Abstract
We previously showed that a noncoding subgenomic flavivirus RNA (sfRNA) is required for viral pathogenicity, as a mutant West Nile virus (WNV) deficient in sfRNA production replicated poorly in wild-type mice. To investigate the possible immunomodulatory or immune evasive functions of sfRNA, we utilized mice and cells deficient in elements of the type I interferon (IFN) response. Replication of the sfRNA mutant WNV was rescued in mice and cells lacking interferon regulatory factor 3 (IRF-3) and IRF-7 and in mice lacking the type I alpha/beta interferon receptor (IFNAR), suggesting a contribution for sfRNA in overcoming the antiviral response mediated by type I IFN. This was confirmed by demonstrating rescue of mutant virus replication in the presence of IFNAR neutralizing antibodies, greater sensitivity of mutant virus replication to IFN-α pretreatment, partial rescue of its infectivity in cells deficient in RNase L, and direct effects of transfected sfRNA on rescuing replication of unrelated Semliki Forest virus in cells pretreated with IFN-α. The results define a novel function of sfRNA in flavivirus pathogenesis via its contribution to viral evasion of the type I interferon response.
INTRODUCTION
West Nile virus (WNV) is a member of the Flavivirus genus of the Flaviviridae family of RNA viruses and is closely related to a number of human pathogens of global concern, including dengue (DENV), yellow fever (YFV), tick-borne encephalitis (TBEV), and Japanese encephalitis (JEV) viruses. Many flaviviruses cause fatal disease in humans, and outbreaks affect 50 to 100 million people every year (20, 36). Since 1999, highly pathogenic North American strains of WNV have caused more than 30,000 clinical cases of meningitis, encephalitis, and acute flaccid paralysis in the United States alone. In comparison, the Australian strains of WNV circulating prior to 2011, which are referred to as Kunjin virus (WNVKUN), are closely related (∼97% homology at the amino acid level) but do not cause disease in immunocompetent adult animals and humans (22).
The flavivirus genome is a single-stranded, positive-polarity RNA of ∼11 kb. It contains one open reading frame flanked by 5′ and 3′ untranslated regions (UTRs) and encodes 10 viral proteins that are required for the complete viral life cycle (30–34, 58, 59). The UTRs play essential functions in the initiation of RNA replication, translation, and genome packaging (40). In addition to full-length genomic RNA (gRNA), an abundant RNA species of about 0.5 kb derived from the 3′ UTR of gRNA was previously detected in flavivirus-infected cells (29, 44, 49, 57) and termed subgenomic flavivirus RNA (sfRNA). Recent studies demonstrated that the sfRNA of WNVKUN and YFV is generated as a product of degradation by a host enzyme, presumably the 5′-3′ exoribonuclease XRN1 (44, 51). XRN1-mediated degradation of gRNA likely stalls due to the rigid and conserved secondary and tertiary RNA structures in the 5′ end of the 3′ UTR (18, 51). Thus, incomplete degradation of WNVKUN RNA results in a 525-nucleotide (nt) RNA remnant that forms the sfRNA. The sfRNA contributes to virus-induced cytopathic effect in cell culture and to virulence in weanling mice highly sensitive to flavivirus infections (44) although the mechanism(s) that explain these outcomes remain unknown. Because of the requirement of sfRNA for virulence in mice but not for replication in BHK-21 or Vero cells that lack intact cell-intrinsic antiviral immune pathways, we hypothesized that sfRNA modulates the host antiviral response.
Viral infection of host cells results in the induction of cell-intrinsic and cell-extrinsic antiviral responses that limit replication and spread. Pathogen recognition receptors (PRRs), including the cytoplasmic receptors retinoid acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene-5 (MDA5), or membrane-bound Toll-like receptors (TLR 3, 7, or 8) serve as initial sensors of pathogen-associated molecular patterns (PAMPs) after RNA virus infection and trigger signal transduction cascades that induce the expression of genes with specific inhibitory functions. For RNA viruses, double-stranded RNA (dsRNA) intermediates of gRNA replication are believed to be the primary PAMPs. Activation of PRRs results in signaling through distinct adaptor molecules. RIG-I and MDA5 signal through beta interferon (IFN-β) promoter stimulator 1 (IPS-1), whereas TLR3 and TLR7/TLR8 signal through Trif and MyD88, respectively. Ultimately, these pathways result in phosphorylation and activation of transcription factors (e.g., interferon regulatory factor 3 [IRF-3], and IRF-7), which, together with NF-κB and ATF-2/c-jun, induce transcription of antiviral cytokines such as IFN-α4 (39) and IFN-β (54). Secreted type I IFN binds to the IFN-α/β receptor (IFNAR) in an autocrine and paracrine manner and activates the Janus kinase/signal transducers and activators of transcription (JAK/STAT) signaling cascade. This leads to formation of the IFN-stimulated gene factor 3 (ISGF3) complex (STAT1, STAT2, and IRF9/p48), which translocates into the nucleus and induces expression of several hundred IFN-stimulated genes (ISGs), many of which likely have antiviral functions. A number of ISGs, including RNase L, PKR, IFIT-1, IFIT-2, ISG20, IFITM3, viperin, and other genes, are believed to possess antiviral activity against flaviviruses (4, 14, 25, 47, 49).
To investigate whether sfRNA modulates virus-host interactions, we compared replication of wild type (wt) WNVKUN virus to a mutant virus incapable of producing sfRNA utilizing cells and mice deficient in various components of the innate antiviral response. Using this approach, we show that sfRNA contributes to viral evasion of the type I IFN-mediated antiviral response.
MATERIALS AND METHODS
Viruses and cells.
Wild type (wt) WNVKUN full-length cDNA clone FLSDX(pro)HDVr (designated FLSDX; where HDVr is hepatitis delta virus ribozyme) and sfRNA-deficient virus FL-IRAΔCS3 were described previously (30, 44). For virus reconstitution, viral RNA was in vitro transcribed from the corresponding full-length cDNA clones, electroporated into BHK-21 cells, and subsequently passaged once on Vero76 cells to grow a high-titer stock. Titers of passage 2 virus were determined by standard plaque assay on BHK-21 cells and used for experiments. Semliki Forest virus (SFV) was grown and titers were determined on Vero76 cells. WNV RNA for in vitro RNase L assays was transcribed from C20DXrep, a cDNA construct containing the FLSDX sequence but lacking the structural C, prM, and E genes (27).
For WNVKUN infection experiments, all murine embryonic fibroblasts (MEFs) with the exception of RNase L−/− MEFs were low-passage-number primary cultures (n = 2 to 10) generated from wt and deficient mice. RNase L−/− and corresponding control MEFs were immortalized after transformation with simian virus 40 (SV40) T antigen (61). IRF-3−/− × IRF-7−/− MEFs for IFN sensitivity experiments and control wt MEFs for sfRNA electroporation and SFV rescue experiments were immortalized spontaneously by iterative passaging.
