown. The glycopeptide from the mmp1081 mutant strain (b) was modified with a trisaccharide composed of the linking GalNAc (■), the GlcNAc3NacA (●), and a terminal sugar that is likely to be di-N-acetyl-mannuronic acid (ManNAc3NAcA [✚). In normal media, both the mmp1082 and mmp1083 mutant strains expressed predominantly the wild-type glycan. However, in nitrogen-free medium supplemented with alanine, both strains expressed predominantly the Δmmp1081 glycan modification (c and d).
Ammonia-limiting growth experiments.
The mmp1081-mmp1082-mmp1083 operon is homologous to wbuX-wbuY-wbuZ from E. coli, an operon that encodes the N-acetyl sugar amidotransferase WbuX (MMP1081 homolog) and two accessory proteins, WbuY and WbuZ (MMP1082 and MMP1083 homologs, respectively) (16). The two accessory proteins in the E. coli system were proposed to be involved in the generation and transfer of ammonia to the amidotransferase, but effects of deletion of the two accessory genes were observed only under ammonia-limiting conditions. To test if deletion of mmp1082 and mmp1083 had effects on the N-linked glycan structure that were observable only under ammonia-limiting conditions, strains harboring mutations in these genes were grown in nitrogen-free medium supplemented with either alanine (ammonia limiting) or NH4Cl as the sole nitrogen source. The nitrogen source under these conditions had no effect on flagellin molecular mass in wild-type cells or the mutant carrying a deletion of mmp1081. However, a clear shift in the migration of the flagellins was seen in both the Δmmp1082 and Δmmp1083 mutant strains, depending on the nitrogen source used (Fig. 3B). Under ammonia-limiting conditions (alanine-grown cells), the MMP1082 and MMP1083 flagellins migrated faster than when cells were grown in NH4Cl-supplemented nitrogen-free medium, suggesting a change in glycan composition. This was confirmed by MS analysis of the flagellin tryptic digests.
In medium with sufficient ammonia, about 93% of the glycan on the flagellins from either the mmp1082 or mmp1083 deletion strain was wild type. However, when cells were grown in ammonia-free medium supplemented with alanine, the relative amounts of wild-type and truncated glycans were altered significantly, with the truncated glycan now predominating (Table 2). In the ammonia-depleted medium, the flagellin glycans of both the mmp1082 and mmp1083 deletion strains were identical in structure to that found in the mmp1081 mutant, i.e., lacking the terminal sugar as well as the acetamidino and threonine groups on the third sugar (Fig. 4c and d). In both of these cases, however, some wild-type glycan, representing about 17% of the total, was still present (Fig. 5; Table 2).
Table 2.
Variation in glycan composition on flagellins from cells grown in different media
| Strain | Growth medium | % Glycan on flagellins |
|
|---|---|---|---|
| Wild type | Mutant | ||
| Wild type | Balch III | 98.79 | 1.21 |
| Δmmp1081 mutant | Balch III | 0 | 100 |
| Δmmp1082 mutant | Balch III | 93.86 | 6.14 |
| Δmmp1083 mutant | Balch III | 93.0 | 7.0 |
| Δmmp1082 mutant | N-free + alanine | 16.47 | 83.53 |
| Δmmp1083 mutant | N-free + alanine | 18.18 | 81.81 |
Fig 5.
Extracted ion chromatograms (EICs) of the wild-type and mutant glycan versions of the FlaB2 tryptic glycopeptide T53–81 isolated from the mmp1082 and mmp1083 mutant strains grown in Balch medium and under ammonia-limiting conditions. The EICs for triply protonated ions for the wild-type and mutant T53–81 glycopeptides are plotted together for the mmp1082 strain grown in Balch (a) and nitrogen-limiting (b) media. The same ions are shown for the mmp1083 strain (c and d). The relative peak areas (expressed as percentages) of the two glycopeptide ions in all the strains examined in this study are presented in Table 2. The glycan symbols used here are explained in the legend for Fig. 4. Though both peptide glycoforms were expressed in the Δmmp1082 and Δmmp1083 strains under the two conditions, the wild-type glycopeptide predominated (>90%) in the Balch-grown strains, while the reverse was true under ammonia-limiting conditions.
