Abstract
All positive-strand RNA viruses reorganize host intracellular membranes to assemble their replication complexes. Similarly, brome mosaic virus (BMV) induces two alternate forms of membrane-bound RNA replication complexes: vesicular spherules and stacks of appressed double-membrane layers. The mechanisms by which these membrane rearrangements are induced, however, remain unclear. We report here that host ACB1-encoded acyl coenzyme A (acyl-CoA) binding protein (ACBP) is required for the assembly and activity of both BMV RNA replication complexes. ACBP is highly conserved among eukaryotes, specifically binds to long-chain fatty acyl-CoA, and promotes general lipid synthesis. Deleting ACB1 inhibited BMV RNA replication up to 30-fold and resulted in formation of spherules that were ∼50% smaller but ∼4-fold more abundant than those in wild-type (wt) cells, consistent with the idea that BMV 1a invaginates and maintains viral spherules by coating the inner spherule membrane. Furthermore, smaller and more frequent spherules were preferentially formed under conditions that induce layer formation in wt cells. Conversely, cellular karmella structures, which are arrays of endoplasmic reticulum (ER) membranes formed upon overexpression of certain cellular ER membrane proteins, were formed normally, indicating a selective inhibition of 1a-induced membrane rearrangements. Restoring altered lipid composition largely complemented the BMV RNA replication defect, suggesting that ACBP was required for maintaining lipid homeostasis. Smaller and more frequent spherules are also induced by 1a mutants with specific substitutions in a membrane-anchoring amphipathic α-helix, implying that the 1a-lipid interactions play critical roles in viral replication complex assembly.
INTRODUCTION
All positive-strand RNA viruses assemble their replication complexes on host intracellular membranes, which are usually rearranged by viral proteins as single- or double-membrane vesicles, convoluted membrane webs, or other membrane rearrangements (13, 52, 67). Recent three-dimensional (3-D) electron microscope tomography has revealed critical aspects of the ultrastructure and organization of the membrane-bound viral replication complexes of Flock House virus (FHV) (39), severe acute respiratory syndrome (SARS) coronavirus (35), and dengue virus (DENV) (79). However, the mechanisms by which membranes are remodeled and the roles that host factors play in this process are not well understood. Moreover, the lipid microenvironment of the viral RNA replication complexes is not well defined.
Lipids are the major components of cellular membranes and thus play critical roles in viral RNA replications. The entry, replication, and secretion of hepatitis C virus (HCV) require cholesterol synthesis (84), which is also necessary for replication of tomato bushy stunt virus (TBSV) (70). Continuous fatty acid (FA) synthesis has been demonstrated to be necessary for replication of multiple positive-strand RNA viruses, including poliovirus (22), Semliki Forest virus (SFV) (55), HCV (33), Drosophila C virus (11), and cowpea mosaic virus (6). In addition, viruses stimulate lipid synthesis to accommodate formation of their replication complexes. Increased phosphatidylcholine (PC) synthesis is induced upon FHV replication (7) and poliovirus infection (53, 74). Similarly, DENV infection promotes a 3-fold increase of total FA synthesis by recruiting host fatty acid synthase (FASN) to viral replication complexes via an interaction between DENV nonstructural protein 3 (NS3) and FASN (25). Better understanding of the interaction between lipid synthesis/composition and viral replication complex assembly and function should provide insights into the mechanisms of membrane rearrangements and identify novel host targets to develop critical antiviral strategies.
Brome mosaic virus (BMV) is a representative member of the alphavirus-like superfamily of human, animal, and plant viruses and has served as a model to study viral replication mechanisms, virus-host interactions, and many other aspects of positive-strand RNA virus infection (reviewed in reference 76). BMV has a tripartite genome and a subgenomic RNA, RNA4. Genomic RNA1 and RNA2 encode the viral proteins required for BMV RNA replication, 1a and 2apol, respectively. BMV 1a has an N-terminal RNA capping domain (1, 38) and a C-terminal nucleoside triphosphatase (NTPase)/helicase-like domain (78). BMV 2apol contains a central RdRp domain and an N-terminal domain that interacts with 1a's NTPase/helicase-like domain (10, 32, 54). RNA3 and subgenomic RNA4 encode the 3a protein and coat protein, respectively, which are required for systemic movement but not for replication (76).
BMV replication induces formation of viral RNA replication compartments, termed spherules, that are the invaginations of the outer perinuclear endoplasmic reticulum (ER) membranes into the ER lumen (68). Spherules are ∼60 to 80 nm in diameter with an ∼10-nm neck connecting the interior of the spherules to the cytoplasm and are the sites where BMV RNA replication takes place. BMV 1a is the only viral component responsible for inducing the formation of viral spherules (68). BMV 1a's membrane association is primarily mediated by an amphipathic α-helix termed helix A, which binds to lipid membrane-mimicking SDS micelles as a stable α-helix (48). BMV 1a also directs 2apol and viral RNA replication templates to viral spherules via the 1a-2apol interaction and by recognizing the recruitment element (RE) cis element present only in BMV genomic RNAs, respectively (10, 14, 40, 62, 63, 68, 73). While viral spherules are the dominant form of the viral replication complexes, increased expression levels of 2apol switch vesicular spherules to appressed stacks of double-membrane layers surrounding the nucleus with an intermembrane distance of ∼50 to 70 nm (69). While morphologically distinct, spherule and layer replication complexes support comparable levels of BMV RNA replication (69).
Lipid synthesis and composition play pivotal roles in BMV replication as well. Expression of 1a enhances total FA accumulation in Saccharomyces cerevisiae yeast by ∼30% per cell, reflecting the presence of numerous spherules (45). Furthermore, a single substitution in OLE1-encoded Δ9 fatty acid desaturase, which converts saturated FA (SFA) to unsaturated FA (UFA), blocks BMV RNA replication by up to 95% (46) and yet only mildly reduces UFA content (by ∼12%) and has no effect on either cell growth or morphology (45). The decreased levels of UFA preferentially affect the lipid composition of membranes surrounding BMV spherules (45), explaining the fact that BMV RNA replication is more sensitive to the altered lipid composition than is host growth.
A screen of a yeast deletion library showed that multiple host genes involved in membrane synthesis and trafficking are required for BMV RNA replication (41). This report focuses on host ACB1, which encodes Acb1p, the yeast ortholog of acyl coenzyme A (acyl-CoA) binding protein (ACBP). ACBP is ubiquitously expressed in all tissues of higher eukaryotes and is highly conserved in humans, plants, and yeast (21, 36). ACBP selectively binds to C14-C22 long-chain fatty acyl-CoAs (LCFA-CoAs) with a remarkably high affinity and thus regulates their stability, metabolism, and transport (36, 43, 59). By binding to LCFA-CoAs, ACBP plays crucial roles in lipid transport, regulation, and synthesis (21, 36). In in vitro assays, ACBP stimulates the activities of many enzymes involved in the synthesis of fatty acids, cholesterols, and phospholipids, including acetyl-CoA carboxylase and FASN (18, 60), acyl-CoA cholesterol acyltransferase II (9), and glycerol-3-phosphate acyltransferase (8), respectively. ACBP is essential in mammals since knocking out hc ACBP gene causes embryonic lethality in transgenic mice (43). Moreover, transfecting several human cell lines with ACBP-specific small interfering RNA (siRNA) leads to cell death (17).
The role of ACBP in viral infection is not well known. ACBP levels are reduced slightly in HIV-infected cells compared to controls (58). In hepatitis B virus-transgenic mice, ACBP protein levels are higher than those in healthy ones (82). Nevertheless, the role that ACBP plays in both virus infections is not clear. Additionally, the requirement for ACBP in genomic replication of any virus has not been documented. We report here that deleting ACB1 specifically altered the proper formation and function of BMV-induced spherule and layer replication complexes but not formation of the cellular structures karmellae. BMV RNA replication phenotypes in cells lacking ACB1, including smaller but significantly more abundant spherules and reduced 2apol accumulation, are similar to those caused by specific amino acid substitutions in 1a's membrane-interacting helix A (48), suggesting that deletion of ACB1 and certain substitutions in 1a affect similar requirements that are necessary for BMV RNA replication. Our lipid complementation data indicate that altered lipid composition in cells lacking ACB1 plays a major role in inhibited BMV RNA replication, suggesting that the 1a-lipid interaction plays important roles in both cases.
MATERIALS AND METHODS
Yeast strain and cell growth.
