Abstract
C-type lectin SIGNR1 directly recognizes Candida albicans and zymosan and has been considered to share properties of polysaccharide recognition with human DC-SIGN (hDC-SIGN). However, the precise specificity of SIGNR1 and the difference from that of hDC-SIGN remain to be elucidated. We prepared soluble forms of SIGNR1 and hDC-SIGN and conducted experiments to examine their respective specificities. Soluble SIGNR1 (sSIGNR1) bound several types of live C. albicans clinical isolate strains in an EDTA-sensitive manner. Inhibition analyses of sSIGNR1 binding by glycans from various yeast strains demonstrated that SIGNR1 preferentially recognizes N-glycan α-mannose side chains in Candida mannoproteins, as reported in hDC-SIGN. Unlike shDC-SIGN, however, sSIGNR1 recognized not only Saccharomyces cerevisiae, but also C. albicans J-1012 glycan, even after α-mannosidase treatment that leaves only β1,2-mannose-capped α-mannose side chains. In addition, glycomicroarray analyses showed that sSIGNR1 binds mannans from C. albicans and S. cerevisiae but does not recognize Lewisa/b/x/y antigen polysaccharides as in shDC-SIGN. Consistent with these results, RAW264.7 cells expressing hDC-SIGN in which the carbohydrate recognition domain (CRD) was replaced with that of SIGNR1 (RAW-chimera) produced comparable amounts of interleukin 10 (IL-10) in response to glycans from C. albicans and S. cerevisiae, but those expressing hDC-SIGN produced less IL-10 in response to S. cerevisiae than C. albicans. Furthermore, RAW–hDC-SIGN cells remarkably reduced IL-10 production after α-mannosidase treatment compared with RAW-chimera cells. These results indicate that SIGNR1 recognizes C. albicans/yeast through a specificity partly distinct from that of its homologue hDC-SIGN.
INTRODUCTION
Numerous microbes are covered with polysaccharides. Recognition of the polysaccharides by pattern recognition receptors (PRRs), including lectins, is vital in order to recognize pathogens, since recognition of the outermost components is the first interactive step with immune cells during infection to evoke innate and adaptive immune responses.
Candida albicans is an opportunistic agent of infection in immunocompromised patients. In the host innate immune system, several types of receptors for sensing ligands on the microbe have been defined, e.g., C-type lectins and Toll-like receptors (TLRs) (18). Ligands for these receptors are present in the outer structure of microbes. However, some ligands are sequestered by the outermost polysaccharides, which consist of mannoproteins, as reported in the case of β-glucan, a ligand for Dectin-1 (6). Mannoproteins are rich in polysaccharides composed mostly of α- and β-mannose and recognized by mannose/mannan-type lectins.
The C-type lectin human DC-SIGN (hDC-SIGN) (CD209) has been shown to interact with a wide range of pathogens, including microbes, viruses, and protozoa (11), via mannose and fucose moieties on the surfaces of the pathogens. Microbes, such as Mycobacterium tuberculosis and C. albicans, are endocytosed and processed for antigen presentation to induce the subsequent T cell-mediated immune responses. However, the recognition also induces immunosuppressive responses in cooperation with TLRs (8).
Mice have eight hDC-SIGN homologues (19, 20). One of these homologues, SIGNR1, is structurally related to hDC-SIGN based on its long neck domain. SIGNR1 is expressed on particular subsets of macrophages (Mϕ)/dendritic cells (DCs) in the marginal zones of the spleen, the resident peritoneal cavity, the medulla of lymph nodes, the skin, and the lamina propria (4, 10, 17, 30). SIGNR1 on these cells plays a role as a sentinel in the recognition of pathogens through capsular polysaccharides. In fact, SIGNR1 on marginal-zone Mϕ recognizes Streptococcus pneumoniae (10), leading to efficient activation of the complement system in situ (9).
Previously, we reported that SIGNR1 recognizes Gram-negative bacteria (Salmonella enterica serovar Typhimurium and Escherichia coli) and C. albicans (27). The former are recognized through the nonreductive end of the lipopolysaccharide (LPS) core sequence by SIGNR1 (17). This is also the case for hDC-SIGN in recognition of E. coli (13). Recently, hDC-SIGN has been reported to strongly recognize the α-mannose structure of N-glycan side chains of C. albicans but to weakly recognize that of Saccharomyces cerevisiae (2). However, the recognition motif for SIGNR1 on C. albicans is not clear at present.
Therefore, we aimed to elucidate the properties of SIGNR1 in the recognition of polysaccharide on C. albicans. To this end, we prepared soluble forms of SIGNR1 (sSIGNR1) and hDC-SIGN (shDC-SIGN) and used structurally distinguished glycans purified from various types of C. albicans and S. cerevisiae, as well as the respective microbes. The results indicate that sSIGNR1 binds equally well to glycans from S. cerevisiae and those from C. albicans. Furthermore, sSIGNR1, but not shDC-SIGN, was shown to readily recognize C. albicans glycan treated with α-mannosidase. In addition, a glycomicroarray based on an evanescent-field fluorescence detection method clearly revealed that sSIGNR1 binds α-mannose monosaccharide and mannans from C. albicans and S. cerevisiae but does not recognize Lewisa/b/x/y antigen polysaccharides as in shDC-SIGN. Different properties in recognition of yeast glycans between the SIGNR1 and hDC-SIGN CRDs were also observed in induction of interleukin 10 (IL-10) from RAW264.7 cells.
MATERIALS AND METHODS
Cells and cultures.
