Abstract
Background
MicroRNAs can play an important role in tumorigenesis through post-transcriptional regulation of gene expression, and are not well characterized in follicular lymphoma.
Design and Methods
MicroRNA profiles of enriched follicular lymphoma tumor cells from 16 patients were generated by assaying 851 human microRNAs. Tandem gene expression profiles were obtained for predicting microRNA targets.
Results
The expression of 133 microRNAs was significantly different (> 2-fold; P<0.05) between follicular lymphoma and follicular hyperplasia. Forty-four microRNAs in three groups generated a unique follicular lymphoma signature. Of these, ten microRNAs were increased (miR-193a-5p, -193b*, -345, -513b, -574-3p, -584, -663, -1287, -1295, and -1471), 11 microRNAs were decreased (miR-17*, -30a, -33a, -106a*, -141, -202, -205, -222, -301b, -431*, and -570), and 23 microRNAs formed a group that was increased in most cases of follicular lymphoma but showed lower expression in a subset of cases (let-7a, let-7f, miR-7-1*, -9, -9*, -20a, -20b, -30b, -96, -98, -194, -195, -221*, -374a, -374b, -451, -454, -502-3p, -532-3p, -664*, -1274a, -1274b, and -1260). Higher expression of this last group was associated with improved response to chemotherapy. Gene expression analysis revealed increased expression of MAPK1, AKT1, PRKCE, IL4R and DROSHA and decreased expression of CDKN1A/p21, SOCS2, CHEK1, RAD51, KLF4, BLIMP1 and IRF4 in follicular lymphoma. Functional studies indicated that CDKN1A/p21 and SOCS2 expression is directly regulated by miR-20a/-20b and miR-194, respectively.
Conclusions
Follicular lymphoma is characterized by a unique microRNA signature, containing a subset of microRNAs whose expression correlate with response to chemotherapy. miR-20a/b and miR-194 target CDKN1A and SOCS2 in follicular lymphoma, potentially contributing to tumor cell proliferation and survival.
Keywords: follicular lymphoma, miRNA, lymphomagenesis, tumor response
Introduction
Follicular lymphoma (FL) is the second most common type of non-Hodgkin’s lymphoma and comprises over 20% of such lymphomas in the adult population. FL is an indolent disease which becomes resistant to most conventional chemotherapies and, in up to 60% of cases, ultimately transforms into the more aggressive diffuse large B-cell lymphoma (DLBCL).1–3 Constitutive expression of the anti-apoptotic protein BCL2 in FL is a consequence of the t(14;18) chromosomal translocation, which is observed in approximately 70–95% of FL. While constitutive BCL2 expression contributes to the survival advantage of FL cells, the t(14;18) alone is not sufficient for lymphomagenesis, as cells harboring the t(14;18) can be isolated from the peripheral blood of healthy individuals.4 Additional molecular lesions or secondary hits are required to cooperate with BCL2 in the evolution of the malignant phenotype, although the precise mechanisms of lymphomagenesis remain to be elucidated. Gene expression profile studies of FL further suggest that the lymph node microenvironment may play a role in the biology of FL through tumor infiltrating T cells and macrophages,5–9 possibly involving cytokines10,11 or other immunomodulatory mechanisms.
MicroRNAs (miRNAs or miRs) employ a post-transcriptional gene regulation mechanism that has been shown to play a role in development, differentiation, disease, and tumorigenesis. miRNAs are evolutionarily conserved small non-coding RNAs, which regulate gene expression by binding to the 3′ untranslated region of mRNAs, leading to mRNA degradation or translational repression.12–14 Numerous studies have demonstrated the ability of miRNA profiling to discriminate correctly between cancer and normal cells, as well as to discriminate sub-classifications of tumors.15,16 miRNA studies in B-cell development with selected panels of B-cell lymphomas indicate that many B-cell lymphomas continue to express B-cell lineage and maturation stage-specific miRNAs.17 Aberrant miRNA expression has been associated with several B-lineage malignancies including chronic lymphocytic leukemia,18 DLBCL17,19–23 and multiple myeloma.24 However, miRNA profiling studies of FL are rare20 and there are virtually no miRNA studies performed on sorted, enriched populations of FL tumor cells. Given the high number of tumor-infiltrating T cells present in FL lymph node samples, it may be critical to enrich FL B cells prior to analysis in order to identify FL tumor-specific miRNA profiles.
