Abstract
LANA is essential for tethering the Kaposi's sarcoma-associated herpesvirus (KSHV) genome to metaphase chromosomes and for modulating host-cell gene expression, but the binding sites in the host-chromosome remain unknown. Here, we use LANA-specific chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-Seq) to identify LANA binding sites in the viral and host-cell genomes of a latently infected pleural effusion lymphoma cell line BCBL1. LANA bound with high occupancy to the KSHV genome terminal repeats (TR) and to a few minor binding sites in the KSHV genome, including the LANA promoter region. We identified 256 putative LANA binding site peaks with P < 0.01 and overlap in two independent ChIP-Seq experiments. We validated several of the high-occupancy binding sites by conventional ChIP assays and quantitative PCR. Candidate cellular LANA binding motifs were identified and assayed for binding to purified recombinant LANA protein in vitro but bound with low affinity compared to the viral TR binding site. More than half of the LANA binding sites (170/256) could be mapped to within 2.5 kb of a cellular gene transcript. Pathways and Gene Ontogeny (GO) analysis revealed that LANA binds to genes within the p53 and tumor necrosis factor (TNF) regulatory network. Further analysis revealed partial overlap of LANA and STAT1 binding sites in several gamma interferon (IFN-γ)-regulated genes. We show that ectopic expression of LANA can downmodulate IFN-γ-mediated activation of a subset of genes, including the TAP1 peptide transporter and proteasome subunit beta type 9 (PSMB9), both of which are required for class I antigen presentation. Our data provide a potential mechanism through which LANA may regulate several host cell pathways by direct binding to gene regulatory elements.
INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV) is a causative agent of Kaposi's sarcoma (KS), pleural effusion lymphomas (PEL), and multicentric Castleman's disease (17, 36, 49). KSHV is a member of the gammaherpesvirus family and can establish long-term latent infection in human B lymphocytes. Latent infection is also detected in endothelial cells associated with KS lesions. During latent infection, only a small portion of viral genes are expressed, and these genes are known to promote host-cell survival and viral genome maintenance. Among these KSHV latency genes, the latency-associated nuclear antigen (LANA) is consistently expressed in all forms of KSHV-positive tissues and cell lines (24, 25, 44, 56).
LANA is essential for maintaining the episomal form of the viral genome during latency (4, 12, 56). This maintenance function is mediated by direct and indirect DNA-binding of LANA to the viral and cellular chromosomes (19, 21). The LANA C-terminal domain binds directly to two LANA-binding sites (LBS) in the KSHV terminal repeats (TR) that confer DNA replication origin function for the latent viral episome (5, 18, 19, 22, 53). LANA maintains stable episome copy number by tethering KSHV episomes to host metaphase chromosomes (4, 5). This function is mediated through the LANA N-terminal chromatin binding domain that interacts with metaphase chromosome constituents, including the core histones H2A and H2B (6, 42). LANA also maintains KSHV latency by repressing transcription of the viral immediate-early gene ORF50, which activates the switch from latency to lytic replication (28, 32, 60).
In addition to regulating viral genome copy number and gene expression, LANA modulates host cell gene expression. LANA interacts with various transcription factors and chromatin regulatory proteins, including BRD2 (57, 58), BRD4 (40, 61), HP1 (30), ORC (31, 53), PARP1 (39), p53 (9, 15), Rb (51), mSin3 (26), DNMT3a (50), and RBP-Jκ (28). Many of these interactions have profound effects on host-cell physiology, including enhanced proliferation and survival. For example, LANA binding to p53 inhibits p53 transcriptional activity and prevents p53-mediated apoptosis (9, 15). LANA can also alter host-cell interactions with the immune system and attenuate the antiviral response. For example, LANA inhibits interleukin-4-mediated STAT4 phosphorylation to inhibit apoptosis and maintain latency (7). LANA also inhibits beta interferon (IFN-β) expression by competing with IFN regulatory factor-3 for binding to IFNB promoter to attenuate the innate antiviral response of the host (10).
Global gene expression profiling studies have revealed that LANA modulates a spectrum of cellular genes when ectopically expressed (2). Among these genes were several members of the p16/Ink4 pathway, as well as components of the immunomodulatory response. Whether LANA binds directly or indirectly to the regulatory regions of these genes remains largely unknown. It is also unknown whether LANA binds to specific regions of the cellular chromosome to support episome tethering during cell division. To identify cellular binding sites for LANA, we applied Illumina/Solexa-based deep sequencing methods to analyze the genome-wide interaction sites of LANA in KSHV latently infected BCBL1 PEL cells. Our result revealed numerous LANA binding sites throughout the host genome and a subset of those binding sites located close to known transcription start sites. Our data also suggest that LANA has the potential to interfere with STAT1-dependent IFN-γ regulation to modulate host cell antiviral immune response.
MATERIALS AND METHODS
Cells and plasmids.
KSHV-positive PEL BCBL1, BC-1, and JSC-1 cells and KSHV-negative Burkitt lymphoma DG75 and BJAB cells were maintained in RPMI-1640 containing 10% fetal bovine serum and supplemented with GlutaMAX (Invitrogen) and antibiotics (penicillin and streptomycin). pCMV-FLAG-LANA-FL was previously described (32). Recombinant LANA-DNA binding domain (DBD) was constructed by PCR amplification of LANA DBD (amino acids [aa] 936 to 1162) with primers introducing a 5′ BamHI and 3′ XhoI site cloned in frame to a baculovirus expression vector pFASTBACHTb. LANA-DBD was expressed with an N-terminal hexahistidine tag from baculovirus-infected Sf9 cells and purified using Ni-NTA agarose.
ChIP-Seq.
Chromatin immunoprecipitation for high-throughput sequencing (ChIP-Seq) experiments were performed with 107 BCBL1 cells per assay with either LANA rat monoclonal antibody (Advanced Biotechnologies, Inc., Gaithersburg, MD) or control mouse IgG (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). The ChIP methods were described previously (33) except that protein G-Sepharose was replaced by Dynabeads protein A (Invitrogen). DNA fragments in the range of ca. 150 to 300 bp were isolated by agarose gel purification, ligated to primers, and then subjected to Solexa-based sequencing using manufacturer's recommendations (Illumina, Inc.).
ChIP assays.
ChIP assays were performed as described previously (8). Quantification of precipitated DNA was determined using real-time PCR and the threshold cycle (CT) method for relative quantification (ABI 7900HT Fast Real-Time PCR System; Applied Biosystems). The primers used for the ChIP assays are listed in Table S1 in the supplemental material. The following antibodies were used for ChIP assays: anti-LANA (rat monoclonal antibody 13-210-100; Advanced Biotechnology), anti-IgG (mouse sc-2025 and rabbit sc-2027 antibodies; Santa Cruz Biotechnology), and anti-STAT1 (rabbit polyclonal sc-345; Santa Cruz Biotechnology).
