Abstract
The monoclonal antibody (MAb) VRC01 was isolated from a slowly progressing HIV-1-infected donor and was shown to neutralize diverse HIV-1 strains by binding to the conserved CD4 binding site (CD4bs) of gp120. To better understand the virologic factors associated with such antibody development, we characterized HIV-1 envelope (Env) variants from this donor and five other donors who developed broadly neutralizing antibodies. A total of 473 env sequences were obtained by single-genome amplification, and 100 representative env clones were expressed and tested for entry and neutralization sensitivity. While VRC01 neutralizes about 90% of the genetically diverse heterologous HIV-1 strains tested, only selective archival Env variants from the VRC01 donor were sensitive to VRC01 and all of the Env variants derived from the donor plasma were resistant, indicating strong antibody-based selection pressure. Despite their resistance to this broadly reactive MAb that partially mimics CD4, all Env variants required CD4 for entry. Three other CD4bs MAbs from the same donor were able to neutralize some VRC01 escape variants, suggesting that CD4bs antibodies continued to evolve in response to viral escape. We also observed a relatively high percentage of VRC01-resistant Env clones in the plasma of four of five additional broadly neutralizing donors, suggesting the presence of CD4bs-directed neutralizing antibodies in these donors. In total, these data indicate that the CD4bs-directed neutralizing antibodies exert ongoing selection pressure on the conserved CD4bs epitope of HIV-1 Env.
INTRODUCTION
Approximately 25% of HIV-1-infected individuals develop cross-reactive neutralizing antibodies, and the serum of some donors potently neutralizes most HIV-1 strains (11, 12, 24, 43, 53, 62, 66, 67, 71). Serum mapping studies and subsequent monoclonal antibody (MAb) isolation have defined several conserved neutralization epitopes on the HIV-1 Env trimer (2, 8, 11, 24, 27, 43, 45, 63, 69–71, 75, 77, 80, 81). One such epitope is the initial site of gp120 attachment to the cellular receptor CD4 (38, 81). We previously described the CD4 binding site (CD4bs) MAb VRC01, isolated from a slowly progressing subtype B-infected donor, which neutralized 91% of the genetically diverse HIV-1 isolates tested (75). Biophysical characterization of VRC01 suggested that it partially mimics the interaction of CD4 with gp120, and the liganded crystal structures of VRC01 defined specific similarities in the heavy chain of VRC01 and domain 1 of CD4 as related to their binding interaction with gp120 (80). Recently, additional potently neutralizing CD4bs antibodies were identified in a total of six donors (63, 77), indicating that other HIV-1-infected donors can make similarly potent CD4bs antibodies.
Previous studies have shown that HIV-1 encounters a genetic bottleneck during transmission, often resulting in a genetically homogenous population of initial plasma viremia (10, 33, 46, 47, 61, 73). Partially due to the host adaptive immunity, the early circulating virus evolves into a diverse population over time, with genetic diversity of up to 10% in the env gene within an infected individual (36, 65). Autologous virus-neutralizing antibodies generally develop within the first months or year of infection, and there is a well-documented antibody-based selection process that results in ongoing viral escape from autologous neutralizing antibodies (1, 3, 4, 18–20, 26, 40, 50, 51, 54, 57, 58, 72). Thus, circulating plasma viral Env variants are poorly neutralized by concurrent serum but are often potently neutralized by serum samples from later time points. Importantly, the early autologous neutralizing antibody response is usually highly strain specific; these antibodies do not appear to target conserved regions of Env and do not neutralize most heterologous viral isolates (25, 48, 52, 54, 58). Until recently, few broadly reactive HIV-1-neutralizing antibodies had been isolated (5, 55, 68, 82); thus, the Env quasispecies in donors with such antibodies have not been characterized.
The isolation of VRC01 and other related MAbs from donor 45 provided the opportunity to study the interaction of these MAbs with the circulating viral quasispecies. It was not clear if circulating virus would escape from a neutralizing antibody targeting a relatively conserved epitope or if broadly reactive antibodies would evolve in relation to viral escape. To better understand these events, we performed detailed studies of the plasma viral quasispecies obtained from donor 45 at three time points (2001, 2006, and 2009). For comparison, we also studied the viral quasispecies in five additional donors with broadly reactive serum neutralizing antibodies. We used single-genome amplification (SGA) to derive viral Env sequences and expressed representative sequences as Env pseudoviruses to assess their in vitro neutralization sensitivity to VRC01, as well as several other MAbs isolated from donor 45. In total, this analysis revealed strong selection pressure by VRC01 on the circulating viral quasispecies and ongoing viral escape and evolution of the antibody response to the CD4bs of HIV-1 gp120.
MATERIALS AND METHODS
Study subjects.
The plasma, serum, and peripheral blood mononuclear cell (PBMC) samples described in this study were from HIV-1-infected individuals enrolled in clinical protocols approved by the appropriate institutional review board of the National Institute of Allergy and Infectious Diseases or the University of Pennsylvania. Donor 45 has never been on antiretroviral treatment (ART), and all other donor samples were from time points prior to or after withdrawal from ART. All donors except B7B5 were considered slow progressors with peripheral blood CD4 T-cell levels of >350 cells/μl and plasma viral RNA levels of <36,000 copies/ml. The clinical status of donor B7B5 was not fully characterized, but he was in the clinically asymptomatic phase of HIV-1 infection.
Antibodies and cells.
The anti-CD4bs MAbs VRC01 and VRC03 have been described previously (75). The CD4-Ig plasmid construct was provided by Joseph Sodroski (Dana-Farber Cancer Institute), and the fusion protein was expressed and purified as described previously (75). The anti-CD4bs MAb b12 was provided by Dennis Burton and Ralph Pantophlet (The Scripps Research Institute, La Jolla, CA). The donor 45 serum IgG was purified using protein G UltraLink columns (Thermo Scientific, Rockford, IL) and following the manufacturer's instructions. The concentrations of purified serum IgG or IgG MAbs were determined using NanoDrop (Thermo Scientific) with 1.40 as the extinction coefficient. The TZM-bl cells were obtained from the NIH AIDS Research and Reference Reagent Program, as contributed by John Kappes and Xiaoyun Wu. The Cf2th-CD4.CCR5 and Cf2th-synCCR5 cell lines, canine thymocyte lines stably transfected to express both human CD4 and CCR5 or only human CCR5, respectively, were obtained from the NIH AIDS Research and Reference Reagent Program, as contributed by Tajib Mizabekov and Joseph Sodroski (Dana-Farber Cancer Institute).
Viral RNA extraction, cDNA synthesis, and genomic DNA extraction.
A 140-μl volume of each plasma or serum sample was used to extract viral RNA using the QIAamp viral RNA minikit (Qiagen, Valencia, CA). The RNA was eluted in 50 μl of elution buffer and subjected to first-strand cDNA synthesis immediately using the SuperScript III reverse transcriptase (Invitrogen Life Technologies, Grand Island, NY). cDNA synthesis was done in a final volume of 100 μl including 50 μl viral RNA, 5 μl of a deoxynucleoside triphosphate (dNTP) mixture (each at 10 mM), 1.25 μl antisense primer envB3out (5′-TTGCTACTTGTGATTGCTCCATGT-3′) at 20 μM, 20 μl 5× first-strand buffer, 5 μl dithiothreitol at 100 mM, 5 μl RNaseOUT (Invitrogen), and 5 μl SuperScript III reverse transcriptase. RNA, primers, and dNTPs were heated at 65°C for 5 min and then chilled on ice for 1 min, and then the entire reaction mixture was incubated at 50°C for 60 min, followed by 55°C for an additional 60 min. Finally, the reaction was heat inactivated at 70°C for 15 min and then treated with 1 μl RNase H at 37°C for 20 min. The QIAamp DNA Blood minikit (Qiagen) was used to extract the donor 45 PBMC genomic DNA. The synthesized cDNA and the extracted genomic DNA were subjected to the first-round PCR immediately or stored frozen at −80°C.
SGA.