FL is a germinal center B-cell tumor. The lineage-specific appropriate control population for FL tumor cells is normal germinal center B cells, which are abundant in follicular hyperplasia (FH). In this study we generated miRNA profiles of enriched sorted populations of FL tumor cells and germinal center B cells from FH. We found significant differences between FL and FH and identified a unique discriminating miRNA signature for FL. We also found that higher expression of a group of miRNAs was associated with improved response to chemotherapy. miRNA profiles were analyzed in tandem with the mRNA expression of a panel of 199 lymphomagenesis-associated genes in order to identify potential miRNA targets. This strategy identified the potential targets CDKN1A and SOCS2 whose expression was found to be directly regulated by miR-20a/-20b and miR-194, respectively, using miRNA mimics in the t(14;18)-positive cell line OCI-Ly8. These findings suggest that aberrant expression of this set of miRNAs contributes to tumor cell proliferation and survival in FL.
Design and Methods
Patients and staging
The samples examined in this study were obtained from patients undergoing evaluation for treatment in clinical trials of idiotype vaccination at the National Institutes of Health. The patients were enrolled on National Cancer Institute, Institutional Review Board-approved protocols and provided written informed consent before biopsy samples were obtained. The 17 patients included in this study were diagnosed with FL grade 1 or 2. Fifteen patients were treated with PACE chemotherapy (cyclophosphamide, doxorubicin, etoposide and prednisone) as previously described,25,26 and two patients received no chemotherapy. Pre-treatment lymph node biopsies were performed in each of the 17 patients; in addition, post-chemotherapy biopsies were taken from two of these patients at the time of disease progression. Response to therapy was evaluated 1 month after six to eight cycles of chemotherapy by clinical history, physical examination, computed tomography scans of the chest, abdomen and pelvis, and bone marrow biopsies. Complete response was defined as the disappearance of all clinical and laboratory (excluding polymerase chain reaction) signs and symptoms of active disease. Patients with minimal stable residual radiographic lesions between the last two cycles of evaluation were considered to be in complete remission. Partial response was defined as a 50% or greater reduction in the size of lesions. Progressive disease was present if there was a 25% or greater increase in disease burden.
B-cell and T-cell separation in samples of follicular lymphoma and follicular hyperplasia
Nineteen viable frozen cell suspensions of FL tumor samples from 17 patients were used in this study. Seventeen of the samples were obtained at pre-treatment time points; and two samples were obtained from two patients after chemotherapy at the time of progressive or recurrent disease. Anonymous samples of tonsillar FH samples were obtained from The Children’s National Medical Center (Washington, DC, USA) and processed as viable frozen cell suspensions. Prior to cell separation, samples were thawed and dead cells from tonsil samples were removed with a Dead Cell Removal kit (Miltenyi Biotech, Auburn, CA, USA). B and T cells from 19 FL and seven FH samples were separated using a Human CD3 Positive Selection kit (Stem Cell Technologies, Vancouver, Canada). The CD3-negative fraction was collected as the B-cell fraction. The purity of the B-cell fraction was verified and contained over 90% CD20-positive B cells in pilot studies with ten FH samples and one FL sample by flow cytometry analysis using a BD FACSCanto II (BD Biosciences, San Jose, CA, USA) and fluorescently labeled (FITC, PE, PERCP and APC) antibodies against CD20, CD10, CD3, CD19, and kappa and lambda.