Yeast strain BY4743 (MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0) was used in the experiments shown in Fig. 1 to 3, and strain YPH500 (MATα ura3-52 lys2-801 ada2-101 trp1-Δ63 his3-Δ200 leu2-Δ1) was used in all experiments. To make the ACB1 deletion mutant in a YPH500 background, the KanMX6 cassette from BY4743 acb1::kanMX6 genomic DNA flanked by 5′ and 3′ homologous recombination regions was amplified and used to delete ACB1 in the YPH500 strain. Cultures were grown at 30°C in defined synthetic medium containing 2% galactose as a carbon source. Leucine, uracil, histidine, or combinations thereof were omitted to maintain selection for different plasmids. Cells were grown in galactose medium for 2 passages (24 to 48 h) and harvested when the optical density at 600 nm (OD600) reached between 0.4 and 1.0. Tergitol NP-40 was added to a final concentration of 1% to solubilize the fatty acids into the growth medium. Palmitoleic acid (16:1), oleic acid (18:1), palmitic acid (16:0), and stearic acid (18:0) were added to NP-40-containing medium to the specified concentrations (72).
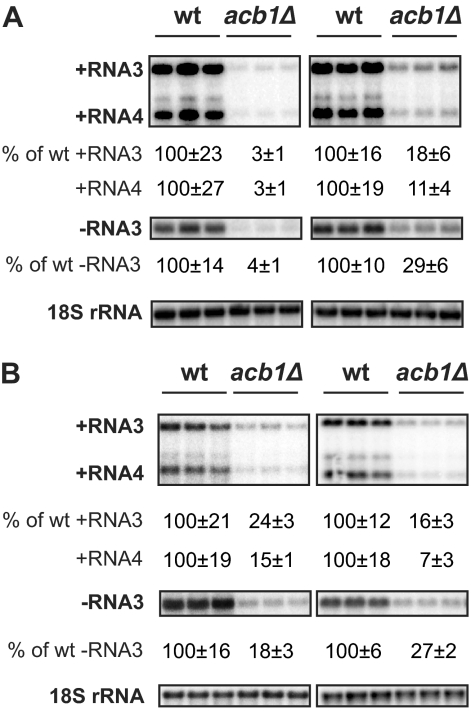
Fig 1.
Deleting host ACB1 inhibits BMV RNA replication. BMV components were expressed under conditions that induced viral spherules (left panel) or layer replication complexes (right panel) in wt cells. (A) BMV replication was assayed in BY4743, the strain in which the yeast deletion library was generated. (B) BMV RNA replication in strains derived from YPH500. BMV positive- and negative-strand RNAs were detected by using probes specific to BMV RNAs. 18S rRNA was detected by using an 18S rRNA probe to serve as loading control. BMV RNA signals were normalized to that of 18S rRNA to eliminate loading variations. Negative-strand RNA blots in all figures were exposed longer than were those for positive-strand RNAs for comparison purposes. All experiments here and in the following figures were repeated multiple times in duplicate or triplicate each time. A representative blot is shown in each figure.
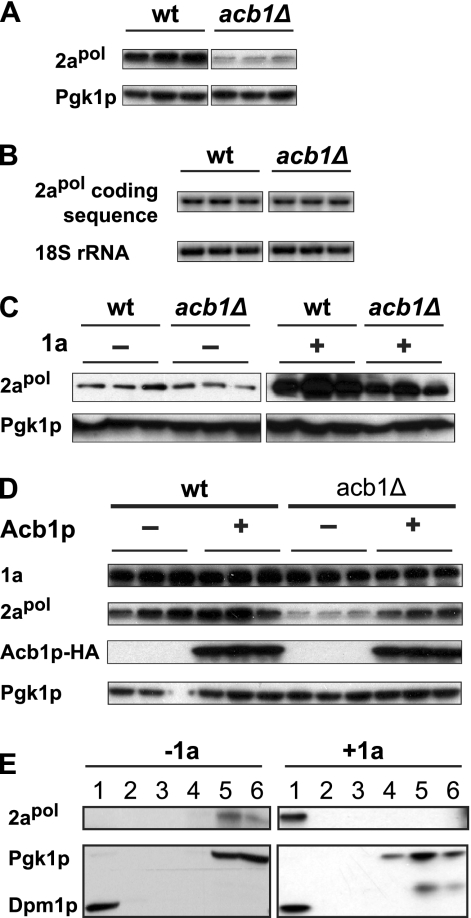
Fig 3.
Accumulation of 2apol is affected by deleting ACB1. (A) Accumulation of 2apol in wt and acb1Δ cells. Total proteins were extracted from equal numbers of yeast cells expressing all BMV components. Accumulation of 2apol and Pgk1p was detected as described for Fig. 2A by using anti-2a and anti-Pgk1p antibodies. (B) Accumulation of 2apol transcripts. In cells expressing 2apol and 1a, 2apol transcripts were detected by using a probe specific to 2apol coding sequences. (C) 1a-mediated 2apol stabilization. 2apol under the control of the GAL1 promoter was expressed on its own (left panel) or along with 1a (right panel). Pgk1p and 2apol were detected as described for panel A. (D) Reduced 2apol accumulation was complemented by expressing HA-tagged Acb1p. Detection of 1a, 2apol, and Pgk1p was performed as described for panel A. Acb1p was detected using an anti-HA antibody. (E) Membrane association of 2apol in the absence and presence of 1a in acb1Δ cells. 2apol was expressed alone or along with 1a and detected in membrane fractions by a membrane flotation assay as described for Fig. 2B. To detect 2apol in the absence of 1a, larger lysate volumes were loaded and the blot was exposed longer than in the presence of 1a. Assays in panels A and D were performed under spherule-forming conditions, and those in panels B, C, and E were performed under layer-forming conditions.
Plasmids and plasmid construction.
BMV RNA3 was launched from pB3VG128-H, in which RNA3 is driven by the CUP1 promoter, for BMV RNA replication without addition of copper (77). To analyze 1a-mediated RNA3 stability, we used plasmid pB3MS82-H, which expresses BMV RNA3 from the GAL1 promoter (77). To assay BMV RNA replication under spherule-forming conditions, 1a and 2apol were expressed from pB12VG1 (41). 1a and 2apol were expressed from pB1YT3 and pB2YT5, respectively, for BMV RNA replication under layer-forming conditions. N-terminally tagged Myc-Mga2p was expressed from YEpLac181-mycMGA2 (57). Constructs overexpressing hemagglutinin (HA)-tagged Acb1p (Acb1p-HA) and Hmg1p were purchased from Open Biosystems, and both open reading frames (ORFs) were inserted into pBG1805, a 2μ plasmid, and were under the control of the GAL1 promoter.
RNA extraction and analysis.
Yeast cells were harvested at OD600 values of 0.4 to 1.0, and total RNA was extracted using the hot phenol method (37). Equal amounts of total RNA were used for Northern blotting. BMV positive- and negative-strand RNAs and host OLE1 transcripts were detected using probes specific to BMV RNAs and OLE1, respectively. 18S rRNA was detected using an 18S rRNA probe to serve as a loading control. BMV RNA signals were normalized to that of 18S rRNA to eliminate loading variations. Negative-strand RNA blots in all figures were exposed longer than those for positive-strand RNAs for comparison purposes.
Western blotting.
Yeast cells were grown to OD600 values of 0.4 to 1.0, and 2 OD600 units of cells were harvested. Total proteins were extracted as described previously (46), and equal volumes of extracted total proteins were used for electrophoresis and transferred to a polyvinylidene difluoride (PVDF) membrane. Expression of target proteins was detected with the following antibodies and dilutions: rabbit anti-BMV 1a at 1:10,000, mouse anti-BMV 2apol at 1:4,000, mouse anti-Pgk1p (Molecular Probes; A6457) at 1:10,000, mouse anti-Dpm1p (Molecular Probes; A6429) at 1:1,000, mouse anti-Myc antibody at 1:3,000 (Calbiochem; OP10), and mouse anti-HA (Invitrogen; 32-6700) at 1:2,000; detection used horseradish peroxidase (HRP)-conjugated secondary antibodies at 1:6,000 and Supersignal West Femto substrate (Thermo Scientific).
Membrane flotation assay.
Yeast spheroplasts prepared from 10 OD600 units of cells were lysed at 4°C in TNM buffer (50 mM Tris-Cl [pH 7.4], 150 mM NaCl, 5 mM MgCl2, and 1:200 dilution of protease inhibitor cocktail [Sigma P8215]). The resulting cell lysate was centrifuged for 5 min at 500 × g to remove cell debris. Supernatant was adjusted to 40% iodixanol by addition of 60% iodixanol (Optiprep; Sigma D1556). An 0.6-ml amount of the mixture was placed at the bottom of a Beckman TLS55 centrifuge tube and overlaid with 1.4 ml of 30% iodixanol in TNM and 100 μl of TNM. The gradients were centrifuged at 55,000 rpm (201,000 × g) at 4°C for 5 h. The gradients were divided into 6 fractions and analyzed by Western blotting for specific proteins.
Fatty acid analysis.