Maintenance of human embryonic kidney HEK293T cells and macrophage-like RAW264.7 cells and preparation of RAW264.7 cells expressing SIGNR1 (RAW-SIGNR1) were as described previously (27). In order to prepare RAW–hDC-SIGN, cDNA-encoded hDC-SIGN (kindly provided by R. M. Steinman, Rockefeller University) was cloned into pMX-IRES-puromycin (12). RAW264.7 cells were transfected by the plasmid with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) in accordance with the manufacturer's protocol. RAW264.7 cells expressing hDC-SIGN (RAW–hDC-SIGN) were maintained in the presence of 4 μg/ml puromycin (InvivoGen, San Diego, CA). To prepare RAW264.7 cells expressing chimeric lectin consisting of hDC-SIGN and SIGNR1 (RAW-chimera), cDNA fragments encoding amino acids 1 to 253 (corresponding to the cytosolic-neck domain of hDC-SIGN) and 193 to 325 (corresponding to the carbohydrate recognition domain [CRD] of SIGNR1) were amplified using KOD polymerase (Toyobo, Tokyo, Japan) and primer pairs 5′-GGTGGTACGGGAATTCATGAGTGACTCCAAGGAACCAAGAC-3′ /5′-GGCACAGGCGTTCCAC-3′ and 5′-TGGAACGCCTGTGCCGACTCTGCCCCTGGGACTGGACATTC-3′/5′-ATTTACGTAGCGGCCGCCTAGCCTTCAGTGCATGGGGTTGC-3′, respectively. They were introduced into the EcoRI-NotI site of pMX-IRES-puromycin using the In-Fusion PCR Cloning System (Clontech, Mountain View, CA). RAW264.7 cells were transfected by the plasmid as described above.
Reagents and yeast strains.
Alexa 647-coupled hamster anti-SIGNR1 monoclonal antibody (MAb) 22D1 and rabbit anti-hDC-SIGN Ab (H-200) were purchased from eBioscience (San Diego, CA) and Santa Cruz Biotechnology (Santa Cruz, CA), respectively. Anti-hDC-SIGN MAb (DCS-8C1; eBioscience) was labeled with Alexa-555 (Invitrogen) in accordance with the manufacturer's protocol. Glycogen of bovine liver type IX (G0885) and Jack bean (Canavalia ensiformis) α-mannosidase (EC 3.2.1.24) were from Sigma-Aldrich (Irvine, CA). C. albicans J-1012 (serotype A; NBRC1060) and NIH B-792 (serotype B; NBRC10108), Candida lusitaniae NBRC1019, and S. cerevisiae X2180-1A (wild type [WT]) (BY21559) were obtained from the Biological Resource Center of the National Institute of Technology and Evaluation (Tokyo, Japan). C. albicans JCM1542 was from the Japan Collection of Microorganisms, RIKEN (Saitama, Japan).
Preparation of mannan from yeast strains.
Glycan was prepared from mannoprotein of the blastospore (yeast) form using Fehling's solution, as previously described (24). The glycans used in this study were purified from strains of C. albicans J-1012 (25), C. albicans NIH B-792 (22), Candida stellatoidea (24), Candida parapsilosis (22), C. lusitaniae (23), S. cerevisiae (WT) (1), S. cerevisiae (mnn1/mnn4) (1), and S. cerevisiae (mnn2) (21). The α-mannosidase treatment of C. albicans J-1012 mannan was carried out in 50 mM sodium acetate buffer (pH 4.6) containing 20 units of α-mannosidase at 37°C for 48 h.
Preparation of sSIGNR1 and shDC-SIGN and analyses of binding to microbes.
Soluble lectin (sLectin) tetramers, such as sSIGNR1 and shDC-SIGN, were prepared as described previously (26). Briefly, cDNA fragments encoding their extracellular domains were cloned into pEXPR-IBA44 (IBA, Göttingen, Germany) to add N-terminal BM40 secretion signal and Strep-Tag II sequences, followed by transfer into pEF6/V5-His (Invitrogen). HEK293T cells were then transfected with each plasmid using the calcium phosphate method (3) and cultured in serum-free medium (293 SFM II; Invitrogen) for the last 48 h. sSIGNR1 and shDC-SIGN in the supernatant were purified using Strep-Tactin Sepharose (IBA) in accordance with the manufacturer's protocol (>95% purity by SDS-PAGE).
Purified sLectins (2.5 μg/ml) were incubated with phycoerythrin (PE)-labeled Strep-Tactin (7.5 μg/ml) in 18 μl of Hanks' balanced salt solution (pH 8.3) (binding buffer) for 2 h at 4°C and for a further 10 min at 37°C. The tetramers thus formed were incubated with 5 × 106 live microbes for 4 h at 4°C in the presence of 1% bovine serum albumin (BSA) (total volume, 25 μl). After washing with the binding buffer, the amount of bound PE–Strep-Tactin was measured with a Gemini EM (Molecular Devices, Sunnyvale, CA). The direct binding of sSIGNR1 is shown as arbitrary units of fluorescence intensity. The percent inhibition was calculated using the following formula: [1 − (fluorescence intensity of C. albicans by staining with sLectin plus inhibitor − that without sLectin)/(that with sLectin without inhibitor − that without sLectin)] × 100.
Lectin enzyme-linked immunosorbent assay (ELISA).
sSIGNR1 and shDC-SIGN were formed by incubating sLectins (62.5 ng) with horseradish peroxidase (HRP)–Strep-Tactin (12.5 ng) in 20 μl of the solution described above. Microtiter plates were coated with 50 μl of mannan/glycan (5 mg/ml) in 50 mM sodium bicarbonate buffer (pH 9.6) for 12 h at 4°C, followed by incubation with 2.5% BSA at room temperature for 2 h after washing with 25 mM Tris-HCl, pH 8.3, plus 150 mM NaCl. The plates were then incubated with sSIGNR1 or shDC-SIGN in the presence of 1% BSA for 2 h at 4°C. For inhibition experiments, sLectin tetramer was preincubated with inhibitors for 1 h at 4°C before being added to the plates. After washing, binding of sLectin tetramer was measured as the absorbance of TMB (eBioscience) at 450 nm by Versamax (Molecular Devices). The percent inhibition was calculated as described above.
