Abstract
The Epstein-Barr virus (EBV) proteins latent membrane proteins 1 and 2 (LMP1 and LMP2) are frequently expressed in EBV-associated lymphoid and epithelial cancers and have complex effects on cell signaling and growth. The effects of these proteins on epithelial cell growth were assessed in vivo using transgenic mice driven by the keratin 14 promoter (K14). The development of papillomas and carcinomas was determined in the tumor initiator and promoter model using dimethyl benzanthracene (DMBA), followed by repeated treatments of 12-O-tetradecanoyl phorbol 13-acetate (TPA). In these assays, LMP1 functioned as a weak tumor promoter and increased papilloma formation. In contrast, mice expressing LMP2A did not induce or promote papilloma formation. Transgenic LMP1 mice had slightly increased development of squamous cell carcinoma; however, the development of carcinoma was significantly increased in the doubly transgenic mice expressing both LMP1 and LMP2A. DMBA treatment induces an activating mutation in the Harvey-ras (H-ras61) oncogene, and this mutation was identified in most papillomas and carcinomas although several papillomas and carcinomas in K14-LMP1 and K14-LMP1/LMP2A mice lacked the mutation. Analysis of signaling pathways that are known to be activated by LMP1 and/or LMP2 indicated that all genotypes had high levels of activated extracellular signal-regulated kinase (ERK) and Stat3 in carcinomas with significantly higher activation in the doubly transgenic carcinomas. These findings suggest that, in combination, LMP1 and LMP2 contribute to carcinoma progression and that this may reflect the combined effects of the proteins on activation of multiple signaling pathways. This study is the first to characterize the effects of LMP2 on tumor initiation and promotion and to identify an effect of the combined expression of LMP1 and LMP2 on the increase of carcinoma development.
INTRODUCTION
Epstein-Barr virus (EBV) is an important human pathogen that is closely linked to major malignancies that develop in both epithelial and lymphoid cells and include nasopharyngeal carcinoma (NPC), gastric cancer, Hodgkin lymphoma, Burkitt lymphoma, and lymphomas that develop in the immunocompromised, including both AIDS and posttransplant lymphomas (38, 54). The EBV latent membrane proteins 1 and 2 (LMP1 and LMP2) are frequently expressed in these cancers and have profound effects on cellular signaling networks and growth properties in vitro (31, 37). In epithelial cells, LMP1 and LMP2 have been shown to affect differentiation, migration, anchorage independence, and tumorigenicity.
LMP1 is required for transformation of B lymphocytes by EBV infection and can transform rodent and human fibroblasts to form foci and colonies in soft agar and tumors in nude mice (38). LMP1 functionally resembles a ligand-independent constitutively active member of the tumor necrosis factor receptor (TNFR) superfamily and binds the same TNFR-associated factors (TRAFs) as CD40. Using transgenic mice, it was shown that, in vivo, LMP1 can partially substitute for CD40 in activating B cell responses (51). Similarly to other members of the TNFR family, LMP1 activates many signaling pathways, including NF-κB, PI3K/Akt, AP-1, Jak/Stat, Jun N-terminal protein kinase (JNK), and the p38 and extracellular signal-regulated kinase (ERK) mitogen-activated protein kinases (MAPKs) (31, 37, 50). Activation of NF-κB is required for B cell transformation by EBV and its inhibition results in cell death through apoptosis (2). However, in rodent fibroblast transformation, activation of NF-κB by LMP1 is not required, whereas activation of both PI3K and ERK is essential (22). Expression of LMP1 in some epithelial cell lines results in epithelial/mesenchymal transition (EMT) and a cadherin switch, an increasingly recognized phenomenon responsible for the metastasis of melanomas, breast, and prostate cancers (46).
Although LMP2 is not strictly required for B cell transformation, it has major effects on B cell signaling and growth, as it can both block and substitute for signaling from the B cell receptor (29, 30). In transgenic mice, where LMP2 is expressed under the control of the immunoglobulin heavy chain promoter and enhancer (Ig-LMP2), LMP2 provides prosurvival signals to allow for the survival and colonization of aberrant B cells lacking a functional B cell receptor in peripheral lymphoid organs (3, 4). Expression of LMP2 in epithelial cell lines inhibits differentiation and induces migration. LMP2 expression also results in transformation of some epithelial cell lines to anchorage independence and tumorigenicity through effects on Syk, PI3K/Akt, and Ras activation (18, 33, 34, 41). LMP1 and LMP2 have also been shown to be secreted in exosomes, and exposure to LMP1 exosomes induces activation of ERK and PI3K (11, 26). These potential paracrine effects may be more apparent in vivo than in in vitro cell cultures.
EBV efficiently infects and transforms B lymphocytes in vitro; however, epithelial infection is inefficient and persistent infection and transformation do not result (55). Thus, many of the properties of LMP1 and LMP2 on epithelial cells have been identified by expression of the proteins in cell lines in vitro and their individual or combined effects have not been tested in vivo. Transgenic mice are a powerful tool to identify and characterize the transforming properties of viral and cellular genes in vivo. For example, the Ig-LMP2 transgenic mice revealed prosurvival signals that were not apparent in cultured cells (3, 4). Similarly, LMP1 cannot transform B lymphocytes by itself; however, Ig promoter transgenic LMP1 mice do have increased development of B cell lymphomas. These lymphomas develop in approximately 50% of elderly mice (51). The low penetrance and advanced age suggest that additional genetic changes occur that synergize with LMP1 to contribute to lymphoma development.
A limited number of studies in epithelial cells in transgenic mice have shown that LMP1 expressed under the control of the polyomavirus early promoter resulted in hyperproliferation of the basal epithelium and in classic initiation-promotion studies showed enhanced carcinogen-induced papilloma formation (8, 52). In these transgenic mice, LMP1 could partially substitute for treatment with the tumor promoter, 12-O-tetradecanoyl phorbol 13-acetate (TPA), in the multistage carcinogenesis model. LMP1 transgenic mice with expression controlled by the ED-L2 EBV early lytic promoter, which is highly active in stratified epithelial cells, had high levels of LMP1 expression particularly evident in the ears, where highly inflamed corrosive lesions developed that could progress to carcinomas (7, 48). Transgenic mice, where expression of LMP2 was targeted to the basal epithelium using the K14 keratin promoter, lacked an apparent phenotype on epithelial cell differentiation or spontaneous cancerous growth (17).
In this study, transgenic mice that express LMP1 and LMP2 using the K14 promoter were evaluated for their ability to function in tumor initiation, promotion, or progression in classical skin painting experiments. K14-LMP1 transgenic mice had increased and prolonged development of papillomas after dimethyl benzanthracene (DMBA) and TPA treatment, indicating that it functioned as a weak promoter. In contrast, K14-LMP2 mice were similar to nontransgenic controls. Additionally, the doubly transgenic mice developed papillomas comparably to the singly transgenic K14-LMP1 mice, indicating the lack of effect of LMP2 on LMP1-enhanced papilloma formation. Importantly, the development of squamous cell carcinoma (SCC) was significantly increased in the doubly transgenic animals. Identification of pathways known to be activated by LMP1 and/or LMP2 revealed that all tumors had high levels of activated ERK and Stat3, with the highest levels in the doubly transgenic carcinomas. This is the first analysis of LMP1 and LMP2 expressed in combination in epithelial cells in transgenic mice. These findings indicate that expression of LMP1 and LMP2 increases susceptibility to tumor development.
MATERIALS AND METHODS
Ethics statement.
Animals were housed at The University of North Carolina at Chapel Hill in facilities accredited by the international Association for the Assessment and Accreditation of Laboratory Animal Care. All protocols were approved by University of North Carolina Institutional Animal Care and Use Committee (under IACUC protocol identification no. 09-247, 06-244, 03-278), compliant with the United States Public Health Service Policy on Humane Care and Use of Laboratory Animals, the Amended Animal Welfare Act of 1985, and the regulations of the United States Department of Agriculture (USDA) (Animal Welfare Assurance no. A3410-01; USDA registration no. 55-R-004). This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. To minimize suffering, animals were monitored daily and subjected to a maximum of 3 skin biopsies. Mice carrying tumor lesions reaching 1.3 cm were euthanized.
Generation of transgenic mice.