Cell culture, transfection and cell proliferation assay
OCI-Ly8 cells (generously provided by Dr. Mark Minden, Ontario Cancer Institute, Toronto, Ontario, Canada)27 were grown in RPMI 1640 medium with 5% CO2. Transient transfection of OCI-Ly8 cells was performed using Amaxa Nucleofector apparatus and Nucleofector solution V (Lonza, Frederick, MD, USA) and program X-001 according to the manufacturer’s instructions. Briefly, 1×107 cells and 125 pmole of mirVana miRNA mimic (miR-20a, miR-20b and miR-194) and negative control oligonucleotides (Ambion, Austin, TX, USA) were separately used in each transfection. After transfection, cells were added to pre-warmed media and immediately placed in an incubator at 37°C in a 5% CO2 atmosphere. Cells were harvested at 24, 36, and 48 hours (h) after transfection. Cell proliferation was assessed at 24 h and 48 h after transfection by counting total cell number using a Scepter hand-held automated cell counter (Millipore, Billerica, MA, USA) and calculating the percentage of dead cells using trypan blue staining (Invitrogen, Carlsbad, CA, USA).
Isolation of RNA
Total RNA was isolated from enriched FL tumor cells, FH B cells, and OCI-Ly8 cells using a miRNeasy Mini Kit (Qiagen, Germantown, MD, USA) according to the manufacturer’s recommendations. RNA concentration and quality were determined using an Agilent Bioanalyzer 2100 (Agilent, Wilmington, DE, USA) and a NanoDrop 2000 instrument (Thermo Scientific, Wilmington, DE, USA).
Agilent human microRNA array
Total RNA (100 ng) from 19 enriched FL tumor cell samples and seven FH B-cell samples was reverse transcribed and labeled with cyanine3-pCp using the Agilent microRNA labeling kit according to the manufacturer’s protocol. Labeled cRNA was purified with Micro Bio-Spin 6 columns (BioRad, Hercules, CA), dried and redissolved in hybridization buffer and hybridized on an Agilent high density human microRNA array (v.3, Rel.12, with 851 human miRNAs on the array) at 56ºC and 20 rpm for 20 h. After washing, array images were scanned on an Agilent G2505C scanner and processed using Agilent Feature Extraction software (v.10.7.3.1). One of the pre-treatment FL samples showed suboptimal hybridization and results from this sample were not included in the miRNA analysis.
NanoString nCounter assay
Gene expression profiling was conducted on 19 enriched FL tumor cell samples and five FH B-cell samples using a NanoString nCounter assay (NanoString Technologies, Seattle, WA, USA)28 with a panel of custom-compiled gene probes related to B-cell differentiation, lymphoma, and/or tumor development. The custom array contained 199 gene probes with four house-keeping genes (GAPDH, GUSB, HPRT1, and PGK1) used as an internal control (Online Supplementary Table S1). The total RNA (4 μL) from enriched FL and FH B-cell samples was hybridized with the capture and reporter probes and incubated overnight at 65°C according to the manufacturer’s recommended protocol. The target and probe complexes were washed and immobilized in the cartridge, and the amount of mRNA was quantified in the nCounter Digital Analyzer. The data were normalized to the spike-in controls and to the geometrical mean of the four house-keeping genes in each hybridization reaction.
Quantitative real-time polymerase chain reaction
Reverse transcription was performed using a High Capacity cDNA Reverse Transcription Kit on a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA). Quantitative real-time polymerase chain reaction (PCR) was performed in triplicate using TaqMan universal PCR master mix and FAM dye-labeled TaqMan MGB probes (CDKN1A and SOCS2 for gene expression; miR-20a, miR-20b, miR-194, RNU66, RNU38B, RNU6B, and Z30 for miR expression) and VIC dye-labeled TaqMan MGB probes (GAPDH and ACTIN) on a 7500 Real-Time PCR System (Applied Biosystems). The ΔCt value was calculated as the Ct value of the target probe subtracted from the geometrical mean of Ct values of GAPDH and ACTIN probes for gene expression; and the geometrical mean of Ct values of RNU66, RNU38b, RNU6B and Z30 probes for miRNA expression. The relative gene or miRNA expression level was calculated as 2-ΔCt.