Fatty acid methyl esters (FAME) were prepared as specified in the work of Lightner et al. (47) with minor modifications. Five OD600 units of cells growing in 1% NP-40 was harvested and washed once with H2O. The pellets were heated at 80°C for 2 h in 2 ml of 2.5% H2SO4 in methanol in screw-cap glass tubes with 100 μg sabic acid as an internal control. In the above mixtures, 1.5 ml of H2O and 0.5 ml hexane were added, vortexed vigorously, and spun for 5 min at low speed to separate the FAME-containing hexane fraction from the rest of the mixture. The FAME preparations were then analyzed by gas chromatography (Agilent 6890).
Electron microscopy.
Yeast cell fixation, dehydration, and embedding were performed as previously described (68). Yeast cells were first fixed with 4% paraformaldehyde and 2% glutaraldehyde for 1 h and then fixed with 1% osmium tetroxide. After dehydration through a gradient of ethanol, samples were embedded in Spurr's resin (Electron Microscopy Sciences). Thin sections (70 nm) were stained with uranyl acetate and lead citrate. Images were obtained using a Philips CM120 transmission electron microscope at the Medical School Electron Microscopy Facility of the University of Wisconsin—Madison. The diameter of spherules was measured using ITEM analysis (Soft Imaging Systems).
RESULTS
Deletion of host ACB1 inhibits BMV RNA replication.
A previous genome-wide yeast deletion mutant screen used a BMV RNA3 derivative expressing Renilla luciferase (RLuc) as a viral replication reporter. The BMV RNA3 (RLuc) was expressed along with 1a and low levels of 2apol, a combination that leads to the formation of spherular viral replication complexes. BMV RNA replication-dependent expression of RLuc decreased 10-fold in cells lacking ACB1 (acb1Δ) (41). To validate this large-scale screening result and to eliminate the possible complications introduced by RLuc, we expressed the wild-type (wt) RNA3, along with 1a and low levels of 2apol, in both wt and acb1Δ cells. BMV RNA-specific probes detected strong signals for negative-strand RNA3 and dramatically amplified positive-strand RNA4 and RNA3 in wt cells (Fig. 1), indicating full BMV RNA replication. In acb1Δ cells, BMV negative-strand RNA3 accumulated to only 4 to 10% of wt levels, indicating that BMV RNA replication was severely inhibited as early as negative-strand RNA synthesis. BMV positive-strand RNA3 and RNA4 accumulation was detected at only 3 to 10% of wt levels (Fig. 1A).
BMV RNA replication induces two forms of the viral replication complexes, spherules and layers, depending on the levels of 2apol expressed in yeast cells (68, 69). Expressing 2apol under the control of the strong GAL1 promoter (GAL1 2apol hereafter) in the presence of 1a leads to the formation of double-membrane layer replication complexes. In acb1Δ cells, inhibition of BMV RNA replication under layer-forming conditions was not as severe as that under spherule-forming conditions. While negative-strand RNA3 was inhibited to 30% of wt levels, accumulation of positive-strand RNAs was much reduced compared to that of negative-strand RNA synthesis, as RNA4 was reduced to 10% while RNA3 was reduced to 20% of wt levels (Fig. 1A).
The yeast deletion library used in a previous screen was based on yeast strain BY4743 (41). Under BMV RNA replication conditions, the doubling time of acb1Δ yeast from a BY4743 background was 4.8 ± 0.2 h compared to 4.4 ± 0.3 h in wt cells. A previous study reported that acb1Δ cells from certain genetic backgrounds adapted from a slow-growth phenotype to a high growth rate at a frequency of >10−5 (19). To confirm that the inhibition in BMV RNA replication was due to a direct function of ACBP and not to a growth phenotype, we deleted ACB1 in strain YPH500, a strain background that has been used in the majority of previous BMV studies (45, 46, 48, 68, 69). During BMV RNA replication, cell growth was significantly affected in YPH500 yeast cells lacking ACB1, resulting in an ∼40% increase in doubling time (12). Moreover, the slow-growth phenotype is stable, as we have not observed any growth adaptations. In YPH500 cells lacking ACB1, BMV RNA replication was also significantly inhibited (Fig. 1B; see also Fig. 7B and C). While negative-strand RNA3 decreased 4- to 5-fold under both spherule- and layer-forming conditions, BMV positive-strand RNA4 accumulation was reduced by 13-fold under layer-forming conditions and by 7-fold under spherule-forming conditions (Fig. 1B). Of note, the inhibition in BMV RNA replication levels under layer-forming conditions in acb1Δ cells from a BY4743 background and that in cells from a YPH500 background were very similar (Fig. 1). In contrast, under spherule-forming conditions, deleting ACB1 inhibited BMV RNA replication 5-fold more in terms of positive-strand RNA4 in the BY4743 background than in the YPH500 background (Fig. 1).
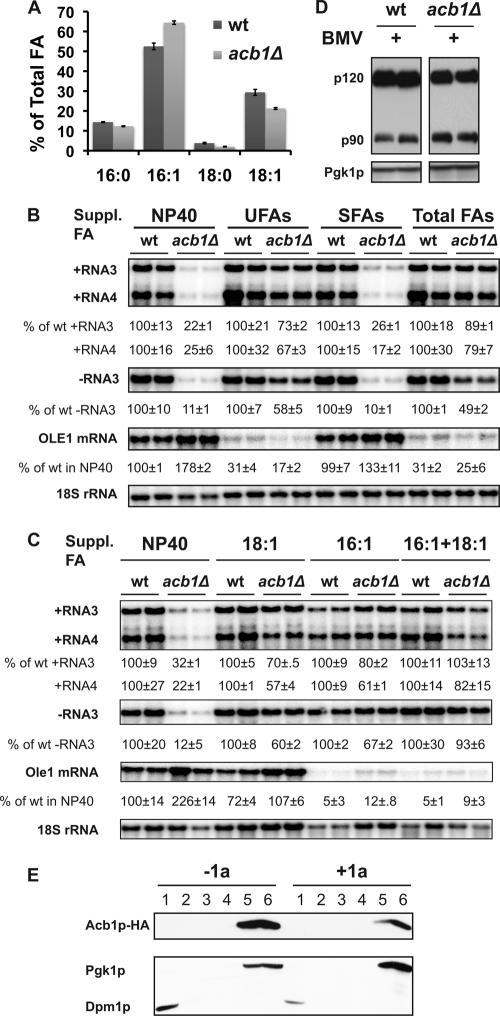
Fig 7.
The BMV RNA replication defect is largely complemented by supplementation with long-chain unsaturated fatty acids. (A) Fatty acid composition of wt and acb1Δ cells grown in Tergitol NP-40 with components supporting BMV RNA replication in spherules. (B) Supplementation with UFAs complemented the BMV RNA replication defect in acb1Δ cells. An 0.25 mM concentration of the specified FAs was provided in yeast growth medium with Tergitol NP-40 as a solvent. NP-40, no FA; UFAs, 16:1- plus 18:1-FA; SFAs, 16:0- plus 18:0-FA; Total FAs, all 4 FAs. BMV negative- and positive-strand RNAs as well as OLE1 transcripts were detected using specific probes. (C) Supplementation with 16:1- and/or 18:1-FA largely complemented the BMV RNA replication defect. To account for loading differences between the samples, viral RNAs and OLE1 transcript levels were normalized to 18S rRNA. (D) Activation of Mga2p in wt and acb1Δ cells. In wt and acb1Δ cells expressing BMV components and Myc-tagged Mga2p, the active and inactive forms of Mga2p, p90 and p120, respectively, were detected with an anti-Myc antibody. (E) Membrane association of Acb1p in the absence and presence of BMV 1a. Acb1p-HA was expressed alone or along with 1a and detected in soluble protein fractions by a membrane flotation assay as described for Fig. 2B.
1a-stimulated 2apol accumulation is reduced in acb1Δ cells.
To identify the specific step(s) of BMV RNA replication affected by deleting ACB1, we checked for the accumulation, interaction, and membrane association of 1a and 2apol as well as 1a-mediated RNA3 protection, steps that occur before negative-strand RNA synthesis during BMV RNA replication.
Accumulation of 1a was not altered in the mutant cells compared to that in wt (Fig. 2A), indicating that deleting ACB1 did not affect the stability of 1a. Upon translation, 1a targets itself to the perinuclear ER membranes and invaginates the outer perinuclear ER membranes to induce spherule formation (68). To determine whether 1a is membrane associated in the absence of ACB1, we performed a membrane flotation assay in which cell lysates from wt or acb1Δ cells were loaded at the bottom of a density gradient. Upon centrifugation, membrane-integral and -associated proteins, such as Dpm1p, float to the top fractions along with low-density membranes while soluble proteins, e.g., Pgk1p, remain in the bottom fractions (Fig. 2B). When expressed alone or along with low (data not shown) or high (Fig. 2B) levels of 2apol, 1a was detected in the top two fractions in both wt and acb1Δ cells, indicating that 1a is membrane associated.