The K14-LMP1 mice were generated from EBV genomic sequences (169570 to 166620) encoding the LMP1 gene cloned into the BamHI site of pGEM2 (Promega). EcoRI linkers were added to the HindIII fragment containing the K14 promoter, a gift from Miroslav Blumenberg of NYU Langone Medical Center, and this fragment was subcloned into the EcoRI site in the multiple cloning site upstream of LMP1. The constructs were confirmed by sequencing. The K14-LMP1 construct was linearized with PvuI and injected into fertilized oocytes from (C57BL/6 × SJL)F1 mice. Founders were crossed initially to BALB/c. Animals were screened for the presence of the transgene by Southern blot analysis of tail DNA and hybridization to the XhoI fragment within LMP1 (13) or by PCR with primers 168821R (5′AGAGTCCACCAGTTTTGTTG-3′) and 169251L (5′ACCTTCTCTGTCCACTTGGA-3′). Transgenic mice expressing LMP2A (K14-LMP2) were a kind gift from Richard Longnecker of Northwestern University (17) and genotyped by PCR with primers LMP2-741R (5′CAGGGGGCCTAGGTACTCTT-3′) and LMP2-951L (5′GTGCCCAAAATCAGTGACGC-3′). Transgenic mice were maintained, and DMBA/TPA treatment studies were performed with the assistance of the Animal Studies Core (University of North Carolina at Chapel Hill).
DMBA/TPA treatment.
The backs of 6- to 8-week-old mice were shaved and painted once with 100 μg of DMBA on one side and acetone as the vehicle control on the other side. One week later, mice were painted with TPA twice weekly with 10 nmol of TPA or acetone for 20 weeks. Mice were shaved every 2 weeks and monitored for benign growths (papillomas) and malignant lesions (carcinomas) every week. Papillomas were categorized and counted by size (small [0 to 0.2 cm], medium [0.2 to 0.4 cm], large [>0.4 cm]). Mice with large growths and lesions were biopsied weekly for histology and were kept for a maximum of 3 skin biopsies. All remaining mice were sacrificed 1 year post-DMBA treatment. At the time of sacrifice, tissues harvested included acetone-painted skin, DMBA/TPA-painted skin, enlarged lymph nodes, growths, and lesions.
Histopathology analysis.
Skin tumors and enlarged lymph nodes were harvested and fixed in buffered formalin. Tissues were embedded in paraffin blocks, sectioned, and stained with hematoxylin and eosin (H&E) by the UNC Histopathology Facility. Tissue sections were examined in a blinded fashion by the same pathologist (J.N.N., UNC Department of Pathology and Laboratory Medicine) and classified as papilloma, carcinoma in situ (CIS), and SCC. The following criteria were used in this classification. Papillomas consisted of well-differentiated epithelial cells forming finger-like projections from the epithelial surfaces, often with a central, connective tissue stalk. Hyperplasia and hyperkeratosis of the surface epithelium, with retention of nuclei in cells of the keratinized layers, was common. Carcinoma in situ was identified by an intact basement membrane of the squamous epithelium; however, the epithelium was thickened and there was failure of normal differentiation. Cells demonstrated pleomorphism, variable nuclear size and shape, abnormal nuclear morphology, and abnormal or misplaced mitotic figures near the epithelial surface. Invasive squamous cell carcinomas had masses of recognizable squamous epithelial cells, arising from the squamous epithelium, but cells were pleomorphic in size and shape, had variable nuclear sizes, and had abnormal nuclear morphology (hyperchromatic dark-staining DNA, coarsely clumped chromatin, large, prominent, often multiple nucleoli identifiable within the nuclei) and variable nuclear/cytoplasmic ratios. Mitoses could be identified and were sometimes atypical. The squamous epithelial cells demonstrated loss of polarity. Orderly differentiation of squamous cells was lost, and large, immature appearing cells or cells containing mitotic figures were present within the middle and superficial layers of epithelium; keratinized cells were present within the middle or deep layers of epithelium. The basal membrane was breached, and atypical cells extended into the dermis, sometimes forming sheets or large masses of cells, occasionally with keratinized cells (keratin pearls) at the center.
Statistical analysis.
For endpoint comparisons, carcinoma, and H-ras61 mutation incidences (see Tables 2 to 5) were analyzed using Fisher's exact test. The Kaplan-Meier (or product-limit) method was used to estimate the time to carcinoma development, and the log rank test was used to test for both overall and pairwise differences (see Fig. 2C). To compare papilloma numbers (Fig. 2A and B), a method known as “response features analysis” was used to analyze the repeated measure of the number of papillomas at each time point (24). This method considers the growth profile of each animal by calculating the change (slope) in papilloma counts over time and is then compared between genotypes using the nonparametric one-way analysis of variance (Kruskal-Wallis test using Van der Waerden normal scores). The Wilcoxon two-group method (also using Van der Waerden normal scores) was then used to test for significant (pairwise) differences between genotypes. Reported P values are for individual tests, unadjusted for multiple comparisons. Statistical analyses were performed using SAS statistical software, version 9.2 (SAS Institute Inc., Cary, NC). For comparing immunoblot densitometry values, the Student t test (two-sided, unpaired) was applied, using the F test to determine variance (equal or unequal).
Table 2.
Carcinoma incidence in the combined DMBA/TPA treatment study
| LMP1/LMP2 genotype | Total mice entered into studya | No. of mice with SCC | No. of mice with CIS | % of mice with carcinoma (SCC or CIS)b |
|---|---|---|---|---|
| −/− | 30 | 2 | 2 | 13 |
| +/− | 28 | 5 | 1 | 21 |
| −/+ | 17 | 3 | 0 | 18 |
| +/+ | 35 | 8 | 4 | 34c |
The total number of mice entered into the study did not necessarily survive the entire study before they got a chance to develop a carcinoma. Mice that completed the study include mice that either survived the entire 1 year study or were sacrificed before the end of the study but had already developed a diagnosed carcinoma.
Percentage of mice with carcinomas represented as total number of mice entered into the study.
P = 0.05, statistically significant between K14-LMP1/2 (+/+) and nontransgenic (−/−) mice.
Table 5.
H-ras61 mutation analysis of papillomasa
| LMP1/LMP2 genotype | No. of mice biopsied for papilloma | No. of mice without H-ras61 mutation/no. of mice analyzed | No. of papillomas without H-ras61 mutation/no. of papillomas analyzed |
|---|---|---|---|
| −/− | 24 | 0/24 | 0/24 |
| +/− | 19 | 1/19 | 1/19 |
| −/+ | 12 | 0/12 | 0/12 |
| +/+ | 19 | 2/19 | 2/19 |
Samples were prescreened for genomic DNA quality by PCR for GAPDH followed by analysis for H-ras61.
Fig 2.
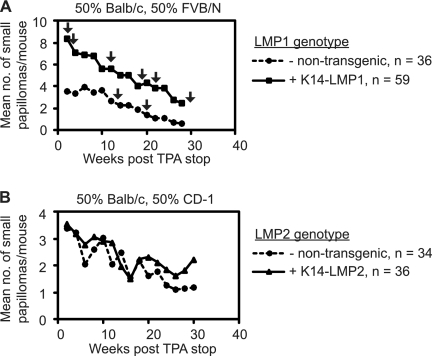
Papilloma and carcinoma incidence in DMBA/TPA-treated singly and doubly transgenic LMP1 and LMP2 mice. Under the common strain background (50% FVB/N, 25% CD-1, 25% BALB/c), mice were treated with DMBA/TPA and observed for papilloma and carcinoma development. The number of mice per study group (n) is indicated on the right, and the mean number of small papillomas per mouse is calculated from mice remaining in the study at each time interval. The number of small papillomas was counted at 2-week intervals for the 20 weeks during TPA treatment (A) and thereafter for 36 weeks post-TPA stop (B). (A) During TPA treatment, no mice were sacrificed and the data are represented as total mice entered into the study. (B) Postcessation of TPA treatment, mice were periodically sacrificed and the data are represented as mice remaining in the study. The Wilcoxon two-group method was used to test for significant differences between genotypes. (C) Kaplan-Meier plot of DMBA/TPA-treated mice depicting carcinoma-free state as a function of time post-TPA start. This comparison was performed on the subset of mice that completed the 1-year carcinoma study. Log rank test indicate differences between the four genotypes (P = 0.01), and pairwise comparisons indicate significant differences between K14-LMP1/LMP2 compared to nontransgenic controls and K14-LMP2 mice (P = 0.005 and P = 0.01, respectively).
Analysis of LMP1 and LMP2 expression by RT-PCR and immunoblot analysis.
Tissues were homogenized using the TissueLyser II (Qiagen). Protein was harvested in RIPA buffer followed by immunoblotting. Total RNA was harvested using the RNeasy minikit (Qiagen), and contaminating genomic DNA was removed with the DNA-free kit (Ambion). Reverse transcriptase (RT)-PCR was performed using Brilliant II quantitative RT-PCR (Stratagene), 100 ng of total RNA and the same primers used for genotyping for LMP1 and LMP2 detection, and primers qGAPDH5′ (5′ TGCACCACCAACTGCTTAGC 3′)/qGAPDH3′ (5′ GAGGGGCCATCCACAGTCTT 3′) for GAPDH (glyceraldehyde-3-phosphate dehydrogenase).
