Abstract
Superinfection exclusion, a phenomenon in which a preexisting viral infection prevents a secondary infection with the same or a closely related virus, has been described for various viruses, including important pathogens of humans, animals, and plants. The phenomenon was initially used to test the relatedness of plant viruses. Subsequently, purposeful infection with a mild isolate has been implemented as a protective measure against virus isolates that cause severe disease. In the medical and veterinary fields, superinfection exclusion was found to interfere with repeated applications of virus-based vaccines to individuals with persistent infections and with the introduction of multicomponent vaccines. In spite of its significance, our understanding of this phenomenon is surprisingly incomplete. Recently, it was demonstrated that superinfection exclusion of Citrus tristeza virus (CTV), a positive-sense RNA closterovirus, occurs only between isolates of the same strain, but not between isolates of different strains of the virus. In this study, I show that superinfection exclusion by CTV requires production of a specific viral protein, the p33 protein. Lack of the functional p33 protein completely eliminated the ability of the virus to exclude superinfection by the same or a closely related virus. Remarkably, the protein appeared to function only in a homology-dependent manner. A cognate protein from a heterologous strain failed to confer the exclusion, suggesting the existence of precise interactions of the p33 protein with other factors involved in this complex phenomenon.
INTRODUCTION
Superinfection exclusion or homologous interference is defined as the ability of an established virus infection to interfere with a secondary infection by the same or a closely related virus. The phenomenon has been described for various virus-host systems, including viruses that cause serious diseases in humans, animals, and plants (2, 9, 10, 11, 14, 19–21, 23, 28, 29, 31, 35, 37, 38, 55, 61–64, 74–77). From an evolutionary standpoint, superinfection exclusion can be a powerful strategy that determines the genetic structure of the virus population. Superinfection exclusion protects the virus from a related competing secondary virus targeting the cell that has been successfully infected by the primary virus. Besides elimination of competition for host resources, the phenomenon could function as a means to maintain the stability of viral sequences because it prevents replication of two or more viral genomes in the same cell, thus reducing the likelihood of the recombination or reassortment of viral genes, with the latter event of particular importance for the evolution of segmented viruses (18, 27). From a practical standpoint, superinfection exclusion can have both positive and negative attributes. Referred to as cross-protection, this phenomenon has been implemented as an agricultural practice in which purposeful infection with a mild isolate was used as a protective measure against isolates of the virus that cause severe disease (reviewed in references 21 and 29). On the other hand, in the medical and veterinary fields, superinfection exclusion was found to interfere with repeated applications of virus-based vaccines to individuals with persistent infections and with the introduction of multicomponent vaccines (14, 64).
I and members of my laboratory are examining superinfection exclusion by Citrus tristeza virus (CTV). CTV is the largest and most complex member of the Closteroviridae family, which contains viruses with mono-, bi-, and tripartite genomes that are transmitted by a range of insect vectors including aphids, whiteflies, and mealybugs and cause severe economic losses in crops including vegetables, grains, grapes, fruit trees, and others (3, 5, 12, 13, 34). CTV has long flexuous virions (2,000 nm by 10 to 12 nm) encapsidated by two coat proteins and a single-stranded RNA genome of approximately 19.3 kb. The RNA genome of CTV encodes 12 open reading frames (ORFs) (32, 47) (Fig. 1). ORFs 1a and 1b are expressed from the genomic RNA and encode polyproteins required for virus replication. ORF 1a encodes a 349-kDa polyprotein containing two papain-like protease domains plus methyltransferase-like and helicase-like domains. Translation of the polyprotein is thought to occasionally continue through the polymerase-like domain (ORF 1b) by a +1 frameshift. Ten 3′-end ORFs are expressed by 3′ coterminal subgenomic RNAs (sgRNAs) (25, 33) and encode the following proteins: major (CP) and minor (CPm) coat proteins, p65 (HSP70 homolog), and p61, which are involved in assembly of virions (57); a hydrophobic p6 protein with a proposed role in virus movement (13, 65); p20 and p23, which along with CP are suppressors of RNA silencing (42); and p33, p13, and p18, which function in extending the virus host range (66). Remarkably, trees of most citrus varieties can be infected with mutants with three genes deleted: p33, p18, and p13 (65).
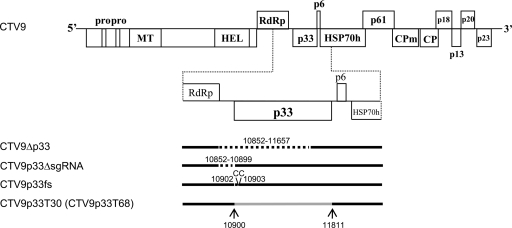
Fig 1.
Schematic diagram of the genome organization of wild-type CTV (CTV9). The open boxes represent ORFs and their translation products. PRO, papain-like protease domain; MT, methyltransferase; HEL, helicase; RdRp, an RNA-dependent RNA polymerase; HSP70h, HSP70 homolog; CPm, minor coat protein; CP, major coat protein. The enlarged view of the region containing the p33 ORF and schematic diagrams of CTV mutants are shown below. The sequences deleted in mutants are indicated by dotted lines with corresponding nucleotide numbers. Solid lines represent sequences present in the genomes of mutants. “CC” indicates two extra cytidylates inserted in CTV9p33fs construct. Sequences substituted from the genomes of T30-1 or T68-1 isolates are shown in gray.
The host range of CTV is limited to citrus and citrus relatives in which the virus infects only phloem-associated cells. CTV has numerous isolates with distinctive biological and genetic characteristics (24, 26, 36, 43, 46, 51–53, 70, 71) that are classified into six major genotype groups, which are referred to as strains T36, T3, T30, T68, VT, and RB, with some isolates remaining undefined (4, 17, 24, 26, 36, 51, 53). The classification strategy of CTV isolates is based upon analysis of nucleotide sequences of the 1a ORF, which shows high genetic diversity between CTV variants, with levels of sequence identity ranging between 72.3 and 90.3% for isolates representing different strains (26, 36, 40, 52, 53). Isolates within the same strain group show minor sequence divergence, generally less than 5% throughout the genome.