WNVKUN infection, growth kinetics, and plaque assays.
MEFs were infected at different multiplicities of infection (MOI) for 2 h at 37°C, washed three times with phosphate-buffered saline (PBS), and incubated with Dulbecco's modified Eagle's medium (DMEM) containing 5% fetal bovine serum (FBS). For viral growth kinetics in Fig. 1 and 5, cell culture supernatant was harvested at the indicated times to determine virus titers by standard plaque assay on BHK-21 cells (44).
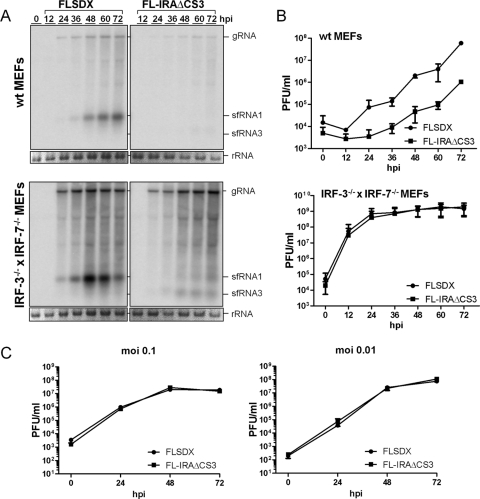
Fig 1.
Replication of sfRNA-deficient WNVKUN is rescued in IRF-3−/− × IRF-7−/− MEFs. (A) Northern blot of viral RNA using a 3′ UTR-specific probe in wt and IRF-3−/− × IRF-7−/− MEFs infected at an MOI of 1 with wt (FLSDX) and sfRNA-deficient (FL-IRAΔCS3) viruses. Ethidium bromide staining of rRNA is included as a loading control. (B) Corresponding titers of infectious virus in the supernatant of the infected cells. The results are the average of two (wt MEFs) or four (IRF-3−/− × IRF-7−/− MEFs) independent experiments. Error bars indicate standard deviations. (C) Virus titers in the supernatants of IRF-3−/− × IRF-7−/− MEFs infected at an MOI of 0.1 and 0.01 with wt (FLSDX) or sfRNA-deficient (FL-IRAΔCS3) virus.
Fig 5.
Replication of sfRNA mutant WNVKUN is partially rescued in RNase L−/− but not in PKR−/− MEFs. (A) Northern blot of viral RNA in wt and PKR−/− MEFs infected with wt (FLSDX) and sfRNA-deficient (FL-IRAΔCS3) viruses. (B) Corresponding titers of infectious virus in the supernatant of the infected cells. The data are the average of two independent experiments. Error bars indicate standard deviations. (C) Northern blot of viral RNA in wt MEFs or RNase L−/− MEFs infected with wt (FLSDX) and sfRNA-deficient (FL-IRAΔCS3) viruses. (D) Corresponding titers of infectious virus in the supernatant of the infected cells. The data are the average of two independent experiments performed in duplicate. Error bars indicate standard deviations. Viral RNA was detected in all Northern blots with a 3′ UTR-specific probe. Ethidium bromide staining of rRNA is included as a loading control for each blot.
RNA isolation and Northern blotting.
RNA from infected cells was isolated with TRIzol reagent (Invitrogen) following the manufacturer's recommendations, and Northern blots were performed using 32P-labeled cDNA probes specific for the WNVKUN 3′ UTR and SFV nsp4 gene as described previously (44).
In vitro sfRNA transcription.
The template for in vitro transcription was generated by PCR from plasmids pBS3′XX (containing T7 promoter and the last 813 nucleotides of viral cDNA including part of WNVKUN NS5 and the entire 3′ UTR) and pBS3′XX-IRAΔCS3 (as above, but containing IRAΔCS3 mutations in the 3′ UTR) (44). The PCR product was purified with a Wizard SV Gel and PCR Clean-Up System (Promega), and 1 μg was used for in vitro transcription with GMP and T7 polymerase according to the manufacturer's instructions (Promega).
RNA electroporation, IFN treatment, and SFV infection.
Wild-type MEFs were trypsinized and resuspended at 2 × 106 cells/ml in OptiMEM (Invitrogen). A 400-μl cell suspension was mixed with ∼10 μg of in vitro transcribed RNA and electroporated in a 4-mm gap cuvette with a single pulse at 400 V for 5 ms using an ECM 830 square wave electroporator (BTX/Harvard Apparatus). Cells were seeded in six-well plates and left to attach for 1 to 2 h before treatment with 1,000 IU/ml mouse IFN-α (Hycult Biotechnology) for 8 h. The IFN solution was removed, and cells were infected with SFV at an MOI of 1 for 1 h. At 12 h postinfection (hpi), cells were lysed with TriReagent (Sigma) for RNA extraction, and supernatants were harvested for plaque assay on Vero76 cells.
Virulence in mice.
All animal studies were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (assurance number A3381-01). All inoculation and experimental manipulation were performed under anesthesia that was induced and maintained with ketamine hydrochloride and xylazine, and all efforts were made to minimize suffering. Wild-type C57BL/6 mice, 17 to 20 days old, were inoculated subcutaneously into the footpad with 103 PFU of WNVKUN FLSDX or FL-IRAΔCS3 in Hanks balanced salt solution (HBSS) supplemented with 1% FBS, monitored for the signs of illness, and sacrificed when signs of encephalitis became apparent. Mice were bled at days 2 and 3 after infection, and viral genomic RNA was determined by real-time reverse transcription-PCR (RT-PCR), as described previously (47).
Eight- to 12-week old IRF-3−/− × IRF-7−/− mice and IFNAR−/− mice (13), both on the C57BL/6 genetic background, were inoculated via footpad with various doses (101, 102, and 103 PFU) of wt (FLSDX) or sfRNA-deficient virus (FL-IRAΔCS3).
In vitro RNase L assays.