The nested PCR of HIV-1 env SGA was described previously (33, 41, 61). Briefly, the synthesized cDNA or genomic DNA was serially diluted and distributed in replicates of 12 to 16 PCRs in ThermoGrid 96-well plates (Denville Scientific, Metuchen, NJ) to identify a dilution where PCR-positive wells constituted about 30% of the total number of reactions. At this dilution, most of the wells contain amplicons derived from a single cDNA or DNA molecule. Additional PCR amplifications were performed using this dilution in full 96-well plates. PCR amplification was carried out using the Platinum Taq High Fidelity PCR system (Invitrogen). The final 20-μl reaction volume was composed of 2 μl 10× buffer, 0.8 μl MgSO4, 0.4 μl dNTP mixture (each at 10 mM), 0.2 μl each primer at 20 μM, 0.1 μl Platinum Taq High Fidelity polymerase, and 1 μl template DNA. The primers for the first-round PCR were envB5out (5′-TAGAGCCCTGGAAGCATCCAGGAAG-3′) and envB3out (5′-TTGCTACTTGTGATTGCTCCATGT-3′). The primers for the second-round PCR were envB5in (5′-CACCTTAGGCATCTCCTATGGCAGGAAGAAG-3′) and envB3in (5′-GTCTCGAGATACTGCTCCCACCC-3′). The cycler parameters were 94°C for 2 min, followed by 35 cycles of 94°C for 15 s, 55°C for 30 s, and 68°C for 4 min and by a final extension of 68°C for 10 min. The product of the first-round PCR (1 μl) was subsequently used as the template in the second-round PCR under the same conditions but with a total of 45 cycles. The amplicons were inspected on a precast 1% agarose gel (Embi Tec, San Diego, CA). All PCR procedures were carried out in a designated PCR clean hood using procedural safeguards against sample contamination.
DNA sequencing.
Amplicons were directly sequenced by BigDye Terminator chemistry (Applied Biosystems, Foster City, CA) by ACGT, Inc. (Wheeling, IL). Both DNA strands were sequenced using partially overlapping fragments. Individual sequence fragments for each amplicon were assembled and edited using Sequencher 5.0 (Gene Codes, Ann Arbor, MI). All chromatograms were inspected for sites of mixed bases (double peaks), which would be evidence of priming from more than one template or the introduction of PCR error in early cycles. Any sequence with evidence of double peaks was excluded from further analysis.
Sequence alignments, diversity, and phylogenetic analysis.
The env sequences containing unproductive mutations (such as those that cause stop codons and frameshifts) or large deletions (>24 nucleotides) in conserved regions were removed from subsequent sequence analysis and env cloning. The 28 PBMC-derived donor 45 env sequences were checked for hypermutation using Hypermut (59) at the Los Alamos HIV sequence database (http://www.hiv.lanl.gov/content/sequence/HYPERMUT/hypermut.html), and none was hypermutated using the intradonor plasma-derived sequence 45_01A14 as a reference. A total of 473 env sequences from the study donors suitable for full analysis were used to generate a neighbor-joining tree as follows. The 473 donor gp160 protein sequences together with the subtype B reference sequence HXB2 were aligned using MUSCLE for multiple-sequence comparison by log expectation (13, 14). The protein distance matrix was calculated by protdist using the Jones-Taylor-Thornton model (32). The tree was constructed by Neighbor using the neighbor-joining method (37). The nucleotide sequences were analyzed as follows. The donor gp160 nucleotide sequences, together with HXB2, were initially aligned with ClustalW (39) and then hand checked using BioEdit (http://www.mbio.ncsu.edu/bioedit/bioedit.html) to improve the alignments according to the codon translation. Each donor nucleotide sequence alignment was submitted to dnadist for calculation of the pairwise nucleotide sequence distance. The F84 model (17, 34) of nucleotide substitution was used for these calculations. After gap stripping, each donor nucleotide sequence alignment was evaluated for potential natural recombination using the recombination analysis tool at http://cbr.jic.ac.uk/dicks/software/RAT/ (15). Of the total of 473 sequences evaluated, none was scored as recombinant using a lower threshold of 80% and an upper threshold of 92% sequence identity. Each donor nucleotide sequence alignment with HXB2 was also submitted to PAUP-easy with built-in MODELTEST (56) for maximum-likelihood analysis. For the 186 donor 45 gp160 sequences aligned with HXB2, the likelihood model selected was GTR+G+I and the settings were as follows. There were six substitution types. The substitution rates were as follows: A ↔ C, 1.2361; A ↔ G, 2.9959; A ↔ T, 0.4754; C ↔ G, 0.3293; C ↔ T, 2.9959; G ↔ T, 1.0000. The nucleotide frequencies were as follows: A, 0.35390; C, 0.18020; G, 0.25000; T, 0.21590. The among-site rate variation was as follows: proportion of invariable sites, 0.5152; distribution of rates at variable sites, gamma (discrete approximation); shape parameter (alpha), 0.5707; number of rate categories, 4; representation of average rate for each category, mean; molecular clock, not enforced. All phylogenetic analysis was performed at using software programs implemented at the NIAID Biocluster. The trees were displayed with Dendroscope (30).
V1V2 length and gp120 glycosylation analysis.
The V1V2 length and the number of putative N-linked glycosylation sites (PNGS) were determined for all of the 473 Env sequences from the six study donors. For comparison, a set of prealigned 897 subtype B Env protein sequences (B_database) was downloaded (with the file name HIV1_FLT_2010_env_PRO) from the Los Alamos HIV database (http://www.hiv.lanl.gov/content/sequence/HIV/mainpage.html). The criteria for this data set were subtype B, intact gp120 sequences, and one sequence per donor. Because all of the study donors were chronically infected, another set of 2,121 chronic Env sequences (B_chronic) isolated by SGA and used in a recent study (21) at the Los Alamos National Laboratory (LANL) was also included, with one sequence removed because of V1V2 deletion (GenBank accession number HQ217183). These sequences were downloaded from the journal website http://www.plospathogens.org/article/info%3Adoi%2F10.1371%2Fjournal.ppat.1002209#s5 (with the file names journal.ppat.1002209.s012.doc and journal.ppat.1002209.s013.doc). The criteria for this data set were subtype B, isolated by SGA, not on ART, and a minimum of 2 years of infection time.
Cloning of HIV-1 env genes.
Representative env sequences from the study donors were selected for cloning from the env phylogenetic trees. Because only 12 env sequences were amplified from donor B7B5, we cloned all 12 env sequences from that donor. The second-round env PCR products containing full-length rev and env genes were directionally cloned into the expression vector pcDNA3.1D (Invitrogen Life Technologies) under the control of the T7 promoter. Each rev and env expression plasmid was maxiprepped (Qiagen), and its sequence was verified. Of a total of 33 env sequences cloned from donor 45, 29 (88%) were functional in mediating virus entry and all 71 env sequences cloned from other donors were functional in mediating virus entry.
Viral stocks and neutralization assay.
The HIV-1 Env pseudovirus stocks were generated and titrated as described previously (42). Briefly, viral stocks were prepared by transfecting 293T cells (6 × 106 in 50 ml growth medium in a T-175 culture flask) with 10 μg of rev and env expression plasmid and 30 μg of an env-deficient HIV-1 backbone vector (pSG3Δenv) using Fugene 6 transfection reagents (Invitrogen). The pseudovirus-containing culture supernatants were harvested 2 days after transfection, filtered (0.45 μm), and stored at −80°C. The 50% tissue culture infectious dose (TCID50) of a single thawed aliquot of each batch of pseudovirus was determined with TZM-bl cells. Briefly, 11 serial 5-fold dilutions of pseudovirus were made in quadruplicate wells in 96-well culture plates to infect TZM-bl cells. Virus infectivity was measured 2 days later by luciferase activity in cell lysates (Promega, Madison, WI). Wells producing luminescence (measured in relative luminescence units [RLU]) >3× the background were scored as positive. The TCID50 was calculated as described previously (31).
Neutralization was measured using HIV-1 Env pseudoviruses to infect TZM-bl cells as described previously (42, 64, 76). Briefly, 40 μl of virus was incubated for 30 min at 37°C with 10 μl of serially diluted test antibody or CD4-Ig in duplicate wells of a 96-well flat-bottom culture plate. To keep assay conditions constant, sham medium was used in place of antibody in specified control wells. The antibody and CD4-Ig concentrations were defined at the point of incubation with virus supernatant. The virus input was set at a multiplicity of infection of 0.01 to 0.1, which generally results in 100,000 to 400,000 RLU in the luciferase assay. Neutralization curves were fitted by nonlinear regression using a five-parameter Hill slope equation programmed into JMP 5.1 statistical software (SAS Institute Inc., Cary, NC). The 50% inhibitory concentration (IC50) was reported as the antibody concentration required to inhibit infection by 50%. The IC50 cutoff was 50 μg/ml for MAbs and CD4-Ig and 1,000 μg/ml for serum IgG because these were the highest concentrations tested.
CD4-independent entry assay.
