Abstract
Generation of G-strand overhangs at Saccharomyces cerevisiae yeast telomeres depends primarily on the MRX (Mre11-Rad50-Xrs2) complex, which is also necessary to maintain telomere length by recruiting the Tel1 kinase. MRX physically interacts with Rif2, which inhibits both resection and elongation of telomeres. We provide evidence that regulation of telomere processing and elongation relies on a balance between Tel1 and Rif2 activities. Tel1 regulates telomere nucleolytic processing by promoting MRX activity. In fact, the lack of Tel1 impairs MRX-dependent telomere resection, which is instead enhanced by the Tel1-hy909 mutant variant, which causes telomerase-dependent telomere overelongation. The Tel1-hy909 variant is more robustly associated than wild-type Tel1 to double-strand-break (DSB) ends carrying telomeric repeat sequences. Furthermore, it increases the persistence at a DSB adjacent to telomeric repeats of both MRX and Est1, which in turn likely account for the increased telomere resection and elongation in TEL1-hy909 cells. Strikingly, Rif2 is unable to negatively regulate processing and lengthening at TEL1-hy909 telomeres, indicating that the Tel1-hy909 variant overcomes the inhibitory activity exerted by Rif2 on MRX. Altogether, these findings highlight a primary role of Tel1 in overcoming Rif2-dependent negative regulation of MRX activity in telomere resection and elongation.
INTRODUCTION
A highly ordered nucleoprotein complex called the telomere prevents the ends of linear chromosomes from being recognized as DNA double-strand breaks (DSBs) (reviewed in reference 30). Another key function of telomeres is to compensate for the incomplete replication of chromosome ends caused by discontinuous DNA synthesis. In most eukaryotes, telomeric DNA comprises tandemly repeated G-rich sequences (TG1-3 repeats in Saccharomyces cerevisiae yeast and T2AG3 in vertebrates), ending in a single-stranded 3′ overhang (G tail) (reviewed in reference 20). The addition of telomeric repeats depends on the action of telomerase, a specialized reverse transcriptase that extends the TG-rich strand of chromosome ends (23). The yeast telomerase complex consists of a reverse transcriptase subunit (Est2), a template RNA (TLC1), and two accessory proteins (Est1 and Est3), which are required for telomerase activity in vivo but not in vitro.
The single-stranded G tails of budding yeast telomeres are short (about 10 to 15 nucleotides [nt]) for most of the cell cycle, but their length increases transiently to 50 to 100 nucleotides in late S phase (15, 50). While telomeric G tails can be generated during lagging-strand replication by removal of the last RNA primer, the 5′ C strand of the telomere generated by leading-strand synthesis must be nucleolytically processed (resected) to generate 3′ overhangs (32, 51). Cyclin-dependent kinase (Cdk1 in S. cerevisiae) activity is required for this telomeric C-strand resection (17, 49), which occurs only during S and G2 cell cycle phases, when Cdk1 activity is high and telomeres are elongated by telomerase (36). The MRX (Mre11-Rad50-Xrs2) complex and Sae2 have been shown to be important for resection of telomeric ends, with MRX playing the major role (8, 14, 28, 31). Moreover, Exo1 and Sgs1-Dna2 can provide a backup mechanism for telomere resection when Sae2-MRX activity is compromised (8).
In the yeast Saccharomyces cerevisiae, telomerase lengthens telomeres only in late S/G2 phase (36). Although chromatin immunoprecipitation (ChIP) studies have found that Est2 is associated to telomeres from G1 to late S phase (44), the use of live-cell imaging has shown that a subset of TLC1 molecules clusters and stably associates with telomeres only in late S phase (18). This clustering has been proposed to represent actively elongating telomeres (18), suggesting that regulation of telomerase activity is achieved at the level of its association with the telomere. In agreement with this hypothesis, budding yeast telomerase is preferentially enriched at short telomeres (4, 43), which are its preferred substrate (35, 45).
Telomere length maintenance depends on the checkpoint kinase Tel1, whose localization to chromosome ends requires the MRX complex (39). The function of Tel1 at telomeres relies on its kinase activity, since tel1 kinase-dead cells have short telomeres like tel1Δ cells (22, 33). However, how Tel1 regulates telomere length is still unknown. Cells lacking Tel1 have a decreased frequency of telomerase-mediated telomere elongation (2). Furthermore, Tel1 has been shown to specifically associate with short telomeres (4, 25, 43) and is needed for preferential binding of Est1 and Est2 to them (21). These findings suggest that length-dependent binding of Tel1 to telomeres is a critical step in the regulation of telomerase association with telomeres in S phase. In budding yeast, telomerase recruitment at telomeres relies on a direct interaction between Est1 and the telomere end-binding protein Cdc13 (5, 9, 16, 40). It has been proposed that Tel1 promotes Est1-Cdc13 interaction by phosphorylating the telomerase recruitment domain of Cdc13 (46), although this model has recently been questioned (19). Tel1 is also important for increasing telomerase processivity at critically short telomeres (10). However, since the occurrence of critically short telomeres (<125 bp in length) should be relatively infrequent, the primary cause of the short tel1Δ telomeres has been proposed to be a reduced frequency of elongation by telomerase, rather than a reduced telomerase processivity (10).
The identity of S. cerevisiae telomeres also relies on a protein complex formed by the Rap1, Rif1, and Rif2 proteins, which inhibit telomerase-dependent telomere elongation (12, 24, 29, 52). Rif2 physically interacts with MRX in vitro (26) and inhibits MRX-dependent 5′-end resection at telomeres (6, 7). Furthermore, the lack of Rif2 enhances MRX and Tel1 association at telomeres (7, 26), suggesting that Rif2 regulates nucleolytic processing of telomeres by inhibiting MRX recruitment. However, the artificial tethering of Rif2 at DNA ends leads to decreased Tel1 binding, but not MRX binding, to these ends (26), indicating that Rif2 counteracts Tel1 association to DNA ends. This observation, together with the finding that MRX localization at telomeres is reduced in tel1Δ cells compared to wild-type cells (26), raises the possibility that Tel1, after its MRX-dependent loading onto telomeric ends, can enhance MRX activity. As MRX is required to generate telomeric single-stranded DNA (ssDNA), this Tel1-dependent regulation of MRX might influence resection of telomeric ends. Consistent with this hypothesis, loss of Tel1 in telomerase-negative cells attenuates the onset of senescence (19, 42), whose primary signal has been proposed to be telomeric ssDNA (1).
To study the physiological consequences of the Tel1-mediated feedback loop on MRX, as well as the role of Rif2 in counteracting Tel1/MRX function, we took advantage of the TEL1-hy909 mutant allele, previously identified to be a dominant suppressor of the hypersensitivity to genotoxic agents and checkpoint defects of Mec1-deficient cells (3). Here we provide evidence that Tel1 regulates G-tail generation at telomeres by promoting MRX function. In fact, the lack of Tel1 impairs MRX-dependent generation of ssDNA at telomeres, whereas the Tel1-hy909 variant enhances both resection and elongation of telomeric ends. Moreover, Tel1-hy909 is more robustly associated to DSBs adjacent to telomeric tracts than wild-type Tel1 and also enhances MRX and Est1 binding at these DNA ends. These findings, together with the observation that TEL1-hy909 telomeres escape the negative regulation exerted by Rif2 on both processing and elongation, indicate that Rif2-mediated inhibition of Tel1/MRX activity is important to regulate nucleolytic processing and elongation of telomeres.
MATERIALS AND METHODS
Strains.