Recently, we demonstrated that superinfection exclusion occurs only between isolates of the same strain, but not between isolates of different CTV strains (17). Remarkably, exclusion among isolates of the same strain was absolute. No trace of the challenging virus was found. On the other hand, isolates from different strains demonstrated a complete lack of exclusion. The isolates of heterologous strains that were established initially appeared to have no effect on infection, movement, and replication of the challenge virus. The amount of the challenge virus in the plants preinfected with isolates of heterologous strains was identical to that found in plants that had no primary infection. Moreover, the exclusion phenomenon appeared to be systemic and functioned not only in cells infected with the primary virus but also in cells that were not infected. Upon invasion in a host, CTV infects only a portion of the phloem-associated cells, usually less than one-third of the cells even in the most susceptible hosts, leaving many cells uninfected (16). However, even though the majority of cells were not infected by the primary isolate, exclusion of a challenging isolate of the same strain was absolute. Not only was the one-third of the cells that contained the primary virus protected, but the other two-thirds of the cells that were not infected became “immune” to the challenging virus (17).
In spite of the significance of superinfection exclusion in viral pathogenesis and in the evolution of virus populations, our understanding of how virus variants exclude each other is surprisingly incomplete. Several mechanisms acting at various stages of the viral life cycle have been postulated to explain superinfection exclusion. However, each of the proposed mechanisms can explain only some instances of superinfection exclusion, and often, in those cases in which a particular mechanism appears to play a role it cannot completely explain all the aspects of this phenomenon. Superinfection exclusion of animal viruses has been related to several mechanisms that explained exclusion by interference with receptor-mediated attachment and penetration into cells, translation, or replication of the secondary virus (2, 35, 38, 59, 62, 63). For plant viruses, initial explanations included competition between primary and challenging viruses for host factors or intracellular replication sites and interference with disassembly of the secondary virus resulting from the excessive production of the coat protein by the primary virus (1, 41, 60; reviewed in references 7, 8, 21, 29, and 78). These mechanisms would be effective only in cells that were infected with the primary virus, while uninfected cells would remain susceptible to the secondary virus. Another mechanism that was implicated in the superinfection exclusion of several plant viruses is based on the induction of RNA silencing by the protector virus that leads to sequence-specific degradation of the challenge virus RNA (48, 49; reviewed in reference 29) and can be induced systemically in both infected and uninfected cells (44, 69, 72, 73). These properties of RNA silencing could explain some of our observations with CTV, leading to the hypothesis that this mechanism could account for CTV superinfection exclusion. However, the results obtained from examination of superinfection exclusion ability of hybrids of different CTV strains in our previous work appeared not to support this hypothesis (17) and further argued for the intriguing complexity of CTV superinfection exclusion phenomenon.
Here, I show that superinfection exclusion by CTV is due to a mechanism that requires production of a specific viral protein, the p33 protein. p33 appears to be a nonconserved protein which shows no significant homology with other known proteins and is not essential for CTV infection in most citrus hosts (65). Lack of functional p33 completely eliminated the ability of the virus to exclude superinfection by the same or a closely related virus. Furthermore, a cognate protein from a heterologous strain failed to confer the exclusion, suggesting the existence of precise interactions of the p33 protein with other factors involved in this phenomenon.
MATERIALS AND METHODS
Virus isolates and inoculation of citrus trees.
The wild-type cloned virus CTV9 (56, 58), deletion mutant viruses CTV9Δp33, CTV9Δp18, CTV9Δp13, CTV9Δp33Δp18, CTV9Δp33Δp13, CTV9Δp33Δp18Δp13, and CTV9Δp18Δp13 (65), green fluorescent protein (GFP)-tagged wild-type virus CTV9-GFP and GFP-tagged p33 deletion mutant CTV9Δp33-GFP (CTV9-GFPC3 and CTV9Δp33-GFPC3 in reference 65), CTV isolates T30-1 (4) and T68-1 (17), and a set of mutant viruses engineered in this work as described below (CTV9p33ΔsgRNA, CTV9p33fs, CTV9p33T30, and CTV9p33T68) (Fig. 1) have been maintained in citrus plants under greenhouse conditions. These plants were used as sources of virus for subsequent graft inoculations of young trees.
To assess superinfection exclusion, 9- to 12-month-old trees of Citrus macrophylla Wester (usually 5 plants per treatment) were initially inoculated by grafting of virus-infected tissue from individual source plants. At 6 weeks after inoculation, systemic tissue was assayed by enzyme-linked immunosorbent assay (ELISA) to confirm the establishment of infection. Secondary (challenge) inoculation of preinfected plants was done by inserting a second graft of bark tissue infected with a challenge virus. When the graft healed, the upper flushes of leaves were trimmed to induce growth of a new flush, which was then evaluated for the ability of the challenge virus to establish systemic infection in plants that were previously infected with a primary virus.
Generation of mutant virus constructs pCTV9p33ΔsgRNA, pCTV9p33fs, pCTV9p33T30, and pCTV9p33T68.
The full-length cDNA clone of CTV T36, pCTV9 (56, 58), was the basis of all constructs in this study. pCTV9p33ΔsgRNA was created by amplifying a PCR product with oligonucleotides C-1493 (corresponding to nucleotides [nt] 4397 to 4424 in the CTV genome) and C-1938 (complementary to nt 10923 to 10900 and 10851 to 10828) and a PCR product with oligonucleotides C-1937 (corresponding to nt 10828 to 10851 and 10900 to 10923) and C-204 (complementary to nt 11949 to 11924). The two products were used to generate an overlap PCR product using C-1493 and C-204. The resulting product was digested with Bsu36I and PmeI restriction endonucleases and substituted for the corresponding fragment into pCTV9 digested with the same enzymes (nt positions 4430 and 11869, respectively). pCTV9p33fs was obtained by amplifying a PCR product with oligonucleotides C-1493 and C-1916 (complementary to nt 10926 to 10877 and containing two extra G's between nt 10903 and 10902) and a PCR product with oligonucleotides C-1915 (corresponding to nt 10877 to 10926 and containing two extra C's between nt 10902 and 10903) and C-204. The two products were used to generate an overlap PCR product using C-1493 and C-204, which was digested with Bsu36I and PmeI and substituted into pCTV9 digested with the same enzymes. To generate pCTV9p33T30 and pCTV9p33T68, double-stranded CTV RNA was extracted from C. macrophylla trees infected with T30-1 or T68-1 as described previously (45). This double-stranded RNA was used for cDNA synthesis and subsequent PCR amplification using a pair of primers, one corresponding to the 21 nt preceding the p33 ORF in CTV9 (nt positions 10879 to 10899) plus 21 nt of the 5′ end of the p33 ORF sequence in the T30-1 or the T68-1 genome (C-1912 or C-1889, respectively) and the other being complementary to the sequence downstream from the stop codon of the p33 ORF in CTV9 containing the PmeI site and the 21 nt of the 3′ end of the T30-1 or T68-1 p33 ORF (C-1888 or C-1891, respectively). A second PCR was carried out using CTV9 as a template and C-1493 and C-1913 complementary to C-1912 for the generation of pCTV9p33T30 or C-1890 complementary to C-1889 for the generation of pCTV9p33T68. The products of the first and second PCRs were used for overlap PCR using C-1493 and C-1888 or C-1891 for the T30 p33 ORF- or the T68 p33 ORF-containing construct. The resulting products were digested with Bsu36I and PmeI and substituted into pCTV9 digested with the same enzymes.