Inhibition of FITC-dextran binding to RAW-SIGNR1 with glycans.
RAW-SIGNR1 cells (2 × 105) were preincubated with various types of glycan and EDTA (25 mM) for 30 min at 4°C and then mixed with 80 μg/ml of fluorescein isothiocyanate (FITC)-dextran (2,000 kDa; Sigma-Aldrich) for 4 h at 4°C. Binding of FITC-dextran was analyzed with a flow cytometer. The percent inhibition was calculated using the following formula: [1 − (mean fluorescence intensity {MFI} of RAW-SIGNR1 cells with FITC-dextran plus inhibitor − that without FITC-dextran)/(that with FITC-dextran without inhibitor − that without FITC-dextran)] × 100.
Glycomicroarray analyses of sLectins by evanescent-field fluorescence detection.
Glycomicroarray analysis was performed as described previously (28). To form immune complexes, sSIGNR1 and shDC-SIGN (10 μg/ml) were preincubated with Alexa 647–anti-SIGNR1 (22D1) and Alexa 555–anti-hDC-SIGN (DCS-8C1; 1 μg/ml) for 15 min at room temperature in 25 mM Tris-HCl buffer (pH 7.4) containing 0.8% NaCl, 1% (vol/vol) Triton X-100, and 2 mM CaCl2 with or without 10 mM EDTA. This complex was directly added to the array immobilized with multivalent glycan ligands (see Fig. S1 in the supplemental material), followed by incubation overnight at 20°C. Binding was then detected using an evanescent-field fluorescence-assisted scanner. The data were analyzed with the Array Pro analyzer ver. 4.5 (Media Cybernetics, Bethesda, MD).
IL-10 production of RAW264.7 transfectants by stimulation in microplates coated with glycan.
Nontreated plates were precoated with 600 μg/ml glycan in phosphate-buffered saline (PBS) for 12 h. After blocking with RPMI1640 containing 10% fetal calf serum (FCS) for 30 min, RAW264.7 transfectants (5 × 104 cells) were cultured in the presence of 100 ng/ml ultrapure LPS (Invitrogen) for 24 h. IL-10 in the supernatants was analyzed using the Cytometric Bead Array (CBA) for mouse inflammation kit (BD Biosciences, Franklin Lakes, NJ).
Statistical analysis.
Data are expressed as the means ± standard deviations (SD) of triplicate assays. Statistical significance was determined by the two-tailed Student t test or by multiple comparisons with Tukey's multiple-range test. All experiments were performed two or more times, and representative results are shown.
RESULTS AND DISCUSSION
SIGNR1 recognizes various types of Candida strains.
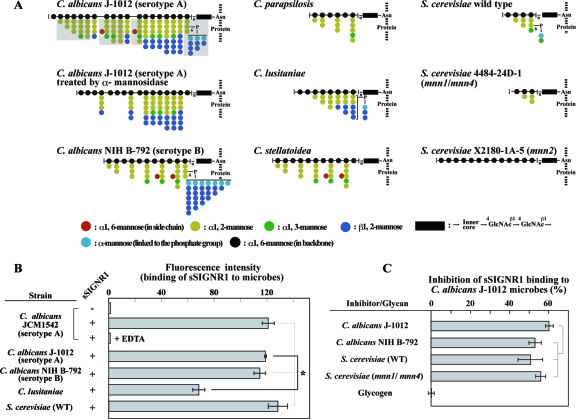
Each Candida strain has a unique set of oligomannose side chains, generating a great diversity of N-glycans (Fig. 1A) compared with that of O-glycans. Moreover, N-glycans account for more than 95% of the glycans in the surface mannoproteins.
Fig 1.
sSIGNR1 binds various types of yeast strains. (A) Structural diagrams of N-glycans used in this study. The structures of N-glycans of C. albicans J-1012 (25), C. albicans NIH B-792 (22), C. stellatoidea (24), C. parapsilosis (22), and C. lusitaniae (23) are adopted from our structural analyses using nuclear magnetic resonance (NMR). The structures of S. cerevisiae wild type, S. cerevisiae 4484-24D-1 (mnn1/mnn4), and S. cerevisiae X2180-1-A-5 (mnn2) are based on previous reports (1, 21). Side chains that are digested by treatment with α-mannosidase in C. albicans J-1012 N-glycan are shaded. The side chain sequence is not specified. (B) Binding of sSIGNR1 to Candida and S. cerevisiae strains. PE–Strep-Tactin alone (−) or PE-sSIGNR1 (+) was incubated with the indicated live yeast strains with or without EDTA (25 mM). (C) Inhibition of sSIGNR1 binding to C. albicans J-1012 by glycans purified from the C. albicans and S. cerevisiae strains indicated. sSIGNR1 was preincubated with 50 μg/ml of glycans before mixing with microbes. Glycogen was used as a negative control. Inhibition is indicated as the percent decrease of fluorescence intensity in experimental groups compared with the control without inhibitor. The results are shown as the means ± SD of triplicate assays. *, P < 0.05 (solid line) by Tukey's multiple-range test. The gray lines indicate no significant differences.