Complementation of mmp1081, mmp1082, and mmp1083.
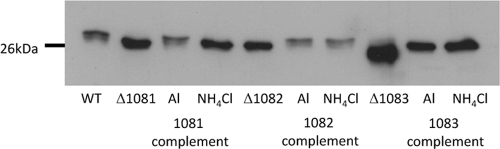
To confirm the role of the deleted genes in the glycosylation pathway, mutants were complemented with plasmids bearing a wild-type copy of the respective genes. The plasmid construct allowed the expression of the cloned genes under the control of the inducible nif promoter (26). For both the mmp1081 and mmp1082 mutants, complementation resulted in the restoration of the flagellin molecular mass, as determined by Western blotting, to wild-type size under ammonia-limiting conditions (Fig. 6). In the case of the mmp1083 mutant, however, complementation was unable to restore the flagellin molecular mass to wild-type size because one of the glycosylation sites of FlaB2 was lost due to a mutation. In this case, complementation led to an increase in flagellin molecular mass equal to that of the mmp1083 mutant grown in Balch III medium or in nitrogen-free medium supplemented with ammonia, i.e., conditions shown to result in a wild-type glycan attached to all sites in the mmp1083 flagellin occupied by glycan in the wild-type flagellin except for the lost mutated site.
Fig 6.
Analysis of complementation experiments with the mmp1081, mmp1082, and mmp1083 deletion strains. Western blots of total cell lysates of the deletion strains containing the respective complementation plasmids and grown in nitrogen-free medium supplemented with either l-alanine (transcription of complementation gene on) or NH4Cl (transcription of complementation gene off) were probed with anti-FlaB2 specific antiserum. Mm900 cells (WT) and the mmp1081 strain grown in nitrogen-free medium supplemented with NH4Cl were used as controls.
Deletions in the mmp1081-mmp1082-mmp1083 operon do not affect flagellum assembly.
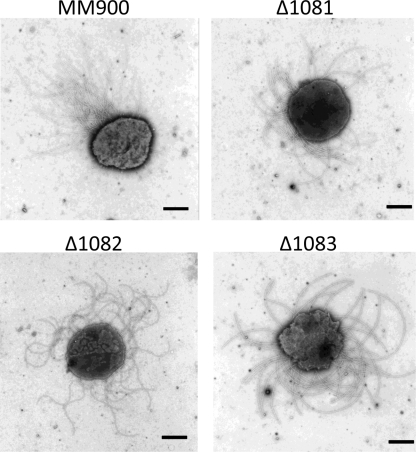
The wild-type and mmp1081, mmp1082, and mmp1083 deletion strains were examined by electron microscopy, and all strains were found to be well flagellated, indicating that none of the gene deletions grossly affected flagellum assembly compared to that in wild-type cells (Fig. 7). In addition, all mutant strains were shown to be motile by light microscopy and semisolid swarm plate motility assays (data not shown).
Fig 7.
Electron micrographs of wild-type and mutant strains. All of the mmp1081, mmp1082, and mmp1083 deletion strains are flagellated like Mm900 (WT) cells. Bars, 0.5 μm.
DISCUSSION
This study identified an operon containing three genes involved in the biosynthesis of the third sugar residue of the N-linked glycan attached to flagellins of M. maripaludis, specifically in the acetamidino group modification. The three genes in the operon, mmp1081, mmp1082, and mmp1083, are located adjacent to genes for glycosyltransferases already identified as having a role in the assembly of the N-linked glycan. MMP1081 shows homology with other known N-acetyl sugar amidotransferases, including WbuX from E. coli (16), LfnA from Pseudomonas aeruginosa (28), and PseA from Campylobacter jejuni (32), which are all involved in the generation of sugars with acetamidino group modifications. WbuX and LfnA have been reported to be required for the biosynthesis of 2,6-dideoxy-2-acetamidino-l-galactose (l-FucNAm), found to be part of the O antigen in several pathogenic strains, including E. coli O145 and P. aeruginosa O12 (28). PseA has been implicated in the attachment of an acetamidino functional group to the glycan O-linked to flagellin of C. jejuni (36). Each of these genes has been found associated with two accessory genes, homologous to the histidine biosynthesis genes hisH and hisF (16, 36). Following the same pattern observed in bacteria, the archaeal genes mmp1082 and mmp1083 share homology with hisH and hisF, respectively. In bacteria with the proposed ammonia tunnel genes, the hisH and hisF homologs found as accessory genes to the amidotransferase gene are not thought to play a role in histidine biosynthesis, as a histidine biosynthesis operon is found elsewhere in the genome, complete with hisF and hisH genes. In M. maripaludis, second copies of both hisF and hisH are also found, and these are likely involved in histidine biosynthesis. However, in M. maripaludis, many genes for amino acid biosynthesis are unlinked (20). In the case of histidine biosynthesis, the various his genes are scattered around the chromosome and not found in a single operon.