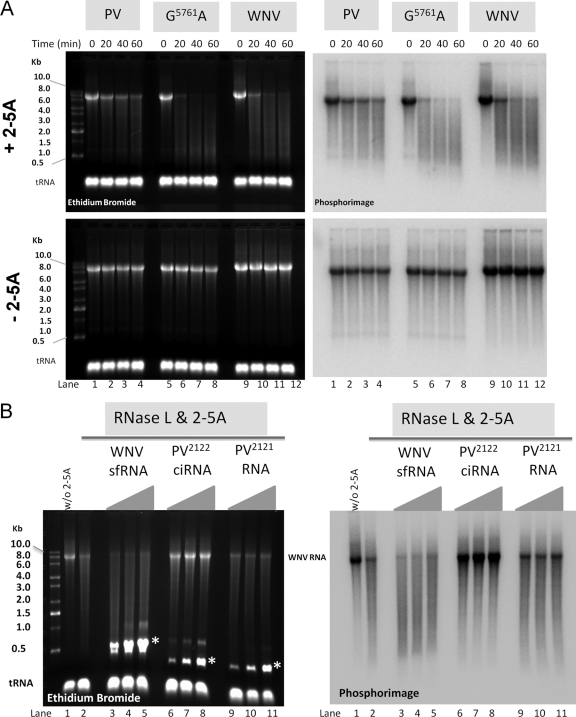
Radiolabeled in vitro transcribed WNV RNA (C20DXrep), RNase L-resistant poliovirus (PV) RNA, and RNase L-sensitive PV RNA containing the mutation G5761A were incubated at a concentration of 150 nM (each) for specified times with 20 nM RNase L and 20 nM 2-5A (a negative control was incubated without 2-5A). For inhibition experiments, radiolabeled in vitro transcribed WNV RNA was incubated for 60 min in reaction mixtures containing 20 nM RNase L and 20 nM 2-5A or 20 nM RNase L without 2-5A. The reaction mixtures included 500, 1,000, and 2,000 nM sfRNA, ORF 2122 of PV competitive inhibitor RNA (PV2122 ciRNA), and PV2121 RNA. All reactions were terminated by addition of SDS buffer, and products were phenol-chloroform extracted, ethanol precipitated with tRNA carrier, and fractionated by electrophoresis in agarose. The integrity of the RNAs was analyzed by ethidium bromide staining and phosphorimaging.
Statistical analyses.
Statistical significance of IFN sensitivity experiments was analyzed by regression analysis using the R program (45). All other data were analyzed using Prism software (GraphPad Prism). A two-way analysis of variance (ANOVA) was used to analyze differences in viral burden in mice. Kaplan-Meier survival curves were analyzed by the log rank test. Fold decrease of SFV titers after electroporation and IFN treatment was analyzed by an unpaired t test.
RESULTS
Antiviral responses mediated by IRF-3 and IRF-7 restrict replication of sfRNA-deficient virus in MEFs.
Our previous studies have shown that sfRNA is required for virulence in vivo as an sfRNA-deficient mutant virus (FL-IRAΔCS3) failed to cause mortality in wild-type (wt) weanling Swiss mice after intraperitoneal infection even at high virus doses (104 PFU) (17, 44). In contrast, sfRNA-deficient mutants replicated like wild-type virus (FLSDX) in cells (e.g., BHK-21 clone 15 and Vero) lacking intact cell-intrinsic antiviral response pathways. Thus, we hypothesized that sfRNA modulates the host antiviral response. To investigate this, we performed viral growth analyses in MEFs deficient in various components of the type I interferon response. IRF-3 and IRF-7 are key transcriptional regulators of IFN induction and ISG expression in response to viral infections (1, 24, 48). Using IRF-3−/− × IRF-7−/− MEFs, we investigated whether IRF-3 and IRF-7 are required for induction of the antiviral response that limits replication of sfRNA-deficient virus. While the sfRNA-deficient mutant FL-IRAΔCS3 replicated poorly in wt MEFs (Fig. 1A and B, upper panels), infection at an MOI of 1 of IRF-3−/− ×IRF-7−/− cells with FL-IRAΔCS3 virus resulted in a substantial increase in RNA replication and virus production, particularly during the first 2 days after infection (Fig. 1A and B, lower panels). Rescue of viral RNA replication and virus production in the IRF-3−/− × IRF-7−/− MEFs was rapid and approached levels of the wt virus as early as 12 to 24 hpi. A similar rescue was observed in IRF-3−/− × IRF-7−/− MEFs when lower levels (MOI of 0.1 or 0.01) of sfRNA-deficient FL-IRAΔCS3 virus were used (Fig. 1C). Again, replication of mutant virus at lower MOIs was as efficient as that of the wt virus. These results demonstrate that the IRAΔCS3 mutation did not affect virus replication in the absence of IRF-3 and IRF-7, thus confirming our prior data in BHK-21 cells, another cell line known to be deficient in IFN response (17), and establish that the mutations that abolish sfRNA production do not inherently attenuate virus replication. The results also suggest that IRF-3/IRF-7-dependent transcription is a primary factor limiting replication of viruses deficient in sfRNA production and imply a role for sfRNA in counteracting this antiviral pathway.
sfRNA contributes to higher viral resistance to type I IFN in MEFs.
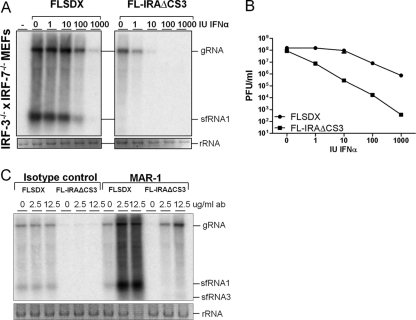
To begin to test whether sfRNA contributes to viral evasion of the IFN response, we compared the sensitivity of wt and mutant viruses to IFN-α pretreatment in IRF-3−/− × IRF-7−/− MEFs. Replication levels of wt and sfRNA-deficient viruses are equivalent in these cells (Fig. 1A), and they produce little, if any, type I IFN after WNV infection (13); so the antiviral effects will reflect only exogenously administered IFN. Cells were pretreated with increasing concentrations of IFN-α for 8 h, infected with wt or sfRNA-deficient virus at an MOI of 1, and incubated for 48 h before viral RNA was harvested from cells and titers of extracellular virus were determined. Although starting at slightly lower levels, viral RNA amounts for sfRNA-deficient virus were reduced to a greater extent than those for wt virus in cells treated with increasing concentrations of IFN-α. While RNA of wt virus could still be detected in cells treated with 1,000 IU/ml IFN-α, RNA of the sfRNA-deficient virus was undetectable after treatment with as little as 100 IU/ml (Fig. 2A). To further quantify this reduction, viral titers were measured by plaque assay and showed a significantly larger reduction in virus yield for the sfRNA mutant virus, ranging from a reduction of ∼10 to 1,000-fold for IFN-α concentrations from 1 to 1,000 IU/ml (P < 0.01), respectively (Fig. 2 B). To confirm the role of type I IFN as the primary factor limiting replication of the mutant virus, wt MEFs were treated with different concentrations of a neutralizing IFNAR-1 monoclonal antibody (MAR1-5A3), which binds to the IFNAR-1 subunit and prevents IFN signaling (50). Treatment of wt MEFs with MAR1-5A3 but not the isotype control antibody rescued replication of sfRNA-deficient virus in wt MEFs and also increased wt virus replication (Fig. 2C). These experiments indicate that the type I IFN response downstream of IFNAR signaling has a primary role in restricting replication of sfRNA-deficient virus in MEFs.