To assess CD4-dependent and -independent viral entry, Env pseudoviruses were generated by small-scale 293T transfections using the HIV-1 pNL4-3 ΔEnv backbone containing a luciferase reporter gene. Briefly, 2 × 105 293T cells were plated in 3 ml growth medium in a 6-well culture plate 1 day before transfection. Transfection was carried out with 1 μg of rev and env expression plasmid and 3 μg of pNL4-3 ΔEnv luciferase backbone using Fugene 6 transfection reagents (Invitrogen). The culture medium was replaced with 2 ml fresh medium on the following day. The pseudovirus-containing culture supernatants were harvested 1 day later and used freshly to infect Cf2th-CD4.CCR5 and Cf2th-synCCR5 cells. Briefly, 100 μl each of undiluted or serially 2-fold diluted viral stock in triplicate wells was incubated with 20 μl medium containing 1 × 104 Cf2th-CD4.CCR5 or Cf2th-synCCR5 cells at 37°C. The culture was fed with 80 μl fresh complete medium on the following day. Virus entry was measured 1 day later by determining the luciferase activity in cell lysates. The previously reported CD4-independent env mutant ADA.Δ197 (35) and its parental wild-type env ADA were used as controls.
Statistical analysis.
To compare the V1V2 lengths and the numbers of gp120 PNGS of Env sequences, a one-way analysis of variance (ANOVA) was used, followed by Dunnett's multiple-comparison test using either B_database or B_chronic as the control group. The nonparametric equivalent tests (Kruskal-Wallis one-way ANOVA followed by Dunn's multiple-comparison test) gave the same statistical significance. Unpaired t test was used to compare the means of two groups. The Pearson and Spearman tests were used to evaluate potential correlations between two variables. All statistical analyses were performed using GraphPad Prism version 5.0 (GraphPad Software Inc.).
Nucleotide sequence accession numbers.
The 473 full-length env nucleotide sequences determined in this study are available in GenBank under accession numbers JQ609684 to JQ610156.
RESULTS
Study subjects.
The main goal of this study was to assess the viral quasispecies of donor 45, from whom the broadly neutralizing MAbs VRC01 and VRC03 were isolated (75). We had previously shown that a major fraction of donor 45 serum neutralizing activity was directed to the CD4bs (43), and this was confirmed by the isolation of the VRC01 and VRC03 CD4bs neutralizing MAbs. We also isolated two additional MAbs from donor 45 (VRC06 and VRC06b) that are clonal relatives of VRC03. Donor 45 plasma samples from three time points (2001, 2006, and 2009) were used for Env isolation. For comparison to donor 45, five additional donors with potent cross-reactive serum neutralizing activity were studied (Table 1). The neutralizing activity of these serum samples has been previously described (12, 43, 45). The serum samples from donors 1 and B7B5 were previously shown to contain a major fraction of neutralizing antibodies directed to the CD4bs of gp120 (43, 45); hence, these two donors were chosen as being generally similar to donor 45. Donor 1 plasma samples from 1995, 2001, and 2006 were studied. Donor B7B5 serum from only one time point (1988) was available for study. The neutralizing antibody specificities of serum samples 18, N26, and N90 could not be clearly defined, but most of their neutralizing activity did not appear to be directed to the CD4bs. A sample taken at one time point from each of these three donors was studied.
Table 1.
Characteristics of the donor samples included in this study
| Donor (yr of diagnosis) and sample date | Sample type | Type of analysis | ARTa | No. of CD4 cells/μl | No. of viral RNA copies/ml | No. of env sequences | Mean env distance ± SD (%) | Maximum env distance (%) | No. of functional env clones |
|---|---|---|---|---|---|---|---|---|---|
| 45 (1990) | |||||||||
| 2/8/2001 | Plasma | env SGA, cloning | Naive | 727 | 11,416 | 71 | 3.81 ± 1.37 | 7.39 | 9 |
| 7/30/2001 | PBMC | env SGA, cloning | Naive | 656 | 9,858 | 28 | 4.87 ± 1.27 | 7.75 | 3 |
| 7/14/2006 | Plasma | env SGA, cloning | Naive | 638 | 9,129 | 32 | 3.93 ± 1.59 | 7.24 | 8 |
| 8/19/2008 | PBMC | MAb isolation | Naive | 530 | 8,588 | Not applicable | Not applicable | Not applicable | Not applicable |
| 6/2/2009 | Plasma | env SGA, cloning | Naive | 686 | 5,153 | 55 | 2.83 ± 1.25 | 6.21 | 9 |
| 1 (1985) | |||||||||
| 8/16/1995 | Plasma | env SGA, cloning | Naive | 1,160 | 14,650 | 39 | 1.53 ± 0.95 | 3.43 | 8 |
| 7/11/2001 | Plasma | env SGA, cloning | Naive | 568 | 10,526 | 25 | 2.49 ± 1.58 | 6.61 | 6 |
| 11/8/2006 | Plasma | env SGA, cloning | Off | 545 | 19,260 | 27 | 2.69 ± 1.25 | 5.09 | 7 |
| B7B5 (1983), 6/20/1988 | Serum | env SGA, cloning | Naive | Unknown | Unknown | 12 | 5.07 ± 1.86 | 7.24 | 12 |
| 18 (1991), 4/20/1999 | Plasma | env SGA, cloning | Naive | 462 | 24,648 | 80 | 2.07 ± 0.79 | 3.66 | 13 |
| N26 (1986), 9/27/2007 | Plasma | env SGA, cloning | Naive | 387 | 35,912 | 67 | 5.44 ± 2.37 | 9.42 | 14 |
| N90 (1985), 5/29/2008 | Plasma | env SGA, cloning | Naive | 912 | 8,216 | 37 | 3.22 ± 1.25 | 5.34 | 11 |
All donors were either ART naïve or off ART at the time that blood specimens were collected.
Analysis of env diversity.
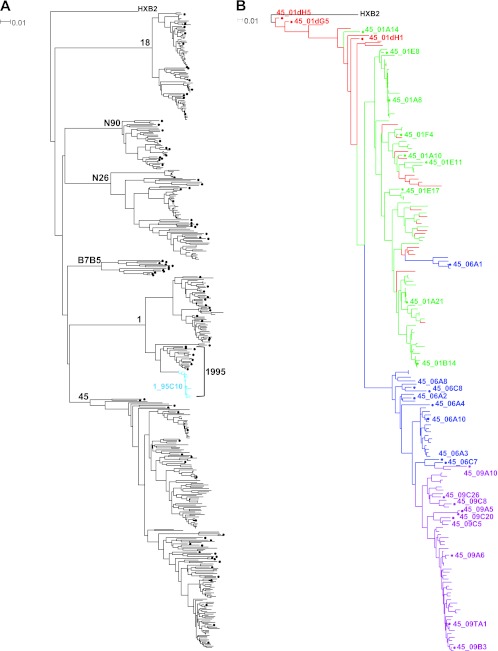
A total of 473 full-length env genes encoding gp160 were amplified and sequenced from viral RNA present in plasma or serum or from proviral DNA integrated in PBMC genomic DNA (median of 73 sequences per donor; range, 12 to 186). All sequences belonged to HIV-1 subtype B. The relationship of these sequences was examined by phylogenetic analysis. In a neighbor-joining phylogenetic tree (Fig. 1A), the viral Env protein sequences formed distinct donor-specific lineages with no evidence of cross-donor contamination. The maximum within-donor env distance at any time point ranged from 3.4% to 9.4% or from 0.23 to 1.45%/year based on the estimated infection time (Table 1). These values reflect the env sequence diversity and diversification rate in these donors and are within the range reported previously (65). In donor 1, a predominant env sequence, representing 20 (51%) of the 39 sequences in 1995, was identified (light blue in Fig. 1A), but this sequence was not found at later time points. The viral quasispecies from donor 45 PBMC obtained in 2001 and from plasma samples obtained in 2001, 2006, and 2009 was studied. A maximum-likelihood tree with a total of 186 env sequences from donor 45 was constructed (Fig. 1B). Most of the sequences from the two 2001 time points (red and green in Fig. 1B) intermingled, with a few integrated proviral DNA sequences (shown in red) clustering toward HXB2, which was displayed as the outgroup root. Most of the sequences derived from the 2006 plasma (shown in blue) formed a separate major branch of the tree. The sequences derived from the 2009 plasma sample (shown in purple) were within the same major tree branch as those from 2006, but most of these formed a distinct subcluster. These data indicate a continuous env evolutionary pattern in donor 45 during the 9-year time span between 2001 and 2009.
Fig 1.