Strain genotypes are listed in Table S1 in the supplemental material. The strains used for monitoring resection at the HO-induced DSB adjacent to telomeric repeats were derivatives of strain UCC5913, kindly provided by D. Gottschling (Fred Hutchinson Cancer Research Center, Seattle, WA). The strains used for monitoring resection at the HO-induced DSB were derivatives of strain RMY169, kindly provided by T. Weinert (University of Arizona, Tucson, AZ), and JKM139, kindly provided by J. Haber (Brandeis University, Waltham, MA). Strain RMY169 was created by replacing the ADE2-TG cassette of strain UCC5913 with the TRP1 gene (38). The strains used for monitoring telomere addition at an HO-induced DSB adjacent to telomeric repeats were derivatives of strain YSN645, kindly provided by D. Shore (University of Geneva, Geneva, Switzerland). Strains UCC5913, RMY169, YSN645, and JKM139 were used to replace the TEL1 chromosomal copy with the TEL1-hy909 allele by PCR, giving rise to strains YLL2836, YLL3066, YLL2255, and YLL2880, respectively. To induce a persistent G1 arrest with α-factor, the BAR1 gene, which encodes a protease that degrades the α-factor, was deleted. Deletions of the YKU70, EXO1, MRE11, TEL1, RIF1, RIF2, RAD51, RAD52, EST2, and BAR1 genes were generated by a one-step PCR disruption method. A TEL1-hy909 xrs2-11 strain was obtained by crossing strain KSC1563 (MATa-inc ADH4cs::HIS2 ade1 his2 leu2 trp1 ura3 xrs2-11::KANMX4 sml1::LEU2), kindly provided by K. Sugimoto (University of New Jersey, Newark, NJ), with a TEL1-hy909 strain. Strain YLL3068, carrying a fully functional TEL1-HA allele at the TEL1 chromosomal locus (the hemagglutinin [HA] tag coding sequence was located 2,394 bp downstream of the TEL1 start codon), was obtained by transforming strain YLL2599 with a PCR product obtained using genomic DNA of strain YLL3001 as the template together with primers PRP1297 (5′-GGC CAC CGT TAA GAA GGG TAA GCC AGA A-3′) and PRP1298 (5′-GTT GCA AAG ACT CTC TGC TTC CCA CCT C-3′). Strain YLL3001 was a derivative of strain KRY22, kindly provided by T. Petes (Duke University School of Medicine, Durham, NC), in which the NATMX cassette was inserted upstream of the TEL1 start codon. Strain YLL3003, carrying the HA-tagged TEL1-hy909 allele at the TEL1 chromosomal locus, was obtained by transforming strain YLL2836 with a PCR product obtained using genomic DNA of strain YLL3001 as the template together with primers PRP1297 and PRP1298. PCR one-step tagging was used to obtain strains carrying fully functional MYC-tagged MRE11 and MYC-tagged EST1 alleles. The TEL1-hy909kd allele, encoding a kinase-defective Tel1-hy909 variant carrying the amino acid changes G2611D, D2612A, N2616K, and D2631E, was obtained by site-directed mutagenesis. The accuracy of all gene replacements and integrations was verified by Southern blot analysis or PCR. Cells were grown in YEP medium (1% yeast extract, 2% Bacto peptone, 50 mg/liter adenine) supplemented with 2% glucose (YEPD) or 3% glycerol, 2% lactic acid, and 0.05% glucose or with 3% glycerol, 2% lactic acid, and 2% galactose.
Resection assay.
Visualization of the single-stranded overhangs at native telomeres was done as described previously (15). The same gel was denatured and hybridized with the end-labeled C-rich oligonucleotide for loading control. Resection of the 5′ strand at the HO-induced DSB adjacent to telomeric repeat sequences was monitored as previously described (7). To monitor resection at the HO-induced DSB, SspI- and XbaI-digested genomic DNA was hybridized with a single-stranded riboprobe, which anneals to the 5′ strand to a site located 248 nt from the HO cutting site. For quantitative analysis of resection, the ratios between the intensities of the cut 5′ strand and loading control bands were calculated by using the NIH image program. For quantitative analysis of G-tail signals at native telomeres, the ratios between the intensities of the TG-ssDNA signals on the native gels and the total amount of TG repeats revealed by the same probe after denaturation of the gels were calculated by using the NIH image program.
ChIP analysis.
ChIP analysis was performed as previously described (47). After exposure to formaldehyde, chromatin samples were immunoprecipitated with appropriate antibodies. Quantification of immunoprecipitated DNA was achieved by quantitative real-time PCR (qPCR) on a Bio-Rad MiniOpticon apparatus using primer pairs located at the nontelomeric ARO1 fragment of chromosome IV (CON) and 640 bp centromere proximal (TG-HO) or 550 bp centromere distal (HO) to the HO cutting site at chromosome VII, and the amount was normalized to the input signal for each primer set; data are expressed as the fold enrichment of TG-HO or HO over the amount of CON in the immunoprecipitates.
RESULTS
Tel1-hy909 causes MRX- and telomerase-dependent telomere overelongation.
In a screening for TEL1 mutations that suppressed the hypersensitivity to genotoxic agents of mec1Δ cells, we previously identified 7 TEL1-hy alleles which can partially bypass the Mec1 requirement for checkpoint activation (3). Most of the Tel1-hy variants displayed enhanced kinase activity in vitro compared to wild-type Tel1, but only the Tel1-hy909 variant causes an impressive telomere elongation (3) (Fig. 1).
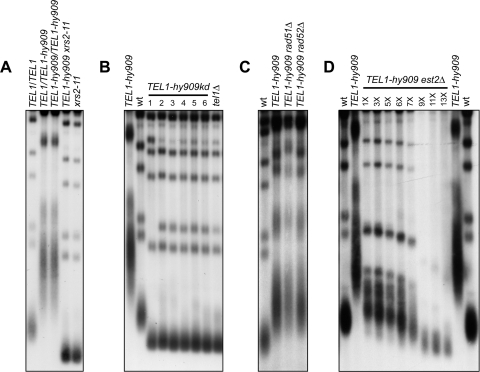
Fig 1.
Telomere overelongation in TEL1-hy909 cells. (A to C) XhoI-cut genomic DNA from exponentially growing cells (YEPD at 25°C) was subjected to Southern blot analysis using a radiolabeled poly(GT) telomere-specific probe. For panel B, 6 independent TEL1-hy909kd transformant clones were analyzed (lanes 1 to 6). (D) Meiotic tetrads from EST2/est2Δ TEL1/TEL1-hy909 diploid cells were dissected on YEPD plates. After ∼25 generations, TEL1-hy909 est2Δ spore clones were streaked for successive times (1 to 13 times) and aliquots of cells from the indicated streaks were propagated in YEPD liquid medium for 5 h to prepare genomic DNA for telomere length detection, as in panels A to C. Each subsequent streak represents ∼25 generations of growth. wt, wild type.
The overelongated telomere phenotype of TEL1-hy909 cells is dominant and requires Tel1 kinase activity. In fact, TEL1-hy909 heterozygous and homozygous diploid cells exhibited very similar telomere lengths (Fig. 1A). Furthermore, cells expressing a Tel1-hy909 variant carrying the G2611D, D2612A, N2616K, and D2631E amino acid changes (Tel1-hy909kd in Fig. 1B), which were shown to destroy Tel1 kinase activity (33), had telomeres as short as those of tel1Δ cells (Fig. 1B). These data further support previous findings that Tel1 acts as a kinase to maintain telomere length (33).
The MRX complex is required to load Tel1 onto DNA ends (39). MRX is properly assembled and binds DSBs in xrs2-11 cells, which express truncated Xrs2 lacking the C-terminal 162 amino acids, but Tel1 binding to DSBs is compromised in the same cells (39). As shown in Fig. 1A, telomeres in xrs2-11 TEL1-hy909 double mutant cells were as short as in xrs2-11 single mutant cells, indicating that Tel1-hy909 action at telomeres does not bypass MRX requirement.
Telomere elongation is primarily accomplished by telomerase and occasionally by homologous recombination, but the latter does not seem to have a role in Tel1-hy909-induced telomere overelongation. In fact, the lack of the recombination protein Rad51 or Rad52 in TEL1-hy909 cells did not modify telomere length (Fig. 1C). We then assessed the contribution of the telomerase enzyme to this TEL1-hy909 phenotype by dissecting meiotic tetrads from a diploid strain heterozygous for the est2Δ and TEL1-hy909 alleles. After 2 days of incubation at 25°C (approximately 25 generations), spore clones were streaked for successive times, and aliquots of cells from the indicated streaks were propagated in YEPD liquid medium for 5 h to prepare genomic DNA for telomere length analysis (Fig. 1D). Telomeres underwent progressive shortening in TEL1-hy909 est2Δ clones (Fig. 1D), indicating that TEL1-hy909-dependent telomere overelongation requires telomerase activity.
The tel1Δ and TEL1-hy909 mutations exert opposite effects on ssDNA generation at a DSB adjacent to telomeric repeat sequences.