Therefore, we first examined the direct binding of sSIGNR1 to several Candida strains (Fig. 1B). We used sSIGNR1 tetramerized with Strep-Tactin, because the affinity of the sSIGNR1 monomer is weak (20). This method not only helps to increase the affinity of sSIGNR1, but also helps to avoid the formation of large complexes using Fc fusion lectins polymerized with anti-Fc polyclonal Ab. Before using this sSIGNR1 tetramer for experiments, we confirmed that sSIGNR1 tetramer bound to mannan-agarose was eluted with EDTA (data not shown), although the yield was less than half the amounts applied. sSIGNR1 bound live C. albicans clinical isolate JCM1542 (serotype A) in an EDTA-sensitive manner, indicating that sSIGNR1 binding occurs via the CRD. The other clinical isolate strains, C. albicans J-1012 (serotype A) and NIH B-792 (serotype B), were also recognized by sSIGNR1, although sSIGNR1 binding was much less to the C. lusitaniae nosocomial strain.
It is notable that sSIGNR1 binds to S. cerevisiae comparably to its binding to a C. albicans clinical isolate (Fig. 1B), and its binding to C. albicans J-1012 microbes was as inhibited by N-glycans from S. cerevisiae (WT and mnn1/mnn4) and C. albicans NIH B-792 as by those from C. albicans J-1012 (Fig. 1C).
SIGNR1 possibly recognizes α-mannose in side chains of C. albicans N-glycan.
To delineate the polysaccharide structure recognized by SIGNR1, inhibition experiments with lectin ELISA were conducted using N-glycans from the various yeast strains listed in Fig. 1A and Table 1.
Table 1.
Compositions of side chains and properties of N-glycans used in the study
| N-glycan | Presence of side chaina |
||||
|---|---|---|---|---|---|
| α1,3-Mannose | α1,2-Mannose | α1,6-Mannoseb | β1,2-mannose | PM | |
| C. albicans J-1012 (serotype A) | + | + | + | + | + |
| C. albicans NIH B-792 (serotype B) | + | + | + | + | + |
| S. cerevisiae (WT) | − | + | − | − | + |
| S. cerevisiae 4484-24D-1 (mnn1/mnn4) | − | + | − | − | − |
| S. cerevisiae X2180-1A-5 (mnn2) | − | − | − | − | − |
| C. stellatoidea | + | + | + | − | − |
| C. parapsilosis | + | + | − | − | − |
| C. lusitaniae | − | + | − | + | + |
| C. albicans J-1012 (α-mannosidase treated) | + | + | − | + | − |
PM, phosphorylated mannose. Side chain structure presence and absence in the N-glycan are indicated by + and −, respectively.
Branching in side chain.
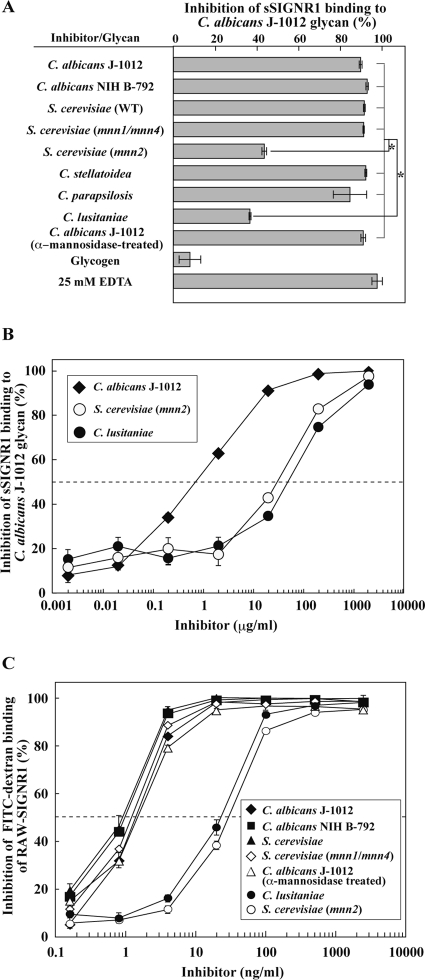
The direct binding of sSIGNR1 to C. albicans J-1012 N-glycan, which is composed of complexed side chains, was inhibited by glycans from several types of microbes: C. albicans NIH B-792 and C. stellatoidea glycans lacking β1,2-mannose, C. parapsilosis glycan lacking β1,2-mannose and α1,6-branched mannose, and S. cerevisiae WT glycan composed of short side chains lacking β1,2-mannose and α1,6-branched mannose (Fig. 2A). These glycans share the α1,2-mannose side chain structure, suggesting that SIGNR1 recognizes a moiety in N-glycan similar to that recognized by hDC-SIGN (2). Moreover, S. cerevisiae 4484-24D-1 (mnn1/mnn4) glycan, which lacks phosphorylated mannose (PM), β1,2-mannose, α1,3-mannose, and α1,6-branched mannose but possesses the short (mono- or di-) mannose side chain structure, appeared to be recognized by SIGNR1. However, S. cerevisiae X2180-1A-5 (mnn2) glycan, which lacks all side chains, was not effective (Fig. 2A and B), suggesting the crucial involvement of the side chain structure. In addition, C. lusitaniae glycan, the side chain of which is composed of more than 75% β1,2-mannose (mono- to tri-β-mannose)-capped side chain (23), was also ineffective in inhibiting sSIGNR1 binding (Fig. 2A and B), implying that these β1,2-mannoses disturb the access of SIGNR1 to the α-dimannose to some extent. Glycogen itself had no effect. As in the case of sSIGNR1 binding to microbes, that to C. albicans J-1012-derived N-glycan was EDTA sensitive (Fig. 2A).
Fig 2.