Mutants lacking mmp1081 produced flagellins that migrated as lower-molecular-mass proteins than the wild-type flagellins in Western blots, which was suggestive of an altered glycan (45). These cells were still able to produce flagella and were motile. Mass spectrometry analysis of purified flagella confirmed that the glycan was composed of only three sugars, with the third sugar lacking the acetamidino group and the threonine moiety, while the terminal sugar found in the wild-type glycan was missing completely. Based on its homology to known amidotransferases and the MS data, it is most likely that MMP1081 is responsible for the addition of the acetamidino group to the third sugar and that the glycosyltransferase for the fourth sugar, MMP1088 (45), is therefore unable to attach the terminal sugar to the defective glycan structure. A less likely possibility is that MMP1081 may be responsible for the attachment of the threonine group. However, it has previously been reported that deletion of mmp1088 resulted in both the absence of the terminal sugar and, unexpectedly, the loss of the threonine group from the third residue (45), thus suggesting that the addition of the threonine may occur only after the fourth sugar is attached. In addition, we recently identified a gene that appears to be essential for the threonine attachment (unpublished results). Complementation of mmp1081 in trans restored the flagellin molecular mass to that in the wild type, thus discounting disruption effects of the deletion on downstream genes.
The flagellins of the mmp1082 mutant did not appear to be of lower molecular mass in Western blots than those of Mm900 when these cells were grown in Balch III medium, while a difference could be seen with the mmp1083 mutant. Both of these mutant strains were flagellated and motile. MS analysis of flagella from the mmp1082 and mmp1083 mutants did not demonstrate any difference in flagellin glycan structure compared to that in wild-type cells. The wild-type nature of the glycan on the mmp1083 flagellins that had a lower molecular mass than wild-type flagellins in Western blots was initially puzzling. However, it was subsequently discovered that the mmp1083 mutant contains a single amino acid mutation that eliminates one of the N-glycosylation consensus sequons on FlaB2. This loss of one of the attached glycans explains the decrease in molecular mass of flagellins observed in Western blots developed with antiserum specific to FlaB2 for this mutant, since a wild-type glycan is added at all of the remaining sites. The mmp1083 mutant also had a mutation in FlaB1 detected by MS analysis that also resulted in the loss of a single N-linked sequon.
HisH and HisF are known to interact (6), and it was proposed that HisH is a glutaminase which acts by hydrolyzing glutamine to produce ammonia and passing it through a molecular tunnel to HisF, which is then able to amidate a precursor molecule of histidine (7, 15, 40). It is thought that the HisH and HisF homologs WbuY and WbuZ accomplish a similar task, with WbuY as the glutaminase generating ammonia and WbuZ acting as the tunnel to deliver the ammonia to the amidotransferase, WbuX, allowing it to modify the target sugar residue (16). Deletions of the hisH and hisF homologs in the C. jejuni system were demonstrated to have an effect on biosynthetic intermediates within the glycan biosynthetic pathway (36).