Fig 2.
sfRNA-deficient virus is more sensitive to IFN-α, and its replication is rescued by neutralizing antibodies to IFNAR. (A) Northern blot of viral RNA in IRF-3−/− × IRF-7−/− MEFs infected with wt (FLSDX) or sfRNA-deficient (FL-IRAΔCS3) virus after pretreatment with IFN-α (0 to 1,000 IU/ml for 8 h prior to infection at an MOI of 1). Supernatants and cells were harvested at 48 hpi for plaque assay and Northern blotting with a 3′ UTR-specific probe. Ethidium bromide staining of rRNA is included as a loading control. (B) Corresponding titers of infectious virus in the supernatant of the infected cells. Results are representative of two independent experiments. (C) Northern blot of viral RNA in wt MEFs infected with wt (FLSDX) and sfRNA-deficient (FL-IRAΔCS3) viruses in the presence of indicated concentrations of IFNAR-neutralizing antibody (ab) MAR1-5A3 or isotype control antibody. wt MEFs were infected with the wt and mutant viruses for 2 h at an MOI of 1, after which MAR1-5A3 or an isotype control antibody was added. Cells were harvested 4 days later, and Northern blotting was performed to detect viral RNA using a 3′ UTR-specific probe. Ethidium bromide staining of rRNA is included as a loading control.
Virulence of sfRNA-deficient virus is partially rescued in IRF-3−/− × IRF-7−/− mice and IFNAR−/− mice.
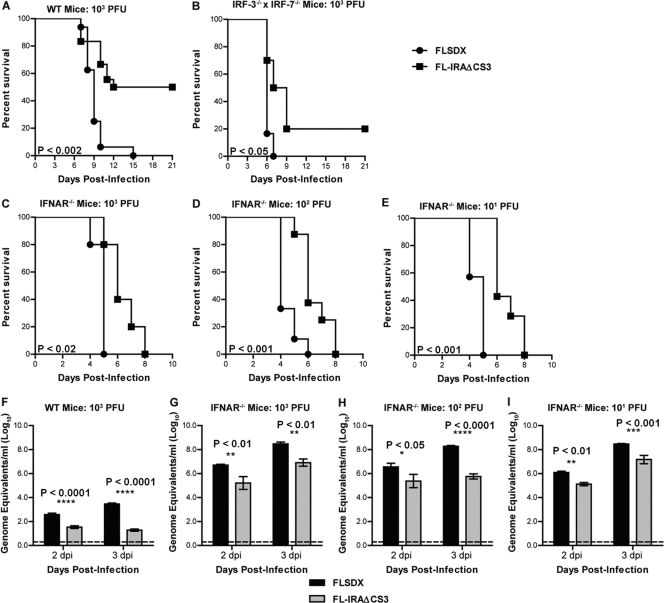
To investigate whether the findings in MEFs translated into an effect on virulence of sfRNA-deficient virus in mice, wt and IRF-3−/− × IRF-7−/− mice were initially infected via footpad injection with 103 PFU of FLSDX and FL-IRAΔCS3 viruses. As adult wt mice are not susceptible to infection with a wt WNVKUN virus (9), weanling (17- to 20-day-old) wt mice were used. In contrast, adult IRF-3−/− × IRF-7−/− mice were used for infections as they are highly susceptible to infection with WNVKUN virus (9). All wt weanling mice succumbed to infection with wt virus by day 15 after infection (n = 16), whereas 50% of animals survived infection with sfRNA-deficient virus (Fig. 3A) (P < 0.002, n = 18). Virulence of the sfRNA-deficient virus was largely rescued in IRF-3−/− × IRF-7−/− mice and approached that of the wt virus, with only 20% of animals surviving the infection (Fig. 3B) (P = 0.02, n = 10). Our previous studies had indicated that wt virus replicated more efficiently in IFNAR−/− mice than in IRF-3−/− × IRF-7−/− mice (9). To examine whether virulence of sfRNA-deficient virus was rescued more efficiently in IFNAR−/− mice, we infected them with the same dose (103 PFU) of wt and sfRNA-deficient virus. All animals died by day 8 after infection with mutant virus, whereas wt virus killed all IFNAR−/− mice by day 5 after infection (Fig. 3C). Quantitative RT-PCR analysis of viral RNA in serum showed a 137-fold decrease in serum virus load at 3 days postinfection (dpi) for wt mice infected with the sfRNA-deficient virus compared to wt virus but only a 6-fold decrease for the mutant virus in IFNAR−/− mice (Fig. 3F and G). As IFNAR−/− mice are highly sensitive to infection, it may be difficult to discern phenotypes with a high (103 PFU) dose of input virus. To address this, we also infected IFNAR−/− mice with 10- and 100-fold smaller amounts of virus. However, lower viral doses resulted in similar kinetics of pathogenesis (Fig. 3D and E) and viral burden in serum (Fig. 3H and I), thus demonstrating that rescue of virulence of sfRNA-deficient virus in the absence of IFNAR-mediated antiviral response was independent of viral dose. For all viral doses tested, wt virus was more virulent, suggesting that a type I IFN-independent pathway also contributed to restricting replication of the sfRNA-deficient virus in mice.
Fig 3.
Rescue of pathogenesis of sfRNA-deficient virus in mice deficient in IFN induction or signaling. (A) Weanling (1- to 20-day-old) wt C57BL/6 mice were inoculated with 103 PFU of wt (FLSDX) or sfRNA-deficient (FL-IRAΔCS3) virus by footpad injection and followed for mortality for 21 days. Survival data were combined from three independent experiments with a total of 16 to 18 mice per group (P < 0.002). (B) Adult (8- to 10-week-old) IRF-3−/− × IRF-7−/− mice were inoculated with 103 PFU of wt (FLSDX) or sfRNA-deficient (FL-IRAΔCS3) virus by footpad injection and followed for mortality for 21 days. Survival data were combined from two independent experiments with a total of 6 to 10 mice per group (P < 0.05). (C to E) Adult (8- to 10-week-old) IFNAR−/− mice were inoculated with 103 (C), 102 (D), or 101 (E) PFU of wt (FLSDX) or sfRNA-deficient (FL-IRAΔCS3) virus by footpad injection and followed for mortality. Survival data were combined from three independent experiments with a total of 5 to 9 mice per group (103 PFU, P < 0.02; 102 and 101 PFU, P < 0.001). (F to I) Viral RNA levels were determined from serum samples harvested on the indicated days after infection of weanling mice with 103 PFU or from adult IFNAR−/− mice infected with 103, 102, or 101 PFU of wt (FLSDX) or sfRNA-deficient (FL-IRAΔCS3) virus using qRT-PCR. Data are shown as log10 viral RNA equivalents per ml from 11 to 18 (wt) or 4 to 9 (IFNAR−/−) mice per time point. The error bar indicates standard error of the mean, and the dotted line represents the limit of sensitivity of the assay. Asterisks and corresponding P values shown represent differences that are statistically significant by two-way ANOVA.
sfRNA partially rescues replication of Semliki Forest virus in IFN-treated MEFs.