EMSA.
DNA fragments were labeled using T4 polynucleotide kinase (NEB) in the presence of [γ-32P]ATP and purified using G-25 spin columns (GE Healthcare). Purified LANA-DBD was incubated with probes at room temperature for 30 min in a total volume of 20 μl of buffer containing 10 mM Tris-HCl (pH 8.0), 100 mM KCl, 1 mM EDTA, 10 mM MgCl2, 0.05 μg of poly(dI-dC)/μl, 0.5 μg of bovine serum albumin/μl, 0.05% NP-40, 35 mM β-mercaptoethanol, and 10% glycerol. For competition assay, 50-pmol portions of cold competitors were mixed with LANA-DBD, and then 0.05 pmol of labeled LBS1/2 probe was added to each reaction, followed by incubation for 30 min. The samples were then separated by electrophoresis on a native 5% polyacrylamide gel. The gels were dried and analyzed using a Typhoon PhosphorImager system. The probe and competitor oligonucleotide sequences are listed in Table S2 in the supplemental material.
Quantitative RT-PCR.
Briefly, 5 × 106 DG75 cells were centrifuged and resuspended in 100 μl of nucleofection solution T (Lonza) containing 5 μg of FLAG-LANA or vector control and 0.5 μg of pmaxGFP (Lonza) and then electroporated on a Lonza nucleofector using the program A-23. After nucleofection, the cells were transferred to a flask containing 20 ml of fresh, complete RPMI 1640 medium, incubated at 37°C for 24 h, and induced with or without 5 ng of human recombinant IFN-γ/ml for another 24 h. The green fluorescent protein (GFP)-positive cells were then sorted and used in reverse transcription-PCR (RT-PCR) analyses. RNA was isolated from 2 × 105 cells using an RNeasy kit (Qiagen) and then further treated with DNase I. RT-PCR was performed as previously described (32). Real-time PCR was performed with SYBR green probe in an ABI Prism 7900 according to the manufacturer's specified parameters. Primer sequences for RT-PCR are listed in Table S3 in the supplemental material.
Bioinformatics analysis of ChIP-Seq sequencing.
We followed published procedures for the bioinformatics analysis of ChIP-Seq data (29, 46, 63). Briefly, image analysis and base calling of ChIP-Seq data was performed using Illumina pipeline software. Sequence alignment to the human genome hg18 was performed using Bowtie. Only uniquely aligned sequence tags, with up to two mismatches, were considered for further analysis. Tags aligned to blacklisted areas (the ENCODE consortium blacklist was downloaded [http://genome.ucsc.edu/cgi-bin/hgFileUi?db=hg19&g=wgEncodeMapability]), and the multiple tags that aligned to the exact same position were also removed before the peak calling step.
Peak calling.
Significant peaks were identified using a two-step procedure, which we have successfully applied in our recent studies (41, 54). In the first step, statistically significant enriched genomic regions (length, 1 kb) were identified. A region is defined as statistically significant if the difference in the number of reads between experiment (LANA) and control IgG samples within the region is higher than a given cutoff read count calculated using a P value of 0.01. Each ChIP-Seq read distribution in the genome can be considered the Poisson distribution, and the difference of two Poisson distributions is given by the Skellam distribution (52). The Skellam probability mass function is given by the following equation:
where n represents number of reads, λ1 and λ2 are mean number of reads for two different samples and In() is a modified Bessel function. A region is considered statistically significant if the difference in the number of reads between experiment (LANA) and control IgG samples within the region is higher than a given cutoff read count calculated using a P value of 0.01 based on the Skellam distribution.
For each of the statistically significant enriched regions, ChIP-Seq read overlapping profiles were created by extending the sequence reads from the 5′ end to the 3′ end of the reads up to 250 bp (the average length of the ChIP-DNA fragment sequenced from the Solexa GA with an Illumina standard ChIP-Seq protocol) for the experiment sample. Then, within each enriched region, the significant enriched peaks in experimental data are identified based on a threshold read count obtained from a second cutoff P value of ≤0.01, using a Poisson distribution on the experimental sample only.
For the KSHV genome, the peak calling method (29, 46, 63) was modified to accommodate the smaller size of the genome, as the KSHV genome is too small to build a robust Poisson distribution model. Also, unlimited multiple mapping was allowed to find the ChIP-Seq peaks in terminal repeat regions. After mapping read tags to the KSHV genome, reads were extended to the average read length. In the case of multiple mapped sequence reads, each read was counted as the fraction −1/n to determine the read counts at each base, where n is the total number of multiple maps of the sequence in the genome. In the next step, the number of reads per base was calculated for moving windows of 250 bp, at every 5-bp interval, by dividing the total read count in each window by its length. Then, the reads per base counts were scaled by using a factor with total reads in the KSHV genome (104/total reads). The normalized scores were assigned to the middle base of the corresponding moving window. After applying the above method for the two samples, average score was obtained as final score at every moving window. Finally, peaks were obtained using the definition of mathematical maximum on the average scores using the predefined cutoff described above. All reads were mapped to the BCBL1 reference genome (NCBI no. HQ404500) with 35 TRs (NC_009333, nucleotides [nt]137169 to 137969).
Annotating the ChIP-Seq LANA peaks to known genes.
LANA peaks that overlap from the two independent ChIP-Seq samples were annotated to genes if they were located within 2,500 bp up- or downstream of known gene open reading frames. Any two consecutive peaks that are within 250 bp of each other are considered overlapping since the average tag length is around 250 to 350 bp.
Ingenuity analysis.
For pathway analysis, we listed all genes that are annotated to ChIP-Seq peaks with a cutoff of peak score of >5. This gene list was then analyzed using Ingenuity Systems IPA software (IPA version 8.7, content version 3202).
LANA binding motif identification.
We selected enriched genomic regions (P < 0.01) from the two independent ChIP-Seq samples (overlapped peaks only) for motif identification. Computational motif prediction was performed by submitting a set of DNA sequences, each 100 bp in length (50 bp around each peak), to the MEME web application version 4.3 (3) (http://meme.nbcr.net/meme/intro.html) to find statistically enriched sequence motifs within the ChIP-Seq peak regions. Possible LANA-binding motifs were predicted based on the highest number of occurrences with the lowest P value, with the parameters “Any number of repetitions” of motifs per sequence, and motif length limited to ca. 6 to 30 nt. Position-weighted matrixes (PWMs) generated by MEME were then represented in the logo format by using WebLogo (13) (http://weblogo.berkeley.edu/logo.cgi) to generate consensus sequences for multiple LANA cellular binding sites. To assess the similarity of the two motifs found from our ChIP-Seq with the published LBS1 and LBS2 sequences (18), we utilized the STAMP DNA motif comparison tool-kit (http://www.benoslab.pitt.edu/stamp/) (35). Motif 1 and motif 2 (as shown in Fig. 4A) were submitted to STAMP as PWMs, along with the LBS1 and LBS2 sequences (5′-CGCCCGGGCATGGGGC-3′ and 5′-CGCCGCCGGGGCCTGCGG-3′, respectively). The STAMP setup parameters were as follows: column comparison was done by using the Pearson correlation coefficient, and local alignment was done by using the ungapped Smith-Waterman method with a preference against the edges of the motif.