Immunohistochemistry staining.
Paraffin-embedded sections were stained for phospho-Stat3 (Tyr705) according to the manufacturer's recommendation (Cell Signaling). For detection, goat anti-rabbit IgG poly-horseradish peroxidase (HRP) immunohistochemistry amplification reagent (Millipore) was used and developed with enhanced 3,3′-diaminobenzidine (DAB), counterstained with Mayer's hematoxylin.
Antibodies.
Rabbit anti-pAkt (Ser473 and Thr308), anti-pStat3 α/β (Tyr705), and Akt isoform antibody sampler kit (Akt1/2/3, monoclonal pan-Akt) were purchased from Cell Signaling. Mouse anti-pERK1/2 (Tyr204) (E-4); rabbit anti-ERK2 (C-14), anti-Stat3 α/β (H-190), and anti-GAPDH (FL-335) were purchased from Santa Cruz. Rabbit polyclonal pan-Akt was purchased from Stressgen. Rat anti-LMP1 (clones 8G3, 1G6, 7E10, and 7G8) was purchased from Ascenion. Rat anti-LMP2 (clone 14B7) was purchased from Abcam.
Analysis of H-ras and K-ras mutations at codons 12, 13, and 61.
Genomic DNA was harvested from homogenized tissue using the DNeasy kit with RNase treatment (Qiagen). DNA quality was assessed by PCR for the GAPDH gene using the same primers for RT-PCR. PCR was performed using the PCR 2× master mix (Fermentas), 100 ng of template DNA, and the primers MH12A1 (5′ ATGACAGAATACAAGCTTGTGGTG 3′)/MH61A2 (5′ GGCAAATACACAGAGGAAGCCCTC 3′) for the H-ras gene (spanning exons 1 to 2). For the K-ras gene, nested PCR was performed on exons 1 and 2. For exon 1 (containing codons 12 and 13), external primers K-ras exon1-5′-3 (5′ TGATAATCTTGTGTGAGACA 3′)/K-ras exon1-3′ (5′ ATATCTTTTTCAAAGCGGCT 3′) were used for the first PCR, and internal primers K-ras exon1-5′ (5′ CTTGTGTGAGACATGTTCTAAT 3′)/K-ras exon1-3′-3 (5′ CTCTATCGTAGGGTCGTACT 3′) were used for the second PCR. For exon 2 (containing codon 61) primers, K-ras exon2-5′ (5′ AGCTGTTTACATCACCTTGT 3′)/K-ras exon 2-3′ (5′ GGCATAACAATTAGCAAAGA 3′) were used for both PCR cycles. For all PCRs, no template served as a negative control. The PCR products were purified using the QIAquick PCR purification kit (Qiagen) and sent to sequencing for codons 12, 13, and 61 or digested with XbaI to confirm the mutation at H-ras61.
RESULTS
LMP1 increases formation and retention of papillomas after tumor promotion and initiation.
Chemical carcinogenesis using a classical regimen of skin painting with a tumor initiator (DMBA) and promoter (TPA) has been extensively used to study multistage tumor development (1, 56). The stages of tumor development involve tumor initiation, promotion to benign papilloma, and progression to malignant and invasive squamous cell carcinoma. In this study, initial experiments tested the K14-LMP1 transgenic mice in the BALB/c mouse strain that were treated with TPA or treated with DMBA, followed by repeated treatments with TPA. Papillomas did not develop in any of the treated mice (Table 1). BALB/c mice are relatively resistant to papilloma formation and carcinoma development and less than strong effects would not be apparent (53). A previous study had characterized LMP1 transgenic mice in the FVB/N mouse strain that is more susceptible to initiation/promotion treatment (8, 21, 52, 53). A considerable number of papillomas developed in all treated mice; however, despite the high background of papilloma formation in the control mice, the studies were able to discern that LMP1 had a subtle effect on numbers of papillomas and cancer development. To decrease the influence of strain-specific effects, and determine if the effects of LMP1 may be more apparent in a less responsive background than the full FVB/N, the K14-LMP1 BALB/c transgenic mice were crossed with FVB/N and the F1 offspring were treated with (i) DMBA one time (1×), (ii) DMBA two times (2×), (iii) TPA alone for 20 weeks, or (iv) DMBA 1× followed by TPA for 20 weeks. Few papillomas developed in the mice treated only with DMBA in LMP1-positive or -negative littermates (Table 1). In the combined DMBA/TPA treatment study, approximately twice as many papillomas developed in the LMP1-positive mice (Fig. 1A). At the first week postcessation of TPA treatment, the LMP1 mice had an average of 8.5 papillomas while the negative mice averaged 3.5 papillomas. By 12 weeks postcessation of TPA treatment, papilloma development decreased in the LMP1 mice to 5.6 papillomas and in the negative controls to 2.6 papillomas per mouse (Table 1). By 28 weeks postcessation of TPA treatment, LMP1 mice had decreased to 2.4 papillomas while the negative mice had an average of less than 1 papilloma (Fig. 1A). Additionally, 6/59 (10%) LMP1 mice developed carcinomas compared to 2/36 (6%) negative littermates. These findings confirmed the previous studies that identified LMP1 to function as a tumor promoter.
Table 1.
Comparison of papilloma and carcinoma incidence in various mouse strains receiving DMBA and/or TPA treatment
| Mouse strain | Treatment | Avg no. of papillomas/mouse at wk 12 post-TPA stop (total no. of mice in study) |
|||
|---|---|---|---|---|---|
| LMP1+ | LMP1− | ||||
| 100% BALB/c | TPAb | 0 (14) | 0 (10) | ||
| DMBA/TPAc | 0 (4) | 0 (4) | |||
| 50% BALB/c, 50% FVB/N | DMBAa 1× | 0 (13) | 0 (16) | ||
| DMBAa 2× | 0.66 (12) | 0.2 (15) | |||
| TPAb | 0 (5) | 0 (8) | |||
| DMBA/TPAc | 5.6 (59) | 2.6 (36) | |||
| LMP2+ | LMP2− | ||||
| 50% BALB/c, 50% CD1 | DMBAa 1× | 0 (9) | 0 (12) | ||
| DMBAa 2× | 0 (15) | 0 (15) | |||
| TPAb | 0 (14) | 0 (13) | |||
| DMBA/TPAc | 2.86 (36) | 2.06 (34) | |||
| 50% BALB/c, 50% FVB/N | DMBA/TPAc | 3.3 (40) | 3.4 (31) | ||
| LMP1+ | LMP2+ | LMP1+/2+ | LMP1−/2− | ||
| 25% BALB/c, 25% CD1, 50% FVB/N | DMBA/TPAc | 6.8 (28) | 1.4 (17) | 7.9 (35) | 2.8 (30) |
100 μg DMBA.
10 nmol TPA.
100 μg/10 nmol DMBA/TPA.
Fig 1.
Papilloma and carcinoma incidence in DMBA/TPA-treated singly transgenic K14-LMP1 (A) and K14-LMP2 (B) mice. Small papillomas were tabulated at 2-week intervals, and carcinomas appeared during the 30-week interval postcessation of TPA treatment. Plots represent papilloma retention following cessation of TPA treatment, and arrows indicate the first appearance of carcinomas. The number of mice per study group (n) is indicated on the right, and the mean number of small papillomas per mouse is calculated from mice remaining in the study at each time interval.
LMP2 does not promote papilloma formation and retention.
LMP2 transgenic mice expressing LMP2A, the isoform containing the N-terminal cytoplasmic signaling domain that is missing from LMP2B, here referred to simply as LMP2, was obtained on the CD1 background and was crossed with BALB/c for initial studies. LMP2-positive and LMP2-negative mice treated with DMBA alone (1× or 2×) or TPA alone did not develop papillomas (Table 1). Papillomas did develop after combined DMBA/TPA treatment with approximately equal numbers in LMP2-positive and -negative mice (Fig. 1B). At the first week postcessation of TPA treatment, both mice had an average of 3.5 papillomas, which decreased to 2 to 3 papillomas by 12 weeks postcessation of treatment (Fig. 1B; Table 1).
To evaluate the effects of LMP2 in the same background as the LMP1 mice, the LMP2 mice were bred onto a BALB/c background and then crossed with FVB/N and the 50% FVB/N F1 generation treated with 100 μg DMBA 1× followed by 20 weeks of TPA treatment. The first week post-TPA. LMP2 mice averaged 4.6 papillomas/mouse while the negative mice averaged 4.3 papillomas/mouse (data not shown). By week 12, the papilloma numbers decreased to 3.3 and 3.4 papillomas/mouse, respectively, for positive and negative mice (Table 1). Several keratoacanthomas developed in both genotypes, and each genotype developed 2 carcinomas. The K14-LMP2 mice have been previously described to lack a discernible phenotype (17). The data presented here indicate that in this transgenic system, LMP2 does not function as a tumor initiator or promoter.