It is predicted that MMP1082 and MMP1083 perform similar tasks in the M. maripaludis system, with MMP1082 acting to generate ammonia which is transferred via a tunnel created by MMP1083 to MMP1081, the actual amidotransferase responsible for the generation of the acetamidino group on the di-N-acetyl mannuronic acid substrate, in an ammonia-dependent fashion. MMP1083 has the conserved TIM phosphate binding domain found in WbuZ and HisF proteins; these proteins have an eight-beta/alpha-closed-barrel structure thought to act as the ammonia tunnel. Presumably, in rich medium such as Balch III medium, there is adequate ammonia available for MMP1081 to function normally. However, under ammonia-limiting conditions, such as growth in nitrogen-free medium with alanine added, the activity of MMP1081 becomes dependent on the ammonia generated by the glutaminase activity of MMP1082 and funneled by MMP1083. It has been reported that effects of deletions in other glutaminase systems are typically not detected when cells are grown in rich medium with sufficient ammonia (27) but become evident when cells are grown in minimal medium without ammonia addition. This was shown to be the case with the mmp1082 and mmp1083 deletion strains in this study. The difference in growth medium had no effect on flagellin molecular mass in Western blots of wild-type cells or the mmp1081 deletion mutant. However, a difference in flagellin molecular mass was seen in the mmp1082 and mmp1083 mutants, depending on whether NH4+ was present or absent in the growth medium. In addition, mass spectrometry analysis of flagellins of the mmp1082 and mmp1083 deletion strains showed a mostly wild-type glycan when the cells were grown with ammonia and a mostly Δmmp1081 mutant-like glycan when the cells were grown under ammonia-deficient conditions. In agreement with this, it has also been reported that the mmp1082 homolog hisH is not required during histidine biosynthesis if ammonium ions are available (10, 29). Complementation of mmp1082 resulted in flagellins that migrated similarly to those of the wild type in both ammonia-sufficient and ammonia-deficient growth medium. Complementation of mmp1083 also resulted in an increased flagellin molecular mass under ammonia-deficient growth conditions, but not to the size of wild-type flagellins, only because the mmp1083 deletion strain also had a mutation that resulted in the loss of a single glycosylation site in the FlaB2 protein.
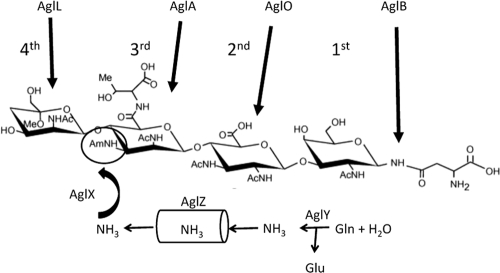
The data presented in this study describe an operon involved in the biosynthesis of the third sugar present in the M. maripaludis flagellin N-linked glycan. The data presented strongly suggest that mmp1081, mmp1082, and mmp1083 are homologous to the wbuX, wbuY, and wbuZ genes of E. coli O145 responsible for acetamidation of l-FucNAc in LPS biosynthesis and for which an ammonia tunneling mechanism is predicted, though without biochemical evidence (16). The archaeal operon encodes a predicted amidotransferase (MMP1081), a predicted ammonia-generating glutaminase-like enzyme (MMP1082), and a predicted ammonia tunnel component (MMP1083), although, as with the bacterial system, biochemical support is still lacking. The incorporation of these findings into the genetic and mass spectrometry data previously published on the assembly of the N-linked glycan is presented in Fig. 8. The glycan is assembled via addition of individual nucleotide-activated sugars to a dolichol lipid carrier. The glycosyltransferase needed for the transfer of the first sugar (N-acetylgalactosamine) is unknown. The remaining three sugars are added through the activities of AglO (MMP1079), AglA (MMP1080), and AglL (MMP1088). Following flipping of the completed glycan across the cytoplasmic membrane, the glycan is transferred to the target protein at designated asparagine residues by the oligosaccharyltransferase, AglB. The identification of MMP1081, MMP1082, and MMP1083 as the gene products involved in generation of the acetamidino group of sugar 3 is a beginning to identifying the pathways of biosynthesis of the four sugars of the N-linked glycan. This modification of the third sugar is a critical step, since subsequent addition of the terminal sugar does not occur without it. However, it is also clear that addition of the third sugar to the glycan can occur without the acetamidino modification. Whether this modification normally occurs before or after the third sugar is added to the glycan is not presently known.
Fig 8.
Model of assembly of the flagellin N-linked glycan. The previously identified glycosyltransferases (AglO, AglA, and AglL) as well as the oligosaccharyltransferase (AglB) are shown. The postulated functions of AglX, AglY, and AglZ in generating ammonia via the glutaminase activity of AglY and its subsequent transfer via the ammonia tunnel of AglZ to the amidotransferase AglX to form the acetamidino group on sugar 3 are depicted.