Statistical analysis
Data from the Agilent microRNA array and Nanostring nCounter assay were analyzed using Agilent GeneSpringGX (v.11.5.1) software. The array data were normalized to the data point of the 75th percentile signal strength and to a set of spike-in and control gene probes on the array, respectively. The differences between the means of experimental groups were analyzed by a two-tailed Mann-Whitney rank sum test (with the Benjamini-Hochberg false discovery rate multiple testing correction method). The miRNAs with statistically significant P values (P≤0.05) and fold change (≥ 2-fold) with both normalization methods (75th percentile and control gene probes) were selected for further analysis. The signal intensity values and fold changes presented in figures and online supplementary tables are from data normalized by control gene probes. Raw data and data normalized by control gene probes were deposited in the GEO database with accession number GSE28090. The differences between the means of experimental groups of normalized Nanostring nCounter assay data were also analyzed by a two-tailed Mann-Whitney rank sum test. P values of less than 0.05 were considered statistically significant. Hierarchical clustering analysis was performed using JMP software (SAS, Cary, NC, USA). Scatter plots were generated using GraphPad Prism software (GraphPad Software, La Jolla, CA, USA). The correlation between miRNA and gene expression was generated using GeneSpringGX (v.11.5.1) software overlapping the list of potential target genes of significant miRNAs (>2-fold difference in expression; P<0.05), generated by TargetScan, with the list of 47 mRNA transcripts significantly differently expressed between FL and FH B cells (>2-fold; P<0.05) in the NanoString nCounter assay. Pathway prediction was done using GeneSpringGX (v.11.5.1) software.
Results
MicroRNAs are differentially expressed in follicular lymphoma tumor cells and follicular hyperplasia germinal center B cells
FL is rich in non-malignant tumor-infiltrating T cells. In order to identify unique miRNA profiles of FL tumor cells, viable cell suspensions from 18 FL tumor biopsies (16 pre-treatment and two post-chemotherapy) and seven control FH samples were sorted and depleted of CD3-positive T cells, resulting in enriched populations of FL tumor cells and FH B cells. The purity of the B-cell fraction was verified and, as evaluated by flow cytometry in pilot studies, contained over 90% CD20-positive B cells. RNA from sorted populations of FL tumor cells and FH B cells was analyzed on a hybridization based Agilent high density microRNA array platform containing probes for 851 human miRNAs. Based on a 2-fold or greater difference in mean values in FL samples relative to FH (P<0.05), we found that 134 miRNAs were significantly differentially expressed in FL tumor cells (Online Supplementary Table S2). Eighty-nine miRNAs were down-regulated and 45 miRNAs showed up-regulation in FL.
A subset of 44 microRNAs discriminates follicular lymphoma from follicular hyperplasia exhibiting a unique follicular lymphoma microRNA signature
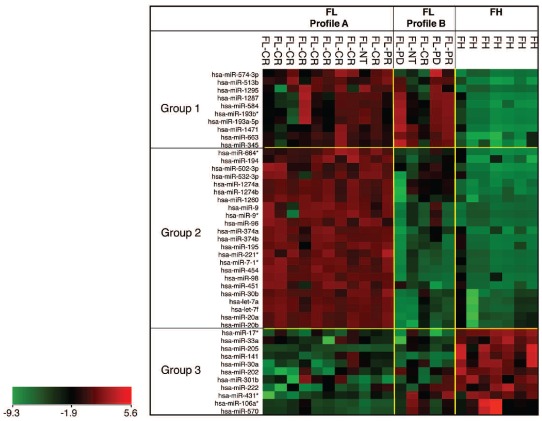
Hierarchical clustering analysis of pre-treatment FL biopsies revealed a subgroup of 44 miRNAs capable of distinguishing FL tumor cells from FH B cells (Figure 1), comprising a unique FL miRNA signature. The 44 miRNAs clustered into three groups. Ten miRNAs showed increased expression in FL tumor cells and were designated miRNA group 1. miRNA group 1 comprised miR-193a-5p, miR-193b*, miR-345, miR-513b, miR-574-3p, miR-584, miR-663, miR-1287, miR-1295 and miR-1471. MiRNA group 2 contained 23 miRNAs that showed increased expression in the majority of FL cases with a subgroup of FL that showed similar expression to FH. miRNA group 2 consisted of let-7a, let-7f, miR-7-1*, miR-9, miR-9*, miR-20a, miR-20b, miR-30b, miR-96, miR-98, miR-194, miR-195, miR-221*, miR-374a, miR-374b, miR-451, miR-454, miR-502-3p, miR-532-3p, miR-664*, miR-1274a, miR-1274b, and miR-1260. Eleven miRNAs were decreased in FL and were designated miRNA group 3 consisting of miR- 17*, miR-30a, miR- 33a, miR-106a*, miR-141, miR-202, miR-205, miR-222, miR-301b, miR-431*, and miR-570 (Figure 1).