The effects of combined expression of LMP1 and LMP2 on promotion and progression.
To evaluate the combined effects of LMP1 and LMP2, the BALB/c LMP1 mice and CD1 LMP2 mice were both crossed with FVB/N and the LMP1+ and LMP2+ F1 were crossed to produce a mixed genotype of approximately 50% FVB/N. A total of 110 mice genotyped into −/−, +/−, −/+, +/+ K14-LMP1/LMP2 were treated with DMBA followed by TPA treatment. Mice were counted for skin growths and lesions every week, and biopsy samples were taken periodically for histological analysis. Papillomas started appearing between 6 to 9 weeks during TPA treatment (Fig. 2A), but differences were most apparent at the end of the 20-week TPA treatment (Fig. 2B). During TPA treatment, the number of mice remained constant and the mean number of papillomas per mouse was averaged from total mice entered into the study (Fig. 2A). Post-TPA treatment, mice with lesions were biopsied or sacrificed and the mean number of papillomas per mouse was averaged from mice remaining in the study (Fig. 2B). The papillomas were grouped into small (<0.2 cm), medium (0.2 to 0.4 cm), and large (>0.4 cm) categories. The numbers of medium and large papillomas were highly variable due to loss or biopsy, and only small papillomas could be consistently enumerated and graphically tabulated. During TPA treatment, K14-LMP1 and K14-LMP1/LMP2 mice had higher numbers of small papillomas compared to both nontransgenic controls and K14-LMP2 mice (P = 0.004) (Fig. 2A). The K14-LMP1 mice and the K14-LMP1/LMP2 mice had 8 to 10 papillomas/mouse by the end of TPA treatment while the K14-LMP2 mice and negative mice averaged 3 to 5 papillomas/mouse (Fig. 2A). The numbers of papillomas decreased at the same rate for the K14-LMP1 and K14/LMP1/LMP2 mice and for the K14-LMP2 mice and controls (Fig. 2B). Importantly, comparison of the K14-LMP1 and K14-LMP1/LMP2 mice to both nontransgenic controls and K14-LMP2 mice indicated that they were highly distinct (P = 0.004, during TPA treatment; P = 0.001, post-TPA treatment) (Fig. 2A and B). Additionally, there was no statistical difference between K14-LMP1/LMP2 and K14-LMP1 mice. These findings support the previous findings that LMP1 increases the incidence of papilloma formation, while LMP2 does not increase papilloma formation or enhance the promotion effects of LMP1.
Carcinoma development was determined by histopathology of the biopsied samples. Samples diagnosed as carcinomas were tabulated and represented as total number of mice entered into the study (Table 2) or as the number of mice that completed the study (Table 3). Based on the total number of mice, the incidence of carcinoma (SCC/CIS) was higher in K14-LMP1 (21%) and K14-LMP2 (18%) mice compared to nontransgenic controls (13%). With the coexpression of both LMP1 and LMP2 in K14-LMP1/LMP2 mice, carcinoma development was increased (34%) compared to nontransgenic controls (13%, P = 0.05). However, animal numbers decreased during the study due to medical complications and animal regulations limiting the number of biopsies; therefore, the data were also analyzed based on the number of mice that completed the study. K14-LMP1 mice again had slightly higher numbers of carcinomas (50%) compared to the nontransgenic controls (33%) or the K14-LMP2 mice (30%). However, a greater number of carcinomas developed in the K14-LMP1/LMP2 mice (86%) than in the nontransgenic controls, which was significant with higher confidence (P = 0.01). These data indicate that the combined expression of LMP1 and LMP2 increases the progression to carcinoma.
Table 3.
Carcinoma incidence in the combined DMBA/TPA treatment study
| LMP1/LMP2 genotype | Total mice completing the studya | No. of mice with SCC | No. of mice with CIS | % of mice with carcinoma (SCC or CIS)b |
|---|---|---|---|---|
| −/− | 12 | 2 | 2 | 33 |
| +/− | 12 | 5 | 1 | 50 |
| −/+ | 10 | 3 | 0 | 30 |
| +/+ | 14 | 8 | 4 | 86c |
The total number of mice entered into the study did not necessarily survive the entire study before they got a chance to develop a carcinoma. Mice that completed the study include mice that either survived the entire 1 year study or were sacrificed before the end of the study but had already developed a diagnosed carcinoma.
Percentage of mice with carcinomas represented as total number of mice completing the study.
P = 0.01, statistically significant between K14-LMP1/2 (+/+) and nontransgenic (−/−) or K14-LMP2 (−/+) mice.
Kaplan-Meier analysis of the number of mice that remain carcinoma free over time revealed that the time to tumor development was not accelerated in the LMP1 or LMP2 transgenic animals (Fig. 2C). However, this analysis confirmed the differences in carcinoma incidences between the four genotypes (P = 0.01). Pairwise comparisons indicated significant differences in results for K14-LMP1/LMP2 mice compared to nontransgenic controls and K14-LMP2 mice (P = 0.005 and P = 0.01, respectively), with borderline significance compared to K14-LMP1 mice (P = 0.06). These analyses reveal that in these studies of initiation, promotion, and progression, LMP1 functions as a tumor promoter to induce papilloma formation while the combined expression of LMP1 and LMP2 contributes to tumor progression.
Expression of LMP1 and LMP2 in the K14 lineages.
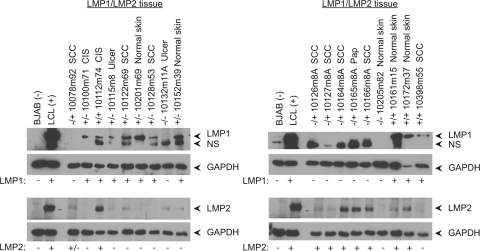
The expression of LMP1 and LMP2 in the K14 lineages was determined using immunoblotting and RT-PCR. Expression of LMP1 and LMP2 protein was determined by immunoblotting using rat monoclonal antibodies in protein extracts from biopsy samples. Samples of normal skin, papilloma, CIS, SCC, and ulcers that result from irritation of skin growths were compared to the levels of expression in an EBV immortalized (type III latency) lymphoblastoid cell line (LCL). LMP1 was detected in all samples and LMP2 expression was detected in most of the sampled tissues. The more frequent and stronger detection of LMP1 likely reflects differences in the antibody reactivities (Fig. 3). The levels of transgene protein expression were variable between samples, reflecting differing amounts of skin epithelial cells and underlying dermis within a biopsy sample. In biopsy samples with weak or undetectable protein levels, expression was confirmed using RT-PCR. These findings revealed that LMP1 and LMP2 were expressed in the normal skin samples, papillomas, and carcinomas of K14-transgenic mice analyzed in the study.
Fig 3.
Detection of LMP1 and LMP2 expression in carcinomas, papillomas, and normal skin samples. Tissues were confirmed for transgene expression by RT-PCR and immunoblot analysis. A representative immunoblot analysis for LMP1 and LMP2 is displayed. GAPDH was used as a loading control, and for LMP1/LMP2 detection controls, an EBV immortalized B cell line (LCL) was used as a positive control and an uninfected B cell line (BJAB) was used as a negative control. Pap, papillomas; NS, nonspecific band.
H-ras61 mutation is induced by DMBA in most papillomas and carcinomas.