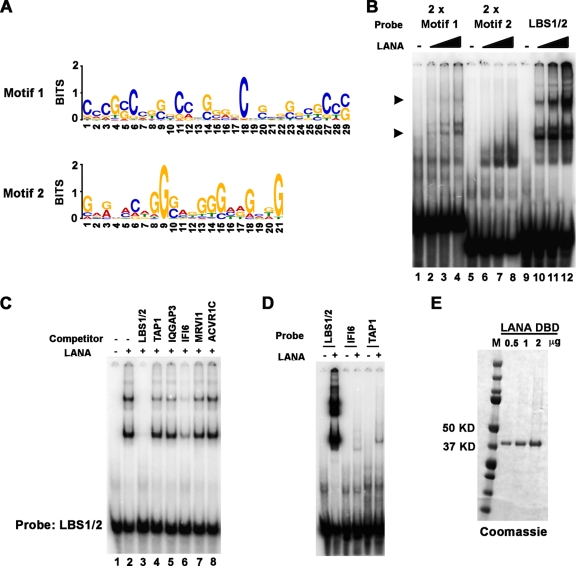
Fig 4.
Consensus binding site of LANA. (A) LANA consensus recognition sequences identified by both ChIP-Seq replicates were identified by MEME analysis and represented in Web Logo format. (B) EMSA analysis of 32P-labeled probes containing 2× motif 1 (lanes 1 to 4), 2× motif 2 (lanes 5 to 8), or LBS1/2 (lanes 9 to 12). Various amounts of LANA DBD protein was used in binding reaction (0, 0.2, 0.6, and 1.8 μg). Arrowheads indicate LANA-specific bound complexes. (C) EMSA with LANA bound to 32P-labeled LBS1/2 and challenged with a 1,000-fold excess of cold competitor oligonucleotides that included LBS1/2 (lane 3), the LANA peak sites in TAP1 (lane 4), IQGAP3 (lane 5), IFI6 (lane 6), MRVI1 (lane 7), and ACVR1C (lane 8). Lane 1 is probe only, and lane 2 is no competitor control. (D) EMSA analysis of 32P-labeled probes containing LBS1/2 or the LANA peak sites in IFI6 or TAP1. (E) Purified LANA-DBD shown on a Coomassie blue-stained gel.
Overlap of LANA and STAT1 bound genomic regions.
The list of LANA-binding enriched genomic regions was matched for overlapping regions with the IFN-γ-stimulated STAT1 ChIP-Seq enrich regions (data set downloaded from the supplementary data of Robertson et al. [46]) by uploading both data sets (in BED format) as custom tracks on to the UCSC genome browser (http://genome.ucsc.edu/) and using the table browser to output the list of all LANA enrich regions that have any overlap with STAT1 enrich regions.
RESULTS
ChIP-Seq analysis of LANA binding to the KSHV genome.
To identify LANA interactions sites on the KSHV and cellular genome, we performed two independent LANA-specific ChIP-Seq experiments using the KSHV-positive PEL cell line BCBL1. BCBL1 cells were selected for LANA ChIP-Seq experiments because they maintain a stable copy number of KSHV episomes and because they are not infected with other gammaherpesviruses, such as Epstein-Barr virus (EBV). Anti-LANA monoclonal antibody and IgG control ChIP DNA was analyzed by Solexa-Illumina-based sequencing methods. A summary of Solexa sequencing and gene mapping statistics is included in Table S4 in the supplemental material. Sequence reads were first mapped to the KSHV genome (NCBI reference sequence HQ404500), and LANA enriched peaks are shown in Fig. 1A. The KSHV genome present in virions consists of a large unique region (LUR) flanked at each end by variable numbers (normally 35 to 45) of terminal repeats (TR). Since HQ404500 contains only the LUR for the KSHV genome derived from BCBL1 cell lines, we therefore attached 35 copies of the TR sequence to the end of the LUR sequence used for mapping. As expected, the major ChIP-Seq peaks for LANA mapped to the TR region (Fig. 1A). We also observed several smaller LANA binding peaks throughout the KSHV genome (Table 1), including a cluster in the latency control region spanning nucleotide (nt) position 120000 to 128000 that contained a major LANA binding peak at the LANA promoter (LANAp) region (marked with red asterisk) as shown in Fig. 1B. To validate these findings, we tested the relative binding of LANA to the site at LANAp compared to TR by conventional ChIP assays. We found that LANA bound TR ∼148-fold more than LANAp (Fig. 1D), indicating that LANAp represents a much less occupied binding site relative to TR. However, LANAp was significantly enriched relative to control R2 region (7-fold enriched) and relative to control IgG at LANAp (9-fold enriched) (Fig. 1E), indicating that LANA does bind to LANAp with specificity. The relatively small peak score at LANAp may reflect weaker or indirect binding of LANA to this region. Previous studies showed LANA associates with KSHV ORF50 promoter through indirect interactions (28, 32), but these binding sites were not observed in our ChIP-Seq data. The failure to detect these additional interactions of LANA with KSHV genome may reflect some of the limitations of our ChIP-Seq methods, as well as the nature of the indirect LANA-DNA interactions at these additional sites. The complete list of LANA binding peaks in KSHV genome with a cutoff of peak score of >2 is included in Table 1. We conclude that LANA binds predominantly to the KSHV TR and to a few additional minor sites throughout the KSHV genome (Table 1).
Fig 1.
LANA binding activity on KSHV genome. The UCSC genome browser was used to map LANA ChIP-Seq enrichment peaks to the BCBL1 KSHV genome (GenBank accession no. HQ404500) with 35 copies of terminal repeat (TR) sequence attached. (A) The peaks represent an average of the results of two independent ChIP-Seq replicates. Zoomed-in views of the latency cluster region and TR region are shown in panels B and C, respectively. ORF50 and LANA transcripts are indicated as colored arrows in panels A and B. LANA and K14 genes are indicated in panel B. The LANA binding peak at the LANA promoter was marked with a red asterisk. The long unique region (LUR) and TR are indicated in panel C. The annealing locations of the primers for ChIP-PCR validation are indicated in panel C, including region 1 (R1), region 2 (R2), region 3 (R3), and LANA binding sites 1 and 2 (LBS1/2). LANA (black bars) or control IgG (gray bars) were assayed by ChIP and quantitative PCR in BCBL1 cells for DNA binding at the LANA promoter compared to LBS1/2 (D) or with R2 (E). (F) LANA (black bars) or control IgG (gray bars) were assayed by ChIP in BCBL1 cells for DNA at R1, R2, R3, or LBS1/2. (G) EMSA analysis of 32P-labeled probes containing R2, R3, or LBS1/2.