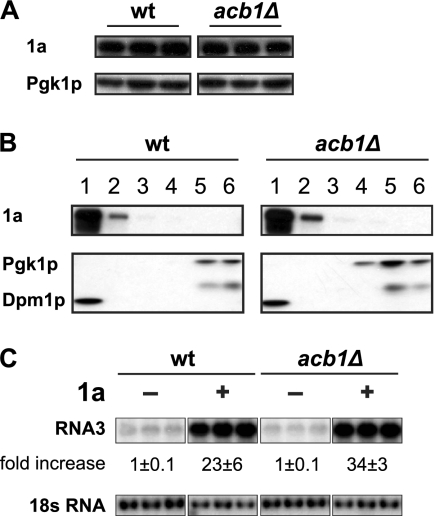
Fig 2.
BMV 1a is associated with membranes and stabilizes RNA3 in acb1Δ cells. (A) BMV 1a accumulation in wild-type (wt) and acb1Δ cells. Total proteins were extracted from equal numbers of yeast cells expressing all BMV components under spherule-forming conditions and analyzed by SDS-PAGE and immunoblot analyses with anti-1a antiserum and anti-Pgk1p antibody. (B) Membrane association of 1a protein. Lysates of wt or acb1Δ cells expressing all BMV components (under layer-forming conditions) were subjected to an iodixanol density gradient and analyzed by SDS-PAGE and Western blotting, using antibodies against ER membrane protein Dpm1p or soluble protein Pgk1p or antiserum against BMV 1a. (C) BMV RNA3 accumulation in the absence or presence of 1a. BMV RNA3 was expressed alone or along with 1a and detected using a BMV positive-strand RNA-specific probe as described for Fig. 1.
BMV RNA3 is unstable when expressed alone, with a half-life of about 5 to 7 min (29). When expressed along with 1a, RNA3 accumulation increases 8- to 20-fold while its half-life increases to >3 h (29, 78). Increased half-life and accumulation of RNA3 are a result of its recruitment into a membrane-associated, RNase-resistant status, with a likely residence inside the viral spherules (68). As shown in Fig. 2C, RNA3 accumulation in acb1Δ cells expressing 1a increased about 34-fold compared to the accumulation in those lacking 1a. The increase in RNA3 accumulation in acb1Δ cells was even higher than that in wt cells, which showed an approximately 23-fold increase, indicating that the recruitment of viral RNA replication templates was enhanced by deleting ACB1 (Fig. 2C).
2apol is a soluble and relatively unstable protein when expressed on its own. 1a recruits 2apol into viral spherule or layer replication complexes, thus protecting 2apol from degradation and increasing its accumulation (Fig. 3C, compare 2apol signals without and with 1a in wt cells) (10). While 1a accumulation was not affected in acb1Δ cells (Fig. 2A), 2apol accumulation was significantly lower than that in wt cells (Fig. 3A). Reduced 2apol accumulation could be a result of inhibited 2apol transcription and/or translation, inhibited 1a-2apol interaction, or other unspecified mechanisms. A BMV RNA2-specific probe detected similar levels of 2apol transcripts in both wt and acb1Δ cells (Fig. 3B), indicating that 2apol transcription was not affected when ACB1 was deleted. When 2apol was expressed on its own, 2apol protein levels were slightly lower in acb1Δ cells than in wt cells (Fig. 3C, left panel), ruling out the possible inhibition of 2apol translation by deleting ACB1.
When coexpressed with 1a, 2apol accumulation increased significantly in wt cells (Fig. 3C) (10). While 2apol accumulation increased with the presence of 1a in acb1Δ cells, the final 2apol level was lower than that in wt cells (Fig. 3C), suggesting that the 1a-2apol interaction might be affected. Decreased 2apol accumulation was ACB1 deletion mediated since HA-tagged Acb1p not only complemented the BMV RNA replication defect (data not shown) but also increased 2apol accumulation (Fig. 3D). To further test whether the mild increase in 2apol was a result of its recruitment onto membranes by 1a, a membrane flotation assay was performed. In both acb1Δ (Fig. 3E) and wt (data not shown) (48) cells, the majority of 2apol was distributed in the bottom two fractions in the absence of 1a. Conversely, in the presence of 1a, all the detected 2apol was found in the top 2 fractions (Fig. 3E) where membrane-integral and -associated proteins were detected (Fig. 3E and 2B). Similar conclusions were reached when performing the above-described assays using acb1Δ cells derived from a BY4743 or a YPH500 background, indicating that cell growth differences do not account for the defective BMV RNA replication phenotypes. Thus, the results described hereafter were based in the YPH500 derivative.
Aberrant viral spherule and layer replication complexes are formed in acb1Δ cells.
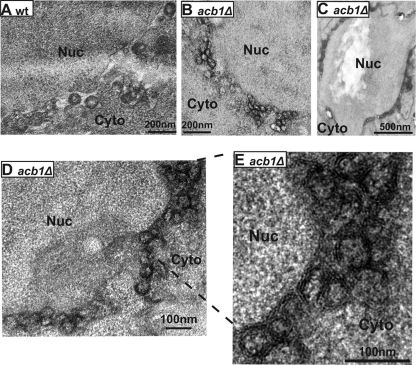
As noted in the introduction, expressing 1a by itself or with low levels of 2apol will induce formation of viral spherules. In wt cells, viral spherules induced by 1a are about ∼60 to 80 nm in diameter with an average number of spherules per cell section of about 6 (Fig. 4A) (15). We found that in cells lacking ACB1, viral spherules were smaller but present at a much higher rate. The average number of spherules per cell section was 23.9 (23.9 ± 7.8, Fig. 4B to D), likely an underestimate since smaller spherules tend to cluster and overlap and thus are sometimes hard to visualize separately (Fig. 4C). Nevertheless, this is an approximately 4-fold increase over that in wt cells. While significantly more spherules were formed in acb1Δ cells than in wt cells, the diameter of the spherules was reduced compared to that in wt cells (Fig. 4B to D). Based on the 281 spherules that we measured, the average spherule diameter was 38.9 ± 11.0 nm, about half the size of those formed in wt cells (74 ± 16 nm) (15). Given the fact that viral RNA replication templates are recruited (Fig. 2C) to spherules and yet the accumulation of both negative- and positive-strand RNAs is substantially lower than that in wt cells (Fig. 1), we conclude that the smaller spherules are inactive or much less efficient than wt spherules in viral RNA synthesis. It should be noted that a very low percentage of cells (<10%) had wt-size spherules, which might account for the low levels of BMV RNA replication seen in this strain.
Fig 4.
Formation of aberrant spherules that are smaller but 4-fold more abundant in cells lacking ACB1. Yeast cells expressing BMV 1a were examined by electron microscopy following osmium staining. (A) Spherules formed in wt cells. (B to E) Smaller (38.9 ± 11.0 nm) but more abundant (23.9 ± 7.8/cell section) spherules were formed in acb1Δ cells. Labels indicate nucleus (Nuc) and cytoplasm (Cyto).
Osmium tetroxide (OsO4), a chemical commonly used in electron microscopy, fixes and stains membranes by reacting with the double bonds of adjacent UFAs, marking the membrane with two parallel, electron-dense lines in high-resolution electron micrographs (24). In cells with decreased UFA levels, membrane lipids are not well cross linked and are subsequently extracted by ethanol during dehydration of the cells, resulting in lipid bilayer-associated lines that are not as distinctive and clear as those in wt yeast. Although the size and frequency of spherules were sharply different from those in wt, two distinct lines representing the outer ER bilayers were clearly observed when spherules in acb1Δ cells were viewed at a higher magnification (Fig. 4E), indicating that UFA levels were not significantly decreased in acb1Δ cells. This is different from the previously characterized ole1w yeast mutant. The ole1w mutation results in a 12% decrease in UFA levels, leading to a 20-fold inhibition in BMV RNA replication (45). In ole1w cells, the bilayer members enveloping the viral spherules are not well fixed and stained by OsO4 due to a local depletion of UFA levels in these membranes (45).
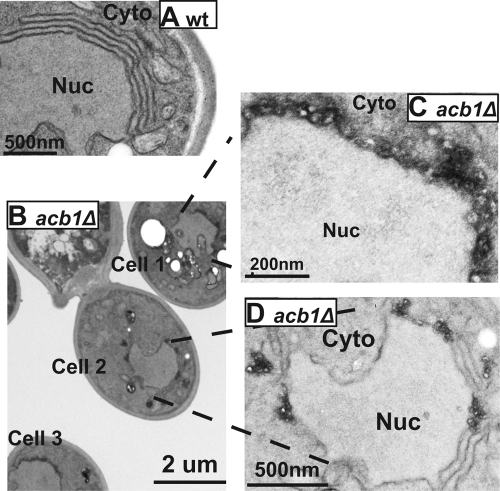
1a, in the presence of high levels of 2apol, induces formation of layer replication complexes in wt cells (69). Accordingly, reduced 2apol accumulation in acb1Δ cells compared to that in wt cells suggested that layer replication complexes might not form or might do so at a very low frequency. In wt cells expressing 1a and GAL1 2apol, 75% of cells containing BMV-induced replication complexes formed layers (Fig. 5A) (48). In contrast, in acb1Δ cells, layer structures were found in ∼30% of cells that had BMV-induced replication complexes (Fig. 5B, cell 2, and Fig. 5D), while the other 70% had vesicles similar in size to those induced by 1a alone (Fig. 5B, cells 1 and 3, and Fig. 5C). Although layer replication complexes formed in wt cells usually have 3 to 7 stacked membranes, layer structures formed in acb1Δ cells usually had no more than 3 stacks and were frequently disrupted by the presence of smaller spherules (Fig. 5D), indicating that smaller spherules were preferentially formed in acb1Δ cells upon overexpression of 1a and 2apol.