Phylogenetic trees of HIV-1 envelope sequences. (A) Neighbor-joining tree showing the phylogenetic clustering of Env protein sequences from six HIV-1-infected donors. A total of 473 gp160 sequences from the six subtype B-infected donors were aligned with the reference sequence HXB2. The tree was constructed based on sequence distance and unrooted and then rooted at HXB2 for visualization. The gp160 sequences were derived at a single time point for donors 18, N90, N26, and B7B5; at three time points for donor 1; and at four time points for donor 45. The donor identification is indicated at each donor's major branch node. Representative sequences indicated by a dot were cloned and expressed. A predominant sequence from donor 1 is shown in light blue from the 1995 time point (indicated by a bracket); one such sequence was cloned as indicated with an identification number. (B) Maximum-likelihood tree of envelope sequences from donor 45. A total of 186 gp160 nucleotide sequences from four temporal samples were aligned with the reference sequence HXB2. The tree was constructed as unrooted and then rooted at HXB2 for visualization. The gp160 sequences are color coded as follows: red, 2001 provirus; green, 2001 plasma; blue, 2006 plasma; purple, 2009 plasma. The major branch colors follow the color of most of the sequences on the branch. Representative sequences indicated by a dot and an identification number were cloned and expressed. The horizontal branch scale is indicated for each tree.
Analysis of gp120 N-linked glycosylation sites and V1V2 lengths.
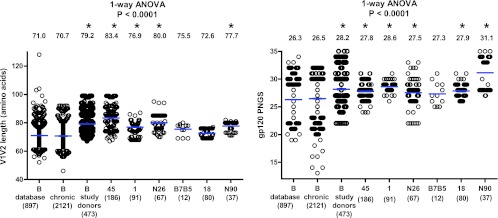
During the course of infection, N-linked glycosylation sites are often added to the variable regions of gp120, sometimes as a consequence of length additions in V1V2 (6, 7, 9, 10, 60). In some cases, these changes in Env were paired with increased viral resistance to autologous or heterologous antibody neutralization (6, 10, 60, 72, 74, 78). Therefore, increased numbers of N-linked glycosylation sites and increased V1V2 lengths have been suggested as part of the viral mechanisms for immune evasion. The V1V2 length and the number of PNGS were also determined for all of the 473 Env sequences from the six study donors (Fig. 2). For comparison, we used a set of 897 subtype B sequences in the HIV database (B_database, described in Materials and Methods). Because this set contained sequences from donors with unknown infection times and included sequences from cultured viruses, we added another set of 2,121 chronic subtype B infection sequences (B_chronic, described in Materials and Methods) isolated by SGA and used in a recent study at the LANL (21). Notably, the average V1V2 length was comparable between the B_database and B_chronic sequences; however, the average V1V2 length of Env variants from four of the six broadly neutralizing study donors was significantly greater than that in both the B_database and B_chronic groups (P < 0.0001, one-way ANOVA, Fig. 2, left). The average number of PNGS was also comparable between the B_database and B_chronic sequences; however, the average number of PNGS among Env variants from five of the six study donors was significantly higher than in both the B_database and B_chronic groups (P < 0.0001, one-way ANOVA, Fig. 2, right). Because an extended V1V2 region with an increased number of glycosylation sites in gp120 has been described as a characteristic of Env sequences from chronically infected individuals (9, 60), our data suggest that donors with broadly reactive serum neutralizing antibodies are further enriched for longer V1V2 regions with more gp120 N-linked glycans.
Fig 2.
Comparisons of V1V2 lengths (left) and numbers of gp120 PNGS (right) among the subtype B sequences from the HIV-1 database (B_database), from chronic infection (B_chronic), and from the six study donors as one group or individually. The blue horizontal bars indicate the group means, and the actual mean values are indicated at the top of the columns. The number of sequences for each group or individual is indicated in parentheses below the group or donor identification. One-way ANOVA P values are indicated, with the asterisks indicating groups that are significantly different (P < 0.05) from both the B_database and B_chronic groups in subsequent Dunnett's multiple-comparison tests.
Neutralization sensitivity of Env variants from the VRC01 donor.
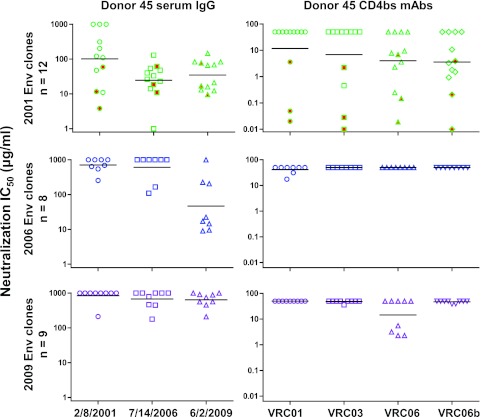
Representative env genes from the VRC01 donor were cloned and expressed as Env pseudoviruses to test for neutralization sensitivity. We first examined the concurrent serum IgG neutralization of Env variants amplified from 2001, 2006, and 2009 plasma samples (Fig. 3, left panels; Table 2). Most of the 2001 Env clones were only weakly neutralized by the concurrent serum IgG (IC50, >100 μg/ml) but were potently neutralized by serum IgGs from 2006 and 2009. Similarly, the 2006 Env clones were poorly neutralized by the concurrent and earlier 2001 serum IgG but were potently neutralized by the 2009 serum IgG. Thus, even in a donor with a broadly reactive serum neutralizing antibody such as VRC01, there is ongoing autologous virus escape from serum neutralizing antibodies, similar to the pattern described for more strain-specific autologous neutralizing antibodies (25, 48, 57, 58, 72).
Fig 3.
Analysis of neutralization sensitivity of Env variants taken from donor 45 at three time points to autologous serum IgG (left panels) and autologous CD4bs MAbs VRC01, VRC03, VRC06, and VRC06b (right panels). The horizontal bars indicate the group geometric means. The three symbols highlighted in red in the 2001 Env plots indicate the archival Env variants derived from proviral DNA.
Table 2.
Neutralization IC50s of autologous serum IgG, autologous CD4bs MAbs, and CD4 surrogate protein CD4-Ig for donor 45 Env variants
| env isolation time, source, and env clone or parameter | Neutralization IC50 (μg/ml) or breadth |
|||||||
|---|---|---|---|---|---|---|---|---|
| Autologous serum IgG |
Autologous CD4bs MAbs |
CD4 surrogate CD4-Ig | ||||||
| 2/8/2001 | 7/14/2006 | 6/2/2009 | VRC01 | VRC03 | VRC06 | VRC06b | ||
| 2001, provirus | ||||||||
| 45_01dG5 | 3.8 | 10.9 | 9.5 | 0.020 | 0.010 | 0.019 | 0.010 | 0.256 |
| 45_01dH1 | 58.6 | 61.4 | 77.0 | 3.6 | 2.2 | 6.9 | 3.9 | 0.870 |
| 45_01dH5 | 11.6 | 18.4 | 16.8 | 0.049 | 0.028 | 0.149 | 0.207 | >50 |
| Breadtha (n = 3) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 2 (67) |
| 2001, plasma | ||||||||
| 45_01A8 | 310 | 47.6 | 13.0 | >50 | >50 | >50 | >50 | 1.3 |
| 45_01A10 | >1,000 | 26.2 | 12.3 | >50 | >50 | 0.248 | 0.925 | 3.6 |
| 45_01A14 | 121 | 39.8 | 21.0 | >50 | 0.453 | >50 | 1.5 | 1.7 |
| 45_01A21 | >1,000 | 22.9 | 65.6 | >50 | >50 | >50 | >50 | 4.4 |
| 45_01B14 | >1,000 | 14.1 | 68.2 | >50 | >50 | >50 | >50 | 5.3 |
| 45_01E8 | 11.0 | 0.823 | 15.9 | >50 | >50 | 9.4 | >50 | 0.265 |
| 45_01E11 | 201 | 129 | 146 | >50 | >50 | 2.3 | 1.8 | 0.698 |
| 45_01E17 | 109 | 33.0 | 83.2 | >50 | >50 | 3.6 | 3.3 | 1.8 |
| 45_01F4 | 47.3 | 54.2 | 83.2 | >50 | >50 | 7.8 | 10.7 | 0.533 |
| Breadth (n = 9) | 6 (67) | 9 (100) | 9 (100) | 0 (0) | 1 (11) | 5 (56) | 5 (56) | 9 (100) |
| 2006, plasma | ||||||||
| 45_06A1 | 689 | >1,000 | 17.2 | >50 | >50 | >50 | >50 | 3.4 |
| 45_06A2 | >1,000 | >1,000 | 9.0 | >50 | >50 | >50 | >50 | 2.8 |
| 45_06A3 | >1,000 | >1,000 | 9.6 | 17.5 | >50 | >50 | >50 | 2.5 |
| 45_06A4 | 542 | 109 | 225 | >50 | >50 | >50 | >50 | 6.0 |
| 45_06A8 | 254 | >1,000 | 22.4 | >50 | >50 | >50 | >50 | 0.614 |
| 45_06A10 | 640 | >1,000 | 14.5 | >50 | >50 | >50 | >50 | 5.3 |
| 45_06C7 | >1,000 | >1,000 | >1,000 | 31.2 | >50 | >50 | >50 | 3.3 |
| 45_06C8 | >1,000 | 165 | 205 | >50 | >50 | >50 | >50 | 1.4 |
| Breadth (n = 8) | 4 (50) | 2 (25) | 7 (88) | 2 (25) | 0 (0) | 0 (0) | 0 (0) | 8 (100) |
| 2009, plasma | ||||||||
| 45_09A5 | >1,000 | 462 | 879 | >50 | >50 | >50 | >50 | >50 |
| 45_09A6 | >1,000 | 447 | 905 | >50 | >50 | 5.5 | 38.4 | >50 |
| 45_09A10 | >1,000 | >1,000 | >1,000 | >50 | >50 | >50 | >50 | 5.2 |
| 45_09B3 | >1,000 | >1,000 | 456 | >50 | 35.7 | 3.2 | >50 | 4.0 |
| 45_09C5 | >1,000 | >1,000 | 564 | >50 | >50 | >50 | >50 | 2.7 |
| 45_09C8 | >1,000 | >1,000 | 743 | >50 | >50 | >50 | >50 | >50 |
| 45_09C20 | 211 | 178 | 209 | >50 | >50 | >50 | >50 | 0.809 |
| 45_09C26 | >1,000 | >1,000 | >1,000 | >50 | >50 | >50 | >50 | 5.5 |
| 45_09TA1 | >1,000 | 785 | 560 | >50 | >50 | 2.3 | >50 | 9.1 |
| Breadth (n = 90) | 1 (11) | 4 (44) | 7 (78) | 0 (0) | 1 (11) | 3 (33) | 1 (11) | 6 (67) |
Breadth is shown as the number (percentage) of Env variants sensitive to the corresponding serum IgG, MAb, or CD4-Ig.