Amplification of virions of engineered virus constructs in Nicotiana benthamiana protoplasts for inoculation of citrus trees.
SP6 RNA polymerase-derived transcripts of CTV cDNAs were used for transfection of N. benthamiana protoplasts as described earlier (56). Passaging of virions in protoplasts for virus amplification was done as described previously (57). The accumulation of virus RNAs was monitored by Northern blot hybridization of the total RNA isolated from protoplasts with a 3′ positive-stranded CTV RNA-specific riboprobe (56). Amplified progeny virions from the final passages in protoplasts were extracted, concentrated by sucrose cushion centrifugation, and used for mechanical “bark flap” inoculation of C. macrophylla (50).
Serological assays.
Triple-antibody sandwich ELISA (TAS-ELISA) was performed as described using antibodies specific to CTV virions (22) to confirm infection in inoculated plants. Plant extracts were prepared at a 1/20 dilution: 0.25 g of citrus bark tissue was ground in 5 ml of the extraction buffer for each sample. Purified IgG from rabbit polyclonal antiserum CTV-908 (1 μg/ml) was used as a coating antibody. ECTV172, a broadly reactive CTV monoclonal antibody, was used to detect antibodies.
Examination of fluorescence in citrus plants infected with GFP-tagged CTV.
Bark tissue from CTV9-GFP- or CTV9Δp33-GFP-inoculated trees was examined for GFP fluorescence beginning at 6 weeks after challenge using a Zeiss Stemi SV 11 UV-fluorescence dissecting microscope (Carl Zeiss Jena, GmbH, Jena, Germany) with an attached Olympus Q-color 5 camera (Olympus America, Inc., Center Valley, PA).
RESULTS
Deletion of the p33 gene eliminates the ability of the virus to exclude superinfection by the parental wild-type virus.
CTV has three genes that appear to be unique to the virus and show no significant homology with other sequences reported in GenBank. Previously, it was demonstrated that CTV mutants with deletions within the p33, p18, or p13 ORF individually or in combination retained the ability to infect, multiply, and spread normally throughout trees of most citrus varieties (65). GFP-tagged CTV variants with deletions in the p33 ORF or the p33, p18, and p13 ORFs demonstrated that the infection levels as well as movement and distribution of these deletion mutants within citrus trees were similar to that of the wild-type virus (65). In this work, we examined how deletions of the p33, p18, or p13 genes from the CTV genome affect superinfection exclusion by assaying the ability of the resulting mutant viruses to prevent superinfection by the GFP-expressing wild-type virus (CTV9-GFP) (15, 65). Young C. macrophylla trees first were inoculated with the mutant viruses CTV9Δp18, CTV9Δp13, CTV9Δp18Δp13, CTV9Δp33 (Fig. 1), CTV9Δp33Δp18, CTV9Δp33Δp13, and CTV9Δp33Δp18Δp13 (65) by grafting virus-infected tissue into stems of receptor trees. As a control for this experiment, a set of plants was inoculated with the parental wild-type CTV (CTV9). Another set of control plants had no primary infection. The upper leaves on the inoculated plants were trimmed to force the growth of a new set of leaves. At 6 weeks after inoculation, systemic infections of the new leaves were confirmed by ELISA using CTV-specific antiserum. Similar ELISA values were obtained for all viruses used for primary inoculations, demonstrating similar levels of accumulation of the viruses in the infected plants (Table 1, experiment A). The plants then were challenged by putting a second graft of bark tissue containing the CTV9-GFP virus. The ability of the challenging virus to superinfect trees was determined by visual observation of GFP fluorescence in the bark tissue of the new flush at 2 and 4 months after challenge inoculation. As a result, the parental CTV9 virus completely prevented superinfection by CTV9-GFP virus: no GFP fluorescence was detected in plants primarily infected with the wild-type virus (Fig. 2; Table 1, experiment A). Similarly to the parental virus, mutants containing a deletion of the p18 or p13 ORF completely excluded the challenge virus, demonstrating that the lack of those genes did not have any impact on the viral superinfection exclusion ability (Fig. 2; Table 1, experiment A). Remarkably, plants that had primary infections with the mutant viruses lacking the p33 gene all displayed GFP fluorescence similar to that observed in plants that had no primary infection and were inoculated only with the challenge virus CTV9-GFP (Fig. 2; Table 1, experiment A). The p33 deletion mutants did not interfere with secondary infection by the GFP-tagged parental virus, indicating that the deletion of this gene removed the ability of the resulting viruses to exclude superinfection by the wild-type virus.
Table 1.