Because the absence of Tel1 attenuates the senescence phenotype of telomerase-negative cells, it was suggested that Tel1 has additional functions at telomeres besides directly affecting telomerase action (19, 42). Tel1 recruitment at telomeres requires the MRX complex (39), which is necessary to generate 3′-ended ssDNA at telomeric ends (8, 14, 28). MRX association to telomeres is decreased in tel1Δ cells (26), suggesting that Tel1 may be involved in stimulating resection of the telomeric ends by promoting MRX activity. To assess whether Tel1 exerts a positive-feedback loop on MRX in vivo, we analyzed resection of telomeric ends in tel1Δ and TEL1-hy909 cells. We used an assay initially developed to examine de novo telomere formation (13, 14), where an 81-bp TG repeat sequence is placed immediately adjacent to an HO endonuclease cut site (Fig. 2A). The strain carries this HO cleavage site inserted into the ADH4 locus on chromosome VII and expresses the HO endonuclease gene from a galactose-inducible promoter. Upon cleavage by HO, the centromere-proximal TG side of the break is “healed” by telomerase and gives rise to a bona fide telomere (13, 14). DNA degradation was assessed by Southern blotting under denaturing conditions using a single-stranded riboprobe that recognizes the 5′ C strand (Fig. 2A). Upon digestion of genomic DNA with EcoRV and RsaI restriction enzymes, the probe reveals an uncut 390-nt DNA fragment (uncut), which is converted by HO cleavage into a 166-nt fragment (cut C strand) (Fig. 2A). Degradation of the 5′ C strand leads to disappearance of the probe signal when resection proceeds beyond the hybridization region.
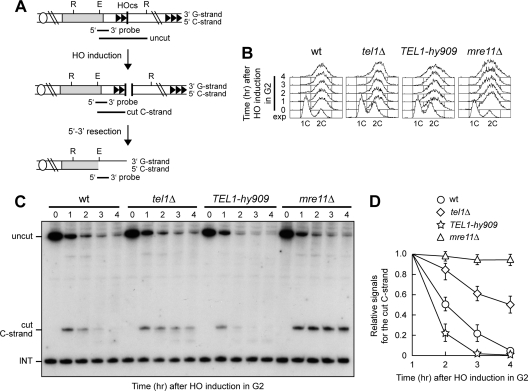
Fig 2.
Effects of the tel1Δ and TEL1-hy909 mutations on ssDNA generation at a DSB end carrying telomeric repeats. (A) Schematic representation of the HO cleavage site (HOcs) with the TG repeat sequences (81 bp; two arrowheads) that were placed centromere proximal to the HO site at the ADH4 locus on chromosome VII-L. The centromere is shown as a circle on the left. The probe used to monitor nucleolytic degradation of the 5′ C strand is also indicated. R, RsaI; E, EcoRV. (B to D) HO expression was induced at time zero by galactose addition to nocodazole-arrested cell cultures that were kept arrested in G2. (B) Fluorescence-activated cell sorter analysis of DNA content. exp, exponentially growing cells. (C) RsaI- and EcoRV-digested genomic DNA was hybridized with a single-stranded riboprobe that anneals to the 5′ C strand to a site located 212 bp from the HO cutting site. The probe reveals an uncut 390-nt DNA fragment (uncut), which is converted by HO cleavage into a 166-nt fragment (cut C strand). Degradation of the 5′ C strand leads to disappearance of the probe signal as resection proceeds beyond the hybridization region. The probe also detects a 138-nt fragment from the ade2-101 locus on chromosome XV (INT), which serves as internal loading control. (D) Densitometric analysis. Plotted values are means ± SDs from three independent experiments, as in panel C.
As generation of ssDNA at telomeres occurs during S and G2/M cell cycle phases, when Cdk1 activity is high (17, 49), we monitored 5′-3′ resection of the TG DSB end by inducing HO expression in tel1Δ, TEL1-hy909, and mre11Δ cells that were kept arrested in G2 with nocodazole (Fig. 2B). Degradation of the band corresponding to the cut 5′ C strand (cut C strand) occurred more slowly in G2-arrested tel1Δ cells than in wild type, and it was completely abolished, as expected (14, 28), in G2-arrested mre11Δ cells (Fig. 2C and D). In contrast, 5′ C-strand degradation was more efficient in TEL1-hy909 cells than in wild type (Fig. 2C and D). Thus, the lack of Tel1 impairs resection at DSB ends adjacent to telomeric DNA, while Tel1-hy909 increases its efficiency. These observations, together with the notion that 5′ C-strand degradation in G2-arrested cells requires the MRX complex (Fig. 2C and D) (14, 28), indicate that Tel1 influences 5′-3′ nucleolytic processing of these DNA ends by promoting MRX activity. Interestingly, MRX function is not completely abrogated by the lack of Tel1, as 5′ C-strand degradation in tel1Δ cells was reduced but not completely abolished as it was in mre11Δ cells (Fig. 2C and D).
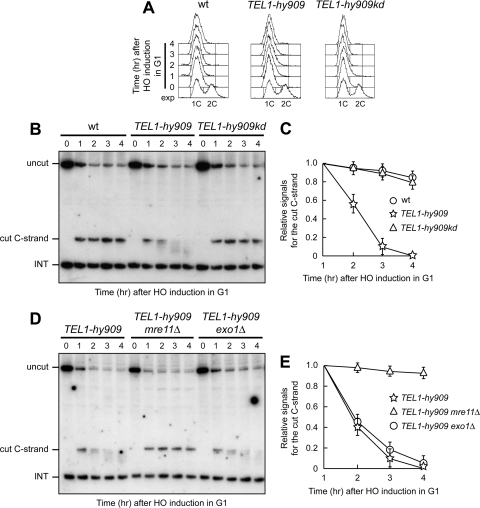
The Tel1-hy909 variant might enhance nucleolytic processing at telomeres by hyperactivating the MRX complex. As nucleolytic degradation at telomeres requires Cdk1 activity (17, 49), which is thought to activate the MRX-dependent resection machinery (6, 7), we asked whether the Tel1-hy909 variant bypasses Cdk1 requirement for resection of a DSB adjacent to telomeric repeat sequences. To this end, the HO cut was induced in G1-arrested wild-type and TEL1-hy909 cells (Fig. 3A) carrying the system described in Fig. 2A. Consistent with the requirement of Cdk1 activity for DNA end resection, the 5′ C-rich strand signal was stable in G1-arrested wild-type cells (Fig. 3B and C). In contrast, it progressively decreased in G1-arrested TEL1-hy909 cells (Fig. 3B and C), indicating that the TEL1-hy909 mutation allows Cdk1-independent nucleolytic processing of a DSB end with telomeric repeats. This ability of Tel1-hy909 to bypass Cdk1 requirement for resection depends on Tel1 kinase activity, as 5′ C-strand degradation was prevented in G1-arrested TEL1-hy909kd cells (Fig. 3A to C). Furthermore, this Tel1-hy909-dependent resection specifically requires MRX, because it was abolished in G1-arrested TEL1-hy909 mre11Δ cells, whereas it took place in TEL1-hy909 cells lacking the Exo1 nuclease (Fig. 3D and E). As Cdk1 has been proposed to induce DSB resection by increasing the efficiency of the MRX-dependent resection machinery (6, 7), these data are consistent with the hypothesis that Tel1-hy909 hyperactivates MRX.
Fig 3.
Tel1-hy909 allows resection in G1 of a DSB adjacent to telomeric repeats. (A to C) HO expression was induced at time zero by galactose addition to α-factor-arrested cells, all carrying the system described in Fig. 2A. Cells were kept arrested in G1. (A) Fluorescence-activated cell sorter analysis of DNA content. (B) RsaI- and EcoRV-digested genomic DNA was analyzed as described in the legend to Fig. 2C. (C) Densitometric analysis. Plotted values are means ± SDs from three independent experiments, as in panel B. (D and E) HO expression was induced at time zero by galactose addition to α-factor-arrested cells that were kept arrested in G1. Cell cycle arrest was verified by fluorescence-activated cell sorter analysis (data not shown). (D) RsaI- and EcoRV-digested genomic DNA was analyzed as described in the legend to Fig. 2C. (E) Densitometric analysis. Plotted values are means ± SDs from three independent experiments, as in panel D.
Tel1-hy909 does not bypass Cdk1 requirement to generate ssDNA at DSB ends lacking telomeric repeats.