We then examined the neutralization sensitivity of donor 45 Env variants to the CD4bs MAbs isolated from this donor (autologous MAbs) (Fig. 3, right panels; Table 2). Almost all of the plasma-derived Env variants from each time point were highly resistant to VRC01, with only two 2006 clones (45_06A3 and 45_06C7) showing moderate sensitivity (IC50s of >10 μg/ml). Three archival Env clones, 45_01dG5, 45_01dH1, and 45_01dH5, derived from proviral DNA of a 2001 PBMC sample and chosen based on phylogenetic analysis (Fig. 1B) were fully sensitive to VRC01. These three sequences were not found in the tested donor plasma by our method of SGA and DNA Sanger sequencing. One 2001 Env clone (45_01A14) was fully sensitive to VRC03, but other Env variants from all time points were resistant to VRC03. In contrast, five of the nine 2001 Env clones were sensitive to VRC06 or VRC06b, suggesting that these closely related MAbs may have evolved after the 2001 time point. By 2006, all tested Env sequences were resistant to VRC06 and VRC06b, with three 2009 sequences sensitive to VRC06. Because the 2009 serum IgG was potent against the 2006 Env clones and this neutralization activity was not recapitulated by the known donor MAbs, these data suggest that there are unidentified antibodies in the 2009 serum that potently neutralized the 2006 Env variants.
Neutralization sensitivity of Env variants from other donors to VRC01.
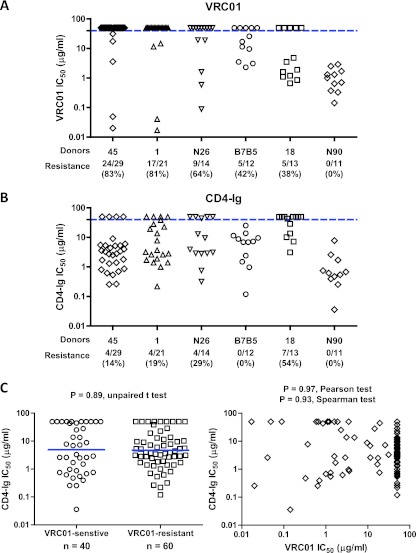
In addition to donor 45, we cloned representative env sequences from five other subtype B-infected donors who developed broadly neutralizing antibodies (Table 1). We expressed representative Env clones as Env pseudoviruses and examined the viral neutralization sensitivity to VRC01 (Fig. 4A) and to VRC03, VRC06, VRC06b, and b12 (Table 3). Prior serum mapping studies indicated that donors 1 and B7B5 had a major fraction of neutralizing antibodies to the CD4bs, and we observed that 81% and 42% of the Env clones, respectively, were resistant to VRC01. In contrast, all of the Env clones from neutralizing donor N90 were VRC01 sensitive. Somewhat surprisingly, a substantial percentage of the sequences from donor N26 (64%) and donor 18 (38%) were also VRC01 resistant.
Fig 4.
Comparisons of VRC01 (A) and CD4-Ig (B) neutralization sensitivities of Env variants derived from the six study donors indicated on the x axis. The blue horizontal dashed line indicates an IC50 of 50 μg/ml, which was used as the cutoff for neutralization sensitivity. The values below the donor designations are the numbers and percentages of Env variants resistant to VRC01 or CD4-Ig neutralization. (C) Plot of log-transformed CD4-Ig neutralization IC50s for VRC01-sensitive (n = 40) and VRC01-resistant (n = 60) Env variants (left). The mean of each group is indicated by a blue horizontal bar. The log-transformed VRC01 and CD4-Ig neutralization IC50s are plotted for each individual Env variant (right, n = 100). The P values and the corresponding statistical analyses are indicated.
Table 3.
Neutralization IC50s of heterologous CD4bs MAbs and CD4 surrogate protein CD4-Ig for Env variants from five donors
| Donor, yr, and env clone or parameter | Neutralization IC50 or breadth |
|||||
|---|---|---|---|---|---|---|
| VRC01 | VRC03 | VRC06 | VRC06b | b12 | CD4-Ig | |
| 1 | ||||||
| 1995 | ||||||
| 1_95A21 | >50 | >50 | >50 | >50 | >50 | >50 |
| 1_95C1 | >50 | >50 | >50 | >50 | >50 | 13.6 |
| 1_95C5 | 0.017 | 0.020 | 3.3 | 0.016 | 0.023 | >50 |
| 1_95C10 | >50 | >50 | >50 | >50 | 0.214 | 3.7 |
| 1_95TC1 | >50 | >50 | >50 | >50 | >50 | >50 |
| 1_95TC6 | >50 | >50 | >50 | >50 | >50 | >50 |
| 1_95TC13 | >50 | >50 | >50 | >50 | 0.773 | 1.4 |
| 1_95TC14 | 14.6 | 5.0 | 26.2 | 7.1 | >50 | 28.0 |
| 2001 | ||||||
| 1_01A10 | >50 | >50 | >50 | >50 | >50 | 38.6 |
| 1_01A19 | >50 | >50 | >50 | >50 | >50 | 1.9 |
| 1_01A20 | 11.5 | 12.0 | >50 | >50 | >50 | 2.6 |
| 1_01B3 | >50 | >50 | >50 | >50 | 0.920 | 1.4 |
| 1_01B5 | 0.041 | 0.111 | >50 | 0.128 | 0.032 | 2.6 |
| 1_01TB5 | >50 | >50 | >50 | >50 | >50 | 3.2 |
| 2006 | ||||||
| 1_06B1 | >50 | >50 | >50 | >50 | 6.1 | 0.995 |
| 1_06B3 | >50 | 0.841 | 13.1 | 2.5 | >50 | 19.3 |
| 1_06B7 | >50 | >50 | >50 | >50 | >50 | 22.8 |
| 1_06B9 | >50 | >50 | >50 | >50 | >50 | 0.218 |
| 1_06C3 | >50 | >50 | >50 | >50 | 0.289 | 7.8 |
| 1_06C5 | >50 | >50 | >50 | >50 | 50.0 | 2.8 |
| 1_06D29 | >50 | >50 | >50 | >50 | 6.3 | 1.7 |
| Breadtha (n = 21) | 4 (19) | 5 (24) | 3 (16) | 4 (19) | 9 (47) | 17 (81) |
| N26, 2007 | ||||||
| N26_07A10 | >50 | >50 | >50 | >50 | >50 | 9.5 |
| N26_07A14 | >50 | >50 | >50 | >50 | >50 | 0.317 |
| N26_07A16 | >50 | >50 | >50 | >50 | >50 | 2.8 |
| N26_07A21 | 1.6 | 1.7 | >50 | 10.9 | 0.640 | 2.9 |
| N26_07A22 | >50 | >50 | >50 | >50 | >50 | >50 |
| N26_07A38 | 0.591 | >50 | >50 | >50 | 1.1 | >50 |
| N26_07B9 | >50 | >50 | >50 | >50 | 7.2 | 3.9 |
| N26_07B10 | >50 | >50 | >50 | >50 | 7.9 | >50 |
| N26_07B18 | 19.0 | >50 | >50 | >50 | >50 | 0.755 |
| N26_07B22 | >50 | >50 | >50 | >50 | >50 | 2.9 |
| N26_07B36 | >50 | >50 | >50 | >50 | >50 | 3.1 |
| N26_07B41 | >50 | >50 | >50 | >50 | >50 | 12.9 |
| N26_07TC39 | 19.3 | >50 | >50 | >50 | >50 | 44.4 |
| N26_07TC40 | 0.089 | >50 | >50 | >50 | 10.4 | >50 |
| Breadth (n = 14) | 5 (36) | 1 (7) | 0 (0) | 1 (7) | 5 (36) | 10 (71) |
| B7B5, 1988 | ||||||