Examination of CTV mutants for the ability to prevent superinfection by GFP-expressing CTV
| Expt group | Primary inoculation | Prechallenge CTV titera | GFPb |
|---|---|---|---|
| Expt A | None | 0.07 ± 0.006 | Yes |
| CTV9 | 3.23 ± 0.046 | No | |
| CTV9Δp18 | 2.98 ± 0.030 | No | |
| CTV9Δp13 | 3.01 ± 0.045 | No | |
| CTV9Δp18Δp13 | 2.99 ± 0.027 | No | |
| CTV9Δp33 | 3.10 ± 0.039 | Yes | |
| CTV9Δp33Δp18 | 3.34 ± 0.060 | Yes | |
| CTV9Δp33Δp13 | 2.88 ± 0.034 | Yes | |
| CTV9Δp33Δp18Δp13 | 3.07 ± 0.066 | Yes | |
| Expt B | None | 0.10 ± 0.005 | Yes |
| CTV9 | 2.81 ± 0.035 | No | |
| CTV9Δp33 | 2.99 ± 0.048 | Yes | |
| Expt C | None | 0.09 ± 0.007 | Yes |
| CTV9 | 3.13 ± 0.028 | No | |
| CTV9Δp33 | 3.33 ± 0.056 | Yes | |
| CTV9p33ΔsgRNA | 2.96 ± 0.042 | Yes | |
| CTV9p33fs | 2.80 ± 0.027 | Yes | |
| CTV9p33T30 | 3.05 ± 0.037 | Yes | |
| CTV9p33T68 | 3.40 ± 0.039 | Yes |
Trees were assayed at 6 weeks after initial inoculation by triple-antibody sandwich ELISA using CTV-specific antibody. ELISA values (A405) are an average of results for 5 plants ± standard deviation.
GFP fluorescence was observed in the bark tissue of trees by using a dissecting fluorescence microscope at 4 months after challenge with CTV9-GFP (experiments A and C) or CTV9Δp33-GFP (experiment B).
Fig 2.
Observation of GFP fluorescence in phloem-associated cells of C. macrophylla trees upon challenge with CTV9-GFP. The left image represents a noninoculated healthy tree. The other images represent trees with no primary infection (second image) or trees preinfected with the wild-type or mutant viruses, which were sequentially challenged with CTV9-GFP. Observations were done on the internal surface of bark at 4 months after challenge inoculation using a dissecting fluorescence microscope. Bars = 0.4 mm.
The p33 deletion mutant fails to exclude superinfection with the same virus.
Next, I examined whether the p33 deletion mutant is able to exclude superinfection by the same mutant virus. I conducted an experiment similar to the one described above, in which the wild-type CTV9 virus and the CTV9Δp33 mutant were used for primary inoculations of citrus trees. Later, upon establishment of the initial infections (Table 1, experiment B), the preinfected plants were challenged with the GFP-expressing CTV9Δp33 virus (65). A set of uninfected plants was also inoculated with the challenge virus as a control in this experiment. Observations of bark tissue of the plants at 2 and 4 months after challenge inoculation revealed that plants preinfected with CTV9 were protected against the challenge virus infection. No GFP fluorescence was detected in those plants (Fig. 3; Table 1, experiment B). However, plants that were initially infected with CTV9Δp33 displayed strong GFP fluorescence similar to that in control plants with no primary infection, indicating that the deletion of the p33 gene resulted in the inability of the virus to exclude itself (Fig. 3; Table 1, experiment B).
Fig 3.
Observation of GFP fluorescence in C. macrophylla trees upon challenge with CTV9Δp33-GFP. The left image represents a noninoculated healthy tree. The other images represent trees with no primary infection (second image) or trees preinfected with the wild type or with CTV9Δp33, which were sequentially challenged with CTV9Δp33-GFP. Observations were done on the internal surface of bark at 4 months after challenge inoculation using a dissecting fluorescence microscope. Bars = 0.4 mm.
p33 protein is a determinant of superinfection exclusion.
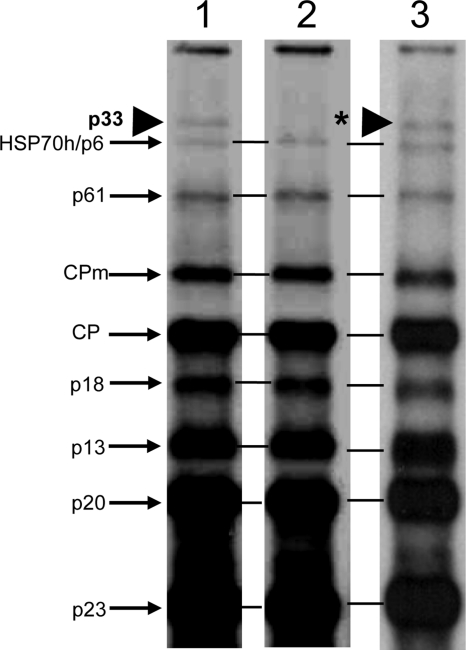
The above findings indicated that deletion of the p33 gene prevented the exclusion of superinfection by the virus. As the next step, I assessed whether exclusion is conferred by the p33 protein or the RNA sequence. I engineered constructs that retain the entire sequence of the p33 ORF but prevent production of the functional protein. One of the constructs (CTV9p33ΔsgRNA) (Fig. 1) was created by removing a 48-nt intergenic region between the stop codon of the 1b ORF and the start codon of the p33 ORF. This region contains a part of the subgenomic mRNA controller element for the p33 sgRNA (65). Removal of this region eliminated the synthesis of the p33 sgRNA, which is used as mRNA for translation of the p33 protein (Fig. 4, lanes 1 and 2). Another construct (CTV9p33fs) (Fig. 1) was engineered by the introduction of two extra cytidylates behind the start codon of the p33 ORF, resulting in a shift of the normal reading frame and the generation of a new stop codon, thus blocking production of the functional p33 protein. The frameshift mutation had no effect on the synthesis of the p33 sgRNA, which was accumulated in amounts similar to that of the wild-type virus (Fig. 4, lane 3). In order to examine how the lack of the p33 protein production affects superinfection exclusion, the constructs were tested for the ability to exclude the parental wild-type virus. The mutant viruses were used for primary inoculation of citrus plants (along with CTV9 and CTV9Δp33, used as controls in this experiment). Upon establishment of primary infections (Table 1, experiment C), the plants were challenged with GFP-tagged CTV. Similarly to the p33 deletion mutant, the virus variant lacking the subgenomic promoter for the p33 sgRNA and the p33 frameshift mutant both failed to exclude the parental wild-type virus. Plants that were preinfected with the mutant viruses and later challenged with CTV9-GFP showed strong GFP fluorescence (Fig. 5; Table 1, experiment C). These results indicated that the lack of p33 protein production eliminated the superinfection exclusion ability of the virus; thus, the p33 protein is a viral factor that functions in the CTV superinfection exclusion phenomenon.