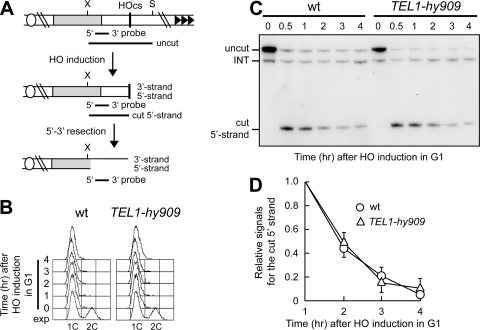
We have previously shown that Tel1-hy909 increases ssDNA generation at intrachromosomal DSBs in G2-arrested cells (3). Thus, we asked whether the Tel1-hy909 variant could bypass Cdk1 requirement for ssDNA generation at an HO-induced DSB devoid of telomeric repeats, as it does when the DSB is adjacent to telomeric tracts (Fig. 3). HO expression was induced in G1-arrested wild-type and TEL1-hy909 cell cultures that carried an HO cut site with no TG repeats (Fig. 4A) and that were kept arrested in G1 with α-factor (Fig. 4B). In contrast to what happens at a DSB end carrying telomeric repeats, whose resection was completely abolished in G1 (Fig. 3B and C), a degradation of the 5′ DSB end with no TG repeats still occurred in G1-arrested wild-type cells (Fig. 4C and D), although it was less efficient than that detected at the same DSB in G2 (data not shown) (27). On the other hand, TEL1-hy909 G1 cells showed a behavior very similar to that of wild type (Fig. 4C and D), indicating that Tel1-hy909 does not promote Cdk1-independent 5′-3′ nucleolytic processing at DSB ends that do not carry telomeric DNA sequences. This hypothesis was further confirmed by the observation that both wild-type and TEL1-hy909 G1 cells generated very similar small amounts of 3′-ended single-stranded resection products at an irreparable HO-induced DSB generated at the MAT locus (see Fig. S1 in the supplemental material). Thus, we can conclude that Tel1-hy909 in G1 exerts its action preferentially at DNA ends adjacent to telomeric repeat sequences.
Fig 4.
Tel1-hy909 does not enhance nucleolytic processing in G1 of a DSB with no telomeric repeats. (A) Schematic representation of the system used to generate the HO-induced DSB on chromosome VII. SspI- and XbaI-digested genomic DNA is hybridized with the indicated single-stranded probe that anneals to the 5′ strand to a site located 248 nt from the HO cutting site, revealing an uncut 897-nt DNA fragment (uncut), which is converted by HO cleavage into a 286-nt fragment (cut 5′ strand). Loss of the 5′ strand beyond the hybridization region of the probe leads to disappearance of the signal generated by the probe. S, SspI; X, XbaI. (B to D) HO expression was induced at time zero by galactose addition to α-factor-arrested wild-type and TEL1-hy909 cells, all carrying the system depicted in panel A. Cells were kept arrested in G1. (B) Fluorescence-activated cell sorter analysis of DNA content. (C) SspI- and XbaI-digested genomic DNA was hybridized with the probe described in panel A. (D) Densitometric analysis. Plotted values are means ± SDs from four independent experiments, as in panel C.
The tel1Δ and TEL1-hy909 mutations exert opposite effects at native telomeres.
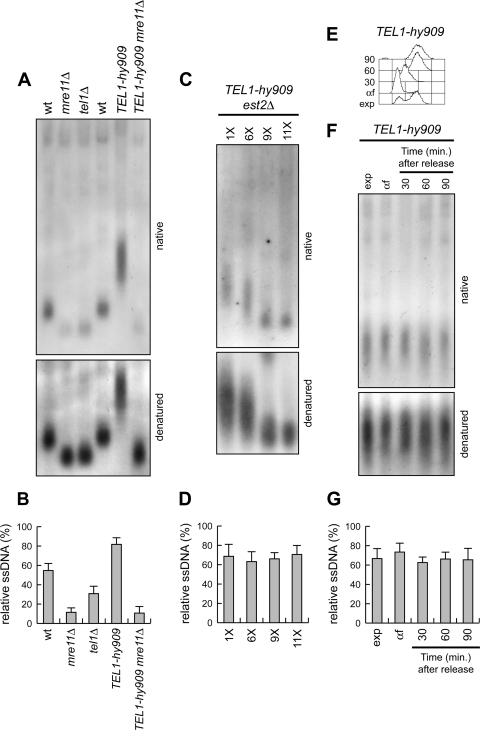
The above-described findings prompted us to investigate whether the role of Tel1 in promoting ssDNA generation at DSB ends carrying TG repeats could be extended to native telomeres. To assess the presence of ssDNA at natural chromosome ends, genomic DNA prepared from exponentially growing cells was analyzed by nondenaturing in-gel hybridization with a C-rich radiolabeled oligonucleotide that detect the G-rich single-stranded telomere overhangs (15). Because tel1Δ telomeres have fewer TG sequences than wild-type telomeres, the TG-ssDNA signals were normalized to the total amount of TG repeats revealed by the same probe after denaturation of the gel. Consistent with a role of Tel1 in promoting single-stranded overhang generation also at native chromosome ends, the amount of ssDNA was lower at tel1Δ telomeres than at wild-type telomeres (Fig. 5A and B). As previously observed (14, 28), mre11Δ telomeres also displayed a reduced amount of telomeric ssDNA compared to wild type (Fig. 5A and B). However, similar to what happens at the DSB ends carrying telomeric repeats (Fig. 2C and D), the single-stranded G-tail signal was lower in mre11Δ than in tel1Δ cells (Fig. 5A and B), indicating that MRX activity at telomeres is not completely abolished by the lack of Tel1.
Fig 5.
Effects of tel1Δ and TEL1-hy909 alleles on ssDNA formation at native telomeres. (A) Genomic DNA prepared from exponentially growing cells was digested with XhoI, and single-stranded G tails were visualized by in-gel hybridization (native) using an end-labeled C-rich oligonucleotide as a probe. The gel was then denatured and hybridized again with the same probe for loading control (denatured). (B) The amount of native TG-ssDNA, as in panel A, was normalized to the total amount of TG sequences detected in each denatured sample. (C and D) Meiotic tetrads from EST2/est2Δ TEL1/tel1-hy909 diploid cells were dissected on YEPD plates. (C) After ∼25 generations, TEL1-hy909 est2Δ spore clones were streaked for successive times (1 to 11 times), and aliquots of cells from the indicated streaks were propagated in YEPD liquid medium for 5 h to prepare genomic DNA for telomeric ssDNA detection, as in panel A. (D) The amount of native TG-ssDNA, as in panel C, was normalized to the total amount of TG sequences detected in each denatured sample. (E to G) Exponentially growing (exp) TEL1-hy909 cells were arrested in G1 with α-factor (αf) and released into the cell cycle. (E) Fluorescence-activated cell sorter analysis of DNA content. (F) Genomic DNA prepared at the indicated time points after release from the α-factor block was analyzed for telomeric ssDNA detection, as in panel A. (G) The amount of native TG-ssDNA, as in panel F, was normalized to the total amount of TG sequences detected in each denatured sample. Plotted values in panels B, D, and G are means ± SDs from three independent experiments.
The same analysis showed a stronger intensity of the G-tail signal in exponentially growing TEL1-hy909 cells than in wild-type cells (Fig. 5A and B). The ssDNA formation at TEL1-hy909 telomeres depends on the MRX complex, as the lack of Mre11 reduced the amount of the G-tail signal in TEL1-hy909 cells to the level observed in mre11Δ cells (Fig. 5A and B). Furthermore, the strong ssDNA signal at TEL1-hy909 telomeres was due to resection rather than to telomerase action, because similar amounts of telomeric ssDNA were detected during successive streaks of TEL1-hy909 cells that shortened telomeres due to the absence of the telomerase subunit Est2 (Fig. 5C and D).
The Tel1-hy909 variant allows resection of a DSB end with telomeric repeats even in G1 (Fig. 3), and this property is extended to native telomeres. In fact, when TEL1-hy909 cells were arrested in G1 with α-factor and released into the cell cycle (Fig. 5E), the intensity of the telomeric ssDNA signal in G1-arrested cells was similar to that observed both during exponential growth and at different times after release (Fig. 5F and G). Thus, the Tel1-hy909 variant appears to bypass Cdk1 requirement for ssDNA generation even at native telomeres.