Figure 1.
Heatmap of 44 miRNA differentially expressed in FL forming a unique FL signature. Signal intensity values used are values normalized by control gene probes and log2-transformed. Sixteen pre-treatment FL tumor cell samples and seven FH B-cell samples are shown in the heatmap. Response to chemotherapy is indicated for 14 patients who received PACE chemotherapy (see text). FL, follicular lymphoma; FH, follicular hyperplasia; CR, complete response; PR, partial response; PD, progressive disease; NT, no treatment.
Follicular lymphoma microRNA profiles and clinical response to chemotherapy
Hierarchical clustering analysis indicated two sub-patterns within the FL signature, which was largely due to differential expression of miRNA group 2 (Figure 1). The first pattern, designated profile A, showed increased expression of miRNA group 2. The second pattern, designated profile B, showed similar expression of miRNA group 2 between FL and FH. The difference between profiles A and B prompted us to correlate miRNA data with available clinical treatment and outcome data. Fourteen of the patients were treated with PACE chemotherapy,25,26 while two of the patients received no chemotherapy. Of the patients treated with chemotherapy, ten achieved a complete response, two achieved a partial response and two had progressive disease. Profile A was found in nine cases of complete response and in one patient with a partial response (Figure 1), while profile B was present in two cases of progressive disease, one partial response and one complete response. The difference between profiles A and B was statistically significant (P<0.0001) and the findings suggest that profile A is associated with an improved response to chemotherapy.
Genes differentially expressed in follicular lymphoma tumor cells
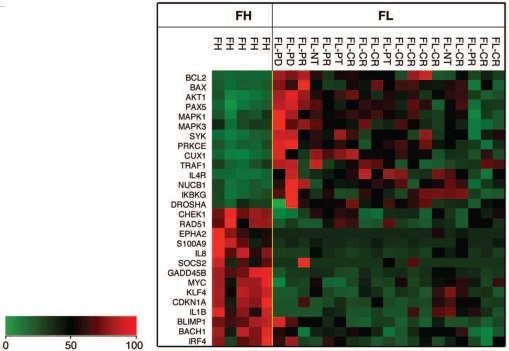
To investigate the potential effect of changes in miRNA expression on gene expression and lymphomagenesis in FL, tandem focused gene expression profiling was performed on 19 sorted, enriched FL tumor samples (17 pre-treatment and 2 post-chemotherapy) and five sorted enriched FH B-cell samples. A panel of 199 genes (Online Supplementary Table S1) related to B-cell development, lymphomagenesis and tumorigenesis was compiled, and the expression of these genes in FL tumor cells and FH B cells was examined using the NanoString nCounter assay platform.28 Forty-seven of the 199 genes in the assay were found to be differentially expressed in FL (P≤0.05); among them, 33 were changed 2-fold or more (Online Supplementary Table S3). Many of these genes were related to cell proliferation, apoptosis or DNA repair. Hierarchical clustering analysis indicated that a subset of genes discriminated between FL and FH. AKT1, BAX, BCL2, BCL6, CUX1, DROSHA, EP300, IKBKG, IL4R, MAPK1, NUCB1, PAX5, PRKCE, and SYK were significantly increased in a proportion of FL; while the expression of BACH1, BRCA1, CDKN1A, CHEK1, EPHA2, GADD45B, IL1B, IL6, IL8, JUN, MKI67, PRDM1/BLIMP1, RAD51, SOCS2, STAT4, TNF and VEGFA was significantly decreased in FL (Figure 2 and Online Supplementary Table S3).
Figure 2.