Table 1.
LANA binding peaks in the KSHV genome with a peak score cutoff of >2a
| Gene | KSHV genome position | Avg peak score |
|---|---|---|
| K7 | 28800 | 2.541 |
| vIRF2 | 92470 | 2.082 |
| ORF71 | 121555 | 2.43 |
| ORF72 | 122615 | 3.055 |
| LANA promoter | 126680 | 4.631 |
| K15 3′UTR | 133655 | 2.277 |
| K15 | 133755 | 2.18 |
| TR | 136125 | 7.724 |
| TR (LBS1/2) | 136955 | 13.167 |
LANA peaks generated from two independent ChIP-Seq analyses were annotated to the KSHV genome (HQ404500). The average peak scores for each LANA binding peak are listed. UTR, untranslated region.
We further investigated the major peaks at the TR region and found that the nucleotides with highest peak scores were positioned at LBS1/2 (Fig. 1C), which were previously identified as high-affinity LANA binding sites at TR. We also found another peak with a relatively lower score located at the border between unique region and TR. We selected three different reads (R1 to R3) from this peak region and examined LANA binding to these three sites relative to LBS1/2. First, we performed an independent ChIP assay using real-time PCR methods. LANA was highly enriched at the LBS1/2, as expected (Fig. 1D and F), and was partially enriched at R3 located at the beginning of the TR but was not detected at sites R1 and R2, which are situated in the unique region of the KSHV genome (Fig. 1F). To determine whether LANA binds directly to the R3 region, we assayed the ability of purified LANA DNA-binding domain (DBD) to bind the candidate sequences in vitro using electrophoretic mobility shift assay (EMSA) (Fig. 1G). As expected, LANA-DBD bound efficiently to the LBS1/2 containing two LANA binding sites LBS1 and LBS2. However, LANA-DBD failed to bind DNA probes containing either R3 or R2 binding sites. This suggests that the R3 ChIP peak is not mediated by direct interaction with the LANA DBD and may represent an indirect interaction between LANA and cellular factors, including higher-order chromatin structure of LANA binding at TR. Alternatively, R3 may represent a shoulder of the LBS1/2 peak, since it is located within ∼150 bp across each iterated TR junction.
ChIP-Seq analysis of LANA binding to the human genome.
To identify highly confident LANA binding sites in the cellular genome, we focused on those peaks with P values of <0.01 (using the Skellam distribution) and overlaps of <250 bp between peak centers for two independent ChIP-Seq experiments. Using these selection criteria, we identified 256 LANA binding peaks. Among the 256 total LANA binding peaks, 170 were mapped to within 2.5 kb of a known gene transcript (see Table S5 in the supplemental material). Inspection of 256 total LANA binding peaks showed that the binding sites were at various positions relative to transcription start sites (Fig. 2A), with 45 peaks (∼17.5% among 256 total LANA binding sites) situated within ±2 kb of the transcription start site (TSS).
Fig 2.
Summary of LANA ChIP-Seq. (A) LANA peaks generated from two independent ChIP-Seq analyses were annotated to genes if they were located within 2,500 bp up- or downstream of known gene open reading frames. The annotated peaks from the two independent ChIP-Seq sets were then scanned for overlapping peaks, where centers within 250 bp of each other are considered overlapping. The overlapped peaks were analyzed for their distance from transcription start sites annotated in the RefSeq database. Peaks were binned into windows of 1,000 bp, as indicated in the x axis. (B) Ingenuity Pathways analysis was performed on the list of gene containing LANA peaks (P < 0.01) generated from overlaps of two independent ChIP-Seq assays. The network in panel B centered around p53 with a score of 26 and annotated as cell death, cellular function and maintenance, and organ morphology. (C) The network shown here centered around TNF with a score of 15 and annotated as cellular function and maintenance, cellular movement, and cell cycle. Genes with LANA binding sites are highlighted with pink. Solid lines represent direct interactions, while dashed lines represent indirect interactions. Lines with a solid arrowhead represent “acts on”; plain lines represent a “binding-only” relationship.
Pathways analysis of genes near LANA binding sites.
To investigate the potential functional significance of LANA association with cellular genes, we performed an Ingenuity analysis with a list of genes selected from LANA ChIP-Seq analysis (see Table S5 in the supplemental material) with a peak score cutoff of 5. Two major biological networks were successfully generated, and included primary nodes linked by p53 (Fig. 2B) and tumor necrosis factor (TNF) (Fig. 2C). The p53 network had a highly significance score of 26, included PTEN and DNMT3A, and was annotated as cell death, cellular function and maintenance, and organ morphology (Fig. 2B). The focus genes with their LANA binding peak scores in the p53 network are listed in Table S6 in the supplemental material. The TNF network had a significance score of 15, included LANA binding sites at the TAP1, PSMB9, and miR21 genes, and was annotated as cellular function and maintenance, cellular movement, and cell cycle (Fig. 2C). The focus genes with their LANA binding peak scores in the TNF network are also listed in Table S7 in the supplemental material.
Validation of LANA ChIP binding sites.
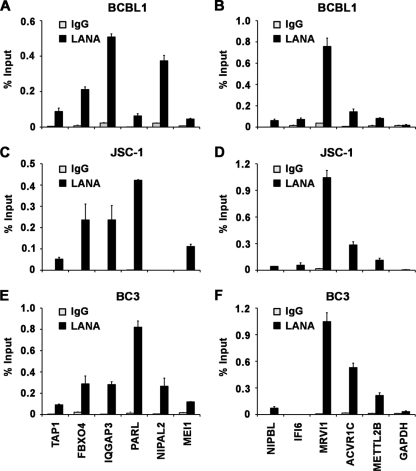
To validate the LANA binding sites determined by ChIP-Seq, we assayed 11 independent loci by traditional ChIP followed by quantitative PCR. Six of these loci were from the 256 peaks that overlap in the two independent ChIP assays (Fig. 3A). Five additional loci were selected from peaks identified in only one of the two ChIP-Seq experiments (Fig. 3B). We also compared LANA binding in three different KSHV positive PEL cell lines, BCBL1 (Fig. 3A and B), JSC-1 (Fig. 3C and D), and BC-3 (Fig. 3E and F). The GAPDH (glyceraldehyde-3-phosphate dehydrogenase) locus was used as a negative control. LANA binding sites derived from the list of 256 overlapping sites observed in both ChIP-Seq experiments (including TAP1, FBXO4, IQGAP3, PARL, NIPAL2, and MEI) were also enriched by real-time PCR relative to IgG in BCBL1 (Fig. 3A). The LANA ChIP-Seq binding sites observed in only one ChIP-Seq experiment (including NIPBL, IFI6, MRVI1, ACVR1C, and METTLE2B) were also enriched by real-time PCR relative to IgG in BCBL1 cells (Fig. 3B). LANA also bound to most of these sites in JSC-1 (Fig. 3C and D) and BC-3 cells (Fig. 3E and F), with some exceptions. LANA failed to bind to NIPAL2 in JSC-1 cells and to IFI6 in BC-3 cells. LANA did not bind to the negative control GAPDH in any cell types. These findings indicate that most, but not all, of the high confidence (P < 0.01) ChIP-Seq peaks can be validated by conventional ChIP assay with site-specific primers.