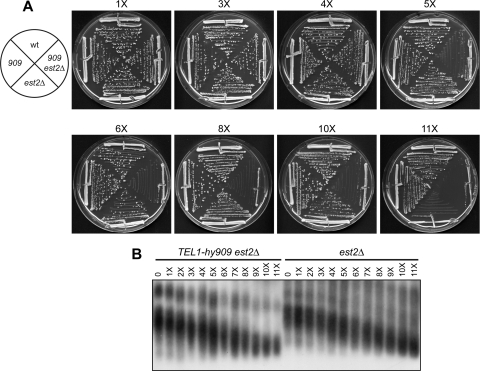
Because ssDNA at telomeres has been proposed to signal the onset of senescence (1), we could expect differences in the decline of growth of est2Δ versus est2Δ TEL1-hy909 cells. Meiotic tetrads were dissected from a diploid strain heterozygous for both est2Δ and TEL1-hy909. After incubation for 2 days at 25°C (approximately 25 generations), spore clones were streaked for successive times (Fig. 6A) and an aliquot of cells from each streak was propagated in YEPD liquid medium for 5 h to prepare genomic DNA for telomere length analysis (Fig. 6B). As the TEL1-hy909 allele also causes telomere overelongation under heterozygous conditions (Fig. 1A), est2Δ and est2Δ TEL1-hy909 spores started with similar very long telomeres, which shortened during the subsequent subculturings (Fig. 6B). Consistent with the enhanced resection promoted by Tel1-hy909, the telomeres shortened slightly faster in est2Δ TEL1-hy909 than in est2Δ cells (Fig. 6B). Furthermore, TEL1-hy909 est2Δ cells showed a senescent phenotype at the fifth passage, whereas est2Δ cells lost viability only at the 11th subculturing (Fig. 6A), indicating that Tel1-hy909 accelerates the onset of senescence in telomerase-negative cells. If ssDNA is the primary signal triggering senescence, as proposed (1), these data suggest that the increased amount of ssDNA in TEL1-hy909 est2Δ compared to est2Δ cells might explain the accelerated senescence phenotype of the TEL1-hy909 est2Δ cells.
Fig 6.
The TEL1-hy909 mutation accelerates the onset of senescence in est2Δ cells. Meiotic tetrads from an EST2/est2Δ TEL1/tel1-hy909 diploid strain were dissected on YEPD plates. (A) After ∼25 generations, spore clones from 15 tetrads were streaked for successive times (1 to 11 times). (B) An aliquot of cells from the indicated streaks was propagated in YEPD liquid medium for 5 h to prepare genomic DNA for telomere length determination by Southern blot analysis. All tetratype tetrads behaved as the one shown in panel A.
TEL1-hy909 telomeres escape Rif2-mediated inhibition of nucleolytic processing and elongation.
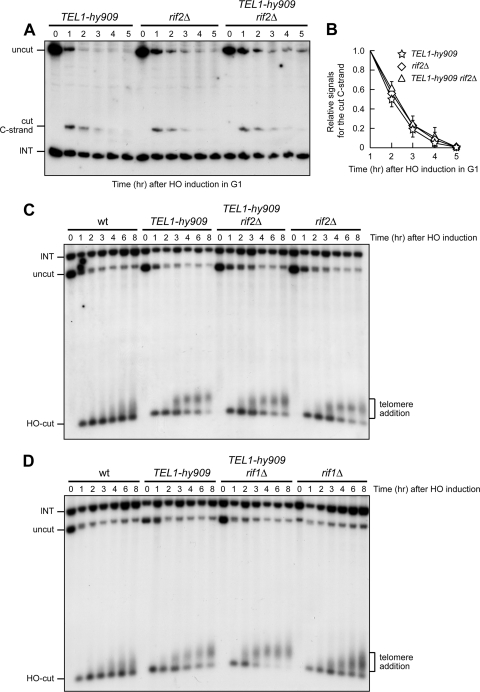
Rif2 inhibits MRX-dependent ssDNA generation at telomeres during both G1 and G2 cell cycle phases (6, 7). Since Rif2 has been proposed to compete with Tel1 for binding to MRX (26), the Tel1-hy909 variant might bypass the Rif2-mediated negative regulation of MRX activity. To address this possibility, we compared resection of the DSB adjacent to telomeric repeats in TEL1-hy909, rif2Δ, and rif2Δ TEL1-hy909 cells. We found that the 5′ C strand at the TG side of the HO cut was degraded with very similar kinetics in G1-arrested TEL1-hy909, rif2Δ, and rif2Δ TEL1-hy909 cells (Fig. 7A and B). Similar results were also obtained when degradation of the 5′ C strand was analyzed in the same strains arrested in G2 (data not shown). Thus, nucleolytic degradation at TEL1-hy909 telomeres is not sensitive to the inhibitory activity of Rif2, indicating that the increased resection of TEL1-hy909 telomeres likely reflects failure of Rif2 to negatively regulate their processing.
Fig 7.
Processing and elongation of a DSB end carrying TG tracts in TEL1-hy909 cells are not affected by RIF2 deletion. (A and B) HO expression was induced at time zero by galactose addition to α-factor-arrested cells, all carrying the system described in Fig. 2A. Cells were kept arrested in G1, and cell cycle arrest was verified by fluorescence-activated cell sorter analysis (data not shown). (A) RsaI- and EcoRV-digested genomic DNA was analyzed as described in the legend to Fig. 2C. (B) Densitometric analysis. Plotted values are means ± SDs from three independent experiments, as in panel A. (C and D) HO expression was induced at time zero by galactose addition to exponentially growing cells. AvaI- and NdeI-digested genomic DNA was subjected to Southern blot analysis using a TRP1 probe, which reveals the ∼800-bp AvaI-HO fragment exposing the TG repeats, whose length progressively increases as new telomere repeats are added. A bracket points out new telomere repeats added to the exposed TG-HO sequence.
As Rif2 inhibits telomerase-dependent telomere elongation (29, 52), we investigated whether Tel1-hy909 also overcomes this Rif2-mediated negative regulation of telomere length. We first used the system described in Fig. 2A to analyze the effect of a lack of Rif2 on the kinetics of telomere addition at the DSB end carrying TG sequences in TEL1-hy909 cells. As a control, we also analyzed the same process in otherwise isogenic TEL1-hy909 cells lacking Rif1, which negatively regulates telomere length by acting in a pathway different from that involving Rif2 (29, 52). When the HO cut was induced in exponentially growing cells, rif2Δ cells elongated the TG DSB end more efficiently than wild-type cells under the same conditions (Fig. 7C), as expected. Sequence addition to this DSB side was also enhanced in TEL1-hy909 cells (Fig. 7C), indicating that the Tel1-hy909 variant also increases the efficiency of telomere elongation in this assay. Interestingly, telomere addition took place with very similar efficiency in TEL1-hy909 rif2Δ double mutant cells and in rif2Δ or TEL1-hy909 single mutants (Fig. 7C), indicating that TEL1-hy909 telomere length escapes the Rif2-dependent negative control. Differently from Rif2, Rif1 still acts as a negative regulator of telomere length in TEL1-hy909 cells. In fact, sequence addition to the telomeric DSB side was enhanced in TEL1-hy909 rif1Δ double mutant cells compared to both TEL1-hy909 and rif1Δ single mutants (Fig. 7D), although RIF1 deletion had a more modest effect than RIF2 deletion in allowing telomere addition (Fig. 7D) (26, 37).
We then analyzed the epistatic relationships between TEL1-hy909, rif2Δ, and rif1Δ alleles in the control of native telomere length. Diploid strains heterozygous for TEL1-hy909 and either rif1Δ or rif2Δ were sporulated, and the resulting tetrads were dissected. Because mutations affecting telomere length often exhibit a phenotypic lag, we examined telomere lengths by Southern blot analysis of genomic DNA prepared from subcultures of the haploid clones derived from the spores. Telomeres of TEL1-hy909 rif1Δ double mutant cells were longer than those of both TEL1-hy909 and rif1Δ single mutants (Fig. 8), whereas RIF2 deletion did not cause further telomere lengthening in TEL1-hy909 cells (Fig. 8). Thus, while Rif1 still negatively controls the length of native telomeres in TEL1-hy909 cells, the same cells also escape the Rif2-mediated negative regulation at native telomeres. Interestingly, TEL1-hy909 cells had longer telomeres than rif2Δ cells (Fig. 8), indicating that TEL1-hy909 telomere overelongation cannot be solely attributed to the lack of Rif2-mediated inhibition of telomerase.
Fig 8.