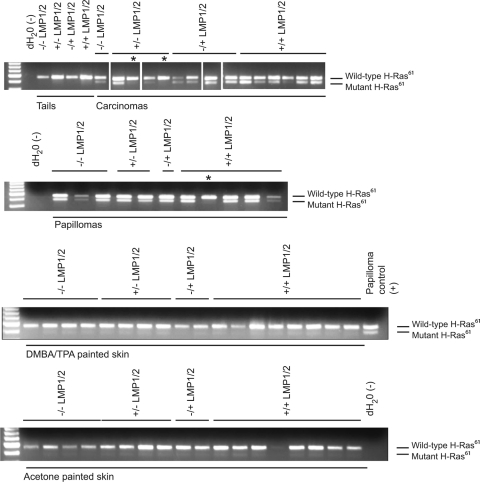
Previous studies have determined that treatment with DMBA results in an activating mutation in codon 61 (A182→T) of the H-ras gene (36). Treatment with other carcinogens such as MNNG and NOR1 induces mutations at other codons in H-ras, including codons 12 and 13 (39, 40). Mutations at these codons are typically mutually exclusive, suggesting that any one activating mutation is sufficient. Biopsy samples were analyzed for mutation at codon 61 of the H-ras gene. Mutation at codon 61 introduces an XbaI site, which can be analyzed by XbaI digestion of the PCR product (spanning exons 1 to 2). A combination of sequencing and XbaI digestion (Fig. 4) of the PCR product identified the mutation in most samples of papillomas and carcinomas from all genotypes. By sequencing, mutations were only detected at codon 61, indicating that LMP1 and/or LMP2 expression does not affect other hot spots (codons 12, 13) in the H-ras gene upon DMBA initiation. However, 2/5 carcinomas from two K14-LMP1 mice did not have detectable mutations in codons 12, 13, or 61 (Table 4 and Fig. 4, indicated by an asterisk). As mutations in K-ras in the absence of a functional H-ras gene have been identified in DMBA-treated H-ras knockout mice, potential mutations in K-ras were also assessed (12, 28). Sequencing of the hot spots in exons 1 and 2 of K-ras (codons 12, 13, and 61) did not identify any mutations that would substitute for the absence of the H-ras mutation at codon 61 in carcinomas that contained or lacked the H-ras61 mutation (22/25 carcinomas analyzed for K-ras exon 1 codons 12, 13; 20/25 carcinomas analyzed for K-ras exon 2 codon 61). Thus, the majority of carcinomas contained the predicted H-ras mutation; however, the absence of mutation in some samples may reflect the ability of LMP1 to activate common effector signals with H-ras.
Fig 4.
Detection of H-ras61mutation in tissues from DMBA/TPA-treated and vehicle control-treated mice. H-ras61 mutation was detected by XbaI digestion (a site introduced through mutation at codon 61) of a PCR product spanning exons 1 and 2. Tissues screened include carcinomas, papillomas, tails, and normal skin. Tails were not treated with carcinogens or vehicle and served as controls for any basal levels of spontaneous H-ras61 mutations. Samples where there were no detectable H-ras61 mutations are indicated (*) and were confirmed by sequencing.
Table 4.
H-ras61 mutation analysis of carcinomasa
| LMP1/LMP2 genotype | No. of mice diagnosed with SCC/CIS | No. of mice without H-ras61 mutation/no. of mice analyzed | No. of SCC/CIS without H-ras61 mutation/no. of SCC/CIS analyzed |
|---|---|---|---|
| −/− | 4 | 0/4 | 0/4 |
| +/− | 6 | 2/4 | 2/5 |
| −/+ | 3 | 0/2 | 0/6 |
| +/+ | 12 | 0/9 | 0/10 |
Samples were prescreened for genomic DNA quality by PCR for GAPDH followed by analysis for H-ras61.
To analyze H-ras61 mutations during earlier stages of tumor progression, papillomas and DMBA/TPA- or vehicle-treated skin samples from the mice that developed carcinomas were evaluated (Fig. 4). H-ras61 mutation was not detected in vehicle-treated or DMBA/TPA-treated skin samples by sequencing or XbaI digestion (Fig. 4). In contrast, H-ras mutation was detected in most papillomas. These findings indicate that mutation in H-ras is the initiating event for papilloma induction. Interestingly, one of the K14-LMP1/LMP2 papillomas lacked the H-ras61 mutation (Fig. 4). Extending the analysis to include a representative papilloma from each mouse harvested identified 1/19 K14-LMP1 papilloma and 2/19 K14-LMP1/LMP2 papillomas that lacked detectable H-ras61 mutations (Table 5). When comparing all growths (papillomas and carcinomas) that lacked the H-ras61 mutation between LMP1-expressing (5/53) and LMP1-nonexpressing (0/46) mice, the difference was borderline significant (P = 0.0593). This suggests that in the presence of LMP1, papillomas and carcinomas from K14-LMP1 or K14-LMP1/LMP2 mice can occasionally develop in the absence of DMBA-induced H-ras61 mutation.
Identification of LMP1- and LMP2-activated signaling pathways in carcinomas.
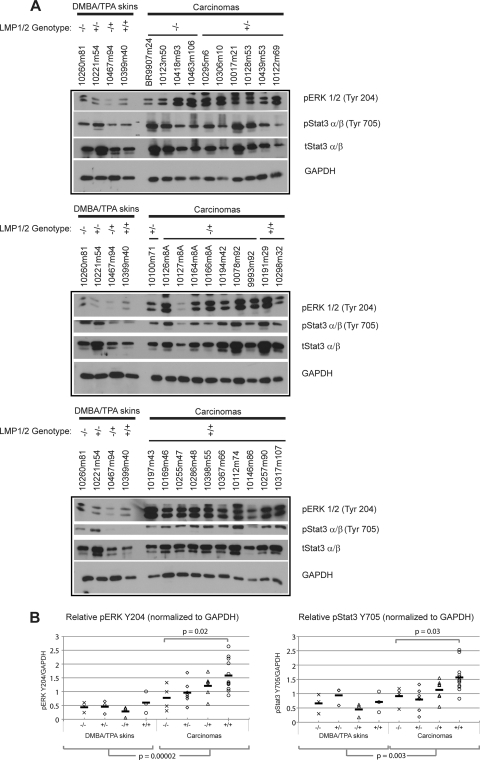
LMP1 is known to activate ERK, STAT3, and Akt, and activation of all three pathways is required for LMP1-mediated rodent fibroblast transformation (10, 14, 22, 45). LMP2 also activates Akt and is linked to Ras activation (33, 35). To assess the activation of these pathways by LMP1 and LMP2 in vivo, normal skin samples and carcinomas were analyzed by immunoblotting using antibodies for phosphorylation activated sites (Fig. 5A). Quantitation of activated ERK relative to GAPDH indicated that all the carcinomas had higher levels (∼2- to 4-fold) of activated ERK compared to DMBA/TPA-treated skin samples (P = 0.00002) (Fig. 5B). Interestingly, comparisons of carcinomas showed that the doubly transgenic K14-LMP1/LMP2 carcinoma samples were significantly more activated than the nontransgenic controls (P = 0.02) (Fig. 5B).
Fig 5.
Immunoblot analysis of normal skin samples and carcinomas for activated ERK and Stat3 in DMBA/TPA-treated mice. (A) Activation was detected with phospho-specific antibodies to ERK1/2 (Y204) and Stat3 (Y705). GAPDH was used as a loading control. (B) ERK and Stat3 activation was normalized to GAPDH and plotted for comparison. The same DMBA/TPA skin samples were run on each gel, allowing for an assessment of variation between gels. The distribution of ERK and Stat3 activation is displayed for each genotype, and the mean value is indicated by a solid line. Student's t test was used to compare between tissue groups and transgenic samples relative to the control. Comparisons with P values of ≤0.05 are displayed.
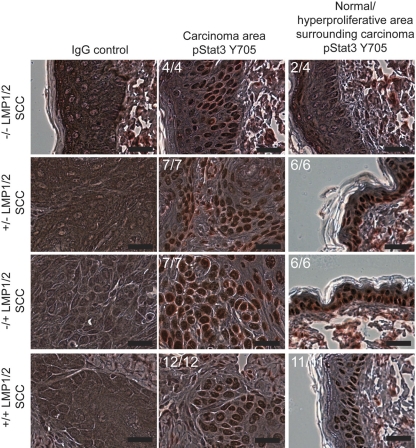
Stat3 is required for both the initiation and promotion of epithelial carcinogenesis by DMBA (6). Activation of Stat3 was determined by blotting for phosphorylation at Tyr705, which is linked to Stat3 nuclear translocation, and quantitated relative to GAPDH (Fig. 5B). Carcinomas from all genotypes had increased levels (∼1.2- to 2.5-fold) of pStat3 above those of nontransgenic DMBA/TPA-treated normal skin samples and, on average, the levels were higher in the carcinoma group than in the DMBA/TPA-treated normal skin samples (P = 0.003) (Fig. 5B). Carcinomas from the doubly transgenic mice had significantly higher levels of pStat3 than carcinomas that developed in the control littermates (P = 0.03). Immunohistochemistry detected nuclear localization of activated Stat3 at Tyr705 in the neoplastic cells of all carcinomas (Fig. 6). Nonmalignant areas surrounding the carcinomas were also analyzed for differences in nuclear p(Tyr705)Stat3. Activated nuclear p(Tyr705)Stat3 was detected in all areas of the LMP1 and LMP2 transgenic skin samples but was not detected in some areas of 2/4 nontransgenic samples (Fig. 6). These observations confirm that ERK and Stat3 activation occurs during initiation and promotion and that their activation is most enhanced in the doubly transgenic group with the highest carcinoma incidence.
Fig 6.
Immunohistochemistry for pStat3 (Y705) in carcinomas and surrounding epithelia of DMBA/TPA-treated mice. The number of samples showing a similar detection to the representative field is indicated. Scale bar, 20 μm.