Recognition of α-mannose side chains in N-glycan by sSIGNR1. (A) Inhibition analysis by lectin ELISA. Binding of sSIGNR1 to microtiter plates coated with C. albicans J-1012 glycan was analyzed in the presence of glycans (25 μg/ml) purified from various types of yeast strains. Blocking activities of inhibitors are shown as the percent inhibition of sSIGNR1 binding. (B) Titration of inhibitory activity of glycans from the indicated yeast strains for sSIGNR1 binding by lectin ELISA. Half of maximal inhibition activity was indicated by the dashed line. (C) Inhibition of FITC binding to RAW-SIGNR1 cells by glycans. Transfectants were incubated with graded doses of glycans as in panel B prior to FITC-dextran. The results are shown as percent inhibition (means ± SD of triplicate assays). *, P < 0.05 (solid lines) by Tukey's multiple-range test. The gray lines indicate no significant differences.
Interestingly, treatment of C. albicans J-1012 mannan with α-mannosidase, which removed the α-mannose side chains other than the β1,2-mannose-capped side chains (Fig. 1B) (14), did not affect the inhibitory activity (Fig. 2A). This result also indicates that β1,2-mannose-capped α-mannose side chains in the N-glycan are sufficient to be recognized by SIGNR1. We also obtained similar results when mannan from S. cerevisiae (M7504; Sigma-Aldrich) was employed to coat plastic plates in lectin ELISA (data not shown). Therefore, it is possible that SIGNR1 recognizes long internal α-mannose (trimannose or greater) capped with β1,2-mannose, in addition to α-mannose side chains.
Since RAW-SIGNR1 cells effectively bind and endocytose high-molecular-weight FITC-dextran in an EDTA-sensitive manner (27), we carried out inhibition analysis using RAW-SIGNR1 cells to bind FITC-dextran instead of lectin ELISA (Fig. 2C). The inhibition activities of glycans from C. albicans J-1012 and NIH B-792 and S. cerevisiae and its mutant (mnn1/mnn4) were again comparably effective. The treatment of C. albicans J-1012 glycan with α-mannosidase was also ineffective in reducing the inhibitory activity. In addition, low efficiencies of glycans from S. cerevisiae (mnn2) and C. lusitaniae were also confirmed in this experimental system. When resident peritoneal Mϕ that express SIGNR1 (27) were used, similar results were obtained (data not shown). Together with the results using N-glycan with only α-mannose in the side chain, these results strengthen the possibility that SIGNR1 recognizes both the α-mannose side chain and β1,2-mannose-capped α-mannose side chains composed of more than trimannoses in the N-glycan.
Specificity of N-glycan recognition by hDC-SIGN.
Cambi et al. previously reported that hDC-SIGN recognizes glycans of S. cerevisiae strains less efficiently than those of Candida strains (2). In contrast, SIGNR1 recognized glycans from both wild-type and mnn1/mnn4 mutant S. cerevisiae equally with those from C. albicans, suggesting that the specificities of hDC-SIGN and SIGNR1 are somehow different from each other. In order to examine this possibility, we prepared shDC-SIGN and compared the sugar specificity with that of sSIGNR1.
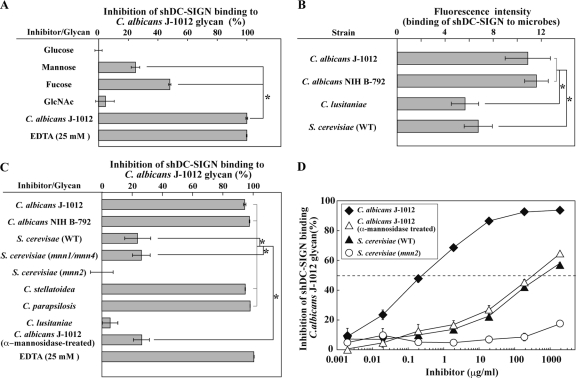
In lectin ELISA, the binding of shDC-SIGN to C. albicans J-1012 glycan was more sensitive to fucose than mannose and less sensitive to glucose and GlcNAc (Fig. 3A), as reported previously (16). Results using this probe showed that shDC-SIGN bound comparably well to C. albicans J-1012 and NIH B-792 (Fig. 3B). Unlike sSIGNR1, shDC-SIGN bound less to S. cerevisiae microbes, as reported previously (2).
Fig 3.
Binding of shDC-SIGN to microbes and inhibition of shDC-SIGN binding by glycans. (A) Inhibition analysis of shDC-SIGN binding by monosaccharides (50 mM) and glycan from C. albicans J-1012 using lectin ELISA. (B) Binding of shDC-SIGN to yeast strains was analyzed as in Fig. 1B. (C) Inhibition analysis using glycans from various yeast strains as in Fig. 2A. (D) Inhibition assay performed in the presence of graded doses of glycans from the indicated yeast strains. Half of maximal inhibition activity was indicated by the dashed line. The results are shown as the means ± SD of triplicate assays. *, P < 0.05 (solid lines) by Tukey's multiple-range test. The gray lines indicate no significant differences.
In addition, shDC-SIGN binding to C. albicans J-1012 glycan was not efficiently blocked by glycans from S. cerevisiae and mnn1/mnn4 mutants (Fig. 3C), indicating different specificities in N-glycan recognition between hDC-SIGN and SIGNR1. It is worth noting that treating C. albicans J-1012 glycan with α-mannosidase dramatically reduced its inhibitory activity (Fig. 3C). We further confirmed the abrogation of inhibitory activity of N-glycan from C. albicans J-1012 by α-mannosidase treatment by titrating its dose (Fig. 3D), showing that the reduced activity is nearly comparable to that of S. cerevisiae. These results suggested that hDC-SIGN recognizes the α1,2-mannose only at the nonreductive end of α-mannose side chains, but not the internal α1,2-mannose capped with β1,2-mannose and short α-mannose chains that are recognized by SIGNR1 of the N-glycan side chains in mannoprotein.
It has been reported that another SIGNR lectin, SIGNR3, also recognizes C. albicans (27) and that its saccharide specificity resembles that of hDC-SIGN (20). Based on the inhibition assay using lectin ELISA, SIGNR3 was shown to bind a little more efficiently to the wild type and the mnn1/mnn4 mutant of S. cerevisiae than hDC-SIGN but less so than SIGNR1 (see Fig. S2 in the supplemental material).