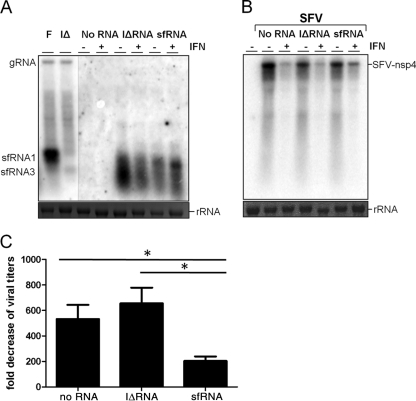
To examine whether sfRNA can inhibit the IFN-induced antiviral response directly in the absence of active WNVKUN replication, an in vitro transcribed 5′ GMP RNA encompassing the last 813 nucleotides of genomic RNA was electroporated into wt MEFs. The cells were then treated with mouse IFN-α for 8 h and infected with Semliki Forest virus (SFV), which is highly sensitive to the antiviral activity of IFN, for 12 h. A longer RNA fragment and incorporation of 5′ GMP during the in vitro transcription reaction were used to ensure proper processing of the electroporated RNA in cells by XRN1 to generate native sfRNA (sfRNA1). An RNA containing IRAΔCS3 mutations (IΔRNA), which rendered it unable to resist XRN1 degradation and produce full-length sfRNA1 (17), was used as a negative control. Indeed, Northern blot hybridization of RNA isolated from electroporated wt MEFs using an sfRNA-specific probe confirmed proper processing of electroporated RNAs into sfRNA1 and sfRNA3 for sfRNA and IΔRNA, respectively (Fig. 4A). Northern blot hybridization of RNA isolated from SFV-infected cells showed a decrease in the level of SFV RNA accumulation in cells pretreated with IFN compared to those not treated with IFN for mock and IΔRNA electroporations; the decrease associated with IFN treatment, however, was not as pronounced in cells electroporated with sfRNA (Fig. 4B). To quantify this effect, SFV titers in the culture supernatant of infected cells were determined. While IFN treatment reduced virus titers in mock or IΔRNA electroporated cells by 533- or 650-fold, respectively, the antiviral effect of IFN was less pronounced in the presence of sfRNA, and titers were reduced on average by only 204-fold (Fig. 4C) (P < 0.05). Thus, sfRNA conferred some resistance to the antiviral activity of IFN-α independently of WNVKUN infection
Fig 4.
sfRNA reduces the inhibitory effect of IFN treatment on SFV replication. Wild-type MEFs were electroporated with in vitro transcribed sfRNA or sfRNA containing IRAΔCS3 mutations (IΔRNA) and treated with 1,000 IU/ml mouse IFN-α for 8 h. Cells were infected subsequently with SFV at an MOI of 1 for 1 h. Samples were harvested at 10 h postelectroporation (before SFV infection) and 12 h after SFV infection. (A) Northern blot of RNA isolated before SFV infection showing different sfRNA species using a 3′ UTR-specific probe. RNA from cells infected with FLSDX (F) and FL-IRAΔCS3 (IΔ) viruses were used as controls for detecting sfRNA1 and sfRNA3, respectively. Ethidium bromide staining of rRNA is included as a loading control. (B) Northern blot detecting SFV gRNA with a probe for the SFV nsp4 gene. Ethidium bromide staining of rRNA is included as a loading control. A representative blot from three independent experiments is shown. (C) Fold reduction of SFV titers after IFN treatment in the presence of sfRNA or IΔRNA. Values are the mean of three independent experiments. Error bars indicate standard errors of the means. Statistical significance as analyzed by unpaired t test is indicated by an asterisk (*, P < 0.05).
RNase L partially restricts replication of sfRNA-deficient virus in MEFs.
To begin to define ISGs responsible for restricting replication of sfRNA-deficient virus, we used MEFs deficient in PKR or RNase L, ISGs which have documented inhibitory activity against WNV in vitro and in vivo (4, 19, 25, 47, 49, 52). Replication of sfRNA-deficient virus was not rescued in PKR−/− MEFs (Fig. 5A and B) but was partially rescued in RNase L−/− MEFs (Fig. 5C and D). However, the extent of rescue in RNase L−/− MEFs was less than that observed in IFNAR−/− MEFs or IRF-3−/− × IRF-7−/− MEFs, suggesting that either additional ISGs or another more general mechanism induced by IFN also contributes to the restriction of replication of sfRNA-deficient virus. Consistent with this, the sfRNA-deficient virus remained attenuated in adult RNase L−/− mice, and no mortality was observed (data not shown).
sfRNA fails to inhibit RNase L activity in vitro.
Poliovirus (PV) competitive inhibitor RNA (ciRNA) is an RNA element in PV genomic RNA (gRNA) that has been shown to inhibit RNase L activity, thereby rendering PV genomic RNA resistant to cleavage by RNase L (23, 55). Because of the partial rescue of the sfRNA-deficient virus in RNase L−/− MEFs, we hypothesized that sfRNA might inhibit RNase L activity in a manner similar to that of PV ciRNA and thus protect the WNV genome from RNase L-mediated degradation. We initially tested the sensitivity of WNV gRNA to RNase L-mediated degradation in an in vitro assay using recombinant RNase L and its activator 2-5A. WNV gRNA and PV RNA containing the G5761A mutation that disables the ciRNA and confers sensitivity to RNase L cleavage (55, 56) were almost completely degraded after 60 min of incubation, whereas RNase L-resistant PV RNA remained mostly intact (Fig. 6A, upper panel). No degradation was observed in reactions without 2-5A, confirming that cleavage was RNase L specific (Fig. 6A, lower panel). To assess the inhibitory potential of sfRNA on RNase L-mediated cleavage of WNV gRNA, we added increasing concentrations of sfRNA to the degradation assay. PV ciRNA and a PV RNA lacking inhibitory capacity (PV2121) were used as positive and negative controls, respectively. As expected, PV ciRNA protected WNV gRNA from RNase L-mediated degradation at concentrations of 500 to 2,000 nM. In contrast, neither PV2121 nor sfRNA was able to protect WNV RNA from degradation by RNase L, even at the highest concentration of 2,000 nM (Fig. 6B). Thus, despite the partial rescue of replication of sfRNA-deficient virus in RNase L−/− MEFs, we did not observe a direct inhibitory effect by sfRNA on RNase L activity in vitro.