Heatmap of 28 genes differentially expressed in FL. Values used are normalized relative mRNA counts scaled to between 0 and 100. Nineteen FL tumor samples (17 pre-treatment biopsies, and 2 post-treatment; see text for details) and five FH B-cell samples are shown. FL: follicular lymphoma; FH: follicular hyperplasia; CR: complete response; PR: partial response; PD: progressive disease; NT: no treatment; PT: post-treatment.
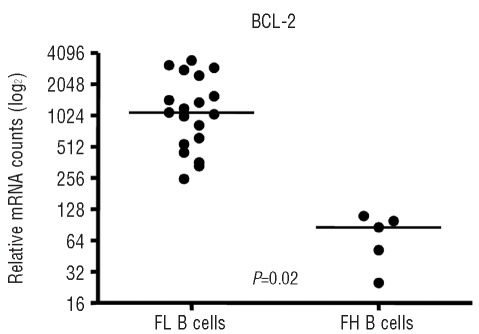
Constitutive expression of the anti-apoptotic protein BCL2 in FL is the consequence of the t(14;18) chromosomal translocation which is present in approximately 70–95% of FL.1–4 We did not have cytogenetic or fluorescence in situ hybridization data on our patients’ samples to document the presence or absence of the t(14;18). However, the enriched FL tumor cells and FH B cells were examined for BCL2 expression. BCL2 expression was 16.3-fold higher in the FL tumor cells than in the FH B cells (P=0.02), suggesting that the majority of the FL samples probably contained the t(14;18) (Figure 3).
Figure 3.
BCL2 is over-expressed in FL tumor cells. The relative mRNA counts of BCL2 were obtained from the NanoString nCounter assay. Lines indicate median values. P=0.02, 16.3-fold difference in BCL2 expression. FL: follicular lymphoma; FH: follicular hyperplasia.
CDKN1A/p21 and SOCS2 are regulated by miR-20a/b and miR-194, contributing to lymphoma cell proliferation
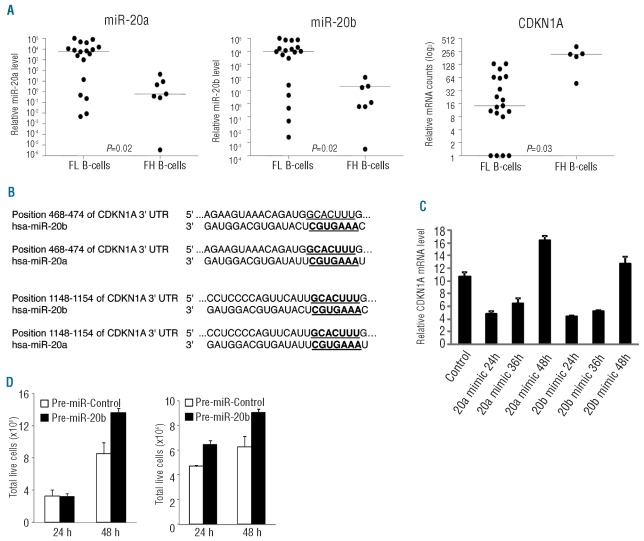
miRNA regulates gene expression by causing degradation of mRNA or by inhibiting mRNA translation.12–14 In order to identify potential targets of miRNAs contained in the FL signature, potential miRNA binding targets were predicted using TargetScan,29 and compared with gene expression profiles of the 47 genes differentially expressed in FL and FH. Multiple putative pathways of gene targets were predicted, including B-cell receptor, interleukins, TNFα/NF-κB, TGFβ receptor, cell cycle control and DNA repair pathways. We did not evaluate mRNAs that were not differently expressed between FL tumor cells and FH B cells, although some of these mRNAs could also be targets of the miRNA in the FL signature. Among many changes in miRNA and gene expression observed, the expression of miR-20a and miR-20b was reversely correlated with the expression of CDKN1A/p21 between FL tumor cells and FH B cells (Figure 4A). The expression of miR-20a was more than 9-fold higher in FL tumor cells than in FH B cells (P=0.02), while CDKN1A was decreased 11-fold in FL (P=0.03). TargetScan analysis predicted that miR-20a and miR-20b potentially regulate the expression of CDKN1A/p21. Sequence comparison revealed that there are two predicted conserved binding sites for miR-20a and miR-20b in the 3′untranslated region of CDKN1A mRNA (Figure 4B). In order to evaluate the validity of the TargetScan predictions, the t(14;18)-containing, germinal center B-cell DLBCL cell line, OCI-Ly8,27 was