| B7B5_88A1 | 17.0 | 38.7 | >50 | >50 | >50 | 1.5 |
| B7B5_88A2 | 2.3 | 6.9 | >50 | 15.5 | 0.452 | 25.3 |
| B7B5_88A3 | 3.1 | 12.6 | >50 | >50 | 0.426 | 13.7 |
| B7B5_88A4 | >50 | >50 | >50 | >50 | 0.731 | 0.119 |
| B7B5_88A5 | >50 | 20.0 | >50 | >50 | >50 | 6.9 |
| B7B5_88A6 | 3.5 | 7.5 | >50 | 19.1 | 0.141 | 0.970 |
| B7B5_88A7 | >50 | >50 | >50 | >50 | 36.1 | 9.6 |
| B7B5_88A10 | 25.9 | >50 | >50 | >50 | >50 | 7.3 |
| B7B5_88TB1 | >50 | >50 | >50 | >50 | >50 | 11.4 |
| B7B5_88TB2 | 9.6 | 2.2 | 5.3 | 1.6 | >50 | 8.8 |
| B7B5_88TB3 | 11.4 | 2.7 | 8.7 | 1.6 | 4.4 | 6.9 |
| B7B5_88TB6 | >50 | >50 | >50 | >50 | >50 | 2.4 |
| Breadth (n = 12) | 7 (58) | 7 (58) | 2 (17) | 4 (33) | 6 (50) | 12 (100) |
| 18, 1999 | ||||||
| 18_99A27 | 0.661 | >50 | >50 | >50 | >50 | >50 |
| 18_99A34 | 3.5 | 1.9 | >50 | >50 | >50 | >50 |
| 18_99A38 | >50 | >50 | >50 | >50 | >50 | >50 |
| 18_99B1 | 4.8 | >50 | >50 | >50 | 9.6 | 13.5 |
| 18_99B15 | 1.9 | 1.5 | >50 | >50 | >50 | >50 |
| 18_99B19 | 0.855 | >50 | >50 | >50 | >50 | 43.1 |
| 18_99B27 | >50 | >50 | >50 | >50 | >50 | 10.2 |
| 18_99B39 | >50 | >50 | >50 | >50 | >50 | 28.8 |
| 18_99B46 | 1.6 | >50 | >50 | >50 | >50 | >50 |
| 18_99B48 | >50 | >50 | >50 | >50 | >50 | 3.1 |
| 18_99B51 | 1.2 | >50 | >50 | >50 | >50 | >50 |
| 18_99TA3 | >50 | >50 | >50 | >50 | >50 | 7.2 |
| 18_99TB13 | 1.1 | >50 | >50 | >50 | >50 | >50 |
| Breadth (n = 13) | 8 (62) | 2 (15) | 0 (0) | 0 (0) | 1 (8) | 6 (46) |
| N90, 2008 | ||||||
| N90_08A13 | 2.5 | 0.364 | 19.4 | 0.349 | 5.8 | 0.618 |
| N90_08A15 | 1.0 | 0.311 | 8.1 | 0.333 | 1.5 | 0.679 |
| N90_08A16 | 1.3 | 0.354 | 11.0 | 0.462 | 3.5 | 0.545 |
| N90_08A19 | 0.372 | 0.128 | 6.2 | 0.354 | >50 | 7.8 |
| N90_08A25 | 1.3 | 0.350 | 5.6 | 0.413 | >50 | 0.752 |
| N90_08B2 | 0.703 | 0.338 | 3.9 | 0.361 | 5.7 | 0.394 |
| N90_08B4 | 2.9 | 0.236 | >50 | 0.380 | 2.5 | 2.6 |
| N90_08B6 | 0.644 | 0.293 | 5.3 | 0.434 | 3.0 | 0.255 |
| N90_08B11 | 0.143 | 0.055 | 4.7 | 0.063 | 1.6 | 0.036 |
| N90_08B16 | 0.328 | 0.196 | 7.0 | 0.259 | 2.5 | 0.472 |
| N90_08B17 | 1.7 | 0.651 | 14.5 | 0.488 | 3.0 | 1.7 |
| Breadth (n = 11) | 11 (100) | 11 (100) | 10 (91) | 11 (100) | 9 (82) | 11 (100) |
Breadth is shown as the number (percentage) of Env variants sensitive to the corresponding corresponding MAb or CD4-Ig or CD4-Ig.
VRC01 escape Env variants are still sensitive to CD4-Ig neutralization and require CD4 for efficient entry.
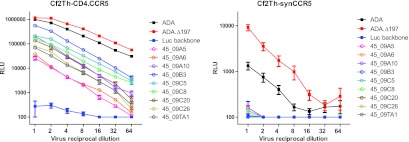
As previously noted, the heavy chain of MAb VRC01 partially mimics the binding interaction of CD4 with viral gp120. The VRC01 binding epitope largely overlaps the binding surface of CD4 in the outer domain of gp120; however, VRC01 makes fewer contacts than CD4 in the inner domain and bridging sheet regions (80). We therefore asked if mutations that confer VRC01 resistance would compromise the CD4-gp120 interaction and lead to reduced CD4-Ig sensitivity or to the evolution of Env clones that could enter cells without CD4, as previous studies have reported rare HIV-1 strains that are capable of CD4-independent entry (29, 35, 79). To test these possibilities, we first looked at the Env variants' sensitivity to CD4-Ig neutralization (Tables 2 and 3; Fig. 4B). Despite high levels of Env resistance to VRC01 in five of the six study donors, most of the donor Env variants were sensitive to CD4-Ig. Comparison of the VRC01-senstive (n = 40) and VRC01-resistant (n = 60) Env variants did not reveal any difference in CD4-Ig sensitivity (P = 0.89, unpaired t test, Fig. 4C, left). In addition, the VRC01 and CD4-Ig neutralization IC50s did not reveal any correlation (P = 0.97, Pearson test; Fig. 4C, right). To test if the VRC01-resistant Env variants can enter cells in a CD4-independent manner, we made luciferase-containing Env pseudoviruses of all 29 donor 45 env clones and examined their entry into CD4− CCR5+ cell line Cf2th-synCCR5 (22) and its CD4+ CCR5+ counterpart Cf2th-CD4.CCR5 (28, 49). All of the Env variants entered the CD4+ cell line Cf2th-CD4.CCR5 efficiently but not the CD4− cell line Cf2th-synCCR5, indicating that these Env variants were able to escape VRC01 neutralization yet retain efficient interaction with the viral cellular receptor CD4. Data from nine env clones from the 2009 plasma of donor 45 are shown in Fig. 5.
Fig 5.
Entry of nine donor 45 Env variants derived from the 2009 plasma into CD4+ cell line Cf2Th-CD4.CCR5 (left) and CD4− cell line Cf2Th-synCCR5 (right).
DISCUSSION
The CD4bs of gp120 is an attractive target for HIV-1 vaccine design because it is functionally conserved and contains epitopes of potently neutralizing antibodies. The isolation of VRC01 and similar CD4bs MAbs demonstrates that the immune system is capable of generating antibodies to this region of gp120. To characterize the circulating Env variants that coexist with VRC01 and to understand how the HIV-1 env gene evolves under the selection pressure of a broadly reactive antibody, we isolated Env variants from the VRC01 donor and studied their neutralization sensitivities. While numerous published studies have shown that autologous plasma virus is often resistant to concurrent serum neutralization (4, 19, 26, 40, 50, 54, 57, 58, 72), there had not previously been a detailed assessment of the Env quasispecies in a donor with a broadly reactive serum neutralizing antibody such as VRC01.