By virtue of their involvement in N-linked glycan synthesis shown here, and considering their homology to wbuX, wbuY, and wbuZ, the mmp1081, mmp1082, and mmp1083 genes have been designated aglX, aglY, and aglZ, respectively, following the naming scheme initially proposed by Chaban et al. (13) for genes involved in N-linked glycosylation in archaea.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the National Research Council of Canada (S.M.L. and J.K.) and by a discovery grant from the Natural Sciences and Engineering Research Council of Canada (NSERC) (K.F.J.).
We thank John Leigh for plasmids and strains.
Footnotes
Published ahead of print 9 March 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Abu-Qarn M, Eichler J. 2007. An analysis of amino acid sequences surrounding archaeal glycoprotein sequons. Archaea 2:73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abu-Qarn M, Eichler J. 2006. Protein N-glycosylation in archaea: defining Haloferax volcanii genes involved in S-layer glycoprotein glycosylation. Mol. Microbiol. 61:511–525 [DOI] [PubMed] [Google Scholar]
- 3. Abu-Qarn M, Eichler J, Sharon N. 2008. Not just for Eukarya anymore: protein glycosylation in Bacteria and Archaea. Curr. Opin. Struct. Biol. 18:544–550 [DOI] [PubMed] [Google Scholar]
- 4. Abu-Qarn M, et al. 2008. Identification of AglE, a second glycosyltransferase involved in N-glycosylation of the Haloferax volcanii S-layer glycoprotein. J. Bacteriol. 190:3140–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abu-Qarn M, et al. 2007. Haloferax volcanii AglB and AglD are involved in N-glycosylation of the S-layer glycoprotein and proper assembly of the surface layer. J. Mol. Biol. 374:1224–1236 [DOI] [PubMed] [Google Scholar]
- 6. Alifano P, et al. 1996. Histidine biosynthetic pathway and genes: structure, regulation, and evolution. Microbiol. Rev. 60:44–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Amaro R, Tajkhorshid E, Luthey-Schulten Z. 2003. Developing an energy landscape for the novel function of a (beta/alpha)8 barrel: ammonia conduction through HisF. Proc. Natl. Acad. Sci. U. S. A. 100:7599–7604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bardy SL, Mori T, Komoriya K, Aizawa S, Jarrell KF. 2002. Identification and localization of flagellins FlaA and FlaB3 within flagella of Methanococcus voltae. J. Bacteriol. 184:5223–5233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Beismann-Driemeyer S, Sterner R. 2001. Imidazole glycerol phosphate synthase from Thermotoga maritima. Quaternary structure, steady-state kinetics, and reaction mechanism of the bienzyme complex. J. Biol. Chem. 276:20387–20396 [DOI] [PubMed] [Google Scholar]
- 11. Blank CE, Kessler PS, Leigh JA. 1995. Genetics in methanogens: transposon insertion mutagenesis of a Methanococcus maripaludis nifH gene. J. Bacteriol. 177:5773–5777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Calo D, Kaminski L, Eichler J. 2010. Protein glycosylation in archaea: sweet and extreme. Glycobiology 20:1065–1076 [DOI] [PubMed] [Google Scholar]
- 13. Chaban B, Voisin S, Kelly J, Logan SM, Jarrell KF. 2006. Identification of genes involved in the biosynthesis and attachment of Methanococcus voltae N-linked glycans: insight into N-linked glycosylation pathways in archaea. Mol. Microbiol. 61:259–268 [DOI] [PubMed] [Google Scholar]
- 14. Chaban B, et al. 2007. Systematic deletion analyses of the fla genes in the flagella operon identify several genes essential for proper assembly and function of flagella in the archaeon, Methanococcus maripaludis. Mol. Microbiol. 66:596–609 [DOI] [PubMed] [Google Scholar]
- 15. Douangamath A, et al. 2002. Structural evidence for ammonia tunneling across the (beta alpha)(8) barrel of the imidazole glycerol phosphate synthase bienzyme complex. Structure 10:185–193 [DOI] [PubMed] [Google Scholar]