utilized in transfection experiments. The transfection efficiency reached approximately 80% as assessed by transfecting Cy3-labeled pre-miRNA negative control oligonucleotides or by transfecting GFP-expressing plasmid in pilot experiments. The endogenous levels of miR-20a, -20b, and -194 in OCI-Ly8 cells were assessed by TaqMan quantitative real-time PCR assays using RNA from seven enriched FH B-cell samples as controls. The miR-20a and miR-194 levels were higher (P=0.006, 2.16-fold for miR-20a; and P=0.006, 1.86-fold for miR-194) and the miR-20b level was lower (P=0.006, 2.16-fold) in OCI-Ly8 cells than in FH B cells (Online Supplementary Figure S1). To examine the effect of miR-20a and miR-20b on CDKN1A transcripts, OCI-Ly8 cells were transiently transfected separately with mirVana miRNA mimics corresponding to miR-20a and miR-20b. The subsequent expression level of CDKN1A was determined by quantitative real-time PCR assay. The mRNA level of CDKN1A was reduced more than 50% by miR-20a and miR-20b mimics 24 and 36 h after the transfection. The inhibitory effect on CDKN1A by miR-20a and -20b was lost 48 h after transfection, likely due to rapid proliferation of OCI-Ly8 cells (Figure 4C). The effect of miR-20a/b transfection on cell proliferation was assessed by counting the total number of live cells 24 and 48 h after transfection. As shown in Figure 4D, transfection of miR-20a and -20b resulted in substantial increases in cell growth 24 and 48 h after transfection in comparison to cell growth in controls.
Figure 4.
CDKN1A is regulated by miR-20a and miR-20b affecting tumor cell growth. (A) CDKN1A, miR-20a and miR-20b expression in patients’ FL tumor cells and control FH B cells. Relative signal intensities of miR-20a and 20b were obtained from a microRNA array; and relative mRNA counts of CDKN1A were obtained from the NanoString nCounter assay. Lines indicate median values. P=0.02, greater than 9-fold difference for miR-20a; P=0.02, greater than 7-fold difference for miR-20b; and P=0.03, 11.8-fold difference in CDKN1A expression. FL, follicular lymphoma; FH, follicular hyperplasia. (B) The predicted conserved miRNA binding sites of miR-20a and miR-20b in the 3′ untranslated region (UTR) of CDKN1A mRNA. The matched putative miRNA binding sequence in 3′UTR of CDKN1A mRNA and miRNA seed sequence are in bold and underlined. (C) and (D), CDKN1A expression is regulated by miR-20a and miR-20b affecting tumor cell growth. OCI-Ly8 cells were separately transfected with miR-20a and-20b mimic oligonucleotides, the changes in CDKN1A/p21 mRNA level at 24, 36 and 48 h after transfection were determined by TaqMan quantitative real-time PCR assay. Mean and standard deviation of triplicate experiments are shown (C). Total live cells were counted 24 and 48 h after transfection using a Scepter hand-held automated cell counter and by trypan blue staining. Mean and standard deviation of triplicate counts are shown (D).