Using SGA followed by DNA Sanger sequencing, we found that plasma-derived Env variants from donor 45 from three time points, spanning from 2001 to 2009, displayed almost uniform resistance to VRC01 neutralization, indicating a strong selection pressure on the viral Env quasispecies by VRC01 and related autologous CD4bs antibodies. While the Env SGA and Sanger sequencing approach has an inherent limitation in the overall depth of the sequences identified, our data show that the vast majority of the viruses circulating in plasma were VRC01 resistant. Donor 45 was first found to be HIV-1 infected in 1990; thus, the time points studied here were in excess of 10 years after HIV-1 infection. The VRC01 MAb was isolated from IgG+ memory B cells from a 2008 PBMC sample (Table 1), but since memory B cells may circulate for a long time, it is not clear when VRC01 first developed. Our finding that all nine plasma Env variants from 2001 were VRC01 resistant suggests that VRC01 arose prior to 2001. It was only through the isolation of HIV-1 env sequences from proviral DNA that we were able to identify archival Env clones that are highly sensitive to VRC01, with IC50s of <1 μg/ml. Thus, it seems unlikely that donor 45 was infected with a VRC01-resistant virus but rather likely that the Env variants evolved to escape from this potently neutralizing antibody.
The neutralization sensitivity of some of the VRC01 escape variants to MAbs VRC03, VRC06, and VRC06b suggests the possibility of continuous evolution of CD4bs neutralizing antibodies in response to viral escape. VRC03 was isolated from the same PBMC samples as VRC01, and its liganded structure has been solved (75, 77). While the VRC03 heavy chain derives from the same IGHV1-2*02 allele as VRC01, it has a different light chain, indicating that it arose from a different B-cell clone. The crystal structure of VRC03 in complex with a gp120 core revealed that the antibody forms contacts with the bridging sheet between the inner and outer domains of gp120, which is part of the coreceptor binding site (77). Therefore, we speculate that the donor immune system responded to VRC01 escape variants by generating antibody specificities extending from the VRC01 epitope toward the coreceptor binding region and resulting in efficient neutralization of some VRC01 escape variants. MAbs VRC06 and VRC06b are clonal relatives of VRC03 and will be described in detail in a separate report (Yuxing Li et al., unpublished data). The sensitivity of some 2001 viral Env clones (which are all resistant to VRC01) to VRC03, VRC06, and VRC06b suggests that these MAbs might have evolved later than VRC01. Indeed, suboptimal 454 pyrosequencing of the IgG+ memory B-cell heavy-chain transcripts from the 2008 PBMC sample identified a population of antibody sequences with >90% identity to VRC03 but not VRC01-related sequences (77), suggesting the presence of greater numbers of memory B cells for VRC03 than VRC01 in 2008. We are currently optimizing PCR and sequencing conditions to analyze donor 45 longitudinal samples to determine the dynamics of these neutralizing antibody clones.
Although MAbs VRC03, VRC06, and VRC06b were able to neutralize VRC01 escape Env variants in donor 45, the neutralization breadth of these MAbs against heterologous viruses (<60%) was less than that of VRC01 (90%) (75). Therefore, the development or maintenance of these MAbs after VRC01 in donor 45 was specific to the relevant autologous viruses but not necessarily associated with better neutralization breadth of “irrelevant” heterologous viruses circulating globally. This highlights the complexity of underlying factors that influence the broadening of antibody neutralization (16, 23). Although most of the donor 45 Env variants from 2001 plasma was neutralized by VRC03, VRC06, and VRC06b, Env variants from 2006 plasma were resistant, suggesting that the virus was able to escape these newly developed antibody variants. Note that the 2006 Env variants remained highly sensitive to the 2009 serum IgG, indicating the presence of unidentified antibody specificities, including the possibility of those outside the CD4bs, in this serum that likely arose in response to the escaped variants.
We also analyzed Env variants from five additional subtype B-infected individuals who developed strong broadly neutralizing antibody responses. We previously reported that the serum samples of donors 1 and B7B5 contained a major fraction of CD4bs-directed neutralizing antibodies (43, 45); thus, we hypothesized that Env sequences from these two donors would contain some VRC01-resistant clones. However, we were surprised to find a majority of Env clones in donor N26 also resistant to VRC01, as our mapping did not definitively indicate CD4bs neutralizing antibodies in this donor. This was in contrast to donor N90, whose Env isolates were fully sensitive to VRC01 neutralization, suggesting a lack of VRC01-like antibody pressure in this donor. Overall, the rather high level of VRC01-resistant clones in four of five donors (other than donor 45) suggests that the CD4bs-directed neutralizing antibodies similar to VRC01 may not be uncommon in donors whose serum samples are broadly neutralizing.
Lastly, we observed that viral variants fully resistant to VRC01 retained their sensitivity to CD4-Ig and required CD4 for entry. These results suggest that VRC01 escape mutations do not substantially impair the gp120 interaction with CD4, at least not enough to have a major impact on viral entry. Our prior studies of VRC01-resistant viruses demonstrated that mutations in the D-loop and V5 and regions of gp120 were often responsible for VRC01 resistance and that some of these mutations were not contact sites for CD4 (44, 80). It remains possible that initial escape from VRC01 is associated with some loss of binding to CD4 and that compensatory mutations restore viral entry and replication. More systematic studies of viral fitness and the affinity of interaction with CD4 are in progress. Donor 45 is a slow progressor, as are several other donors from whom broadly neutralizing CD4bs MAbs have been isolated (63, 77). More systematic and unbiased sampling is required to know if CD4bs neutralizing MAbs are more commonly found in donors with slow progression.
In summary, our data show that neutralizing antibodies to the conserved CD4bs exert selection pressure on HIV-1 Env and that the viruses evolve to escape from such neutralization. The B-cell response to the CD4bs also evolves by generating antibodies that neutralize viral escape mutants. Hence, even for this functionally conserved region of gp120, there is ongoing viral evolution matched by antibody evolution. The facts that escape from VRC01-like CD4bs antibodies appears to occur commonly within a donor but that the large majority of heterologous viral strains are sensitive to VRC01 suggest some constraint on resistance to such CD4bs antibodies, although whether there is any measurable fitness cost to VRC01 escape remains to be determined. Finally, the study of Env species in donors with broadly neutralizing antibodies may reveal clues about the viral antigenic stimulus that led to the development of this antibody. Such conclusions are difficult in the case of donor 45, who had been infected for more than 10 years prior to the time when the samples we studied were taken. Similar studies of known seroconverter donors with longitudinal sampling could reveal the relationship between the early circulating Env sequences and the early B-cell clones that ultimately develop into mature broadly neutralizing antibodies.
ACKNOWLEDGMENTS
We thank Kim-Truc Pham for technical assistance with SGA of env from donor 1 plasma samples.
Support for this work was provided by the Intramural Research Program of the Vaccine Research Center, NIAID, NIH. G.M.S. was supported by grants from the NIH (AI067854) and the Bill & Melinda Gates Foundation Grand Challenges Program (37874).