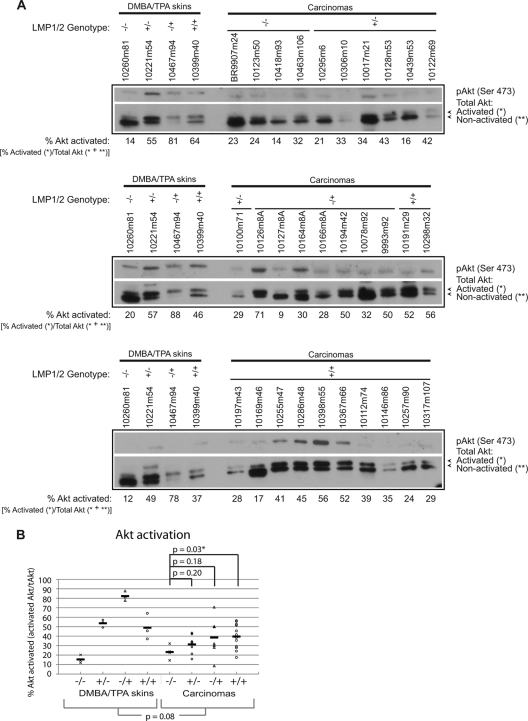
Activation of Akt was also assessed in transgenic skin samples and carcinomas. Immunoblotting for total Akt using polyclonal antisera identified two bands, and the slower migrating upper band was also detected with antibody specific for phosphorylation at Ser473, which is mediated by the mammalian target of rapamycin complex 2 (mTORC2) (Fig. 7A). Quantitation of the upper Akt band to the total amount of Akt revealed that higher levels of activated Akt were detected in the doubly transgenic K14-LMP1/LMP2 carcinomas compared to the nontransgenic carcinomas (P = 0.03) (Fig. 7B). However, Akt activation was not significantly higher in the carcinomas from all genotypes compared to the DMBA/TPA-treated normal skin samples (P = 0.08) (Fig. 7B), suggesting that the effects of LMP1 and LMP2 on Akt may contribute to earlier stages of tumor initiation/promotion, as has been shown in other DMBA/TPA skin tumorigenesis studies (42, 43).
Fig 7.
Immunoblot analysis of normal skin samples and carcinomas for activated Akt in DMBA/TPA-treated mice. (A) Activated Akt was detected using phospho-specific antibodies to Ser-473 and as the upper species of the doublet band detected with the polyclonal total Akt antibody. The percentage of Akt activation was calculated from the average of three exposures of the total Akt immunoblot (activated band/sum of activated and nonactivated bands). One representative exposure from each gel is shown. (B) Percentage of Akt activation is plotted for each sample, and the mean value for each genotype is indicated by a solid line. The same DMBA/TPA skin samples were run on each gel, allowing for an assessment of variation between gels. Student's t test was used to compare between tissue groups and transgenic samples relative to the control. An asterisk denotes significant P values of ≤0.05.
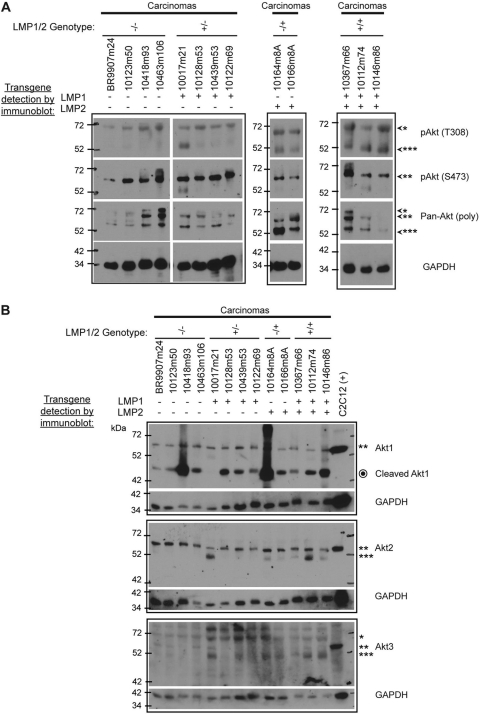
To further evaluate the effects of LMP1 and LMP2 on Akt activation, samples where LMP1 and LMP2 were readily detected by immunoblotting were analyzed using additional antibodies to distinguish different phospho- and isoform-specific forms of Akt (Fig. 8). Immunoblotting for total Akt with the rabbit polyclonal antibody detected three bands in some samples and clearly in two of the control carcinomas (Fig. 8A). Antibody specific for phospho-Ser473 predominantly detected the middle band (**) and weakly detected the fastest migrating band (***). The slowest (*) and fastest (***) migrating bands were equally detected with a phospho-T308 Akt-specific antibody. This residue is thought to be phosphorylated through PI3K activation of PDK1. The fastest migrating (***) band detected with phospho-Thr308 was not detected in the control carcinomas but was detected in 1/4 LMP1+ carcinomas, 2/2 LMP2+ carcinomas, and 3/3 LMP1+/LMP2+ carcinoma samples (Fig. 8A).
Fig 8.
Immunoblot analysis of highly expressing carcinomas for activated pAkt (S473, T308) and Akt isoforms. (A) Various modifications of Akt (*, **, ***, slow, medium, and fast migrating, respectively) were detected with phospho-specific antibodies (pAkt-S473 and -T308) and a polyclonal pan-Akt antibody. (B) The expression of Akt isoforms was detected using isoform-specific peptide antibodies. The corresponding position of the various Akt modification bands is indicated with asterisks (*, **, ***). A cleaved Akt1 product was also detected and is indicated by an enclosed circle ( ). GAPDH was used as a loading control.
). GAPDH was used as a loading control.
The lysates were also analyzed using antibodies to peptides specific for the Akt1, -2, and -3 isoforms to screen for possible differences in isoform expression that could account for phenotypic differences (Fig. 8B). The C2C12 mouse myoblast cell line was included as a positive control for all three isoforms. Akt1 and Akt2 antibodies detected the ** band and Akt2 antibody also detected the *** band. Additionally, Akt1 antibody strongly detected a smaller band of approximately 48 kDa previously described to be a caspase cleavage product (20, 27). The isotype-specific antibodies have been shown to not cross-react; thus the detection of the ** band by both antibodies perhaps indicates that both forms migrate identically. The Akt3 antibody faintly detected multiple bands on long exposure, suggesting that Akt3 is not expressed. Analysis of the vehicle control-treated skin samples identified only Akt2, the cleaved form of Akt1, and no Akt3 (data not shown). These findings indicate that Akt2 is the predominant isoform of Akt that is activated in this model.
DISCUSSION
LMP1 and LMP2 are expressed in NPC and other EBV-associated tumors and have potent effects on cell growth and transformation (38, 54, 55). The DMBA/TPA painting regimen has been a critical tool to determine the contribution of chemical carcinogens and oncogene activation in a model of multistage carcinogenesis that distinguishes effects on tumor initiation, promotion, and progression (1, 56). In combination with transgenic mice, the approach has revealed distinct effects of the human papillomavirus (HPV) E7, E6, and E5 on papilloma promotion, carcinoma progression, and both stages, respectively (25, 47). Animal models are not available for HPV, and these distinct functional contributions could not have been identified using in vitro culture systems. Similarly, EBV also lacks a mouse model and the study of its effects on epithelial cell growth is dependent on cell culture systems. However, interesting transgenic mouse models have revealed a contribution of LMP1 to lymphoma development when expressed in B cells and to papilloma and carcinoma development when expressed using polyoma or EBV promoters (8, 13, 44, 48, 51, 52). This study shows that LMP1 but not LMP2 functions as a tumor promoter when expressed specifically in basal epithelial cells. The K14-LMP2 mice have been previously shown to lack any obvious pathological phenotype (17), and in this current study, LMP2 did not contribute to initiation or promotion but importantly did contribute to cancer progression.