Glycomicroarray analyses of SIGNR1 and hDC-SIGN.
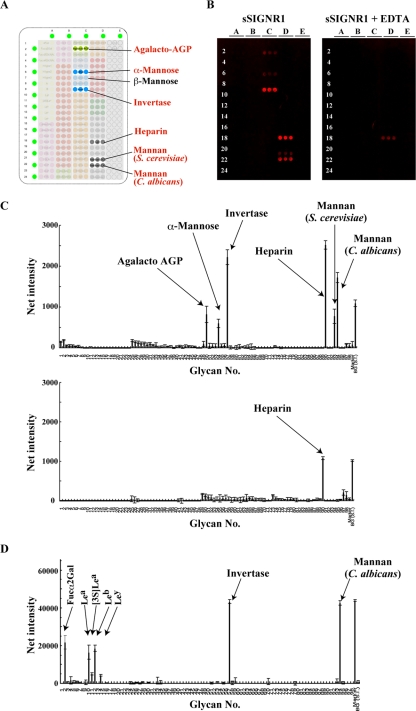
Previously, Powlesland et al. reported that sSIGNR1 did not bind any ligand on an array using the regular method (20), suggesting a weak affinity of SIGNR1. However, a sensitive glycomicroarray based on evanescent-field fluorescence-assisted detection has recently been developed (28). This method, in which analysis is performed in the presence of lectin probe without washing, enabled us to detect weak glycan-lectin interactions in the equilibrium state (15, 29), possibly representing genuine interactions of ligand and cellular lectin.
An array plate immobilized with the glycans indicated in Fig. 4A (see Fig. S1 in the supplemental material) was visualized by staining with Alexa 647-coupled sSIGNR1/anti-SIGNR1 MAb complex (Fig. 4B). The results clearly show that SIGNR1 binds to α-mannose and yeast mannans from C. albicans and S. cerevisiae, but not β-mannose, in an EDTA-sensitive manner (Fig. 4B and C). Of note, SIGNR1 did not recognize fucose-containing moieties, such as Fucα2Gal and Lewisa/b/x/y antigen. The binding of sSIGNR1 to heparin is likely to be false positive because of the insensitivity to EDTA.
Fig 4.
Glycomicroarray analysis of sSIGNR1. (A) Layout of the glycomicroarray. (B) Results of glycomicroarray analyses. Binding of soluble SIGNR1/Alexa 647–anti-SIGNR1 MAb immune complex to the array was performed in the absence (left) or presence (right) of 10 mM EDTA and detected by an evanescent-field fluorescence-assisted scanner. (C) Data analyzed with the Array Pro analyzer ver. 4.5. (D) Glycoarray analysis was performed using an immune complex of soluble hDC-SIGN/Alexa 555–anti-hDC-SIGN MAb as for sSIGNR1.
We previously performed array analyses using an shDC-SIGN–Fc fusion as a probe (28). However, the structural forms of SIGNR1 and shDC-SIGN–Fc were different, and this made it difficult to compare the precise sugar specificities. Therefore, we again performed glycoarray analysis using shDC-SIGN dimer as in sSIGNR1. The results clearly showed the recognition of fucose-containing glycans and C. albicans, but not S. cerevisiae, mannan by shDC-SIGN (Fig. 4D), which was consistent with our results in lectin ELISA and direct binding to microbes.
However, there are some discrepancies in our current observations and previous array results. It has been shown that SIGNR1 can recognize the fucose-containing moiety using glycoarray and solid-phase competition binding assays (5). One of our previous microarray analyses also demonstrated that the hDC-SIGN–Fc fusion was able to bind to glycans from C. albicans and S. cerevisiae (28). In both reports, the lectin CRD was fused with the Fc portion of IgG and polymerized with anti-Fc polyclonal Ab, giving rise to a very large and multivalent complexed probe. In the current glycomicroarray analyses, we utilized dimerized lectin probes, which possibly have lower avidity than those used in previous studies, enabling us to uncover the difference in the binding activities of shDC-SIGN to S. cerevisiae and C. albicans glycan.
Biological significance of the different sugar specificities of SIGNR1 and hDC-SIGN in IL-10 production using RAW264.7 transfectants.