Fig 3.
Real-time PCR validation of ChIP-Seq data for LANA cellular binding sites. ChIP assays with antibody to LANA (black bars) or control IgG (gray bars) were conducted with either BCBL1 cells (A and B), JSC-1 cells (C and D), or BC-3 cells (E and F). LANA peaks annotated to TAP1, FBXO4, IQGAP3, PARL, NIPAL2, and MEI1 (peaks found in two independent ChIP-Seq) were validated in panels A, C and E. NIPBL, ACVR1C, METTL2B, MRVI1, or IFI6 (peaks from a single ChIP-Seq only) or GAPDH (control) were validated in panels B, D, and F.
Identification of LANA-binding motifs in the host chromosome.
To identify consensus LANA-binding sites in the cellular genome, the enriched LANA binding sites from both ChIP-Seq samples were analyzed using the MEME web application (http://meme.nbcr.net/meme4_3_0/cgi-bin/meme.cgi). The top two candidate motifs are shown in Web LOGO format (Fig. 4A). A total of 50 LANA binding sites contribute to the motif 1, and 36 total binding sites contribute to the motif 2. Since the motif 1 sequence appears to be similar to the core sequence of KSHV LBS1, we subjected both motif 1 and motif 2 to computational comparison with the sequences of LBS1 and LBS2 (18). Interestingly, motif 1 aligned to both LBS1 and LBS2, with E-values of 1.1102e–16 and 6.3625e–12, respectively, while motif 2 is relatively less similar to either LBS1 or LBS2 with E-values of 1.7621e–05 and 9.3032e–04 (see Table S8 in the supplemental material). The consensus sequences of each motif were synthesized as 2× repeat oligonucleotides for use in EMSA to compare with LANA binding site LBS1/2 (see Table S2 in the supplemental material). As shown in Fig. 4B, 2× motif 1 probe (lanes 1 to 4) formed both fast and slow mobility complexes with LANA-DBD. The shift pattern was similar to the control LBS1/2 probe (lanes 9 to 12), although the binding affinity was considerably lower than for LBS1/2. LANA-DBD also shifted 2× motif 2 probe but formed only a single fast mobility complex. To determine whether LANA bound to any of the specific sites at cellular loci, we next tested the ability of candidate LANA binding sites to compete for binding to the LBS1/2 probe (Fig. 4C). As a positive control, we assayed the ability of 1,000-fold excess of unlabeled LBS1/2 fragment to compete with 32P-labeled LBS1/2 (Fig. 4C, lanes 3) and, as expected, found that unlabeled LBS1/2 competitor abolished LANA-DBD to labeled LBS1/2. Among five cellular binding sites, only IFI6 (lane 6) showed partial competition, and substantially weaker competition was observed for TAP1 (lane 4) and IQGAP3 (lane 5). Since LANA affinity for LBS1/2 may be orders of magnitude higher than that of other candidate sites, we performed EMSA with direct labeling of several of the candidate binding sites (Fig. 4D, and data not shown). Compared to the LBS1/2 probe, only the TAP1 and IFI6 binding site formed a relatively weak complex with LANA (Fig. 4D). These data suggest LANA DBD may bind weakly to some cellular binding sites, such as those found at the TAP1 promoter region. These findings also indicate that LANA binds LBS1/2 with considerably higher affinity than for most other viral and cellular binding sites tested and that additional cellular factors or modifications may be required for high-affinity binding in vitro.
Overlapping LANA binding peaks with STAT1.
We noted that the LANA binding site upstream of the TAP1 and PSMB9 (LMP2) genes are located in a bidirectional promoter regulatory element known to be IFN-γ-inducible (16, 34, 45). Since IFN-γ signals mainly through the JAK-STAT pathway to achieve transcriptional activation of IFN-γ-inducible genes (23, 55, 62), we compared our LANA ChIP-Seq analysis data with IFN-γ-inducible STAT1 DNA binding ChIP-Seq data from a previously published study (http://www.nature.com/nmeth/journal/v4/n8/suppinfo/nmeth1068_S1.html) (46). We observed that several LANA binding sites either colocalized with or are located adjacent to STAT1 binding sites at promoter regions of several IFN-γ inducible genes, including TAP1/PSMB9, FBXO4, IQGAP3, and PARL (Fig. 5). These observations suggest that LANA may alter STAT1 function at these IFN-γ-inducible genes.
Fig 5.
Example of LANA and STAT1 binding peaks near transcriptional start sites of cellular genes. The UCSC genome browser was used to map LANA peaks (black) and STAT1 IFN-γ-stimulated (blue, top) and unstimulated (blue, bottom) peaks to cellular genes for TAP1 (A), IQGAP3 (B), FBXO4 (C), and PARL (D). RefSeq annotated transcripts are indicated below each ChIP-Seq peak.
Regulation of IFN-γ response gene expression by LANA.
To determine whether LANA could affect transcription regulation of genes with overlapping LANA and STAT1 binding sites, we assayed the ability of LANA to alter transcription in response to IFN-γ treatment (Fig. 6A). We observed that TAP1/PSMB9, FBXO4, IQGAP3, and PARL were IFN-γ inducible in KSHV-positive PEL cells. Unfortunately, we were unable to deplete LANA sufficiently using siRNA or shRNA methods to observe any LANA-dependent changes in this induction (data not shown). Alternatively, we transiently transfected KSHV negative B-cell lymphoma cell lines with expression vectors for FLAG-LANA or control vector and then assayed the changes in IFN-γ-inducible gene expression for LANA bound genes. In the absence of IFN-γ treatment, we found that FLAG-LANA inhibited the expression of TAP1 and PARL mRNA by ∼3-fold but had no measurable effect on other candidate genes (Fig. 6A). IFN-γ treatment led to the transcription activation of TAP1, PSMB9, and PARL, which is consistent with previously published studies (48). Ectopic expression of LANA inhibited the IFN-γ-inducible transcription of TAP1, PSMB9, and PARL by ca. 7- to 5-fold, while not affecting transcription of control gene NIPBL. FBXO4 and IQGAP3 mRNA levels did not change with or without IFN-γ treatment and were not further affected by LANA expression. These findings suggest that LANA can modulate the IFN-γ response of some host cell genes.