Fig 6.
WNV RNA is sensitive to RNase L degradation in vitro, and sfRNA fails to protect WNV RNA from RNase L degradation. (A) Radiolabeled RNase L-resistant PV RNA, RNase L-sensitive PV G5761A RNA, and WNV RNA at 150 nM were incubated for the indicated periods of time in reaction mixtures containing 20 nM RNase L and 20 nM 2-5A or 20 nM RNase L without 2-5A. Reactions were terminated in SDS buffer, and products were phenol-chloroform extracted, ethanol precipitated with tRNA carrier, and fractionated by electrophoresis in 1.2% agarose-morpholinepropanesulfonic acid (MOPS)-formaldehyde. RNA was detected by ethidium bromide staining and UV light (left panels) and by phosphorimaging (right panels). (B) Radiolabeled WNV RNA (50 nM) was incubated for 60 min in reaction mixtures containing 20 nM RNase L and 20 nM 2-5A (lanes 2 to 11) or 20 nM RNase L without 2-5A (lane 1). sfRNA (lanes 3 to 5), PV2122 ciRNA (lanes 6 to 8), and PV2121 RNA (lanes 9 to 11) at 500, 1,000, and 2,000 nM were included in the reaction mixtures. Reactions were terminated in SDS buffer, and products were phenol-chloroform extracted, ethanol precipitated with tRNA carrier, and fractionated by electrophoresis in 1.2% agarose-MOPS-formaldehyde. RNA was detected by ethidium bromide staining and UV light (left panel) and by phosphorimaging (right panel). The mobilities of sfRNA and poliovirus RNAs are indicated with asterisks in the left panel.
DISCUSSION
We along with others have shown previously that a small noncoding RNA (ncRNA) derived from the 3′ UTR of the flavivirus genome (sfRNA) is produced in vitro and in vivo after infection by several members of the genus (29, 44, 49, 57). sfRNA is produced as a result of incomplete degradation of the gRNA by a cellular RNase, presumably XRN1, and is required for efficient replication and virulence in mice (44). Flaviviruses are not unique in their ability to generate small noncoding RNA or RNA structures within the genome that inhibit the host response. Several picornaviruses, including group C enteroviruses and poliovirus, encode structures within their open reading frames that inhibit the endoribonuclease domain of RNase L (23, 55). In addition, a number of viruses produce noncoding RNAs (ncRNAs) including microRNAs (miRNAs) and longer ncRNAs (53). Viral miRNAs function in evasion of the host antiviral response or in regulation of the viral life cycle (8). Longer ncRNAs also modulate host antiviral responses. For example, an adenovirus-associated RNA of ∼160 nt and Epstein-Barr virus-encoded EBER1 and EBER2 ncRNAs of ∼170 nt inhibit PKR (6, 41). Viral ncRNAs also have other functions, including maintenance of ATP levels during human cytomegalovirus infection (46) or activation of T cells during herpesvirus saimiri infection by inhibiting a cellular miRNA (5, 7). Given the role of viral ncRNAs in inhibiting host antiviral responses and the highly attenuated phenotype of a WNVKUN mutant lacking sfRNA, we hypothesized that sfRNA also might counteract the host response against flavivirus infection.
Type I and, to a lesser extent, type II IFNs control WNV and other flavivirus infections (reviewed in reference 12). Nonetheless, WNV has evolved several strategies that counteract IFN-dependent antiviral responses. WNV delays the induction of type I IFNs in infected cells by uncertain mechanisms, which allows for more efficient early replication (16). In addition, WNV encodes proteins with specific inhibitory functions that prevent activation of certain arms of the innate immune response (2, 3, 21, 28, 34, 35, 42, 43, 60). Importantly, the efficiency with which a particular WNV strain inhibits the antiviral activity of IFN is linked to virulence and pathogenicity (26). A contribution of NS5 to this effect recently has been established (28). In contrast to the extensive studies on the role of flavivirus proteins in inhibition of the IFN response, RNA-based evasion mechanisms have only recently begun to be identified (14).
Our results show that a mutant WNVKUN deficient in the production of sfRNA replicated poorly in wt mice and in wt MEFs, whereas it replicated more efficiently in mice and MEFs deficient in major factors involved in the type I IFN response. A mortality rate of 50% in 17- to 20-day-old wt C57BL/6 mice after footpad challenge with mutant virus differs from our previous data that showed no mortality after intraperitoneal challenge of 19- to 21-day-old Swiss outbred mice (44). It is most likely that the difference is due to the difference in the genetic background of the mice; however, the different routes of inoculation and age variations could also contribute to observed differences. Nevertheless, mutant virus was still significantly less virulent than wt virus in wt C57BL/6 mice. In contrast, virulence of the mutant virus was largely rescued in mice with a combined deficiency of the transcription factors IRF-3 and IRF-7, which act downstream of IPS-1 and MyD88, or a deficiency of IFNAR. Both IRF-3 and IRF-7 are master transcriptional regulators of the type I IFN response to WNV infection (10, 11). In IRF-7−/− MEFs, the IFN-α response and positive feedback loop are abolished, whereas the IFN-β response remains intact (11). IRF-3−/− MEFs, in contrast, show a significant reduction in early IFN-α and IFN-β responses to WNV (10) and fail to limit WNV spread (15). A combined deficiency of IRF-3 and IRF-7 rescued sfRNA-deficient mutant virus almost to wt virus levels, which is consistent with the finding that IRF-3−/− × IRF-7−/− MEFs had severely impaired type I IFN responses after WNV infection (13). WNV was more pathogenic in IRF-3−/− × IRF-7−/− mice than in wt mice but was still not as virulent as in IFNAR−/− mice, possibly because of a small residual IFN response (13). Analogously, the virulence of the sfRNA-deficient virus was greater in IFNAR−/− mice than in IRF-3−/− × IRF-7−/− mice. Clearly, replication of the sfRNA-deficient virus was impaired by the IFN-mediated antiviral response although remaining differences in virulence between wt and sfRNA-deficient virus in IFN response-defective mice suggest also some contribution of an IFN-independent antiviral response in restricting virulence of sfRNA-deficient virus.