Fig 4.
Replication of CTV9 (lane 1), CTV9p33ΔsgRNA (lane 2), or CTV9p33fs (lane 3) in N. benthamiana protoplasts at 4 days postinoculation. Northern blot hybridizations were carried out using CTV 3′ positive RNA strand-specific riboprobe. Positions of sgRNAs are shown. A big arrowhead indicates the position of p33 sgRNA; an asterisk shows the absence of the p33 sgRNA band in lane 2.
Fig 5.
Observation of GFP fluorescence in phloem-associated cells of C. macrophylla trees upon challenge with CTV9-GFP. The left image represents a noninoculated healthy tree. The other images represent trees with no primary infection (second image) or trees preinfected with the wild-type or mutant viruses, which were sequentially challenged with CTV9-GFP. Observations were done on the internal surface of bark at 4 months after challenge inoculation using a dissecting fluorescence microscope. Bars = 0.4 mm.
Heterologous p33 protein does not support superinfection exclusion.
Earlier, we demonstrated that modifications in the 3′ portion of the CTV genome, such as substitutions of the 3′ half sequences with the sequences derived from other virus strains, did not affect the superinfection ability of the virus: viruses with substitutions within this genomic region retained the ability to exclude superinfection by the parental virus (17). In the experiments above, I determined that p33 protein is required for superinfection exclusion. Our next question was whether the p33 protein functions in a homology-dependent manner, or in other words, whether a heterologous p33 would support the exclusion of superinfection by the parental wild-type virus. To examine how substitution of the p33 gene with a cognate gene from another strain would affect the ability of the virus to exclude the wild-type virus, I engineered two similar constructs in which the p33 ORF in the T36 cDNA clone pCTV9 was replaced with the p33 ORF sequence from an isolate of the T30 or the T68 strain. The sequences of the p33 ORF of both the T30-1 and T68-1 isolates have 84% identity with the p33 ORF sequence of the T36 isolate. The corresponding p33 proteins of the T30-1 and T68-1 isolates show 85.5 and 82.5% amino acid similarity with the T36 p33 protein, respectively (4, 17). The resulting hybrid viruses, CTV9p33T30 and CTV9p33T68 (Fig. 1), were used for primary inoculation of plants. Sets of control plants were inoculated with CTV9 and CTV9Δp33, with some plants not being inoculated. Upon establishment of the initial infections (Table 1, experiment C), the plants were challenged with the GFP-tagged CTV virus and further observed for GFP fluorescence. The hybrid viruses carrying substitutions of the p33 ORF with the sequences derived from heterologous strains failed to exclude the parental virus (Fig. 5; Table 1, experiment C). Similar to the p33 deletion, the p33 swap removed the ability of the virus to exclude superinfection by the parental virus. Plants that were preinfected with CTV9Δp33, CTV9p33T30, or CTV9p33T68 and challenged with CTV9-GFP virus displayed strong GFP fluorescence, indicating the inability of the mutant viruses to interfere with superinfection by the parental virus (Fig. 5; Table 1, experiment C).
DISCUSSION
The experiments that examined the ability of several mutant viruses to exclude superinfection by the wild-type virus demonstrated that superinfection exclusion by CTV requires production of a specific viral protein, the p33 protein. Modifications of the p33 gene which restricted production of the functional protein prevented superinfection exclusion. The virus mutants that failed to produce p33 failed to exclude superinfection by the parental wild-type virus. Superinfection exclusion was conferred by the p33 protein rather than the RNA sequence: the mutant viruses that retained the entire sequence of the p33 ORF, yet had a deletion of the subgenomic mRNA controller element for the p33 sgRNA or a frameshift mutation within the p33 ORF, failed to exclude the wild-type virus. The distribution and intensity of GFP fluorescence observed in trees preinfected with the p33 mutants and then challenged with the GFP-marked virus were comparable to those found upon inoculation of trees with no primary infection.
Remarkably, not only was the p33 protein required for superinfection exclusion, but the protein functioned in a homology-dependent manner. A cognate protein from a heterologous strain failed to confer the exclusion. Unlike substitutions of the 3′ half genes that had no impact on superinfection exclusion as was demonstrated in our previous work (17), the substitution of the p33 ORF with a cognate sequence from a different virus strain removed the ability of the virus to exclude the parental virus. The hybrid viruses with the p33 substitutions behaved similarly to the mutants that produced no p33: they were unable to interfere with the secondary infection by the wild-type virus. These data suggest that the p33 protein has a precise interaction(s) with some other viral factor(s) involved in this phenomenon.
Superinfection exclusion of viruses has been related to a number of different mechanisms acting at various stages of the viral life cycle, yet the phenomenon still remains obscure. Most of the proposed mechanisms, such as competition between primary and challenging viruses for host factors and intracellular replication sites or interference with disassembly, translation, or replication of the secondary virus (1, 2, 7, 35, 38, 41, 59, 60, 62, 63), could function only “locally,” in cells that were infected with the primary virus, leaving uninfected cells susceptible to the secondary virus. Based on our data, such mechanisms would not be relevant for superinfection exclusion by CTV due to the observed “systemic” nature of this phenomenon. A proposed model that could explain the exclusion of related viruses from uninfected cells is based on RNA silencing (48, 49; reviewed in references 21 and 29). RNA silencing has been considered as the major antiviral defense mechanism in plants and invertebrates (6, 39, 69, 72, 73). It can be induced systemically, not only in cells that contained the primary virus, but also in cells that were not preinfected with the primary virus. The mechanism targets nearly identical RNA sequences; thus, the introduction of homologous sequences, in some cases as short as 23 nucleotides, into genomes of heterologous viruses has been shown to induce degradation of RNA molecules containing those sequences (30, 49, 67, 72). For a number of plant viruses, RNA silencing was suggested as a mechanism that confers homologous interference of viruses (48, 49, 68; reviewed in references 21 and 29). Upon infection, the primary virus would act as an elicitor of the host defense machinery, which then would recognize and destroy homologous sequences, such as those present in the genomes of closely related viruses invading the same host, thus preventing the secondary infection by the latter viruses.