Fig 6.

Ectopic expression of LANA inhibits IFN-γ-induced TAP1 gene expression. (A) KSHV-negative DG75 was transfected with control vector or with FLAG-LANA expression vector and pmaxGFP. At 24 h after transfection, the cells were induced with 5 ng of human recombinant IFN-γ/ml for another 24 h. The GFP-positive cells were then sorted out and assayed by RT-PCR for IFN-γ-induced transcription of TAP1, PSMB9, PARL, FBXO4, IQGAP3, and NIPBL, as indicated. (B) BCBL1 cells were stimulated or unstimulated with IFN-γ for 30 min and then subjected to immunoprecipitation with antibodies to IgG, LANA, or STAT1. Immunoprecipitates were analyzed by Western blotting for LANA and then reprobed for STAT1 or STAT3 as indicated. (C) DG75 cells were transfected with control vector or with FLAG-LANA expression vector and pmaxGFP. At 24 h after transfection, GFP-positive cells were sorted, then treated or not treated with 5 ng of IFN-γ for 30 min/ml, and assayed by ChIP with antibodies to IgG and STAT1 at the TAP1/PSMB9 site.
To further explore the mechanism by which LANA can regulate IFN-γ-mediated transcription, we first assayed the ability of LANA and STAT1 or STAT3 to associate using coimmunoprecipitation with LANA and STAT1 or STAT3 antibodies (Fig. 6B). STAT3 is a common binding partner of STAT1 for many IFN-γ-inducible genes (62). We were unable to detect any stable interaction between LANA and STAT1, but we did observe a weak interaction between LANA and STAT3. As expected, STAT3 was coimmunoprecipitated with STAT1. These findings suggest that LANA may interact weakly with STAT3 to modulate IFN-γ-responsive gene transcription.
We next examined the effect of LANA expression on STAT1 binding to the TAP1 promoter region (Fig. 6C). To this end, we transfected KSHV-negative DG75 cells with control or FLAG-LANA expression vectors and then treated them with or without IFN-γ. STAT1 binding was examined by ChIP assay. We found that STAT1 binding was modestly enriched on the TAP1/PSMB9 promoter when DG75 cells were treated with interferon-γ, but this enrichment was abrogated in cells transfected with FLAG-LANA (Fig. 6C). Similar experiments with STAT3 were also attempted, but STAT3 binding could not be detected by ChIP assay (data not shown). These findings suggest that LANA can interfere with the IFN-γ-induced enrichment of STAT1 on the TAP1/PSMB9 promoter.
DISCUSSION
In this study, we used two independent ChIP-Seq experiments to identify ∼256 high-confidence binding sites for LANA in the cellular chromosome of a human PEL cell line. Several (∼11) of the high-occupancy binding sites identified by ChIP-Seq were validated for binding by conventional ChIP and real-time PCR (Fig. 3) in three different KSHV PEL cell lines. Among these 256 binding sites, we noted that ∼17.5% (45 sites) are located within 2 kb of an annotated or predicted transcription start sites (Fig. 2). Two GC-rich DNA motifs were identified in the high-confidence LANA binding site data set, and position-weighted matrix (PWM) analysis, Web LOGO presentation, and STAMP DNA motif comparison toolkit revealed that motif 1 sequence is similar to LBS1, while motif 2 shows less apparent similarity to LBS1 or LBS2 (Fig. 4A and see Table S8 in the supplemental material). We also observed relatively weak or undetectable direct binding of purified LANA protein to most validated cellular sites, suggesting that the viral binding sites in TR are the primary direct binding sites for LANA throughout the virus and host genomes. The relatively few high-confidence LANA binding sites (∼256) identified in our study suggest that LANA may interact in a dynamic or indirect manner with many different chromatin domains and require additional cellular factors to mediate high-affinity chromosome binding.
LANA binding sites poorly overlap with previously characterized LANA regulated gene targets.
A previous gene expression profiling study identified a list of cellular gene transcripts that were affected by LANA (2). In our study, we identified 170 candidate LANA cellular binding sites that could be mapped to 218 known genes (see Table S5 in the supplemental material). We found that only three of these genes (PITPNB, PTPRC, and PPP2R5E) were common to the previous gene expression profiling study. Our RT-PCR analysis also showed that the transcription of several candidate genes with the LANA binding site on their promoter was not changed by ectopic expression of LANA (data not shown). This suggests that many of the LANA-bound genes are not subject to LANA-dependent gene activation or repression. Since our data did not analyze LANA in the same cells as those analyzed in the expression profiling, it is possible that LANA regulates genes in a cell-type-specific or context-specific manner. Alternatively, it is possible that many LANA binding sites in the cellular genome regulate transcription of genes located outside of the ∼1-kb window that we examined. It is also possible that LANA affects chromosome structure that modulate transcription of genes far from the LANA binding sites.
The P53 and TNF pathway as a target for LANA function.
Network analysis revealed a potential role for LANA in the p53 and TNF pathways (Fig. 2). LANA has been shown to physically and functionally interact with p53 (9, 14), and our pathways analysis indicated that LANA can bind near the TSS of several genes that functionally interact with p53. Thus, LANA may function coordinately with p53 to regulate a common network of cellular genes important for cell cycle control in response to DNA damage or other cellular stress responses. We also found that LANA may have a role in the TNF network (Fig. 2) and bind to the promoter proximal regions of several STAT1-dependent IFN-γ-regulated genes (Fig. 5). One of the most enriched LANA binding sites was in the divergent promoter region of TAP1 and PSMB9. TAP1 plays a major role in assembling peptides into the major histocompatibility complex class I (MHC-I) (43), and PSMB9 (previously designated low-molecular-weight polypeptide 2 [LMP2]) is a component of 20S proteasome, which is responsible for generating the pool of peptides that are presented on the cell surface by MHC-I molecules (47). TAP1 was identified as an IFN-γ-inducible gene with STAT1 binding sites on its promoter (34). PSMB9, which shares a bidirectional promoter with TAP1, was also identified as IFN-γ-inducible gene (16, 45). Our alignment of LANA and STAT1 peaks on TAP1/PSMB9 promoter region revealed a significant overlap, which suggests that LANA and STAT1 are likely to either co-occupy or compete for binding on TAP1/PSMB9 promoter. The RT-PCR analysis revealed the IFN-γ-induced TAP1 and PSMB9 gene expression was downregulated by ectopic expression of LANA in KSHV-negative BL cells. This suggests that LANA may compete with STAT1 for binding to the TAP1/PSMB9 promoter region, and this was partially supported by ChIP assays in IFN-γ-treated DG75 cells transfected with or without LANA (Fig. 6). Thus, LANA can attenuate IFN-γ-inducible activation of a target gene and modulate the host cell antiviral immune response.