Fig 5.
Small spherules are preferentially formed in acb1Δ cells under conditions that promote layer formation in wt cells. (A) Layer replication complexes formed in wt cells expressing 1a and high levels of 2apol. (B to D) Layers and large numbers of small spherules formed in acb1Δ cells. (C and D) Higher-magnification micrographs of cell 1 (C) and cell 2 (D) from panel B. Nuc, nucleus; Cyto, cytoplasm.
Deleting ACB1 does not affect karmella formation.
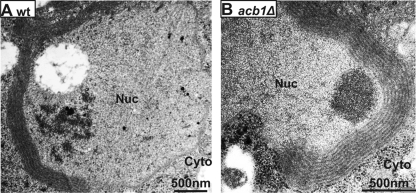
BMV layer replication complexes are similar to karmellae, which are cellular structures formed in yeast and mammalian cells upon overexpression of certain ER membrane-resident proteins, such as 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase 1 (Hmp1p) (80) and cytochrome b (5, 71, 75). Furthermore, interactions between the cytoplasmic domains of the karmella-inducing proteins are required for karmella production (4, 56, 75), similar to the 1a-2apol interaction being a prerequisite for formation of BMV layer replication complexes (69). However, karmellae are more closely spaced, with a cytoplasmic space of ∼8 to 11 nm between the stacked double membranes (4, 56, 75), compared to an intermembrane distance of ∼50 to 70 nm in the BMV layer replication compartments (Fig. 5A) (69).
Hmp1p is one of two isoenzymes in yeast that convert HMG-CoA to mevalonate, the rate-limiting step in sterol synthesis (4). While overproduction of Hmg1p does not increase sterol synthesis in yeast, overexpressed Hmg1p induces karmella formation (80). As expected, expressing Hmg1p from the GAL1 promoter induced membrane proliferations surrounding the nucleus in wt cells (Fig. 6A). Similarly, when Hmg1p was overexpressed in acb1Δ cells, karmellae were formed at a frequency comparable to that in wt cells (Fig. 6B). The number of karmella layers in each cell section was between 5 and 9 in both wt and acb1Δ cells, indicating that the defects caused by deleting ACB1 specifically affected the membrane remodeling induced by BMV 1a.
Fig 6.
Karmellae are formed normally in acb1Δ cells. Electron microscopy images of karmella structures in wt (A) and acb1Δ (B) cells overexpressing yeast Hmg1p. Nuc, nucleus; Cyto, cytoplasm.
The BMV RNA replication defect can be largely complemented by supplementing unsaturated long-chain fatty acids.
Decreased accumulation of long-chain fatty acids, including stearic acid (18:0), oleic acid (18:1), and palmitic acid (16:0), and an increase of the shorter unsaturated fatty acids myristoleic acid (14:1) and palmitoleic acid (16:1) in acb1Δ cells from different genetic backgrounds have been reported by several groups, albeit the magnitude of the reported alterations varied (12, 19, 64). Northern blotting showed 2- to 3-fold increases in accumulated OLE1 transcripts in acb1Δ cells (Fig. 7B and C), agreeing well with other reports (12, 19). We measured FA composition in wt and acb1Δ cells and confirmed a reduction in 16:0, 18:0, and 18:1 levels but an increase in 16:1, which is the major component of total FAs in yeast (Fig. 7A). Increased 16:1 and decreased SFAs resulted in a UFA/SFA ratio of 6.0 in acb1Δ cells compared to a ratio of 4.5 in wt cells, a 33% increase.
To partially complement the altered FA composition, we provided different combinations of individual FAs in the growth medium, each at 0.25 mM. Since acb1Δ cells contain less 16:0 and 18:0, we added SFAs as a supplement in the medium; however, it did not enhance BMV RNA replication (Fig. 7B). In contrast, BMV RNA replication was enhanced when UFAs (16:1 and 18:1) or all four major FAs were added as supplements to the growth medium (Fig. 7B). In both cases, negative-strand RNA3 accumulation increased about 4- to 5-fold over that in untreated medium, resulting in an increase from 11% to 58% and 49%, respectively. In addition, both positive-strand RNA3 and RNA4 increased at least 3-fold in acb1Δ yeast grown in medium supplemented with UFAs or total FAs (Fig. 7B). To identify the specific UFA responsible for restoring BMV RNA replication in acb1Δ cells, 16:1 or 18:1 was added as a supplement individually or in combination in the medium. Figure 7C shows that either 16:1 or 18:1 can individually restore BMV RNA replication to levels similar to those when both were provided.
Transcription of OLE1 and a group of lipid metabolism genes is regulated by two homologous transcription factors, Spt23p and Mga2p (3, 27, 77). Both are initially sequestered as ER membrane-integral proteins, termed p120, and subsequently activated by proteasomes, which degrade the C-terminal membrane-spanning motif and release the N-terminal activation domain from membranes. The active form, p90, is targeted to the nucleus to execute its functions (26, 27, 57). We previously reported that inhibited activation of Spt23p and/or Mga2p resulted in decreased accumulation of OLE1 transcripts and thus inhibited BMV RNA replication (77). The active form of Mga2p, p90, accumulated to higher levels in BMV-replicating acb1Δ cells than in wt cells (Fig. 7D), indicating that ACB1 deletion leads to a better activation of Mga2p, a conclusion supported by the increased accumulation of OLE1 transcripts (Fig. 7B and C) and increased UFA/SFA ratio (Fig. 7A) (12, 19). Activation of Spt23p and Mga2p can be inhibited by exogenously adding UFAs as a supplement via a feedback regulation but not by adding SFAs as a supplement (27, 31). As expected, addition of 16:1, UFAs, or total FAs substantially inhibited accumulation of OLE1 transcripts by approximately 70% to 80% (Fig. 7B) or even more (Fig. 7C) compared to untreated wt cells. 18:1 FA was not as efficient as 16:1 at inhibiting OLE1 transcription at 0.25 mM; nonetheless, it decreased OLE1 transcripts to wt levels in acb1Δ cells (Fig. 7C).
The restoration of BMV RNA replication by supplemental UFAs suggested that Acb1p is functionally required for BMV RNA replication. As a first step to test whether Acb1p is associated with viral spherules and physically required for BMV RNA replication, we performed a membrane flotation assay. The great majority of Acb1p was detected in the soluble protein-containing bottom fractions in the absence and presence of BMV components (Fig. 7E). While we cannot totally rule out the possibility that a small portion of Acb1p was present in the membrane fraction or viral spherules, the membrane flotation and UFA feeding assays collectively suggest that Acb1p is functionally, but not physically, required in BMV RNA replication.
Supplemental unsaturated fatty acids increase spherule size.
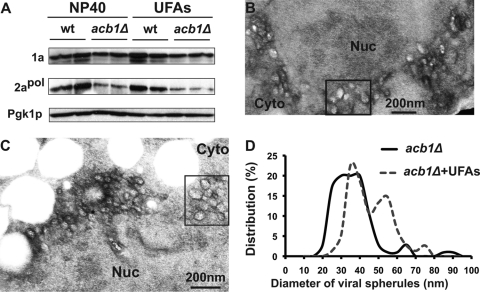
In an attempt to dissect the mechanism by which supplemental UFA largely complemented BMV RNA replication, we measured 2apol accumulation and checked for spherule formation in acb1Δ cells supplemented with UFAs. Compared to untreated cells, 2apol accumulation in the presence of UFAs was not restored in acb1Δ cells (Fig. 8A), even though BMV RNA replication was largely restored. However, previous reports have shown that cells expressing substantially reduced levels of 2apol can still support BMV RNA replication (16). Moreover, despite significant differences in 2apol levels, spherule and layer replication complexes support similar levels of BMV RNA replication (69), suggesting that a decrease in 2apol levels may not be the major contributor to the inhibition of BMV RNA replication.
Fig 8.