The higher sensitivity of sfRNA-deficient virus compared to wt virus to IFN pretreatment in IRF-3−/− × IRF-7−/− MEFs and rescue of its replication in wt MEFs in the presence of IFNAR-neutralizing antibodies suggest a role for sfRNA in modulating an IFN-induced effector function. Consistent with this, sfRNA generated in MEFs (presumably by XRN1) from an in vitro transcribed longer RNA with an additional 5′ 288 nucleotides after electroporation resulted in partial inhibition of the IFN-induced antiviral response against an unrelated virus, SFV. Interestingly, when a shorter in vitro transcribed RNA representing sfRNA with only six additional nucleotides upstream of the sfRNA start was used under the same conditions, no inhibition of the IFN response was observed (data not shown), indicating that inhibition of the IFN response by sfRNA may require prior processing or coprocessing by XRN1. Although the exact mechanism of sfRNA-mediated inhibition of the effector function of IFN is unclear, we hypothesized that sfRNA may directly interact with and antagonize an ISG that binds RNA. Two of these RNA-binding ISGs, PKR and RNase L, have antiviral activity against a number of viruses including WNV (4, 19, 25, 47, 49, 52). In addition to degrading single-stranded RNA, which could have direct antiviral functions, RNase L can generate viral or host-derived small RNAs that amplify the IFN response by generating PAMPs that activate the RIG-I/MDA5 pathway (37, 38). Because of these two crucial direct and indirect antiviral activities, RNase L is targeted by viral evasion mechanisms, including a small poliovirus RNA, termed PV ciRNA (23, 55). In the current study, we observed a partial rescue of sfRNA-deficient virus in RNase L−/− MEFs, suggesting that RNase L could be one of the ISG targets of sfRNA. We hypothesized that sfRNA might inhibit RNase L-mediated cleavage of WNV RNA in a manner similar to PV ciRNA. However, our in vitro experiments showed that WNV RNA was sensitive to degradation by RNase L but that sfRNA failed to inhibit RNase L activity directly. Thus, the mechanism by which sfRNA interferes with RNase L-mediated inhibition of WNVKUN remains uncertain. Further detailed study on how sfRNA interferes with the function of RNase L and/or other yet unidentified ISGs, as well as other IFN-dependent and IFN-independent antiviral pathways, is required.
In summary, we have demonstrated that WNVKUN sfRNA contributes to viral evasion of the IFN-induced antiviral response. In addition to the many prior studies showing that viral proteins can inhibit IFN induction or signaling, flaviviruses, and likely other RNA viruses, independently utilize RNA-based strategies to subvert the antiviral response. An improved understanding of viral evasion strategies may facilitate novel strategies for therapeutic intervention against viral pathogens. Possibly, new classes of drugs that alter production of sfRNA in infected cells could sensitize flaviviruses to the innate antiviral effects of type I IFN.
ACKNOWLEDGMENTS
We thank Ezequiel Balmori Melian and Judy Edmonds for helpful discussions and Sarah Vick for technical assistance.
This work was supported by grants from the National Health and Medical Research Council of Australia and NIH grants UO1 AI066321 (A.A.K.), U54 AI081680 (Pacific Northwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research) and U19 AI083019 (M.S.D), and CA044059 (R.S). D.J.B was supported by Public Health Service grant AI042189 from the NIH. H.M.L was supported by an NIH training grant (T32-AI007172).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 29 February 2012
REFERENCES
- 1. Andersen J, VanScoy S, Cheng TF, Gomez D, Reich NC. 2008. IRF-3-dependent and augmented target genes during viral infection. Genes Immun. 9:168–175 [DOI] [PubMed] [Google Scholar]
- 2. Arjona A, et al. 2007. West Nile virus envelope protein inhibits dsRNA-induced innate immune responses. J. Immunol. 179:8403–8409 [DOI] [PubMed] [Google Scholar]
- 3. Best SM, et al. 2005. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J. Virol. 79:12828–12839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brass AL, et al. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cazalla D, Yario T, Steitz J. 2010. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science 328:1563–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clarke PA, Schwemmle M, Schickinger J, Hilse K, Clemens MJ. 1991. Binding of Epstein-Barr virus small RNA EBER-1 to the double-stranded RNA-activated protein kinase DAI. Nucleic Acids Res. 19:243–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cook HL, et al. 2005. Small nuclear RNAs encoded by Herpesvirus saimiri upregulate the expression of genes linked to T cell activation in virally transformed T cells. Curr. Biol. 15:974–979 [DOI] [PubMed] [Google Scholar]
- 8. Cullen BR. 2011. Viruses and microRNAs: RISCy interactions with serious consequences. Genes Dev. 25:1881–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Daffis S, et al. 2011. The naturally attenuated Kunjin strain of West Nile virus shows enhanced sensitivity to the host type I interferon response. J. Virol. 85:5664–5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daffis S, Samuel MA, Keller BC, Gale M, Jr, Diamond MS. 2007. Cell-specific IRF-3 responses protect against West Nile virus infection by interferon-dependent and -independent mechanisms. PLoS Pathog. 3:e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Daffis S, et al. 2008. Interferon regulatory factor IRF-7 induces the antiviral alpha interferon response and protects against lethal West Nile virus infection. J. Virol. 82:8465–8475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Daffis S, Suthar MS, Gale M, Jr, Diamond MS. 2009. Measure and countermeasure: type I IFN (IFN-alpha/beta) antiviral response against West Nile virus. J. Innate Immun. 1:435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Daffis S, Suthar MS, Szretter KJ, Gale M, Jr, Diamond MS. 2009. Induction of IFN-beta and the innate antiviral response in myeloid cells occurs through an IPS-1-dependent signal that does not require IRF-3 and IRF-7. PLoS Pathog. 5:e1000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Daffis S, et al. 2010. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468:452–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fredericksen BL, Gale M., Jr 2006. West Nile virus evades activation of interferon regulatory factor 3 through RIG-I-dependent and -independent pathways without antagonizing host defense signaling. J. Virol. 80:2913–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fredericksen BL, Smith M, Katze MG, Shi PY, Gale M., Jr 2004. The host response to West Nile Virus infection limits viral spread through the activation of the interferon regulatory factor 3 pathway. J. Virol. 78:7737–7747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Funk A, et al. 2010. RNA structures required for production of subgenomic flavivirus RNA. J. Virol. 84:11407–11417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Funk A, et al. 2010. RNA structures required for production of subgenomic flavivirus RNA. J. Virol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gilfoy FD, Mason PW. 2007. West Nile virus-induced interferon production is mediated by the double-stranded RNA-dependent protein kinase PKR. J. Virol. 81:11148–11158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gubler DJ. 2002. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 10:100–103 [DOI] [PubMed] [Google Scholar]