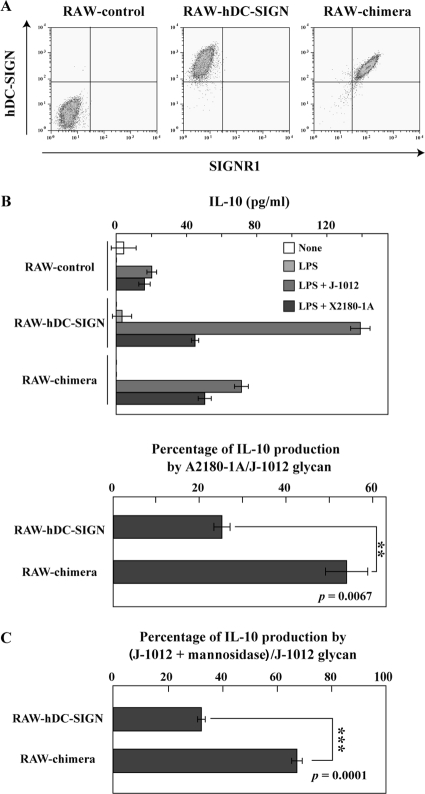
To examine the biological significance of distinct saccharide recognition between SIGNR1 and hDC-SIGN, we measured IL-10 production by RAW-SIGNR1 and RAW–hDC-SIGN cells, because hDC-SIGN on DCs is known to induce IL-10 production by recognizing C. albicans (7), and lamina propria DCs expressing SIGNR1 are also capable of producing IL-10 (30). After stimulation with C. albicans J-1012 microbes and glycan on plastic plates, RAW–hDC-SIGN, but not RAW-SIGNR1, cells produced IL-10 (data not shown). Therefore, we prepared RAW264.7 cells expressing the chimeric hDC-SIGN molecule (RAW-chimera) in which the CRD was replaced with the SIGNR1 CRD to compare glycan recognition specificities in terms of the induction of IL-10 production. The RAW-chimera cells expressed levels of lectin comparable to those of RAW–hDC-SIGN cells (Fig. 5A). Upon stimulation with S. cerevisiae X2180-1A (WT) glycan, RAW–hDC-SIGN and RAW-chimera cells produced equivalent amounts of IL-10 (Fig. 5B, top). RAW–hDC-SIGN cells produced much less IL-10 in response to S. cerevisiae than C. albicans glycan, in comparison with RAW-chimera cells (Fig. 5B, bottom). Interestingly, in the case of C. albicans J-1012 glycan, RAW–hDC-SIGN cells produced more IL-10 than RAW-chimera cells (Fig. 5B, top). However, the former significantly reduced IL-10 production after α-mannosidase treatment compared with the latter (Fig. 5C), which was consistent with the results showing that SIGNR1, but not hDC-SIGN, recognizes β-mannose-capped α-mannose side chains in C. albicans glycan.
Fig 5.
IL-10 production of RAW264.7 transfectants upon stimulation with glycan on a plastic plate. (A) RAW-control, RAW–hDC-SIGN, and RAW-chimera cells were analyzed by flow cytometry using polyclonal anti-hDC-SIGN Ab and anti-SIGNR1 MAb (22D1) specific for the SIGNR1 CRD. (B) The transfectants (5 × 104 cells) were cultured on plates precoated with C. albicans J-1012 and S. cerevisiae X2180-1A (WT) glycan in the presence or absence of LPS (100 ng/ml). After 24 h, IL-10 in the supernatants was analyzed (top). IL-10 production against S. cerevisiae glycan is shown as a percentage of that against C. albicans glycan (bottom). (C) IL-10 production after stimulation with native and α-mannosidase-treated C. albicans J-1012 glycan was analyzed as in panel B. IL-10 production against the treated glycan is shown as a percentage of that against the native glycan. **, P = 0.0067, and ***, P = 0.0001 by Student's t test.
It is possible that C. albicans induces higher IL-10 production than S. cerevisiae via hDC-SIGN on DCs in humans, leading to the immunosuppressive milieu at the site of infection. This may explain why C. albicans is more virulent than S. cerevisiae. Regarding the sugar specificity of SIGNR1, there were no significant differences in the binding specificity to mannose moieties between C. albicans and S. cerevisiae at the molecular level. However, IL-10 production was slightly but significantly higher in response to C. albicans than in response to S. cerevisiae, implying that cellular responses by recognition through lectin receptors is affected by some other, unknown factors. In this study, we used glycans prepared from the blastospore (yeast) form of each yeast strain. However, it should also be kept in mind that the difference in the growth form between C. albicans and S. cerevisiae might modulate cellular activity against microbes in situ. hDC-SIGN in the mouse cells prepared in this study properly transduces signals for IL-10 production. Although it is unknown how hDC-SIGN activates the mouse Src family and subsequent Raf-1 kinase in this situation, hDC-SIGN likely has a certain motif that is lacking in SIGNR1 to work in both human and mouse cells for IL-10 production.
Collectively, SIGNR1 and hDC-SIGN bind polysaccharides in surface mannoprotein on live C. albicans. However, SIGNR1 recognizes both α-mannose and β1,2-mannose-capped α-mannose side chains composed of more than trimannoses, whereas hDC-SIGN recognizes only α-mannose at the nonreductive end of the side chains, but not internal α-mannose capped with β1,2-mannose and short α-mannose chains of N-glycan side chains in mannoprotein. Differential recognition of yeast strains by SIGNR1 and hDC-SIGN may be relevant to the differences in cellular responsiveness.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by Grants-in-Aid for Scientific Research (19590389 to K.T. and 18390121 and 20390109 to K.I.) and a Grant-in-Aid for Scientific Research on Priority Area (19041036 to K.T.) from the Ministry of Education, Science, Sports and Culture of Japan and by the Hokuto Foundation for Bioscience (K.T.) and the Naito Foundation (K.I.).
Footnotes
Published ahead of print 13 February 2012
Supplemental material for this article may be found at http://iai.asm.org.