Supplementation with long-chain UFAs increases viral spherule diameter. (A) Supplementation with UFAs did not restore the accumulation of 2apol in acb1Δ cells. Total proteins were extracted from equal numbers of yeast cells in the absence and presence of UFAs. Western blotting was done as described for Fig. 3. (B and C) Electron micrographs of spherules formed in acb1Δ cells supplemented with UFAs. Boxed areas highlight larger spherules. Nuc, nucleus; Cyto, cytoplasm. (D) Distribution of spherule diameters in acb1Δ cells without (acb1Δ) or with (acb1Δ +UFAs) UFA treatment. Distribution of spherule diameters in acb1Δ cells was based on measurements of 281 spherules, and the spherule diameters from cells with UFA treatment were from 75 spherules. All assays were performed under spherule-forming conditions.
The number of spherules formed in acb1Δ cells in the presence of long-chain UFAs, about 23 spherules per cell section, was not significantly altered (Fig. 8B and C). However, the diameter of the spherules increased from 39 nm to about 43 nm (43.4 nm ± 10.8 nm) in the presence of UFAs, based on measurements of 75 spherules. While the majority of spherules were smaller, groups of larger spherules aggregated in the same vicinity (boxed areas in Fig. 8B and C). The average size of these spherules was approximately 58.1 ± 8.4 nm (Fig. 8B to D). Thus, the distribution in spherule diameter in UFA-treated acb1Δ cells is different from that in untreated cells, where the majority of spherules were between 25 and 45 nm in diameter (Fig. 8D). Despite restoring the BMV RNA replication defect, supplemental UFAs only partially complemented spherule formation, as these spherules were smaller than those in wt cells.
DISCUSSION
In the studies reported here, we further explored the relationship between cellular membrane lipid synthesis and composition and positive-strand RNA virus replication (45, 46, 77). We found that host ACBP (Acb1p in yeast), a protein highly conserved among all eukaryotic species, is required for both the assembly and activity of BMV RNA replication complexes. Deleting ACB1 resulted in formation of spherules that were significantly more abundant but smaller than those formed with ACB1 (Fig. 4 and 5). Our data suggest that Acb1p is required for maintaining host lipid homeostasis, as supplemental long-chain UFA(s) largely restored BMV RNA replication and the great majority of Acb1p was not associated with membranes under BMV RNA replication conditions (Fig. 7B to C and E). As discussed further below, the defective BMV RNA replication phenotypes in acb1Δ cells were very similar to those caused by a group of 1a mutants with specific amino acid substitutions within an amphipathic α-helix, termed helix A, a domain crucial for 1a-ER membrane association and spherule formation (48). Thus, loss of host ACBP may have effects on BMV RNA replication similar to those of mutations within 1a (48), possibly disrupting 1a-lipid interactions required for the assembly of functional replication complexes.
Membrane rearrangements induced by BMV 1a are specifically affected in cells lacking ACB1.
Acb1p plays important roles in the synthesis of cellular lipids and maintenance of cellular membrane structures (19). In cells lacking ACB1, aberrant membrane structures are frequently accumulated, including 50- to 60-nm vesicles in the cytoplasm (19). Based on this, it is possible that the smaller spherules induced by BMV in acb1Δ cells were due to general defects associated with the absence of ACB1. However, two lines of evidence indicate that membrane rearrangements induced by BMV are selectively affected: formation of cellular karmella structures was not affected (Fig. 6) and the spherule phenotype was similarly induced by specific 1a mutants (48).
While the mechanism by which BMV spherules are formed is not fully understood, 1a's helix A plays crucial roles in the process (48). Mutations in helix A result in two distinct phenotypes (48). The class I 1a mutants, in which helix A is deleted or disrupted, fail to associate with the perinuclear ER membranes and induce spherules (48). In contrast, class II 1a mutants not only retain efficient ER membrane association but cause phenotypes similar to those caused by deletion of ACB1, including the formation of smaller but more frequent spherules (Fig. 4), preferential formation of smaller and more frequent spherules under layer-forming conditions (Fig. 5), a decrease in 1a-promoted 2apol accumulation and membrane association (Fig. 3), and an increase in 1a-mediated RNA3 stabilization (Fig. 2C). The class II 1a mutants have specific substitutions in amino acids T397, Y401, K403, and Y404, which are in the major membrane-interacting face of helix A (1a amino acids 392 to 409) (48) and possibly interact with phospholipid head groups. Amphipathic α-helices are often flexible and unstructured in solution and not well resolved in crystal structures but switch to a helical conformation upon binding to membranes (20, 30, 42). Consistent with this, the 1a helix A peptide adopts a helical conformation upon binding to membrane-mimicking SDS micelles (48). A conformational change of 1a upon binding to membranes may favor 1a-1a interactions that facilitate formation of ∼60- to 80-nm spherules. For SFV nonstructural protein 1 (nsP1), an RNA capping enzyme homologous to the capping domain of 1a, binding to anionic phospholipids causes a conformational change which leads to activation of the protein (2). The interactions between head groups of cellular membrane lipids and helix A may determine 1a's affinity for membranes and the depth of 1a penetration into membranes and/or may modulate 1a-1a interactions, which in turn may affect spherule size. Thus, either mutations within helix A or changes in the lipid composition of ER membrane upon deletion of ACB1 might modulate the aforementioned interactions and lead to formation of smaller spherules. Supplementation with 16:1- and 18:1-FA, which are long-chain UFAs, increased the size of a subset of spherules, supporting the notion that altered lipid composition was partially responsible for decreased spherule sizes. However, only a portion of the spherule population increased in size, possibly due to inefficient incorporation of supplemental UFAs into phospholipids in cells lacking ACB1, among others.
Alternatively, altered lipid composition may affect the relocalization or activity of host proteins that are required for spherule formation, such as reticulon-homology proteins (RHPs). RHPs are recruited by 1a from peripheral ER membranes to the interior of viral spherules (15). RHPs are a group of membrane-shaping proteins that induce and stabilize ER membrane tubules (83) and not only regulate spherule diameter but also are required for initiating and/or maintaining BMV spherules (15). Deleting one or two RHP-encoding genes decreased spherule diameters, ranging from 27 nm (in cells lacking RTN2 and YOP1) to 58 nm (in cells lacking RTN2) (15). In acb1Δ cells, similar to 2apol, RHPs may not be efficiently recruited into perinuclear ER membranes, leading to the formation of smaller spherules. However, unlike the small spherules formed by 1a class II mutants or in acb1Δ cells, the smaller spherules formed upon deletion of RHPs support BMV RNA replication (15).
BMV and FHV are among many positive-strand RNA viruses that induce formation of vesicular viral replication complexes (39, 68). FHV protein A, which is the replicase and a membrane integral protein, invaginates outer mitochondrial membranes to form viral spherules (50, 51). Based on three-dimensional (3-D) electron microscope tomography and stoichiometric and biochemical analyses, protein A coats the inside of spherule membranes (39). Similarly, the self-interacting and membrane-bound 1a resides as a shell within the inner spherule membrane to initiate and maintain the spherular structure (68). In acb1Δ cells, 1a accumulated to levels similar to those in wt cells (Fig. 2A). In addition, 1a-induced formation of spherules with half the diameter of those induced in wt cells correlated with a 4-fold increase in spherule frequency (Fig. 4), consistent with the conclusion that 1a occupies the majority, if not all, of the spherule interior.
BMV RNA replication requires a balanced lipid microenvironment.
Deleting ACB1 affects cellular lipid synthesis, including decreased accumulation of long-chain FAs and increased accumulation of OLE1 transcripts as well as shorter and unsaturated FAs (Fig. 7A to C) (12, 19, 64), most likely through enhanced activation of Mga2p and/or Spt23p (Fig. 7D). It is possible that Mga2p- and/or Spt23p-regulated genes, aside from OLE1, are overexpressed and may result in increased accumulation of sterols and/or sphingolipid intermediates and/or final products, which may affect the composition of membranes surrounding the replication complexes and thus the efficiency of viral RNA replication.
In acb1Δ cells, the UFA/SFA ratio increased ∼33% (Fig. 7A). Although we cannot exclude other possibilities, complementation of BMV RNA replication defects by supplemental long-chain UFAs is consistent with the idea that altering the UFA contents or the UFA/SFA ratio affects BMV RNA replication. Consistent with this, BMV RNA replication is inhibited by up to 95% in the ole1w mutant (46). A 12% reduction in UFA levels in this mutant preferentially affects the lipid composition of spherule-enveloping membranes, indicating that the lipid composition of membranes surrounding the spherules is different from that of the rest of perinuclear ER membranes (45). These results suggest that BMV viral replication requires a well-controlled lipid microenvironment that, when mildly altered, sabotages the viral replication more dramatically than host growth and division. Emerging evidence indicates that genomic replication of positive-strand RNA viruses requires a suitable lipid microenvironment. Coxsackievirus B3 (CVB3) selectively recruits phosphatidylinositol-4-kinase III β (PI4KIIIβ) via its 3A protein to the viral replication sites (28). PI4KIIIβ, which produces phosphatidylinositol 4-phosphate (PI4P), creates a PI4P-enriched microenvironment that is required for CVB3 replication, most likely by promoting membrane association and/or the activity of the viral replicase (28). A PI4P-enriched environment is also required for HCV replication (61). HCV nonstructural protein 5A (NS5A) interacts with PI4KIIIα and stimulates its kinase activity to promote PI4P production, consistent with the elevated levels of PI4P in liver tissues from HCV patients (61). Moreover, depleting PI4KIIIα activity dramatically affects HCV RNA replication complex assembly, either by reducing the formation of viral replication complexes (5) or by forming abnormal NS5A “clusters” in cells expressing the NS3-to-NS5B polyprotein fragment (61).