- 16. Feng L, et al. 2005. Structural and genetic characterization of enterohemorrhagic Escherichia coli O145 O antigen and development of an O145 serogroup-specific PCR assay. J. Bacteriol. 187:758–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guan Z, Naparstek S, Calo D, Eichler J. 2011. Protein glycosylation as an adaptive response in archaea: growth at different salt concentrations leads to alterations in Haloferax volcanii S-layer glycoprotein N-glycosylation. Environ. Microbiol. 14:743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guan Z, Eichler J. 2011. Liquid chromatography/tandem mass spectrometry of dolichols and polyprenols, lipid sugar carriers across evolution. Biochim. Biophys. Acta 1811:800–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guan Z, Meyer BH, Albers SV, Eichler J. 2011. The thermoacidophilic archaeon Sulfolobus acidocaldarius contains an unsually short, highly reduced dolichyl phosphate. Biochim. Biophys. Acta 1811:607–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hendrickson EL, et al. 2004. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J. Bacteriol. 186:6956–6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Igura M, et al. 2007. Purification, crystallization and preliminary X-ray diffraction studies of the soluble domain of the oligosaccharyltransferase STT3 subunit from the thermophilic archaeon Pyrococcus furiosus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63:798–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jarrell KF, Jones GM, Nair DB. 2010. Biosynthesis and role of N-linked glycosylation in cell surface structures of archaea with a focus on flagella and S layers. Int. J. Microbiol. 2010:470138 doi:10.1155/2010/470138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jarrell KF, Jones GM, Kandiba L, Nair DB, Eichler J. 2010. S-layer glycoproteins and flagellins: reporters of archaeal posttranslational modifications. Archaea 2010:612948 doi:10.1155/2010/612948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kaminski L, et al. 2010. AglJ adds the first sugar of the N-linked pentasaccharide decorating the Haloferax volcanii S-layer glycoprotein. J. Bacteriol. 192:5572–5579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kelly J, Logan SM, Jarrell KF, Vandyke DJ, Vinogradov E. 2009. A novel N-linked flagellar glycan from Methanococcus maripaludis. Carbohydr. Res. 344:648–653 [DOI] [PubMed] [Google Scholar]
- 26. Kessler PS, Leigh JA. 1999. Genetics of nitrogen regulation in Methanococcus maripaludis. Genetics 152:1343–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. King JD, Vinogradov E, Tran V, Lam JS. 2010. Biosynthesis of uronamide sugars in Pseudomonas aeruginosa O6 and Escherichia coli O121 O antigens. Environ. Microbiol. 12:1531–1544 [DOI] [PubMed] [Google Scholar]
- 28. King JD, et al. 2008. lfnA from Pseudomonas aeruginosa O12 and wbuX from Escherichia coli O145 encode membrane-associated proteins and are required for expression of 2,6-dideoxy-2-acetamidino-l-galactose in lipopolysaccharide O antigen. J. Bacteriol. 190:1671–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Klem TJ, Davisson VJ. 1993. Imidazole glycerol phosphate synthase: the glutamine amidotransferase in histidine biosynthesis. Biochemistry 32:5177–5186 [DOI] [PubMed] [Google Scholar]
- 30. Lie TJ, Wood GE, Leigh JA. 2005. Regulation of nif expression in Methanococcus maripaludis: roles of the euryarchaeal repressor NrpR, 2-oxoglutarate, and two operators. J. Biol. Chem. 280:5236–5241 [DOI] [PubMed] [Google Scholar]
- 31. Linton D, et al. 2005. Functional analysis of the Campylobacter jejuni N-linked protein glycosylation pathway. Mol. Microbiol. 55:1695–1703 [DOI] [PubMed] [Google Scholar]
- 32. Logan SM, Kelly JF, Thibault P, Ewing CP, Guerry P. 2002. Structural heterogeneity of carbohydrate modifications affects serospecificity of Campylobacter flagellins. Mol. Microbiol. 46:587–597 [DOI] [PubMed] [Google Scholar]