The “systemic” nature of superinfection exclusion by CTV parallels characteristics of RNA silencing. Thus, we previously attempted to trigger exclusion between heterologous CTV isolates by substituting various regions in the genome of the protecting virus with the exact cognate sequences (up to 3.7 kb in size) from the genome of the challenging virus. The hybrid viruses did not induce exclusion of the challenging virus: sharing of extended homologous RNA sequences did not confer exclusion of the secondary virus (17). The substituted regions contained 3′ end genes, which amplify large amounts of double-stranded RNAs (25, 45, 46). As has been demonstrated recently, this part of the CTV genome generates production of most viral small RNAs upon CTV infection (54). Yet, the hybrids in which these regions were substituted from the challenge isolate failed to exclude the latter isolate despite the fact that they shared extended identical sequences (17). These results do not appear to support the RNA silencing-based model. With the mutant viruses examined in this work, one could possibly suggest that the lack of superinfection exclusion by the p33 deletion mutant and the mutant containing deletion of the subgenomic promoter for the p33 sgRNA could be due to the absence of the p33 sgRNA, which results in failure to accumulate small RNAs from the p33 ORF region. However, the mutant that contained a frameshift mutation within the p33 ORF would be expected to produce small RNAs corresponding to this region from the p33 sgRNA, which was produced in amounts similar to that of the wild-type CTV. Yet, in contrast to the wild-type virus, the frameshift mutant failed to exclude the parental virus, as did the other two mutants that were also deficient in production of the p33 protein. More studies will be needed to determine whether superinfection exclusion by CTV involves components of the RNA silencing pathway or operates via another novel mechanism.
The data presented here demonstrate that superinfection exclusion by CTV is an active virus-controlled function. It is a powerful process that is capable of completely preventing infection by a challenging virus. It apparently involves a precise interaction between a virus-encoded p33 protein and other unknown factors. At this point, its effectiveness is limited to related viruses. However, with further understanding, it could possibly be deconstructed and reconstructed into a process to manage viruses in unforeseen ways. It is believed that this phenomenon can be further utilized for the development of new effective strategies for controlling diseases caused by this group of viruses and possibly by viruses from other taxonomic groups, including the development of new virus-based therapeutics to reduce the effect of viral diseases in the fields of medicine and agriculture.
ACKNOWLEDGMENTS
I thank William O. Dawson for insightful discussions, encouragement, and critical reading of the manuscript, W. O. Dawson and Satyanarayana Tatineni for providing deletion mutant viruses, Turksen Shilts for excellent technical assistance, and Siddarame Gowda for providing technical advice for engineering some of the constructs.
This research was supported by grant from the National Science Foundation (1050883; to Svetlana Y. Folimonova).
Footnotes
Published ahead of print 7 March 2012
REFERENCES
- 1. Abel PP, et al. 1986. Delay of disease development in transgenic plants that express the tobacco virus coat protein gene. Science 232:738–743 [DOI] [PubMed] [Google Scholar]
- 2. Adams RH, Brown DT. 1985. BHK cells expressing Sindbis virus-induced homologous interference allow the translation of nonstructural genes of superinfecting virus. J. Virol. 54:351–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Agranovsky AA. 1996. Principles of molecular organization, expression, and evolution of closteroviruses: over the barriers. Adv. Virus Res. 47:119–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Albiach-Martí MR, et al. 2000. Sequences of Citrus tristeza virus separated in time and space are essentially identical. J. Virol. 74:6856–6865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bar-Joseph MS, Garnsey SM, Gonsalves D. 1979. The closteroviruses: a distinct group of elongated plant viruses. Adv. Virus Res. 25:93–168 [DOI] [PubMed] [Google Scholar]
- 6. Baulcombe D. 2004. RNA silencing in plants. Nature 431:356–363 [DOI] [PubMed] [Google Scholar]
- 7. Beachy RN. 1999. Coat-protein-mediated resistance to tobacco mosaic virus: discovery mechanisms and exploitation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 354:659–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bendahmane M, Beachy RN. 1999. Control of tobamovirus infection via pathogen-derived resistance. Adv. Virus Res. 53:369–386 [DOI] [PubMed] [Google Scholar]
- 9. Bennett CW. 1953. Interactions between viruses and virus strains. Adv. Virus Res. 1:39–67 [DOI] [PubMed] [Google Scholar]
- 10. Bratt MA, Rubin H. 1968. Specific interference among strains of Newcastle disease virus. 3. Mechanism of interference. Virology 35:395–407 [DOI] [PubMed] [Google Scholar]
- 11. Delwart EL, Panganiban AT. 1989. Role of reticuloendotheliosis virus envelope glycoprotein in superinfection interference. J. Virol. 63:273–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dolja VV, Karasev AV, Koonin EV. 1994. Molecular biology and evolution of closteroviruses: sophisticated build-up of large RNA genomes. Annu. Rev. Phytopathol. 32:261–285 [Google Scholar]
- 13. Dolja VV, Kreuze JF, Valkonen JPT. 2006. Comparative and functional genomics of closteroviruses. Virus Res. 117:38–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ehrengruber MU, Goldin AL. 2007. Semliki Forest virus vectors with mutations in the nonstructural protein 2 gene permit extended superinfection of neuronal and non-neuronal cells. J. Neurovirol. 13:353–363 [DOI] [PubMed] [Google Scholar]
- 15. Folimonov AS, Folimonova SY, Bar-Joseph M, Dawson WO. 2007. A stable RNA virus-based vector for citrus trees. Virology 368:205–216 [DOI] [PubMed] [Google Scholar]