Possible role of ACBP in BMV infection in plants and potential role in infection by other positive-strand RNA viruses.
Many plant positive-strand RNA viruses form vesicular replication complexes in infected host cells. Viruses of the Tombusviridae family form spherular replication complexes in mitochondria (66) and peroxisomes (65), and viruses in the Bromoviridae family replicate in ER (34) and tonoplast (23) membrane-bound vesicles. Our efforts to prove that ACBP is required for or involved in BMV RNA replication in plants were hindered by several issues. For instance, there are six members in Arabidopsis thaliana, AtACBP1 to -6, which are all capable of binding to LCFA-CoAs (81), suggesting that plant ACBPs have redundant functions. This notion is supported by the fact that no growth phenotype is observed upon deletion of individual AtACBP genes (81). In addition, expression of the Arabidopsis and Nicotiana benthamiana orthologs of ACB1 in acb1Δ yeast, AtACBP6 and NbACBP6, as well as a paralog, AtACBP4, complemented BMV RNA replication defects (data not shown), suggesting that ACBPs from different plant species are capable of supporting BMV RNA replication in yeast and likely play a similar role in plants. A better understanding of the roles of Acb1p in BMV RNA replication in yeast should help in designing experiments that can be feasibly tested in whole plants. Human, mouse, bovine, and Caenorhabditis elegans ACBPs complement slow growth of acb1Δ yeast cells (17, 44, 49), indicating that the functions of ACBPs from different species are conserved. As all ACBPs are involved in lipid synthesis, it is possible that ACBPs in some species could be directly or indirectly involved in the replication of positive-strand RNA viruses by maintaining proper lipid homeostasis.
ACKNOWLEDGMENTS
We thank Alejandra Gutierrez, Yanmei Hu, Natali Mejia, and Gabriela Ontiveros for general assistance. We also thank Randall Massey and Benjamin August at the University of Wisconsin Medical School Electron Microscope Facility and Anxiu Kuang at the Microscopy Core Research Facility, University of Texas-Pan American, for assistance with electron microscopy work. We thank Samuel Piña, Jr., and Raul Rivera at the Kika de la Garza Subtropical Agricultural Research Center, USDA, for processing our lipid samples.
This work was supported by NIH grant GM35072 to P.A. and Texas AgriLife Research Start-up Fund and NSF grant IOS-1120598 to X.W. A.D. was partially supported by NIH training grant T32 AI078985. P.A. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print 15 February 2012
REFERENCES
- 1. Ahola T, Ahlquist P. 1999. Putative RNA capping activities encoded by brome mosaic virus: methylation and covalent binding of guanylate by replicase protein 1a. J. Virol. 73:10061–10069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahola T, Lampio A, Auvinen P, Kaariainen L. 1999. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J. 18:3164–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Auld KL, Brown CR, Casolari JM, Komili S, Silver PA. 2006. Genomic association of the proteasome demonstrates overlapping gene regulatory activity with transcription factor substrates. Mol. Cell 21:861–871 [DOI] [PubMed] [Google Scholar]
- 4. Basson ME, Thorsness M, Rine J. 1986. Saccharomyces cerevisiae contains two functional genes encoding 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proc. Natl. Acad. Sci. U. S. A. 83:5563–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berger KL, et al. 2009. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc. Natl. Acad. Sci. U. S. A. 106:7577–7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carette JE, Stuiver M, Van Lent J, Wellink J, Van Kammen A. 2000. Cowpea mosaic virus infection induces a massive proliferation of endoplasmic reticulum but not Golgi membranes and is dependent on de novo membrane synthesis. J. Virol. 74:6556–6563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Castorena KM, Stapleford KA, Miller DJ. 2010. Complementary transcriptomic, lipidomic, and targeted functional genetic analyses in cultured Drosophila cells highlight the role of glycerophospholipid metabolism in Flock House virus RNA replication. BMC Genomics 11:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chao H, et al. 2002. Membrane charge and curvature determine interaction with acyl-CoA binding protein (ACBP) and fatty acyl-CoA targeting. Biochemistry 41:10540–10553 [DOI] [PubMed] [Google Scholar]
- 9. Chao H, Zhou M, McIntosh A, Schroeder F, Kier AB. 2003. ACBP and cholesterol differentially alter fatty acyl CoA utilization by microsomal ACAT. J. Lipid Res. 44:72–83 [DOI] [PubMed] [Google Scholar]
- 10. Chen J, Ahlquist P. 2000. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J. Virol. 74:4310–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cherry S, et al. 2006. COPI activity coupled with fatty acid biosynthesis is required for viral replication. PLoS Pathog. 2:e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Choi JY, Stukey J, Hwang SY, Martin CE. 1996. Regulatory elements that control transcription activation and unsaturated fatty acid-mediated repression of the Saccharomyces cerevisiae OLE1 gene. J. Biol. Chem. 271:3581–3589 [DOI] [PubMed] [Google Scholar]
- 13. den Boon J, Diaz A, Ahlquist 2010. Cytoplasmic viral replication complexes. Cell Host Microbe 8:77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. den Boon JA, Chen J, Ahlquist P. 2001. Identification of sequences in brome mosaic virus replicase protein 1a that mediate association with endoplasmic reticulum membranes. J. Virol. 75:12370–12381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Diaz A, Wang X, Ahlquist P. 2010. Membrane-shaping host reticulon proteins play crucial roles in viral RNA replication compartment formation and function. Proc. Natl. Acad. Sci. U. S. A. 107:16291–16296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dinant S, Janda M, Kroner PA, Ahlquist P. 1993. Bromovirus RNA replication and transcription require compatibility between the polymerase- and helicase-like viral RNA synthesis proteins. J. Virol. 67:7181–7189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Faergeman NJ, Knudsen J. 2002. Acyl-CoA binding protein is an essential protein in mammalian cell lines. Biochem. J. 368:679–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Faergeman NJ, Knudsen J. 1997. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem. J. 323:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gaigg B, et al. 2001. Depletion of acyl-coenzyme A-binding protein affects sphingolipid synthesis and causes vesicle accumulation and membrane defects in Saccharomyces cerevisiae. Mol. Biol. Cell 12:1147–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gallop JL, et al. 2006. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 25:2898–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gossett RE, et al. 1996. Acyl-CoA binding proteins: multiplicity and function. Lipids 31:895–918 [DOI] [PubMed] [Google Scholar]
- 22. Guinea R, Carrasco L. 1990. Phospholipid biosynthesis and poliovirus genome replication, two coupled phenomena. EMBO J. 9:2011–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hatta T, Francki RI. 1981. Cytopathic structures associated with tonoplasts of plant cells infected with cucumber mosaic and tomato aspermy viruses. J. Gen. Virol. 53:343–346 [Google Scholar]
- 24. Hayat MA. 2000. Principles and techniques of electron microscopy: biological applications, 4th ed Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 25. Heaton NS, et al. 2010. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc. Natl. Acad. Sci. U. S. A. 107:17345–17350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hitchcock AL, et al. 2001. The conserved npl4 protein complex mediates proteasome-dependent membrane-bound transcription factor activation. Mol. Biol. Cell 12:3226–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hoppe T, et al. 2000. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell 102:577–586 [DOI] [PubMed] [Google Scholar]
- 28. Hsu NY, et al. 2010. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141:799–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Janda M, Ahlquist P. 1998. Brome mosaic virus RNA replication protein 1a dramatically increases in vivo stability but not translation of viral genomic RNA3. Proc. Natl. Acad. Sci. U. S. A. 95:2227–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jao CC, et al. 2010. Roles of amphipathic helices and the bin/amphiphysin/rvs (BAR) domain of endophilin in membrane curvature generation. J. Biol. Chem. 285:20164–20170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jiang Y, et al. 2002. Mga2p processing by hypoxia and unsaturated fatty acids in Saccharomyces cerevisiae: impact on LORE-dependent gene expression. Eukaryot. Cell 1:481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kao CC, Ahlquist P. 1992. Identification of the domains required for direct interaction of the helicase-like and polymerase-like RNA replication proteins of brome mosaic virus. J. Virol. 66:7293–7302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kapadia SB, Chisari FV. 2005. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc. Natl. Acad. Sci. U. S. A. 102:2561–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim KS. 1977. An ultrastructural study of inclusions and disease development in plant cells infected by cowpea chlorotic mottle virus. J. Gen. Virol. 35:535–543 [Google Scholar]