Footnotes
Published ahead of print 14 March 2012
REFERENCES
- 1. Albert J, et al. 1990. Rapid development of isolate-specific neutralizing antibodies after primary HIV-1 infection and consequent emergence of virus variants which resist neutralization by autologous sera. AIDS 4:107–112 [DOI] [PubMed] [Google Scholar]
- 2. Bonsignori M, et al. 2011. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J. Virol. 85:9998–10009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bosch KA, Rainwater S, Jaoko W, Overbaugh J. 2010. Temporal analysis of HIV envelope sequence evolution and antibody escape in a subtype A-infected individual with a broad neutralizing antibody response. Virology 398:115–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bunnik EM, Pisas L, van Nuenen AC, Schuitemaker H. 2008. Autologous neutralizing humoral immunity and evolution of the viral envelope in the course of subtype B human immunodeficiency virus type 1 infection. J. Virol. 82:7932–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burton DR, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024–1027 [DOI] [PubMed] [Google Scholar]
- 6. Cheng-Mayer C, Brown A, Harouse J, Luciw PA, Mayer AJ. 1999. Selection for neutralization resistance of the simian/human immunodeficiency virus SHIVSF33A variant in vivo by virtue of sequence changes in the extracellular envelope glycoprotein that modify N-linked glycosylation. J. Virol. 73:5294–5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chohan B, et al. 2005. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1-V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J. Virol. 79:6528–6531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Corti D, et al. 2010. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One 5:e8805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Curlin ME, et al. 2010. HIV-1 envelope subregion length variation during disease progression. PLoS Pathog. 6:e1001228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Derdeyn CA, et al. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019–2022 [DOI] [PubMed] [Google Scholar]
- 11. Dhillon AK, et al. 2007. Dissecting the neutralizing antibody specificities of broadly neutralizing sera from human immunodeficiency virus type 1-infected donors. J. Virol. 81:6548–6562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Doria-Rose NA, et al. 2010. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J. Virol. 84:1631–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Etherington GJ, Dicks J, Roberts IN. 2005. Recombination analysis tool (RAT): a program for the high-throughput detection of recombination. Bioinformatics 21:278–281 [DOI] [PubMed] [Google Scholar]
- 16. Euler Z, et al. 2012. Longitudinal analysis of early HIV-1-specific neutralizing activity in an elite neutralizer and in five patients who developed cross-reactive neutralizing activity. J. Virol. 86:2045–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Felsenstein J, Churchill GA. 1996. A hidden Markov model approach to variation among sites in rate of evolution. Mol. Biol. Evol. 13:93–104 [DOI] [PubMed] [Google Scholar]
- 18. Flynn NM, et al. 2005. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J. Infect. Dis. 191:654–665 [DOI] [PubMed] [Google Scholar]
- 19. Frost SD, et al. 2005. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc. Natl. Acad. Sci. U. S. A. 102:18514–18519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gilbert PB, et al. 2005. Correlation between immunologic responses to a recombinant glycoprotein 120 vaccine and incidence of HIV-1 infection in a phase 3 HIV-1 preventive vaccine trial. J. Infect. Dis. 191:666–677 [DOI] [PubMed] [Google Scholar]
- 21. Gnanakaran S, et al. 2011. Recurrent signature patterns in HIV-1 B clade envelope glycoproteins associated with either early or chronic infections. PLoS Pathog. 7:e1002209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gorry PR, et al. 2002. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J. Virol. 76:6277–6292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gray ES, et al. 2011. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J. Virol. 85:4828–4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gray ES, et al. 2009. Broad neutralization of human immunodeficiency virus type 1 mediated by plasma antibodies against the gp41 membrane proximal external region. J. Virol. 83:11265–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gray ES, et al. 2011. Isolation of a monoclonal antibody that targets the alpha-2 helix of gp120 and represents the initial autologous neutralizing-antibody response in an HIV-1 subtype C-infected individual. J. Virol. 85:7719–7729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gray ES, et al. 2007. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J. Virol. 81:6187–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gray ES, et al. 2009. Antibody specificities associated with neutralization breadth in plasma from human immunodeficiency virus type 1 subtype C-infected blood donors. J. Virol. 83:8925–8937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haim H, et al. 2009. Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathog. 5:e1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haim H, et al. 2011. Contribution of intrinsic reactivity of the HIV-1 envelope glycoproteins to CD4-independent infection and global inhibitor sensitivity. PLoS Pathog. 7:e1002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huson DH, et al. 2007. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics 8:460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Johnson VA, Byington RE. 1990. Infectivity assay (virus yield assay), p 71–76 In Aldovani A, Walker BD. (ed), Techniques in HIV research. Stockton Press, New York, NY [Google Scholar]
- 32. Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275–282 [DOI] [PubMed] [Google Scholar]
- 33. Keele BF, et al. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kishino H, Hasegawa M. 1989. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J. Mol. Evol. 29:170–179 [DOI] [PubMed] [Google Scholar]
- 35. Kolchinsky P, Kiprilov E, Bartley P, Rubinstein R, Sodroski J. 2001. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J. Virol. 75:3435–3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Korber B, et al. 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288:1789–1796 [DOI] [PubMed] [Google Scholar]
- 37. Kuhner MK, Felsenstein J. 1994. A simulation comparison of phylogeny algorithms under equal and unequal evolutionary rates. Mol. Biol. Evol. 11:459–468 [DOI] [PubMed] [Google Scholar]
- 38. Kwong PD, et al. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 40. Li B, et al. 2006. Evidence for potent autologous neutralizing antibody titers and compact envelopes in early infection with subtype C human immunodeficiency virus type 1. J. Virol. 80:5211–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li H, et al. 2010. High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog. 6:e1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li M, et al. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li Y, et al. 2007. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat. Med. 13:1032–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li Y, et al. 2011. Mechanism of neutralization by the broadly neutralizing HIV-1 monoclonal antibody VRC01. J. Virol. 85:8954–8967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li Y, et al. 2009. Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J. Virol. 83:1045–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Long EM, et al. 2000. Gender differences in HIV-1 diversity at time of infection. Nat. Med. 6:71–75 [DOI] [PubMed] [Google Scholar]
- 47. Long EM, Rainwater SM, Lavreys L, Mandaliya K, Overbaugh J. 2002. HIV type 1 variants transmitted to women in Kenya require the CCR5 coreceptor for entry, regardless of the genetic complexity of the infecting virus. AIDS Res. Hum. Retroviruses 18:567–576 [DOI] [PubMed] [Google Scholar]
- 48. Lynch RM, et al. 2011. The B cell response is redundant and highly focused on V1V2 during early subtype C infection in a Zambian seroconverter. J. Virol. 85:905–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Madani N, et al. 2008. Small-molecule CD4 mimics interact with a highly conserved pocket on HIV-1 gp120. Structure 16:1689–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mahalanabis M, et al. 2009. Continuous viral escape and selection by autologous neutralizing antibodies in drug-naive human immunodeficiency virus controllers. J. Virol. 83:662–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Montefiori DC, et al. 1991. Homotypic antibody responses to fresh clinical isolates of human immunodeficiency virus. Virology 182:635–643 [DOI] [PubMed] [Google Scholar]
- 52. Moore PL, et al. 2008. The c3-v4 region is a major target of autologous neutralizing antibodies in human immunodeficiency virus type 1 subtype C infection. J. Virol. 82:1860–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Moore PL, et al. 2011. Potent and broad neutralization of HIV-1 subtype C by plasma antibodies targeting a quaternary epitope including residues in the V2 loop. J. Virol. 85:3128–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Moore PL, et al. 2009. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog. 5:e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Muster T, et al. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642–6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818 [DOI] [PubMed] [Google Scholar]
- 57. Richman DD, Wrin T, Little SJ, Petropoulos CJ. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 100:4144–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rong R, et al. 2009. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS Pathog. 5:e1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rose PP, Korber BT. 2000. Detecting hypermutations in viral sequences with an emphasis on G → A hypermutation. Bioinformatics 16:400–401 [DOI] [PubMed] [Google Scholar]
- 60. Sagar M, Wu X, Lee S, Overbaugh J. 2006. Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J. Virol. 80:9586–9598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Salazar-Gonzalez JF, et al. 2008. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 82:3952–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sather DN, et al. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83:757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Scheid JF, et al. 2011. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333:1633–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Seaman MS, et al. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for neutralizing antibody assessment. J. Virol. 84:1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shankarappa R, et al. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489–10502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Simek MD, et al. 2009. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J. Virol. 83:7337–7348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Stamatatos L, Morris L, Burton DR, Mascola JR. 2009. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat. Med. 15:866–870 [DOI] [PubMed] [Google Scholar]
- 68. Trkola A, et al. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Walker LM, et al. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Walker LM, et al. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Walker LM, et al. 2010. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 6:e1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wei X, et al. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312 [DOI] [PubMed] [Google Scholar]
- 73. Wolinsky SM, et al. 1992. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science 255:1134–1137 [DOI] [PubMed] [Google Scholar]
- 74. Wu X, et al. 2006. Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J. Virol. 80:835–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wu X, et al. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wu X, et al. 2009. Mechanism of human immunodeficiency virus type 1 resistance to monoclonal antibody B12 that effectively targets the site of CD4 attachment. J. Virol. 83:10892–10907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wu X, et al. 2011. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333:1593–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wyatt R, Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888 [DOI] [PubMed] [Google Scholar]
- 79. Zhang PF, et al. 2002. A variable region 3 (V3) mutation determines a global neutralization phenotype and CD4-independent infectivity of a human immunodeficiency virus type 1 envelope associated with a broadly cross-reactive, primary virus-neutralizing antibody response. J. Virol. 76:644–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhou T, et al. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhou T, et al. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zwick MB, et al. 2001. Identification and characterization of a peptide that specifically binds the human, broadly neutralizing anti-human immunodeficiency virus type 1 antibody b12. J. Virol. 75:6692–6699 [DOI] [PMC free article] [PubMed] [Google Scholar]