Distinct inbred and outbred strains have highly variable responses to initiation and promotion. In this study, most experiments used an F1 hybrid mouse that was 50% FVB/N. Importantly, the control mice in all experiments had very consistent low levels of papilloma formation, with 2 to 3 papillomas at 12 weeks post-TPA. Regardless of the background, the LMP1-positive mice developed 2 to 3 times as many papillomas as control mice and the papillomas had prolonged persistence. This effect was apparent both in the LMP1-positive transgenic mice and in the doubly positive LMP1/LMP2 mice. In the multistep carcinoma model, treatment with DMBA results in mutation of H-ras while TPA is thought to contribute possibly through the activation of Stat3 (6, 36). Activation of epidermal growth factor receptor (EGFR) signaling has also been identified as an important component in the development of carcinomas, which are distinguished from normal skin by elevated activation of ERK, Stat3, and Akt (5, 19, 43). As LMP1 is known to activate ERK, Stat3, and Akt and to induce constitutively phosphorylated EGFR, it might be expected that expression of LMP1 would have major effects on both papilloma development and progression to carcinoma (9, 10, 14, 15, 22, 45). However, the effects of LMP1, in general, on tumor promotion and induction of papilloma formation were relatively subtle. Interestingly, ras mutation was lacking in 2/5 LMP1 carcinomas, in 1/19 LMP1 papillomas, and in 2/19 LMP1/LMP2 papillomas. The absence of ras mutation in this small subset of carcinomas and papillomas may indicate that the effects of LMP1, potentially on ERK and Stat3, can sometimes substitute for constitutively activated Ras. In EBV-associated tumors, including NPC, ras mutation does not occur. However, ras pathways are activated and one contributing factor is the lack of RASSF1A expression, a ras-binding protein that acts as a tumor suppressor (16, 23).
It has been previously shown that LMP2 induces Akt activation and can block epithelial cell differentiation, and some similarities between the Ig-LMP2 transgenic and activated Ras transgenic mice suggested that LMP2 can directly activate the Ras/PI3K/Akt pathway to mediate B cell survival (33, 35, 41). However, when expressed in epithelial cells, differences in skin development or pathology were not detected in the K14-LMP2A transgenic mice (17). Akt signaling contributes to both tumor promotion and progression during TPA treatment and constitutively activated Akt results in enhanced papilloma formation (19, 42, 43). The data in this study show that even with the treatment of DMBA and TPA, LMP2 did not enhance tumor promotion, as evidenced by the lack of increased or sustained papilloma development. However, the expression of LMP2 in combination with LMP1 did significantly increase carcinoma development (Table 2).
To discern how LMP1 and LMP2 affect cell growth in this model, the activation of the signaling pathways that are known to contribute to promotion and progression was assessed. In confirmation of previous studies, the carcinomas were distinguished from normal skin samples with constitutive, elevated ERK and Stat3 activation, which was highest in the doubly transgenic K14-LMP1/LMP2 carcinomas (Fig. 5) (43). It is known that both LMP1 and LMP2 can activate Akt; however, distinct outcomes of activation have been identified (10, 22, 33, 41, 45, 46). For example, only LMP2 induces β-catenin nuclear translocation in an Akt-dependent manner (33). Analysis of Akt activation in the carcinomas identified multiple bands that had distinct reactivities using phospho-specific antibodies. The predominant forms of Akt that were detected were Akt2 and a cleaved form of Akt1. The pattern of phosphorylation was complex; however, a unique band identified with Thr308-specific pAkt antibody was detected predominantly in the transgenic carcinomas that expressed LMP2. However, the same pathways, overall, were activated in carcinomas that developed in control and transgenic mice. This suggests that the contribution of EBV and its oncogenes, LMP1 and LMP2, to tumor development is likely the enhanced activation of pathways that can also be sporadically activated in the absence of LMP1 or LMP2 and lead to tumor development. This is similar to EBV-associated cancers in vivo where pathways that are activated by LMP1 are also activated in the less prevalent EBV-negative forms of the cancers (32, 49).
Both LMP1 and LMP2 are expressed in NPC, and their cooperation in this carcinogenesis model reveals their combined oncogenic potential in causing disease. As in the carcinogenesis model, genetic changes are likely required in vivo, in addition to LMP1 and LMP2 expression, to result in carcinoma formation, while the signaling events initiated by LMP1 and LMP2 likely provide the critical signals needed for tumor promotion and progression. The further characterization of the specific effects of LMP1 and LMP2 may identify essential pathways and interactions that can be specifically targeted to develop therapeutic strategies against EBV-associated cancers.
ACKNOWLEDGMENTS
We are grateful to Richard Longnecker (Northwestern University) for his generous gift of K14-LMP2A (TgE lineage) transgenic mice.
This work was supported by NIH grants CA19014 and CA32979 to N.R.-T.
Footnotes
Published ahead of print 22 February 2012
REFERENCES
- 1. Balmain A, Pragnell IB. 1983. Mouse skin carcinomas induced in vivo by chemical carcinogens have a transforming Harvey-ras oncogene. Nature 303:72–74 [DOI] [PubMed] [Google Scholar]
- 2. Cahir-McFarland ED, Davidson DM, Schauer SL, Duong J, Kieff E. 2000. NF-kappa B inhibition causes spontaneous apoptosis in Epstein-Barr virus-transformed lymphoblastoid cells. Proc. Natl. Acad. Sci. U. S. A. 97:6055–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Caldwell RG, Brown RC, Longnecker R. 2000. Epstein-Barr virus LMP2A-induced B-cell survival in two unique classes of EmuLMP2A transgenic mice. J. Virol. 74:1101–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Caldwell RG, Wilson JB, Anderson SJ, Longnecker R. 1998. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity 9:405–411 [DOI] [PubMed] [Google Scholar]
- 5. Chan KS, et al. 2004. Epidermal growth factor receptor-mediated activation of Stat3 during multistage skin carcinogenesis. Cancer Res. 64:2382–2389 [DOI] [PubMed] [Google Scholar]
- 6. Chan KS, et al. 2004. Disruption of Stat3 reveals a critical role in both the initiation and the promotion stages of epithelial carcinogenesis. J. Clin. Invest. 114:720–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Charalambous CT, Hannigan A, Tsimbouri P, McPhee GM, Wilson JB. 2007. Latent membrane protein 1-induced EGFR signalling is negatively regulated by TGF alpha prior to neoplasia. Carcinogenesis 28:1839–1848 [DOI] [PubMed] [Google Scholar]
- 8. Curran JA, Laverty FS, Campbell D, Macdiarmid J, Wilson JB. 2001. Epstein-Barr virus encoded latent membrane protein-1 induces epithelial cell proliferation and sensitizes transgenic mice to chemical carcinogenesis. Cancer Res. 61:6730–6738 [PubMed] [Google Scholar]
- 9. Dawson CW, Laverick L, Morris MA, Tramoutanis G, Young LS. 2008. Epstein-Barr virus-encoded LMP1 regulates epithelial cell motility and invasion via the ERK-MAPK pathway. J. Virol. 82:3654–3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dawson CW, Tramountanis G, Eliopoulos AG, Young LS. 2003. Epstein-Barr virus latent membrane protein 1 (LMP1) activates the phosphatidylinositol 3-kinase/Akt pathway to promote cell survival and induce actin filament remodeling. J. Biol. Chem. 278:3694–3704 [DOI] [PubMed] [Google Scholar]
- 11. Ikeda M, Longnecker R. 2007. Cholesterol is critical for Epstein-Barr virus latent membrane protein 2A trafficking and protein stability. Virology 360:461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ise K, et al. 2000. Targeted deletion of the H-ras gene decreases tumor formation in mouse skin carcinogenesis. Oncogene 19:2951–2956 [DOI] [PubMed] [Google Scholar]
- 13. Kulwichit W, et al. 1998. Expression of the Epstein-Barr virus latent membrane protein 1 induces B cell lymphoma in transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 95:11963–11968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kung CP, Meckes DG, Jr, Raab-Traub N. 2011. Epstein-Barr virus LMP1 activates EGFR, STAT3, and ERK through effects on PKCdelta. J. Virol. 85:4399–4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kung CP, Raab-Traub N. 2008. Epstein-Barr virus latent membrane protein 1 induces expression of the epidermal growth factor receptor through effects on Bcl-3 and STAT3. J. Virol. 82:5486–5493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lo PH, et al. 2007. Reduced expression of RASSF1A in esophageal and nasopharyngeal carcinomas significantly correlates with tumor stage. Cancer Lett. 257:199–205 [DOI] [PubMed] [Google Scholar]