Modulation of the innate immune response by viral proteins appears to be an essential and common strategy for virus infection and persistence (62). Several other gammaherpesvirus proteins have been implicated in the regulation of the STAT pathway, including KSHV vIL6, which can transform cells through a STAT-dependent pathway (37, 38), and the ORF50 immediate-early protein RTA, which can relocalize STAT3 to its nuclear targets (20). STAT1-responsive gene expression can also be stimulated by KSHV vFLIP protein during endothelial cell infection (1). The EBV-encoded EBNA1 protein, which is the functional orthologue of KSHV LANA, has also been implicated in regulating STAT1 gene expression, although the mechanism of this regulation is not clear (59). Our studies suggest that LANA interferes with the IFN-γ/STAT1-dependent antigen presentation pathway, since LANA inhibited the IFN-γ-inducible expression of TAP1 and PSMB9 (Fig. 6). Several other KSHV proteins also promote the downregulation of host cell antigen presentation. The virus-encoded proteins MIR1 and MIR2 can restrict MHC protein cycling at the cell membrane (11), and vIRF1 can inhibit MHC-I gene transcription (27). Further attenuation of antigen presentation and IFN-γ signaling by LANA could greatly facilitate KSHV latently infected cells from host T-cell recognition and elimination. It is interesting that LANA bound to genes within the p53 and TNF networks, both of which would help to promote long-term survival of latently infected cells. The identification and further characterization of these LANA targets could reveal a remarkable global strategy for LANA-dependent regulation of host cell processes critical for viral genome maintenance and host-cell survival during latent infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sridhar Hannenhalli, University of Maryland, for bioinformatic advice. We also thank Rolf Renne for sharing related ChIP Seq data prior to publication.
We acknowledge support from a Wistar Institute Cancer Center grant (P30 CA10815) and from the Core Facilities for Genomics, Bioinformatics, and Flow Cytometry. This study was funded by a grant from the National Institutes of Health (RO1 CA117830).
Footnotes
Published ahead of print 14 March 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Alkharsah KR, et al. 2011. Deletion of Kaposi's sarcoma-associated herpesvirus FLICE inhibitory protein, vFLIP, from the viral genome compromises the activation of STAT1-responsive cellular genes and spindle cell formation in endothelial cells. J. Virol. 85:10375–10388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. An FQ, et al. 2005. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus modulates cellular gene expression and protects lymphoid cells from p16 INK4A-induced cell cycle arrest. J. Biol. Chem. 280:3862–3874 [DOI] [PubMed] [Google Scholar]
- 3. Bailey TL, Williams N, Misleh C, Li WW. 2006. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34:W369–W373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ballestas ME, Chatis PA, Kaye KM. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641–644 [DOI] [PubMed] [Google Scholar]
- 5. Ballestas ME, Kaye KM. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barbera AJ, et al. 2006. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311:856–861 [DOI] [PubMed] [Google Scholar]
- 7. Cai Q, Verma SC, Choi JY, Ma M, Robertson ES. 2010. Kaposi's sarcoma-associated herpesvirus inhibits interleukin-4-mediated STAT6 phosphorylation to regulate apoptosis and maintain latency. J. Virol. 84:11134–11144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chau CM, Lieberman PM. 2004. Dynamic chromatin boundaries delineate a latency control region of Epstein-Barr virus. J. Virol. 78:12308–12319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen W, Hilton IB, Staudt MR, Burd CE, Dittmer DP. 2010. Distinct p53, p53:LANA, and LANA complexes in Kaposi's sarcoma-associated herpesvirus lymphomas. J. Virol. 84:3898–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cloutier N, Flamand L. 2010. Kaposi sarcoma-associated herpesvirus latency-associated nuclear antigen inhibits interferon (IFN) beta expression by competing with IFN regulatory factor-3 for binding to IFNB promoter. J. Biol. Chem. 285:7208–7221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coscoy L, Ganem D. 2000. Kaposi's sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc. Natl. Acad. Sci. U. S. A. 97:8051–8056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cotter MA, II, Robertson ES. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254–264 [DOI] [PubMed] [Google Scholar]
- 13. Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fakhari FD, Jeong JH, Kanan Y, Dittmer DP. 2006. The latency-associated nuclear antigen of Kaposi sarcoma-associated herpesvirus induces B cell hyperplasia and lymphoma. J. Clin. Invest. 116:735–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Friborg J, Jr, Kong W, Hottiger MO, Nabel GJ. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889–894 [DOI] [PubMed] [Google Scholar]
- 16. Gaczynska M, Goldberg AL, Tanaka K, Hendil KB, Rock KL. 1996. Proteasome subunits X and Y alter peptidase activities in opposite ways to the interferon-gamma-induced subunits LMP2 and LMP7. J. Biol. Chem. 271:17275–17280 [DOI] [PubMed] [Google Scholar]
- 17. Ganem D. 2006. KSHV infection and the pathogenesis of Kaposi's sarcoma. Annu. Rev. Pathol. 1:273–296 [DOI] [PubMed] [Google Scholar]
- 18. Garber AC, Hu J, Renne R. 2002. Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, and both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication. J. Biol. Chem. 277:27401–27411 [DOI] [PubMed] [Google Scholar]
- 19. Garber AC, Shu MA, Hu J, Renne R. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:7882–7892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gwack Y, et al. 2002. Kaposi's Sarcoma-associated herpesvirus open reading frame 50 stimulates the transcriptional activity of STAT3. J. Biol. Chem. 277:6438–6442 [DOI] [PubMed] [Google Scholar]
- 21. Han SJ, Hu J, Pierce B, Weng Z, Renne R. 2010. Mutational analysis of the latency-associated nuclear antigen DNA-binding domain of Kaposi's sarcoma-associated herpesvirus reveals structural conservation among gammaherpesvirus origin-binding proteins. J. Gen. Virol. 91:2203–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hu J, Renne R. 2005. Characterization of the minimal replicator of Kaposi's sarcoma-associated herpesvirus latent origin. J. Virol. 79:2637–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hu X, Ivashkiv LB. 2009. Cross-regulation of signaling pathways by interferon-gamma: implications for immune responses and autoimmune diseases. Immunity 31:539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kedes DH, Lagunoff M, Renne R, Ganem D. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Invest. 100:2606–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Komatsu T, Ballestas ME, Barbera AJ, Kaye KM. 2002. The KSHV latency-associated nuclear antigen: a multifunctional protein. Front. Biosci. 7:d726–d730 [DOI] [PubMed] [Google Scholar]