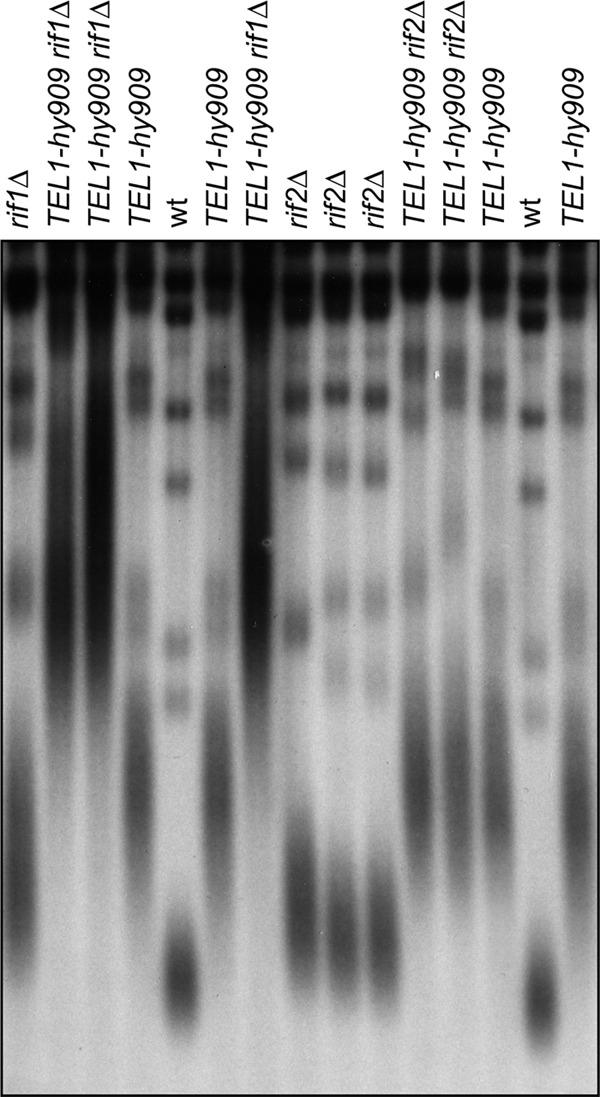
Different effects of RIF1 and RIF2 deletion on the length of native TEL1-hy909 telomeres. XhoI-cut genomic DNA from exponentially growing cell cultures with the indicated genotypes was analyzed as described in the legend to Fig. 1.
Tel1-hy909 is more robustly associated than wild-type Tel1 to DSB ends carrying telomeric repeat sequences.
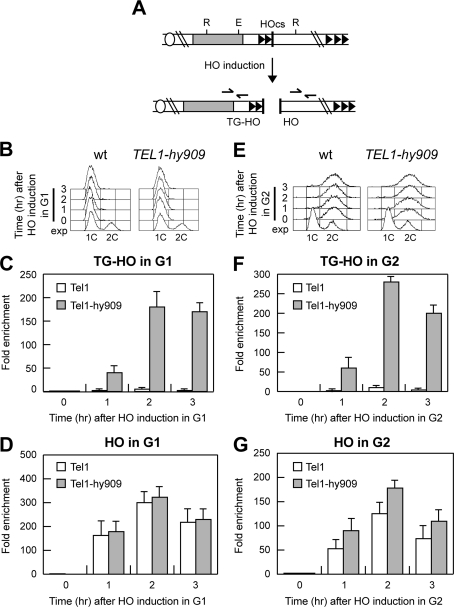
Rif2 is thought to inhibit telomere elongation by counteracting Tel1 association to telomeric DNA (26). Thus, one possibility is that Rif2 is unable to inhibit telomere processing and elongation in TEL1-hy909 cells because Tel1-hy909 is more robustly associated to the telomeric ends than wild-type Tel1. To investigate this hypothesis, we analyzed by ChIP experiments the binding of HA-tagged Tel1 and Tel1-hy909 variants at the telomeric (TG-HO) and nontelomeric (HO) sides of the HO-induced DSB depicted in Fig. 9A. The HO cut was induced by galactose addition in G1- or G2-arrested TEL1-HA and TEL1-hy909-HA cell cultures that were kept arrested in G1 (Fig. 9B) or in G2 (Fig. 9E), respectively. Consistent with the notion that Tel1 association at telomeres is inhibited by Rif2 (26), the amount of wild-type Tel1-HA bound at the TG-HO side of the break was greatly reduced compared to that associated to the nontelomeric (HO) side during both G1 (Fig. 9C and D) and G2 (Fig. 9F and G) arrest. Interestingly, the amount of Tel1-hy909-HA bound at the TG-HO DSB end was dramatically higher than that of wild-type Tel1-HA in both G1 and G2 (Fig. 9C and F), indicating that Tel1-hy909 is more robustly associated than wild-type Tel1 to DNA ends carrying telomeric DNA. In contrast, the amount of Tel1-hy909-HA that associated to the HO end with no telomeric repeats was similar to that of wild-type Tel1-HA in G1 cells (Fig. 9D), whereas it was slightly increased in G2 cells (Fig. 9G), in agreement with the observation that Tel1-hy909 can enhance resection of a nontelomeric DSB end in G2, but not in G1.
Fig 9.
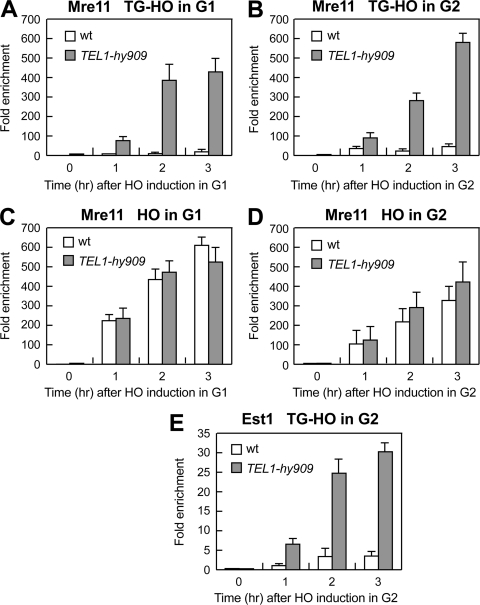
Association of Tel1-HA and Tel1-hy909-HA to DNA ends. (A) The system described in Fig. 2A was used to generate TG-HO and HO DNA ends, and the primers used to detect protein association centromere proximal (TG-HO) or centromere distal (HO) to the HO-induced DSB are represented by arrows in the bottom part of the panel. (B to G) HO expression was induced at time zero by galactose addition to G1-arrested (B to D) or G2-arrested (E to G) TEL1-HA (wild-type) and TEL1-hy909-HA (TEL1-hy909) cells, which were kept arrested in G1 or G2 by α-factor and nocodazole, respectively. (B and E) Fluorescence-activated cell sorter analysis of DNA content. (C to G) Chromatin samples taken at the indicated times after HO induction were immunoprecipitated with anti-HA antibody. (C and F) Coimmunoprecipitated DNA was analyzed by qPCR using primer pairs located 640 bp centromere proximal to the HO cleavage site (HOcs; TG-HO) and at the nontelomeric ARO1 fragment of chromosome IV (CON). (D and G) Coimmunoprecipitated DNA was analyzed by qPCR using primer pairs located 550 bp centromere distal to the HO cleavage site (HOcs; HO) and at the nontelomeric ARO1 fragment of chromosome IV (CON). In all graphs, data are expressed as relative fold enrichment of the TG-HO or HO signal over the CON signal after normalization to input signals for each primer set. The data presented are means ± SDs from three different experiments.
Tel1-hy909 increases MRX and Est1 persistence at DSB ends carrying telomeric repeats.
If Tel1 promotes MRX activity by facilitating MRX persistence onto DNA ends, then the more robust association to telomeric ends of Tel1-hy909 than wild-type Tel1 might lead to increased MRX binding at these ends, thus eventually leading to enhanced telomere resection. We therefore also analyzed G1- or G2-arrested wild-type and TEL1-hy909 cells for the binding of MYC-tagged Mre11 to either the TG-HO telomeric end or the HO nontelomeric end of the HO-induced DSB depicted in Fig. 9A. The association of Mre11 to the TG-HO DSB end was greatly increased in both G1- and G2-arrested TEL1-hy909 cells compared to wild type (Fig. 10A and B). In contrast, Mre11 binding to the nontelomeric DSB end was similar in G1-arrested wild-type and TEL1-hy909 cells (Fig. 10C), whereas it was slightly increased in TEL1-hy909 G2 cells (Fig. 10D), consistent with the finding that the enhanced resection of DSB ends with no TG sequences in TEL1-hy909 cells is restricted to the G2 cell cycle phase (3).
Fig 10.
Mre11 and Est1 association to DNA ends in wild-type and TEL1-hy909 cells. G1- or G2-arrested wild-type and TEL1-hy909 cells carrying the TG-HO system described in Fig. 2A and expressing fully functional MYC-tagged Mre11 (A to D) or MYC-tagged Est1 (E) were treated as described in the legend to Fig. 9, and cell cycle arrest was verified by fluorescence-activated cell sorter analysis (data not shown). Chromatin samples taken at the indicated times after HO induction were immunoprecipitated with anti-MYC antibody. (A, B, and E) Coimmunoprecipitated DNA was analyzed by qPCR with the primer pairs used for Fig. 9C and F. (C and D) Coimmunoprecipitated DNA was analyzed by qPCR with the primer pairs used for Fig. 9D and G. In all graphs, data are expressed as relative fold enrichment of the TG-HO or HO signal over the CON signal after normalization to input signals for each primer set. The data presented are means ± SDs from three different experiments.