- 33. Magidovich H, Eichler J. 2009. Glycosyltransferases and oligosaccharyltransferases in archaea: putative components of the N-glycosylation pathway in the third domain of life. FEMS Microbiol. Lett. 300:122–130 [DOI] [PubMed] [Google Scholar]
- 34. Magidovich H, et al. 2010. AglP is an S-adenosyl-l-methionine-dependent methyltransferase that participates in the N-glycosylation pathway in Haloferax volcanii. Mol. Microbiol. 76:190–199 [DOI] [PubMed] [Google Scholar]
- 35. Maita N, Nyirenda J, Igura M, Kamishikiryo J, Kohda D. 2010. Comparative structural biology of eubacterial and archaeal oligosaccharyltransferases. J. Biol. Chem. 285:4941–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McNally DJ, et al. 2006. Functional characterization of the flagellar glycosylation locus in Campylobacter jejuni 81-176 using a focused metabolomics approach. J. Biol. Chem. 281:18489–18498 [DOI] [PubMed] [Google Scholar]
- 37. Mescher MF, Strominger JL. 1976. Purification and characterization of a prokaryotic glucoprotein from the cell envelope of Halobacterium salinarium. J. Biol. Chem. 251:2005–2014 [PubMed] [Google Scholar]
- 38. Meyer BH, et al. 2011. Sulfoquinovose synthase—an important enzyme in the N-glycosylation pathway of Sulfolobus acidocaldarius. Mol. Microbiol. 82:1150–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moore BC, Leigh JA. 2005. Markerless mutagenesis in Methanococcus maripaludis demonstrates roles for alanine dehydrogenase, alanine racemase, and alanine permease. J. Bacteriol. 187:972–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mouilleron S, Golinelli-Pimpaneau B. 2007. Conformational changes in ammonia-channeling glutamine amidotransferases. Curr. Opin. Struct. Biol. 17:653–664 [DOI] [PubMed] [Google Scholar]
- 41. Namboori SC, Graham DE. 2008. Acetamido sugar biosynthesis in the Euryarchaea. J. Bacteriol. 190:2987–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ng SYM, et al. 2011. Genetic and mass spectrometry analysis of the unusual type IV-like pili of the archaeon Methanococcus maripaludis. J. Bacteriol. 193:804–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nothaft H, Liu X, McNally DJ, Szymanski CM. 2010. N-linked protein glycosylation in a bacterial system. Methods Mol. Biol. 600:227–243 [DOI] [PubMed] [Google Scholar]
- 44. Peyfoon E, et al. 2010. The S-layer glycoprotein of the crenarchaeote Sulfolobus acidocaldarius is glycosylated at multiple sites with the chitobiose-linked N-glycans. Archaea 2010:754101 doi:10.1155/2010/754101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. VanDyke DJ, et al. 2009. Identification of genes involved in the assembly and attachment of a novel flagellin N-linked tetrasaccharide important for motility in the archaeon Methanococcus maripaludis. Mol. Microbiol. 72:633–644 [DOI] [PubMed] [Google Scholar]
- 46. VanDyke DJ, et al. 2008. Identification of putative acetyltransferase gene, MMP0350, which affects proper assembly of both flagella and pili in the archaeon Methanococcus maripaludis. J. Bacteriol. 190:5300–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Voisin S, et al. 2005. Identification and characterization of the unique N-linked glycan common to the flagellins and S-layer glycoprotein of Methanococcus voltae. J. Biol. Chem. 280:16586–16593 [DOI] [PubMed] [Google Scholar]
- 48. Yurist-Doutsch S, Chaban B, VanDyke DJ, Jarrell KF, Eichler J. 2008. Sweet to the extreme: protein glycosylation in archaea. Mol. Microbiol. 68:1079–1084 [DOI] [PubMed] [Google Scholar]
- 49. Yurist-Doutsch S, et al. 2010. N-glycosylation in archaea: on the coordinated actions of Haloferax volcanii AglF and AglM. Mol. Microbiol. 75:1047–1058 [DOI] [PubMed] [Google Scholar]
- 50. Yurist-Doutsch S, et al. 2008. aglF, aglG and aglI, novel members of a gene island involved in the N-glycosylation of the Haloferax volcanii S-layer glycoprotein. Mol. Microbiol. 69:1234–1245 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.