- 26. Krithivas A, Young DB, Liao G, Greene D, Hayward SD. 2000. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J. Virol. 74:9637–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lagos D, et al. 2007. Kaposi sarcoma herpesvirus-encoded vFLIP and vIRF1 regulate antigen presentation in lymphatic endothelial cells. Blood 109:1550–1558 [DOI] [PubMed] [Google Scholar]
- 28. Lan K, Kuppers DA, Robertson ES. 2005. Kaposi's sarcoma-associated herpesvirus reactivation is regulated by interaction of latency-associated nuclear antigen with recombination signal sequence-binding protein Jκ, the major downstream effector of the Notch signaling pathway. J. Virol. 79:3468–3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lim C, Lee D, Seo T, Choi C, Choe J. 2003. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus functionally interacts with heterochromatin protein 1. J. Biol. Chem. 278:7397–7405 [DOI] [PubMed] [Google Scholar]
- 31. Lim C, Sohn H, Lee D, Gwack Y, Choe J. 2002. Functional dissection of latency-associated nuclear antigen 1 of Kaposi's sarcoma-associated herpesvirus involved in latent DNA replication and transcription of terminal repeats of the viral genome. J. Virol. 76:10320–10331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lu F, Day L, Gao SJ, Lieberman PM. 2006. Acetylation of the latency-associated nuclear antigen regulates repression of Kaposi's sarcoma-associated herpesvirus lytic transcription. J. Virol. 80:5273–5282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lu F, et al. 2010. Genome-wide analysis of host-chromosome binding sites for Epstein-Barr virus nuclear antigen 1 (EBNA1). Virol. J 7:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ma W, Lehner PJ, Cresswell P, Pober JS, Johnson DR. 1997. Interferon-gamma rapidly increases peptide transporter (TAP) subunit expression and peptide transport capacity in endothelial cells. J. Biol. Chem. 272:16585–16590 [DOI] [PubMed] [Google Scholar]
- 35. Mahony S, Benos PV. 2007. STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 35:W253–W258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mesri EA, Cesarman E, Boshoff C. 2010. Kaposi's sarcoma and its associated herpesvirus. Nat. Rev. Cancer 10:707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moore PS, Boshoff C, Weiss RA, Chang Y. 1996. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science 274:1739–1744 [DOI] [PubMed] [Google Scholar]
- 38. Nicholas J, et al. 1997. Kaposi's sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat. Med. 3:287–292 [DOI] [PubMed] [Google Scholar]
- 39. Ohsaki E, et al. 2004. Poly(ADP-ribose) polymerase 1 binds to Kaposi's sarcoma-associated herpesvirus (KSHV) terminal repeat sequence and modulates KSHV replication in latency. J. Virol. 78:9936–9946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ottinger M, et al. 2006. Kaposi's sarcoma-associated herpesvirus LANA-1 interacts with the short variant of BRD4 and releases cells from a BRD4- and BRD2/RING3-induced G1 cell cycle arrest. J. Virol. 80:10772–10786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pal S, et al. 2011. Alternative transcription exceeds alternative splicing in generating the transcriptome diversity of cerebellar development. Genome Res. 21:1260–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Piolot T, Tramier M, Coppey M, Nicolas JC, Marechal V. 2001. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 75:3948–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Raghavan M, Del Cid N, Rizvi SM, Peters LR. 2008. MHC class I assembly: out and about. Trends Immunol. 29:436–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rainbow L, et al. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Robek MD, Garcia ML, Boyd BS, Chisari FV. 2007. Role of immunoproteasome catalytic subunits in the immune response to hepatitis B virus. J. Virol. 81:483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Robertson G, et al. 2007. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods 4:651–657 [DOI] [PubMed] [Google Scholar]
- 47. Rock KL, et al. 1994. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78:761–771 [DOI] [PubMed] [Google Scholar]
- 48. Rouyez MC, et al. 2005. IFN regulatory factor-2 cooperates with STAT1 to regulate transporter associated with antigen processing-1 promoter activity. J. Immunol. 174:3948–3958 [DOI] [PubMed] [Google Scholar]
- 49. Schulz TF. 2006. The pleiotropic effects of Kaposi's sarcoma herpesvirus. J. Pathol. 208:187–198 [DOI] [PubMed] [Google Scholar]
- 50. Shamay M, Krithivas A, Zhang J, Hayward SD. 2006. Recruitment of the de novo DNA methyltransferase Dnmt3a by Kaposi's sarcoma-associated herpesvirus LANA. Proc. Natl. Acad. Sci. U. S. A. 103:14554–14559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Si H, Robertson ES. 2006. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen induces chromosomal instability through inhibition of p53 function. J. Virol. 80:697–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Skellam JG. 1946. The frequency distribution of the difference between two Poisson variates belonging to different populations. J. Royal Stat. Soc. Ser. A 109:296. [PubMed] [Google Scholar]
- 53. Stedman W, Deng Z, Lu F, Lieberman PM. 2004. ORC, MCM, and histone hyperacetylation at the Kaposi's sarcoma-associated herpesvirus latent replication origin. J. Virol. 78:12566–12575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sun H, et al. 2011. Genome-wide mapping of RNA Pol-II promoter usage in mouse tissues by ChIP-Seq. Nucleic Acids Res. 39:190–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. van Boxel-Dezaire AH, Stark GR. 2007. Cell type-specific signaling in response to interferon-gamma. Curr. Top. Microbiol. Immunol. 316:119–154 [DOI] [PubMed] [Google Scholar]
- 56. Verma SC, Lan K, Robertson E. 2007. Structure and function of latency-associated nuclear antigen. Curr. Top. Microbiol. Immunol. 312:101–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Viejo-Borbolla A, et al. 2003. A Domain in the C-terminal region of latency-associated nuclear antigen 1 of Kaposi's sarcoma-associated Herpesvirus affects transcriptional activation and binding to nuclear heterochromatin. J. Virol. 77:7093–7100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Viejo-Borbolla A, et al. 2005. Brd2/RING3 interacts with a chromatin-binding domain in the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 (LANA-1) that is required for multiple functions of LANA-1. J. Virol. 79:13618–13629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wood VH, et al. 2007. Epstein-Barr virus-encoded EBNA1 regulates cellular gene transcription and modulates the STAT1 and TGFβ signaling pathways. Oncogene 26:4135–4147 [DOI] [PubMed] [Google Scholar]
- 60. Ye FC, et al. 2004. Disruption of Kaposi's sarcoma-associated herpesvirus latent nuclear antigen leads to abortive episome persistence. J. Virol. 78:11121–11129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. You J, et al. 2006. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes. J. Virol. 80:8909–8919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yu H, Pardoll D, Jove R. 2009. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat. Rev. Cancer 9:798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang ZD, Rozowsky J, Snyder M, Chang J, Gerstein M. 2008. Modeling ChIP sequencing in silico with applications. PLoS Comput. Biol. 4:e1000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.