As Tel1 is thought to regulate telomere length by promoting telomerase recruitment at telomeres, increased association of Tel1-hy909 at telomeric ends might also enhance telomerase association to the same ends. Indeed, the amount of MYC-tagged Est1 bound to the TG-HO side of the HO-induced DSB was much higher in G2-arrested TEL1-hy909 cells than in wild type (Fig. 10E), suggesting that enhanced Est1 association at TEL1-hy909 telomeres may account for their overelongation.
DISCUSSION
The MRX complex is required for the recruitment of Tel1 to DSB ends (39). On the other hand, the lack of Tel1 causes a decrease of MRX binding at telomeres (26), suggesting that Tel1 exerts a positive-feedback loop on MRX. However, the physiological relevance of this control was unknown. By studying the effects on telomere processing and elongation of the lack of Tel1 compared with the effects of the dominant Tel1-hy909 variant, we provide evidence that Tel1 is crucial for counteracting Rif2-dependent negative regulation of telomere resection and elongation.
We show that the lack of Tel1, which is known to cause telomere shortening (22), decreases both MRX-dependent resection at a DSB end carrying telomeric repeats and the amount of ssDNA formation at native telomeres. Together with the notion that MRX association at telomeres is impaired by the lack of Tel1 (26), these data indicate that Tel1, once loaded onto DNA ends by MRX, regulates 5′-3′ nucleolytic degradation of telomere ends by promoting MRX activity. This role of Tel1 is not restricted to telomere ends, as the lack of Tel1 was shown to slightly impair generation of ssDNA also at intrachromosomal DSBs (34).
Consistent with a role of Tel1 in regulating telomere end resection, the TEL1-hy909 mutation, which causes telomerase-dependent telomere overelongation, increases the amount of ssDNA at telomeres. This increase becomes apparent even in the absence of telomerase, indicating that it is due to an enhanced resection rather than to telomerase action. Moreover, the TEL1-hy909 mutation accelerates the onset of senescence of telomerase-negative cells. As telomeric ssDNA has been proposed to trigger senescence (1), this finding further supports the hypothesis that the Tel1-hy909 variant enhances nucleolytic degradation at telomeres. Accordingly, the lack of Tel1, which reduces the amount of telomeric ssDNA, attenuates the senescence phenotype of telomerase-negative cells (19, 42).
How does Tel1-hy909 enhance telomere processing and elongation? Both the enhanced resection and elongation of TEL1-hy909 telomeres require Tel1-hy909 kinase activity, further confirming that Tel1 acts as a kinase at telomeres (33). Like most of the other Tel1-hy variants that we found by virtue of their ability to suppress the hypersensitivity to genotoxic agents of mec1Δ cells, the Tel1-hy909 variant has enhanced kinase activity in vitro compared to wild-type Tel1 (3). However, only the Tel1-hy909 variant confers a striking telomere overelongation phenotype (3), implying that this phenotype cannot be entirely ascribed to the high Tel1-hy909 kinase activity.
Tel1-hy909 is more robustly associated than wild-type Tel1 to DSB ends carrying telomeric repeats during both the G1 and G2 cell cycle phases, likely leading to increased MRX and Est1 persistence at telomeric ends. In fact, the amounts of Mre11 and Est1 bound to telomeric DNA ends are higher in TEL1-hy909 cells than wild-type cells. While enhanced MRX association could explain the increased efficiency of TEL1-hy909 telomere resection, stabilization of Est1 binding to telomeric ends may account for the overelongation of TEL1-hy909 telomeres. Because MRX is required to load Tel1 onto DNA ends (39) and to support Tel1-hy909 activities, this robust Tel1-hy909 association to telomeres may be due to an increased ability of Tel1-hy909 to interact with MRX compared to wild-type Tel1, as also suggested by the dominant effects of the TEL1-hy909 allele. Unfortunately, we have so far been unable to coimmunoprecipitate Tel1 with MRX to assess this possibility.
MRX-dependent generation of ssDNA at telomeres is prevented by Rif2 during both G1 and G2 (7). We show that neither processing nor elongation of TEL1-hy909 telomeres is inhibited by Rif2, indicating a failure of Rif2 to counteract MRX activity in the presence of the Tel1-hy909 variant. As Rif2 has been proposed to compete with Tel1 for binding to MRX (26), the dominant Tel1-hy909 variant might escape the Rif2-mediated negative control by binding to MRX more efficiently than wild-type Tel1. Because Tel1 exerts a positive-feedback loop on MRX, this robust Tel1-hy909 recruitment may in turn stabilize MRX association to telomeric ends, possibly through phosphorylation events.
The action of Tel1-hy909 is not restricted to telomeric ends, as Tel1-hy909 also enhances resection at intrachromosomal DSBs (3). However, this Tel1-hy909 function is restricted to the G2 cell cycle phase, because Tel1-hy909 is not able to enhance resection in G1 (when Cdk1 activity is low) of a DSB end without telomeric sequences, while it does it when the DSB end carries TG repeats. Noteworthy, while Rif2 prevents MRX activity specifically at telomeres, MRX-dependent resection at DSBs devoid of telomeric repeats is counteracted by Yku, which exerts this inhibitory role only in G1 (11). These observations, together with our finding that Tel1 regulates telomere resection by promoting MRX function, suggest that Tel1-hy909 is unable to promote resection in G1 at DSB ends without telomeric repeats because it cannot overcome the inhibitory effect exerted by Yku on MRX. This inability does not prevent Tel1-hy909 from enhancing MRX-dependent resection at telomeres, because Yku was shown to protect them mainly from Exo1 and not from MRX (7, 48). Since Yku-mediated inhibition of MRX activity at intrachromosomal DSBs is relieved in G2 (11), Tel1-hy909 can promote resection of intrachromosomal DSBs only during this cell cycle stage. Consistent with different mechanisms inhibiting MRX activity at telomeres than at DSBs, MRX-dependent resection in G1 is completely abolished at a DSB end with TG repeats, whereas it takes place, although less efficiently than in G2, when the DSB is devoid of telomeric sequences. Interestingly, the resection kinetics of DSB ends with no TG repeats in G1-arrested wild-type cells is similar to that occurring in G1-arrested TEL1-hy909 cells when the DSB end carries TG tracts (compare Fig. 3 and 4). Furthermore, the amount of MRX bound to DSB ends with no telomeric sequences in wild-type cells is similar to the MRX amount that is recruited at DSB ends carrying TG repeats in TEL1-hy909 cells. Thus, the difference in terms of ssDNA generation at DSBs versus telomeric DNA ends appears to rely on Rif2-mediated inhibition of Tel1 and, hence, of MRX activity.
The molecular mechanism underlying Tel1-mediated regulation of telomere length has not yet been solved. The finding that the amount of telomeric G tails is reduced by the lack of Tel1, whereas it is increased in TEL1-hy909 cells, leads to a model, also suggested by Lundblad and colleagues (19), where Tel1 function in telomere length maintenance can be linked to its role in generating telomeric G tails, which are the substrates of telomerase. In this model, the telomere length defect observed in tel1Δ cells might be a consequence of the reduced ssDNA amount at the telomeric ends, whereas the increased ssDNA generation at TEL1-hy909 telomeres might improve telomerase-dependent elongation by increasing the substrates available for the telomerase enzyme. However, the finding that the single-stranded G-tail signal was lower in mre11Δ cells than in tel1Δ cells, which display a similar telomere length defect and are likely defective in the same telomere length maintenance pathway (41), is inconsistent with the idea that Tel1 function in telomere length maintenance is limited to ssDNA generation.
The correlation between telomere overelongation and increased Est1 binding at TEL1-hy909 telomeres provides additional support for a model in which Tel1 is a positive activator of telomerase. Because the kinase activity of Tel1 is needed for its role in telomere maintenance (33), phosphorylation of one or more telomere binding proteins by Tel1 could increase the frequency of elongation by making the telomeric chromatin more accessible to telomerase. In this scenario, the robust association of hyperactive Tel1-hy909 kinase at telomeres may improve the efficiency of this process.
In conclusion, regulation of telomere processing and elongation appears to rely on a balance between Tel1 and Rif2 activities. As Tel1 hyperactivation can improve both telomere resection and telomerase action, Rif2-dependent inhibition of Tel1/MRX function at telomeres is important to ensure the maintenance of telomere identity by limiting ssDNA generation and elongation. In contrast, such a control appears to be dispensable at intrachromosomal DSBs, where generation of ssDNA is necessary to repair the break.