- 17. Longan L, Longnecker R. 2000. Epstein-Barr virus latent membrane protein 2A has no growth-altering effects when expressed in differentiating epithelia. J. Gen. Virol. 81:2245–2252 [DOI] [PubMed] [Google Scholar]
- 18. Lu J, et al. 2006. Syk tyrosine kinase mediates Epstein-Barr virus latent membrane protein 2A-induced cell migration in epithelial cells. J. Biol. Chem. 281:8806–8814 [DOI] [PubMed] [Google Scholar]
- 19. Lu J, Rho O, Wilker E, Beltran L, Digiovanni J. 2007. Activation of epidermal akt by diverse mouse skin tumor promoters. Mol. Cancer Res. 5:1342–1352 [DOI] [PubMed] [Google Scholar]
- 20. Luo HR, et al. 2003. Akt as a mediator of cell death. Proc. Natl. Acad. Sci. U. S. A. 100:11712–11717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Macdiarmid J, Stevenson D, Campbell DH, Wilson JB. 2003. The latent membrane protein 1 of Epstein-Barr virus and loss of the INK4a locus: paradoxes resolve to cooperation in carcinogenesis in vivo. Carcinogenesis 24:1209–1218 [DOI] [PubMed] [Google Scholar]
- 22. Mainou BA, Everly DN, Jr, Raab-Traub N. 2005. Epstein-Barr virus latent membrane protein 1 CTAR1 mediates rodent and human fibroblast transformation through activation of PI3K. Oncogene 24:6917–6924 [DOI] [PubMed] [Google Scholar]
- 23. Man C, et al. 2007. Latent membrane protein 1 suppresses RASSF1A expression, disrupts microtubule structures and induces chromosomal aberrations in human epithelial cells. Oncogene 26:3069–3080 [DOI] [PubMed] [Google Scholar]
- 24. Matthews JN, Altman DG, Campbell MJ, Royston P. 1990. Analysis of serial measurements in medical research. BMJ 300:230–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maufort JP, Williams SM, Pitot HC, Lambert PF. 2007. Human papillomavirus 16 E5 oncogene contributes to two stages of skin carcinogenesis. Cancer Res. 67:6106–6112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meckes DG, Jr, et al. 2010. Human tumor virus utilizes exosomes for intercellular communication. Proc. Natl. Acad. Sci. U. S. A. 107:20370–20375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Medina EA, et al. 2005. Tumor necrosis factor-{alpha} decreases Akt protein levels in 3T3-L1 adipocytes via the caspase-dependent ubiquitination of Akt. Endocrinology 146:2726–2735 [DOI] [PubMed] [Google Scholar]
- 28. Megosh L, Halpern M, Farkash E, O'Brien TG. 1998. Analysis of ras gene mutational spectra in epidermal papillomas from K6/ODC transgenic mice. Mol. Carcinog. 22:145–149 [DOI] [PubMed] [Google Scholar]
- 29. Miller CL, et al. 1995. Integral membrane protein 2 of Epstein-Barr virus regulates reactivation from latency through dominant negative effects on protein-tyrosine kinases. Immunity 2:155–166 [DOI] [PubMed] [Google Scholar]
- 30. Miller CL, Lee JH, Kieff E, Longnecker R. 1994. An integral membrane protein (LMP2) blocks reactivation of Epstein-Barr virus from latency following surface immunoglobulin crosslinking. Proc. Natl. Acad. Sci. U. S. A. 91:772–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morris MA, Dawson CW, Young LS. 2009. Role of the Epstein-Barr virus-encoded latent membrane protein-1, LMP1, in the pathogenesis of nasopharyngeal carcinoma. Future Oncol. 5:811–825 [DOI] [PubMed] [Google Scholar]
- 32. Morrison JA, Gulley ML, Pathmanathan R, Raab-Traub N. 2004. Differential signaling pathways are activated in the Epstein-Barr virus-associated malignancies nasopharyngeal carcinoma and Hodgkin lymphoma. Cancer Res. 64:5251–5260 [DOI] [PubMed] [Google Scholar]
- 33. Morrison JA, Klingelhutz AJ, Raab-Traub N. 2003. Epstein-Barr virus latent membrane protein 2A activates beta-catenin signaling in epithelial cells. J. Virol. 77:12276–12284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pegtel DM, et al. 2005. Epstein-Barr-virus-encoded LMP2A induces primary epithelial cell migration and invasion: possible role in nasopharyngeal carcinoma metastasis. J. Virol. 79:15430–15442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Portis T, Longnecker R. 2004. Epstein-Barr virus (EBV) LMP2A mediates B-lymphocyte survival through constitutive activation of the Ras/PI3K/Akt pathway. Oncogene 23:8619–8628 [DOI] [PubMed] [Google Scholar]
- 36. Quintanilla M, Brown K, Ramsden M, Balmain A. 1986. Carcinogen-specific mutation and amplification of Ha-ras during mouse skin carcinogenesis. Nature 322:78–80 [DOI] [PubMed] [Google Scholar]
- 37. Raab-Traub N. 2002. Epstein-Barr virus in the pathogenesis of NPC. Semin. Cancer Biol. 12:431–441 [DOI] [PubMed] [Google Scholar]
- 38. Raab-Traub N. 1996. Pathogenesis of Epstein-Barr virus and its associated malignancies. Semin. Virol. 7:315–323 [Google Scholar]
- 39. Rehman I, et al. 2000. Frequent codon 12 Ki-ras mutations in mouse skin tumors initiated by N-methyl-N′-nitro-N-nitrosoguanidine and promoted by mezerein. Mol. Carcinog. 27:298–307 [PubMed] [Google Scholar]
- 40. Satomi Y, Bu P, Okuda M, Tokuda H, Nishino H. 2003. H-ras mutations at codon 61 or 13 in tumors initiated with a NO donor in mouse skin. Cancer Lett. 196:17–22 [DOI] [PubMed] [Google Scholar]
- 41. Scholle F, Bendt KM, Raab-Traub N. 2000. Epstein-Barr virus LMP2A transforms epithelial cells, inhibits cell differentiation, and activates Akt. J. Virol. 74:10681–10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Segrelles C, et al. 2007. Deregulated activity of Akt in epithelial basal cells induces spontaneous tumors and heightened sensitivity to skin carcinogenesis. Cancer Res. 67:10879–10888 [DOI] [PubMed] [Google Scholar]
- 43. Segrelles C, et al. 2002. Functional roles of Akt signaling in mouse skin tumorigenesis. Oncogene 21:53–64 [DOI] [PubMed] [Google Scholar]
- 44. Shair KH, et al. 2007. EBV latent membrane protein 1 activates Akt, NFkappaB, and Stat3 in B cell lymphomas. PLoS Pathog. 3:e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shair KH, Schnegg CI, Raab-Traub N. 2008. EBV latent membrane protein 1 effects on plakoglobin, cell growth, and migration. Cancer Res. 68:6997–7005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shair KH, Schnegg CI, Raab-Traub N. 2009. Epstein-Barr virus latent membrane protein-1 effects on junctional plakoglobin and induction of a cadherin switch. Cancer Res. 69:5734–5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Song S, Liem A, Miller JA, Lambert PF. 2000. Human papillomavirus types 16 E6 and E7 contribute differently to carcinogenesis. Virology 267:141–150 [DOI] [PubMed] [Google Scholar]
- 48. Stevenson D, Charalambous C, Wilson JB. 2005. Epstein-Barr virus latent membrane protein 1 (CAO) up-regulates VEGF and TGF alpha concomitant with hyperlasia, with subsequent up-regulation of p16 and MMP9. Cancer Res. 65:8826–8835 [DOI] [PubMed] [Google Scholar]
- 49. Thornburg NJ, Pathmanathan R, Raab-Traub N. 2003. Activation of nuclear factor-kappaB p50 homodimer/Bcl-3 complexes in nasopharyngeal carcinoma. Cancer Res. 63:8293–8301 [PubMed] [Google Scholar]
- 50. Tsao SW, Tramoutanis G, Dawson CW, Lo AK, Huang DP. 2002. The significance of LMP1 expression in nasopharyngeal carcinoma. Semin. Cancer Biol. 12:473–487 [DOI] [PubMed] [Google Scholar]
- 51. Uchida J, et al. 1999. Mimicry of CD40 signals by Epstein-Barr virus LMP1 in B lymphocyte responses. Science 286:300–303 [DOI] [PubMed] [Google Scholar]
- 52. Wilson JB, Weinberg W, Johnson R, Yuspa S, Levine AJ. 1990. Expression of the BNLF-1 oncogene of Epstein-Barr virus in the skin of transgenic mice induces hyperplasia and aberrant expression of keratin 6. Cell 61:1315–1327 [DOI] [PubMed] [Google Scholar]
- 53. Woodworth CD, et al. 2004. Strain-dependent differences in malignant conversion of mouse skin tumors is an inherent property of the epidermal keratinocyte. Carcinogenesis 25:1771–1778 [DOI] [PubMed] [Google Scholar]
- 54. Young LS, Murray PG. 2003. Epstein-Barr virus and oncogenesis: from latent genes to tumours. Oncogene 22:5108–5121 [DOI] [PubMed] [Google Scholar]
- 55. Young LS, Rickinson AB. 2004. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4:757–768 [DOI] [PubMed] [Google Scholar]
- 56. Yuspa SH. 1994. The pathogenesis of squamous cell cancer: lessons learned from studies of skin carcinogenesis—thirty-third G. H. A. Clowes Memorial Award Lecture. Cancer Res. 54:1178–1189 [PubMed] [Google Scholar]