- 35. Knoops K, et al. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6:e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Knudsen J, Burton M, Faergeman NJ. 2004. Long chain acyl-CoA esters and acyl-CoA binding protein (ACBP) in cell function. Adv. Mol. Cell Biol. 33:123–152 [Google Scholar]
- 37. Kohrer K, Domdey H. 1991. Preparation of high molecular weight RNA. Methods Enzymol. 194:398–405 [DOI] [PubMed] [Google Scholar]
- 38. Kong F, Sivakumaran K, Kao C. 1999. The N-terminal half of the brome mosaic virus 1a protein has RNA capping-associated activities: specificity for GTP and S-adenosylmethionine. Virology 259:200–210 [DOI] [PubMed] [Google Scholar]
- 39. Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 5:e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Krol MA, et al. 1999. RNA-controlled polymorphism in the in vivo assembly of 180-subunit and 120-subunit virions from a single capsid protein. Proc. Natl. Acad. Sci. U. S. A. 96:13650–13655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kushner DB, et al. 2003. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc. Natl. Acad. Sci. U. S. A. 100:15764–15769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lampio A, et al. 2000. Membrane binding mechanism of an RNA virus-capping enzyme. J. Biol. Chem. 275:37853–37859 [DOI] [PubMed] [Google Scholar]
- 43. Landrock D, et al. 2010. Acyl-CoA binding protein gene ablation induces pre-implantation embryonic lethality in mice. Lipids 45:567–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Larsen MK, Tuck S, Faergeman NJ, Knudsen J. 2006. MAA-1, a novel acyl-CoA-binding protein involved in endosomal vesicle transport in Caenorhabditis elegans. Mol. Biol. Cell 17:4318–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee WM, Ahlquist P. 2003. Membrane synthesis, specific lipid requirements, and localized lipid composition changes associated with a positive-strand RNA virus RNA replication protein. J. Virol. 77:12819–12828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee WM, Ishikawa M, Ahlquist P. 2001. Mutation of host delta9 fatty acid desaturase inhibits brome mosaic virus RNA replication between template recognition and RNA synthesis. J. Virol. 75:2097–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lightner J, Wu J, Browse J. 1994. A mutant of Arabidopsis with increased levels of stearic acid. Plant Physiol. 106:1443–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu L, et al. 2009. An amphipathic alpha-helix controls multiple roles of brome mosaic virus protein 1a in RNA replication complex assembly and function. PLoS Pathog. 5:e1000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mandrup S, et al. 1993. Effect of heterologous expression of acyl-CoA-binding protein on acyl-CoA level and composition in yeast. Biochem. J. 290:369–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Miller DJ, Ahlquist P. 2002. Flock House virus RNA polymerase is a transmembrane protein with amino-terminal sequences sufficient for mitochondrial localization and membrane insertion. J. Virol. 76:9856–9867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Miller DJ, Schwartz MD, Ahlquist P. 2001. Flock House virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J. Virol. 75:11664–11676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Miller S, Krijnse-Locker J. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6:363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mosser AG, Caliguiri LA, Tamm I. 1972. Incorporation of lipid precursors into cytoplasmic membranes of poliovirus-infected HeLa cells. Virology 47:39–47 [DOI] [PubMed] [Google Scholar]
- 54. O'Reilly EK, Paul JD, Kao CC. 1997. Analysis of the interaction of viral RNA replication proteins by using the yeast two-hybrid assay. J. Virol. 71:7526–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Perez L, Guinea R, Carrasco L. 1991. Synthesis of Semliki Forest virus RNA requires continuous lipid synthesis. Virology 183:74–82 [DOI] [PubMed] [Google Scholar]
- 56. Profant DA, Roberts CJ, Koning AJ, Wright RL. 1999. The role of the 3-hydroxy 3-methylglutaryl coenzyme A reductase cytosolic domain in karmellae biogenesis. Mol. Biol. Cell 10:3409–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rape M, et al. 2001. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell 107:667–677 [DOI] [PubMed] [Google Scholar]
- 58. Rasheed S, Yan JS, Lau A, Chan AS. 2008. HIV replication enhances production of free fatty acids, low density lipoproteins and many key proteins involved in lipid metabolism: a proteomics study. PLoS One 3:e3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rasmussen JT, Faergeman NJ, Kristiansen K, Knudsen J. 1994. Acyl-CoA-binding protein (ACBP) can mediate intermembrane acyl-CoA transport and donate acyl-CoA for beta-oxidation and glycerolipid synthesis. Biochem. J. 299:165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rasmussen JT, Rosendal J, Knudsen J. 1993. Interaction of acyl-CoA binding protein (ACBP) on processes for which acyl-CoA is a substrate, product or inhibitor. Biochem. J. 292:907–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Reiss S, et al. 2011. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 9:32–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Restrepo-Hartwig M, Ahlquist P. 1999. Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J. Virol. 73:10303–10309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Restrepo-Hartwig MA, Ahlquist P. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 70:8908–8916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rijken PJ, et al. 2009. Cardiolipin molecular species with shorter acyl chains accumulate in Saccharomyces cerevisiae mutants lacking the acyl coenzyme A-binding protein Acb1p: new insights into acyl chain remodeling of cardiolipin. J. Biol. Chem. 284:27609–27619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Russo M, Di Franco A, Martelli GP. 1983. The fine structure of Cymbidium ringspot virus infections in host tissues. III. Role of peroxisomes in the genesis of multivesicular bodies. J. Ultrastruct. Res. 82:52–63 [DOI] [PubMed] [Google Scholar]
- 66. Russo M, Martelli GP. 1982. Ultrastructure of turnip crinkle- and saguaro cactus virus-infected tissues. Virology 118:109–116 [DOI] [PubMed] [Google Scholar]
- 67. Salonen A, Ahola T, Kaariainen L. 2005. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285:139–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schwartz M, et al. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505–514 [DOI] [PubMed] [Google Scholar]
- 69. Schwartz M, Chen J, Lee WM, Janda M, Ahlquist P. 2004. Alternate, virus-induced membrane rearrangements support positive-strand RNA virus genome replication. Proc. Natl. Acad. Sci. U. S. A. 101:11263–11268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sharma M, Sasvari Z, Nagy PD. 2010. Inhibition of sterol biosynthesis reduces tombusvirus replication in yeast and plants. J. Virol. 84:2270–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Snapp EL, et al. 2003. Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol. 163:257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Stukey JE, McDonough VM, Martin CE. 1989. Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. J. Biol. Chem. 264:16537–16544 [PubMed] [Google Scholar]
- 73. Sullivan ML, Ahlquist P. 1999. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 73:2622–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Vance DE, Trip EM, Paddon HB. 1980. Poliovirus increases phosphatidylcholine biosynthesis in HeLa cells by stimulation of the rate-limiting reaction catalyzed by CTP: phosphocholine cytidylyltransferase. J. Biol. Chem. 255:1064–1069 [PubMed] [Google Scholar]
- 75. Vergeres G, Yen TS, Aggeler J, Lausier J, Waskell L. 1993. A model system for studying membrane biogenesis. Overexpression of cytochrome b5 in yeast results in marked proliferation of the intracellular membrane. J. Cell Sci. 106:249–259 [DOI] [PubMed] [Google Scholar]
- 76. Wang X, Ahlquist P. 2008. Brome mosaic virus (Bromoviridae), p 381–386 In Mahy BWJ, van Regenmortel MHV. (ed), Encyclopedia of virology, 3rd ed Elsevier, Oxford, United Kingdom [Google Scholar]
- 77. Wang X, et al. 2011. Intersection of the multivesicular body pathway and lipid homeostasis in RNA replication by a positive-strand RNA virus. J. Virol. 85:5494–5503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang X, et al. 2005. Brome mosaic virus 1a nucleoside triphosphatase/helicase domain plays crucial roles in recruiting RNA replication templates. J. Virol. 79:13747–13758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Welsch S, et al. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wright R, Basson M, D'Ari L, Rine J. 1988. Increased amounts of HMG-CoA reductase induce “karmellae”: a proliferation of stacked membrane pairs surrounding the yeast nucleus. J. Cell Biol. 107:101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Xiao S, Chye ML. 2011. New roles for acyl-CoA-binding proteins (ACBPs) in plant development, stress responses and lipid metabolism. Prog. Lipid Res. 50:141–151 [DOI] [PubMed] [Google Scholar]
- 82. Yang F, et al. 2008. Expression of hepatitis B virus proteins in transgenic mice alters lipid metabolism and induces oxidative stress in the liver. J. Hepatol. 48:12–19 [DOI] [PubMed] [Google Scholar]
- 83. Yang YS, Strittmatter SM. 2007. The reticulons: a family of proteins with diverse functions. Genome Biol. 8:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ye J. 2007. Reliance of host cholesterol metabolic pathways for the life cycle of hepatitis C virus. PLoS Pathog. 3:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]