Supplementary Material
ACKNOWLEDGMENTS
We thank D. Gottschling, J. Haber, T. Petes, D. Shore, K. Sugimoto, and T. Weinert for yeast strains. We thank Laura Candelora for the help in some experiments.
This work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro (grant number IG11407), Cofinanziamento 2008 MIUR/Università di Milano-Bicocca to M.P.L., and Cofinanziamento 2009 MIUR/Università di Milano-Bicocca to G.L. M.M. was supported by a fellowship from the Fondazione Confalonieri.
Footnotes
Published ahead of print 21 February 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Abdallah P, et al. 2009. A two-step model for senescence triggered by a single critically short telomere. Nat. Cell Biol. 11:988–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arnerić M, Lingner J. 2007. Tel1 kinase and subtelomere-bound Tbf1 mediate preferential elongation of short telomeres by telomerase in yeast. EMBO Rep. 8:1080–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baldo V, Testoni V, Lucchini G, Longhese MP. 2008. Dominant TEL1-hy mutations compensate for Mec1 lack of functions in the DNA damage response. Mol. Cell. Biol. 28:358–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bianchi A, Shore D. 2007. Increased association of telomerase with short telomeres in yeast. Genes Dev. 21:1726–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bianchi A, Negrini S, Shore D. 2004. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol. Cell 16:139–146 [DOI] [PubMed] [Google Scholar]
- 6. Bonetti D, Clerici M, Manfrini N, Lucchini G, Longhese MP. 2010. The MRX complex plays multiple functions in resection of Yku- and Rif2-protected DNA ends. PLoS One 5:e14142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bonetti D, et al. 2010. Shelterin-like proteins and Yku inhibit nucleolytic processing of Saccharomyces cerevisiae telomeres. PLoS Genet. 6:e1000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bonetti D, Martina M, Clerici M, Lucchini G, Longhese MP. 2009. Multiple pathways regulate 3′ overhang generation at S. cerevisiae telomeres. Mol. Cell 35:70–81 [DOI] [PubMed] [Google Scholar]
- 9. Chan A, Boulé JB, Zakian VA. 2008. Two pathways recruit telomerase to Saccharomyces cerevisiae telomeres. PLoS Genet. 4:e1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chang M, Arneric M, Lingner J. 2007. Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 21:2485–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clerici M, Mantiero D, Guerini I, Lucchini G, Longhese MP. 2008. The Yku70-Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO Rep. 9:810–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Conrad MN, Wright JH, Wolf AJ, Zakian VA. 1990. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell 63:739–750 [DOI] [PubMed] [Google Scholar]
- 13. Diede SJ, Gottschling DE. 1999. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases α and δ. Cell 99:723–733 [DOI] [PubMed] [Google Scholar]
- 14. Diede SJ, Gottschling DE. 2001. Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr. Biol. 11:1336–1340 [DOI] [PubMed] [Google Scholar]
- 15. Dionne I, Wellinger RJ. 1996. Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc. Natl. Acad. Sci. U. S. A. 93:13902–13907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Evans SK, Lundblad V. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286:117–120 [DOI] [PubMed] [Google Scholar]
- 17. Frank CJ, Hyde M, Greider CW. 2006. Regulation of telomere elongation by the cyclin-dependent kinase CDK1. Mol. Cell 24:423–432 [DOI] [PubMed] [Google Scholar]
- 18. Gallardo F, et al. 2011. Live cell imaging of telomerase RNA dynamics reveals cell cycle-dependent clustering of telomerase at elongating telomeres. Mol. Cell 44:819–827 [DOI] [PubMed] [Google Scholar]
- 19. Gao H, et al. 2010. Telomerase recruitment in Saccharomyces cerevisiae is not dependent on Tel1-mediated phosphorylation of Cdc13. Genetics 186:1147–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gomes NM, Shay JW, Wright WE. 2010. Telomere biology in Metazoa. FEBS Lett. 584:3741–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goudsouzian LK, Tuzon CT, Zakian VA. 2006. S. cerevisiae Tel1p and Mre11p are required for normal levels of Est1p and Est2p telomere association. Mol. Cell 24:603–610 [DOI] [PubMed] [Google Scholar]
- 22. Greenwell PW, et al. 1995. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell 82:823–829 [DOI] [PubMed] [Google Scholar]
- 23. Greider CW, Blackburn EH. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43:405–413 [DOI] [PubMed] [Google Scholar]
- 24. Hardy CF, Sussel L, Shore D. 1992. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 6:801–814 [DOI] [PubMed] [Google Scholar]
- 25. Hector RE, et al. 2007. Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol. Cell 27:851–858 [DOI] [PubMed] [Google Scholar]
- 26. Hirano Y, Fukunaga K, Sugimoto K. 2009. Rif1 and Rif2 inhibit localization of Tel1 to DNA ends. Mol. Cell 33:312–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ira G, et al. 2004. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431:1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Larrivée M, LeBel C, Wellinger RJ. 2004. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 18:1391–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Levy DL, Blackburn EH. 2004. Counting of Rif1p and Rif2p on Saccharomyces cerevisiae telomeres regulates telomere length. Mol. Cell. Biol. 24:10857–10867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Longhese MP. 2008. DNA damage response at functional and dysfunctional telomeres. Genes Dev. 22:125–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Longhese MP, Bonetti D, Manfrini N, Clerici M. 2010. Mechanisms and regulation of DNA end resection. EMBO J. 29:2864–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Makarov VL, Hirose Y, Langmore JP. 1997. Long G tails at both ends of human chromosomes suggest a C-strand degradation mechanism for telomere shortening. Cell 88:657–666 [DOI] [PubMed] [Google Scholar]
- 33. Mallory JC, Petes TD. 2000. Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisiae proteins related to the human ATM protein kinase. Proc. Natl. Acad. Sci. U. S. A. 97:13749–13754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mantiero D, Clerici M, Lucchini G, Longhese MP. 2007. Dual role for Saccharomyces cerevisiae Tel1 in the checkpoint response to double-strand breaks. EMBO Rep. 8:380–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marcand S, Brevet V, Gilson E. 1999. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 18:3509–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marcand S, Brevet V, Mann C, Gilson E. 2000. Cell cycle restriction of telomere elongation. Curr. Biol. 10:487–490 [DOI] [PubMed] [Google Scholar]
- 37. McGee JS, et al. 2010. Reduced Rif2 and lack of Mec1 target short telomeres for elongation rather than double-strand break repair. Nat. Struct. Mol. Biol. 17:1438–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Michelson RJ, Rosenstein S, Weinert T. 2005. A telomeric repeat sequence adjacent to a DNA double-stranded break produces an anticheckpoint. Genes Dev. 19:2546–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nakada D, Matsumoto K, Sugimoto K. 2003. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 17:1957–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pennock E, Buckley K, Lundblad V. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104:387–396 [DOI] [PubMed] [Google Scholar]
- 41. Ritchie KB, Petes TD. 2000. The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics 155:475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ritchie KB, Mallory JC, Petes TD. 1999. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6065–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sabourin M, Tuzon CT, Zakian VA. 2007. Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol. Cell 27:550–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Taggart AK, Teng SC, Zakian VA. 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297:1023–1026 [DOI] [PubMed] [Google Scholar]
- 45. Teixeira MT, Arneric M, Sperisen P, Lingner J. 2004. Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117:323–335 [DOI] [PubMed] [Google Scholar]
- 46. Tseng SF, Lin JJ, Teng SC. 2006. The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res. 34:6327–6336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Viscardi V, Bonetti D, Cartagena-Lirola H, Lucchini G, Longhese MP. 2007. MRX-dependent DNA damage response to short telomeres. Mol. Biol. Cell 18:3047–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vodenicharov MD, Laterreur N, Wellinger RJ. 2010. Telomere capping in non-dividing yeast cells requires Yku and Rap1. EMBO J. 29:3007–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vodenicharov MD, Wellinger RJ. 2006. DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell-cycle kinase. Mol. Cell 24:127–137 [DOI] [PubMed] [Google Scholar]
- 50. Wellinger RJ, Wolf AJ, Zakian VA. 1993. Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell 72:51–60 [DOI] [PubMed] [Google Scholar]
- 51. Wellinger RJ, Ethier K, Labrecque P, Zakian VA. 1996. Evidence for a new step in telomere maintenance. Cell 85:423–433 [DOI] [PubMed] [Google Scholar]
- 52. Wotton D, Shore D. 1997. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11:748–760 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.