- 16. Folimonova SY, Folimonov AS, Tatineni S, Dawson WO. 2008. Citrus tristeza virus: survival at the edge of the movement continuum. J. Virol. 82:6546–6556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Folimonova SY, et al. 2010. Infection with strains of Citrus tristeza virus does not exclude superinfection by other strains of the virus. J. Virol. 84:1314–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Formella S, Jehle C, Sauder C, Staeheli P, Schwemmle M. 2000. Sequence variability of Borna disease virus: resistance to superinfection may contribute to high genome stability in persistently infected cells. J. Virol. 74:7878–7883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fulton RW. 1978. Superinfection by strains of tobacco streak virus. Virology 85:1–8 [DOI] [PubMed] [Google Scholar]
- 20. Fulton RW. 1986. Practices and precautions in the use of cross protection for plant virus disease control. Annu. Rev. Phytopathol. 24:67–81 [Google Scholar]
- 21. Gal-On A, Shiboleth YM. 2006. Cross protection, p 261–288 In Loebenstein G, Carr JP. (ed), Natural resistance mechanisms of plants to viruses. Springer, Dordrecht, The Netherlands [Google Scholar]
- 22. Garnsey S, Cambra M. 1991. Enzyme-linked immunosorbent assay (ELISA) for citrus pathogens, p 193–216 In Roistacher CN. (ed), Graft-transmissible diseases of citrus. Handbook for detection and diagnosis, FAO, Rome, Italy [Google Scholar]
- 23. Geib T, et al. 2003. Selective virus resistance conferred by expression of Borna disease virus nucleocapsid components. J. Virol. 77:4283–4290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Harper SJ, Dawson TE, Pearson MN. 2010. Isolates of citrus tristeza virus that overcome Poncirus trifoliata resistance comprise a novel strain. Arch. Virol. 155:471–480 [DOI] [PubMed] [Google Scholar]
- 25. Hilf ME, et al. 1995. Characterization of citrus tristeza virus subgenomic RNAs in infected tissue. Virology 208:576–582 [DOI] [PubMed] [Google Scholar]
- 26. Hilf ME, Mavrodieva VA, Garnsey SM. 2005. Genetic marker analysis of a global collection of isolates of Citrus tristeza virus: characterization and distribution of CTV genotypes and association with symptoms. Phytopathology 95:909–917 [DOI] [PubMed] [Google Scholar]
- 27. Huang I-C, et al. 2008. Influenza A virus neuraminidase limits viral superinfection. J. Virol. 82:4834–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hull R, Plaskitt A. 1970. Electron microscopy on the behaviour of two strains of alfalfa mosaic virus in mixed infections. Virology 42:773–776 [DOI] [PubMed] [Google Scholar]
- 29. Hull R. 2002. Matthews' plant virology. Academic Press, New York, NY [Google Scholar]
- 30. Jan FJ, Fagoaga C, Pang SZ, Gonsalves D. 2000. A minimum length of N gene sequence in transgenic plants is required for RNA-mediated tospovirus resistance. J. Gen. Virol. 81:235–242 [DOI] [PubMed] [Google Scholar]
- 31. Johnston RE, Wan K, Bose HR. 1974. Homologous interference induced by Sindbis virus. J. Virol. 14:1076–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Karasev AV, et al. 1995. Complete sequence of the citrus tristeza virus RNA genome. Virology 208:511–520 [DOI] [PubMed] [Google Scholar]
- 33. Karasev A, Hilf ME, Garnsey SM, Dawson WO. 1997. Transcriptional strategy of closteroviruses: mapping the 5′ termini of the citrus tristeza virus subgenomic RNAs. J. Virol. 71:6233–6236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karasev AV. 2000. Genetic diversity and evolution of closteroviruses. Annu. Rev. Phytopathol. 38:293–324 [DOI] [PubMed] [Google Scholar]
- 35. Karpf AR, Lenches E, Strauss EG, Strauss JH, Brown DT. 1997. Superinfection exclusion of alphaviruses in three mosquito cell lines persistently infected with Sindbis virus. J. Virol. 71:7119–7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kong P, Rubio L, Polek M, Falk BW. 2000. Population structure and genetic diversity within California Citrus tristeza virus (CTV) field isolates. Virus Genes 21:139–145 [DOI] [PubMed] [Google Scholar]
- 37. Lecoq H, Lemaire JM, Wipf-Scheibel C. 1991. Control of zucchini yellow mosaic virus in squash by cross protection. Plant Dis. 75:208–211 [Google Scholar]
- 38. Lee YM, Tscherne DM, Yun SI, Frolov I, Rice CM. 2005. Dual mechanisms of pestiviral superinfection exclusion at entry and RNA replication. J. Virol. 79:3231–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li H-W, Ding S-W. 2005. Antiviral silencing in animals. FEBS Lett. 579:5965–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. López CM, et al. 1998. Molecular variability of the 5′ and 3′ terminal regions of citrus tristeza virus RNA. Phytopathology 88:685–691 [DOI] [PubMed] [Google Scholar]
- 41. Lu B, Stubbs G, Culver JN. 1998. Coat protein interactions involved in tobacco mosaic tobamovirus cross-protection. Virology 248:188–198 [DOI] [PubMed] [Google Scholar]
- 42. Lu R, et al. 2004. Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc. Natl. Acad. Sci. U. S. A. 101:15742–15747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mawassi M, Mietkiewska E, Gofman R, Yang G, Bar-Joseph M. 1996. Unusual sequence relationships between two isolates of citrus tristeza virus. J. Gen. Virol. 77:2359–2364 [DOI] [PubMed] [Google Scholar]
- 44. Mlotshwa S. 2002. RNA silencing and the mobile silencing signal. Plant Cell 14:S289–S301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moreno P, Guerri J, Muñoz N. 1990. Identification of Spanish strains of citrus tristeza virus (CTV) by analysis of double-stranded RNAs (dsRNA). Phytopathology 80:477–482 [Google Scholar]
- 46. Moreno P, Ambros S, Albiach-Marti MR, Guerri J, Pena L. 2008. Citrus tristeza virus: a pathogen that changed the course of the citrus industry. Mol. Plant Pathol. 9:251–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pappu HR, et al. 1994. Nucleotide sequence and organization of eight 3′ open reading frames of the citrus tristeza closterovirus genome. Virology 199:35–46 [DOI] [PubMed] [Google Scholar]