REFERENCES
- 1. Ballou CE, Kern KA, Raschke WC. 1973. Genetic control of yeast mannan structure. Complementation studies and properties of mannan mutants. J. Biol. Chem. 248:4667–4671 [PubMed] [Google Scholar]
- 2. Cambi A, et al. 2008. Dendritic cell interaction with Candida albicans critically depends on N-linked mannan. J. Biol. Chem. 283:20590–20599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen C, Okayama H. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dupasquier M, Stoitzner P, van Oudenaren A, Romani N, Leenen PJ. 2004. Macrophages and dendritic cells constitute a major subpopulation of cells in the mouse dermis. J. Investig. Dermatol. 123:876–879 [DOI] [PubMed] [Google Scholar]
- 5. Galustian C, et al. 2004. High and low affinity carbohydrate ligands revealed for murine SIGN-R1 by carbohydrate array and cell binding approaches, and differing specificities for SIGN-R3 and langerin. Int. Immunol. 16:853–866 [DOI] [PubMed] [Google Scholar]
- 6. Gantner BN, Simmons RM, Underhill DM. 2005. Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. EMBO J. 24:1277–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Geijtenbeek TB, et al. 2003. Mycobacteria target DC-SIGN to suppress dendritic cell function. J. Exp. Med. 197:7–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Geijtenbeek TBH, Gringhuis SI. 2009. Signalling through C-type lectin receptors: shaping immune responses. Nat. Rev. Immunol. 9:465–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kang YS, et al. 2006. A dominant complement fixation pathway for pneumococcal polysaccharides initiated by SIGN-R1 interacting with C1q. Cell 125:47–58 [DOI] [PubMed] [Google Scholar]
- 10. Kang YS, et al. 2003. SIGN-R1, a novel C-type lectin expressed by marginal zone macrophages in spleen, mediates uptake of the polysaccharide dextran. Int. Immunol. 15:177–186 [DOI] [PubMed] [Google Scholar]
- 11. Khoo US, Chan KY, Chan VS, Lin CL. 2008. DC-SIGN and L-SIGN: the SIGNs for infection. J. Mol. Med. 86:861–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kitamura T. 1998. New experimental approaches in retrovirus-mediated expression screening. Int. J. Hematol. 67:351–359 [DOI] [PubMed] [Google Scholar]
- 13. Klena J, Zhang P, Schwartz O, Hull S, Chen T. 2005. The core lipopolysaccharide of Escherichia coli is a ligand for the dendritic-cell-specific intercellular adhesion molecule nonintegrin CD209 receptor. J. Bacteriol. 187:1710–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kobayashi H, Shibata N, Mitobe H, Ohkubo Y, Suzuki S. 1989. Structural study of phosphomannan of yeast-form cells of Candida albicans J-1012 strain with special reference to application of mild acetolysis. Arch. Biochem. Biophys. 272:364–375 [DOI] [PubMed] [Google Scholar]
- 15. Kuno A, et al. 2005. Evanescent-field fluorescence-assisted lectin microarray: a new strategy for glycan profiling. Nat. Methods 2:851–856 [DOI] [PubMed] [Google Scholar]
- 16. Mitchell DA, Fadden AJ, Drickamer K. 2001. A novel mechanism of carbohydrate recognition by the C-type lectins DC-SIGN and DC-SIGNR. Subunit organization and binding to multivalent ligands. J. Biol. Chem. 276:28939–28945 [DOI] [PubMed] [Google Scholar]
- 17. Nagaoka K, et al. 2005. Association of SIGNR1 with TLR4-MD-2 enhances signal transduction by recognition of LPS in gram-negative bacteria. Int. Immunol. 17:827–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Netea MG, Brown GD, Kullberg BJ, Gow NA. 2008. An integrated model of the recognition of Candida albicans by the innate immune system. Nat. Rev. Microbiol. 6:67–78 [DOI] [PubMed] [Google Scholar]
- 19. Park CG, et al. 2001. Five mouse homologues of the human dendritic cell C-type lectin, DC-SIGN. Int. Immunol. 13:1283–1290 [DOI] [PubMed] [Google Scholar]
- 20. Powlesland AS, et al. 2006. Widely divergent biochemical properties of the complete set of mouse DC-SIGN-related proteins. J. Biol. Chem. 281:20440–20449 [DOI] [PubMed] [Google Scholar]
- 21. Raschke WC, Kern KA, Antalis C, Ballou CE. 1973. Genetic control of yeast mannan structure. Isolation and characterization of mannan mutants. J. Biol. Chem. 248:4660–4666 [PubMed] [Google Scholar]
- 22. Shibata N, et al. 1995. Existence of branched side chains in the cell wall mannan of pathogenic yeast, Candida albicans. Structure-antigenicity relationship between the cell wall mannans of Candida albicans and Candida parapsilosis. J. Biol. Chem. 270:1113–1122 [DOI] [PubMed] [Google Scholar]
- 23. Shibata N, Kobayashi H, Okawa Y, Suzuki S. 2003. Existence of novel beta-1,2 linkage-containing side chain in the mannan of Candida lusitaniae, antigenically related to Candida albicans serotype A. Eur. J. Biochem. 270:2565–2575 [DOI] [PubMed] [Google Scholar]
- 24. Shibata N, et al. 1997. Demonstration of the presence of alpha-1,6-branched side chains in the mannan of Candida stellatoidea. Eur. J. Biochem. 246:477–485 [DOI] [PubMed] [Google Scholar]
- 25. Shibata N, Suzuki A, Kobayashi H, Okawa Y. 2007. Chemical structure of the cell-wall mannan of Candida albicans serotype A and its difference in yeast and hyphal forms. Biochem. J. 404:365–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Takahara K, et al. 2011. C-type lectin SIGNR1 enhances cellular oxidative burst response against C. albicans in cooperation with Dectin-1. Eur. J. Immunol. 41:1435–1444 [DOI] [PubMed] [Google Scholar]
- 27. Takahara K, et al. 2004. Functional comparison of the mouse DC-SIGN, SIGNR1, SIGNR3 and Langerin, C-type lectins. Int. Immunol. 16:819–829 [DOI] [PubMed] [Google Scholar]
- 28. Tateno H, et al. 2008. Glycoconjugate microarray based on an evanescent-field fluorescence-assisted detection principle for investigation of glycan-binding proteins. Glycobiology 18:789–798 [DOI] [PubMed] [Google Scholar]
- 29. Uchiyama N, et al. 2008. Optimization of evanescent-field fluorescence-assisted lectin microarray for high-sensitivity detection of monovalent oligosaccharides and glycoproteins. Proteomics 8:3042–3050 [DOI] [PubMed] [Google Scholar]
- 30. Zhou Y, et al. 2010. Oral tolerance to food-induced systemic anaphylaxis mediated by the C-type lectin SIGNR1. Nat. Med. 16:1128–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.