- 21. Guo JT, Hayashi J, Seeger C. 2005. West Nile virus inhibits the signal transduction pathway of alpha interferon. J. Virol. 79:1343–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hall RA, Broom AK, Smith DW, Mackenzie JS. 2002. The ecology and epidemiology of Kunjin virus. Curr. Top. Microbiol. Immunol. 267:253–269 [DOI] [PubMed] [Google Scholar]
- 23. Han JQ, et al. 2007. A phylogenetically conserved RNA structure in the poliovirus open reading frame inhibits the antiviral endoribonuclease RNase L. J. Virol. 81:5561–5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Honda K, et al. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772–777 [DOI] [PubMed] [Google Scholar]
- 25. Jiang D, et al. 2010. Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infection. J. Virol. 84:8332–8341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Keller BC, et al. 2006. Resistance to alpha/beta interferon is a determinant of West Nile virus replication fitness and virulence. J. Virol. 80:9424–9434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Khromykh AA, Varnavski AN, Westaway EG. 1998. Encapsidation of the flavivirus Kunjin replicon RNA by using a complementation system providing Kunjin virus structural proteins in trans. J. Virol. 72:5967–5977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Laurent-Rolle M, et al. 2010. The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. J. Virol. 84:3503–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin KC, Chang HL, Chang RY. 2004. Accumulation of a 3′-terminal genome fragment in Japanese encephalitis virus-infected mammalian and mosquito cells. J. Virol. 78:5133–5138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu WJ, Chen HB, Khromykh AA. 2003. Molecular and functional analyses of Kunjin virus infectious cDNA clones demonstrate the essential roles for NS2A in virus assembly and for a nonconservative residue in NS3 in RNA replication. J. Virol. 77:7804–7813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu WJ, Chen HB, Wang XJ, Huang H, Khromykh AA. 2004. Analysis of adaptive mutations in Kunjin virus replicon RNA reveals a novel role for the flavivirus nonstructural protein NS2A in inhibition of beta interferon promoter-driven transcription. J. Virol. 78:12225–12235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu WJ, Sedlak PL, Kondratieva N, Khromykh AA. 2002. Complementation analysis of the flavivirus Kunjin NS3 and NS5 proteins defines the minimal regions essential for formation of a replication complex and shows a requirement of NS3 in cis for virus assembly. J. Virol. 76:10766–10775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu WJ, et al. 2006. A single amino acid substitution in the West Nile virus nonstructural protein NS2A disables its ability to inhibit alpha/beta interferon induction and attenuates virus virulence in mice. J. Virol. 80:2396–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu WJ, et al. 2005. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J. Virol. 79:1934–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mackenzie JM, Khromykh AA, Parton RG. 2007. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe 2:229–239 [DOI] [PubMed] [Google Scholar]
- 36. Mackenzie JS, Gubler DJ, Petersen LR. 2004. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med. 10:S98–109 [DOI] [PubMed] [Google Scholar]
- 37. Malathi K, Dong B, Gale M, Jr, Silverman RH. 2007. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 448:816–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Malathi K, et al. 2010. RNase L releases a small RNA from HCV RNA that refolds into a potent PAMP. RNA 16:2108–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marie I, Durbin JE, Levy DE. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 17:6660–6669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Markoff L. 2003. 5′- And 3′-noncoding regions in flavivirus RNA. Adv. Virus Res. 59:177–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mathews MB, Shenk T. 1991. Adenovirus virus-associated RNA and translation control. J. Virol. 65:5657–5662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mazzon M, Jones M, Davidson A, Chain B, Jacobs M. 2009. Dengue virus NS5 inhibits interferon-alpha signaling by blocking signal transducer and activator of transcription 2 phosphorylation. J. Infect. Dis. 200:1261–1270 [DOI] [PubMed] [Google Scholar]
- 43. Munoz-Jordan JL, et al. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 79:8004–8013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pijlman GP, et al. 2008. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 4:579–591 [DOI] [PubMed] [Google Scholar]
- 45. R Development Core Team 2010. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 46. Reeves MB, Davies AA, McSharry BP, Wilkinson GW, Sinclair JH. 2007. Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science 316:1345–1348 [DOI] [PubMed] [Google Scholar]
- 47. Samuel MA, et al. 2006. PKR and RNase L contribute to protection against lethal West Nile Virus infection by controlling early viral spread in the periphery and replication in neurons. J. Virol. 80:7009–7019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sato M, et al. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539–548 [DOI] [PubMed] [Google Scholar]
- 49. Scherbik SV, Paranjape JM, Stockman BM, Silverman RH, Brinton MA. 2006. RNase L plays a role in the antiviral response to West Nile virus. J. Virol. 80:2987–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sheehan KC, et al. 2006. Blocking monoclonal antibodies specific for mouse IFN-alpha/beta receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection. J. Interferon Cytokine Res. 26:804–819 [DOI] [PubMed] [Google Scholar]
- 51. Silva PA, Pereira CF, Dalebout TJ, Spaan WJ, Bredenbeek PJ. 2010. An RNA pseudoknot is required for production of yellow fever virus subgenomic RNA by the host nuclease XRN1. J. Virol. 84:11395–11406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Silverman RH. 2007. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J. Virol. 81:12720–12729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sullivan CS. 2008. New roles for large and small viral RNAs in evading host defences. Nat. Rev. Genet. 9:503–507 [DOI] [PubMed] [Google Scholar]
- 54. Takeuchi O, Akira S. 2009. Innate immunity to virus infection. Immunol. Rev. 227:75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Townsend HL, et al. 2008. A viral RNA competitively inhibits the antiviral endoribonuclease domain of RNase L. RNA 14:1026–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Townsend HL, Jha BK, Silverman RH, Barton DJ. 2008. A putative loop E motif and an H-H kissing loop interaction are conserved and functional features in a group C enterovirus RNA that inhibits ribonuclease L. RNA Biol. 5:263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Urosevic N, van Maanen M, Mansfield JP, Mackenzie JS, Shellam GR. 1997. Molecular characterization of virus-specific RNA produced in the brains of flavivirus-susceptible and -resistant mice after challenge with Murray Valley encephalitis virus. J. Gen. Virol. 78:23–29 [DOI] [PubMed] [Google Scholar]
- 58. Westaway EG, Mackenzie JM, Khromykh AA. 2003. Kunjin RNA replication and applications of Kunjin replicons. Adv. Virus Res. 59:99–140 [DOI] [PubMed] [Google Scholar]
- 59. Westaway EG, Mackenzie JM, Khromykh AA. 2002. Replication and gene function in Kunjin virus. Curr. Top. Microbiol. Immunol. 267:323–351 [DOI] [PubMed] [Google Scholar]
- 60. Wilson JR, de Sessions PF, Leon MA, Scholle F. 2008. West Nile virus nonstructural protein 1 inhibits TLR3 signal transduction. J. Virol. 82:8262–8271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhou A, et al. 1997. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 16:6355–6363 [DOI] [PMC free article] [PubMed] [Google Scholar]