- 48. Ratcliff F, Harrison BD, Baulcombe DC. 1997. A similarity between viral defense and gene silencing in plants. Science 276:1558–1560 [DOI] [PubMed] [Google Scholar]
- 49. Ratcliff F, MacFarlane S, Baulcombe DC. 1999. Gene silencing without DNA. RNA-mediated cross-protection between viruses. Plant Cell 11:1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Robertson C, Garnsey SM, Satyanarayana T, Folimonova S, Dawson WO. 2005. Efficient infection of citrus plants with different cloned constructs of Citrus tristeza virus amplified in Nicotiana benthamiana protoplasts, p 187–195 Proc. 16th Conf. Int. Org. Citrus Virol IOCV, Riverside, CA [Google Scholar]
- 51. Rocha-Peña MA, et al. 1995. Citrus tristeza virus and its vector Toxoptera citricida. Plant Dis. 79:437–445 [Google Scholar]
- 52. Roy A, Manjunath KL, Brlansky RH. 2005. Assessment of sequence diversity in the 5′-terminal region of Citrus tristeza virus from India. Virus Res. 113:132–142 [DOI] [PubMed] [Google Scholar]
- 53. Rubio L, et al. 2001. Genetic variation of citrus tristeza virus isolates from California and Spain: evidence for mixed infections and recombination. J. Virol. 75:8054–8062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ruiz-Ruiz S, et al. 2011. Citrus tristeza virus infection induces the accumulation of viral small RNAs (21-24-nt) mapping preferentially at the 3′-terminal region of the genomic RNA and affects the host small RNA profile. Plant Mol. Biol. 75:607–619 [DOI] [PubMed] [Google Scholar]
- 55. Salaman RN. 1933. Protective inoculation against a plant virus. Nature 131:468 [Google Scholar]
- 56. Satyanarayana T, et al. 1999. An engineered closterovirus RNA replicon and analysis of heterologous terminal sequences for replication. Proc. Natl. Acad. Sci. U. S. A. 96:7433–7438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Satyanarayana T, et al. 2000. Closterovirus encoded HSP70 homolog and p61 in addition to both coat proteins function in efficient virion assembly. Virology 1278:253–265 [DOI] [PubMed] [Google Scholar]
- 58. Satyanarayana T, Gowda S, Ayllon MA, Dawson WO. 2003. Frameshift mutations in infectious cDNA clones of Citrus tristeza virus: a strategy to minimize the toxicity of viral sequences to Escherichia coli. Virology 313:481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schaller TN, et al. 2007. Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes. J. Virol. 81:4591–4603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sherwood JL, Fulton RW. 1982. The specific involvement of coat protein in tobacco mosaic virus crossprotection. Virology 119:150–158 [DOI] [PubMed] [Google Scholar]
- 61. Singh IR, Suomalainen M, Varadarajan S, Garoff H, Helenius A. 1997. Mechanisms for the inhibition of entry and uncoating of superinfecting Semliki forest virus. Virology 231:59–71 [DOI] [PubMed] [Google Scholar]
- 62. Steck FT, Rubin H. 1996. The mechanism of interference between an avian leukosis virus and Rous sarcoma virus. I. Establishment of interference. Virology 29:628–641 [DOI] [PubMed] [Google Scholar]
- 63. Steck FT, Rubin H. 1966. The mechanism of interference between an avian leukosis virus and Rous sarcoma virus. II. Early steps of infection by RSV of cells under conditions of interference. Virology 29:642–653 [DOI] [PubMed] [Google Scholar]
- 64. Strauss JH, Strauss EG. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tatineni S, et al. 2008. Three genes of Citrus tristeza virus are dispensable for infection and movement throughout some varieties of citrus trees. Virology 376:297–307 [DOI] [PubMed] [Google Scholar]
- 66. Tatineni S, Robertson CJ, Garnsey SM, Dawson WO. 2011. A plant virus evolved by acquiring multiple nonconserved genes to extend its host range. Proc. Natl. Acad. Sci. U. S. A. 108:17366–17371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Thomas CL, Jones LDC, Maule AJ. 2001. Size constraints for targeting post-transcriptional gene silencing and for using RNA-directed methylation in N. benthamiana using a potato virus X vector. Plant J. 25:417–425 [DOI] [PubMed] [Google Scholar]
- 68. Valkonen JP, Rajamäki ML, Kekarainen T. 2002. Mapping of viral genomic regions important in cross-protection between strains of a potyvirus. Mol. Plant Microbe Interact. 15:683–692 [DOI] [PubMed] [Google Scholar]
- 69. Vance V, Vaucheret H. 2001. RNA silencing in plants defense and counterdefense. Science 292:2277–2280 [DOI] [PubMed] [Google Scholar]
- 70. Vives MC, et al. 1999. The complete genome sequence of the major component of a mild citrus tristeza virus isolate. J. Gen. Virol. 80:811–816 [DOI] [PubMed] [Google Scholar]
- 71. Vives MC, et al. 2005. Evidence of multiple recombination events between two RNA sequence variants within a Citrus tristeza virus isolate. Virology 331:232–237 [DOI] [PubMed] [Google Scholar]
- 72. Voinnet O. 2001. RNA silencing as a plant immune system against viruses. Trends Genet. 17:449–459 [DOI] [PubMed] [Google Scholar]
- 73. Voinnet O. 2005. Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6:206–220 [DOI] [PubMed] [Google Scholar]
- 74. Walkey DGA, Lecoq H, Collier R, Dobson S. 1992. Studies on the control of zucchini yellow mosaic virus in courgettes by mild strain protection. Plant Pathol. 41:762–771 [Google Scholar]
- 75. Wen F, Lister M, Fattouh FA. 1991. Cross-protection among strains of barley yellow dwarf virus. J. Gen. Virol. 72:791–799 [DOI] [PubMed] [Google Scholar]
- 76. Whitaker-Dowling PA, Youngner JS, Widnell CC, Wilcox DK. 1983. Superinfection exclusion by vesicular stomatitis virus I. Virology 131:137–143 [DOI] [PubMed] [Google Scholar]
- 77. Wildum SM, Schindler M, Munch J, Kirchhoff F. 2006. Contribution of Vpu, Env, and Nef to CD4 down-modulation and resistance of human immunodeficiency virus type I-infected T cells to superinfection. J. Virol. 80:8047–8059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ziebell H, Carr JP. 2010. Cross-protection: a century of mystery. Adv. Virus Res. 76:211–264 [DOI] [PubMed] [Google Scholar]