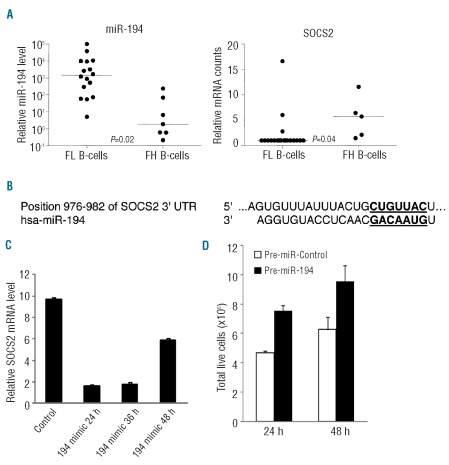
Similarly, the expression of miR-194 was reversely correlated with the expression of SOCS2 (Figure 5A) in patients’ FL tumor cells compared to in FH B cells. The expression of miR-194 was increased more than 5-fold (P=0.2); and the expression of SOCS2 was decreased by 2.9-fold (P=0.04) in patients’ FL tumor cells (Figure 5A). TargetScan analysis predicts that, among many other genes, miR-194 potentially regulates the expression of SOCS2. One predicted conserved binding site of miR-194 was found in the 3′ untranslated region of SOCS2 mRNA (Figure 5B). To test whether SOCS2 may be regulated by miR-194, OCI-Ly8 cells were transiently transfected with miR-194 mimic oligonucleotides. The expression level of SOCS2 was determined by quantitative real-time PCR assay. As shown in Figure 5C, the mRNA level of SOCS2 was reduced by more than 80% at 24 and 36 h and more than 40% at 48 h after transfection. The effect of miR-194 transfection on cell proliferation was also assessed. miR-194 mimic transfection leads to substantial increase in cell growth 24 and 48 h after transfection in comparison to controls (Figure 5D).
Figure 5.
SOCS2 is regulated by miR-194 affecting tumor cell growth. (A), SOCS2 and miR-194 expression in patients’ FL tumor cells and control FH B cells. Relative signal intensity of miR-194 was obtained from a microRNA array and relative mRNA counts of SOCS2 were obtained from a NanoString nCounter assay. Lines indicate median values. P=0.02, 5.8-fold for miR-194; and P=0.04, 2.9-fold for SOCS2. FL, follicular lymphoma; FH, follicular hyperplasia. (B), The predicted conserved miR binding sites in 3′ untranslated (3′ UTR) of SOCS2 mRNA. The matched putative miRNA binding sequence in 3′ UTR of SOCS2 mRNA and miRNA seed sequence are in bold and underlined. (C) and (D), SOCS2 expression is regulated by miR-194 affecting tumor cell growth. OCI-Ly8 cells were transfected with miR-194 mimic oligonucleotides; the changes in SOCS2 mRNA level at 24, 36 and 48 h after transfection were determined by TaqMan quantitative real-time PCR assay. Mean and standard deviation of triplicate experiments are shown (C). Total live cells were counted 24 and 48 h after transfection using a Scepter hand-held automated cell counter and by trypan blue staining. Mean and standard deviation of triplicate counts are shown (D).
Taken together, these findings indicate that miR-20a/b might regulate the expression of CDKN1A and miR-194 regulates the expression of SOCS2 in FL. The decreased expression of the cell cycle inhibitor CDKN1A/p21 and SOCS2 observed in FL patients’ samples may be a consequence of elevated expression of miR-20a, miR-20b, and miR-194. The altered expression of miR-20a/b and miR-194 may contribute to cell proliferation in FL through regulation of protein expression of CDKN1A/p21 and SOCS2 transcripts.
Discussion
In this study we identified a unique miRNA signature of FL comprising 44 miRNAs in three groups. This signature was capable of discriminating FL tumor cells from normal germinal center B cells in FH. Two patterns of miRNA expression were noted within FL patients’ samples and higher expression of group 2 miRNAs was associated with an improved response to chemotherapy. mRNA targets of three miRNAs in group 2 (miR-20a, -20b and -194) were predicted and validated in functional studies; these targets have tumor suppressor properties and are involved in regulation of cell cycle arrest and cytokine signaling.
Both miR-20a and miR-20b were found to be over-expressed in the majority of FL samples (profile A) in our study. MiR-20a is a member of the miR-17-92 cluster, a cluster that encodes six miRNAs (miR-17, miR-18a, mir-19a, miR-20a, miR-19b-1 and miR-92-1). The miR-17-92 cluster is located on chromosome 13 (q31.3) and has been found to be over-expressed due to genomic amplification in several lymphomas including DLBCL and FL.30–32 Expression of this cluster has been shown to be oncogenic in animal models and cell lines.33,34 MiR-20b is derived from the miR-106a-92 cluster on chromosome X, which encodes miR-363, miR-92-2, miR-19b-2, miR-20b, miR-18b and miR-106a. This cluster is homologous to the oncogenic miR-17-92 cluster.35
TargetScan predicted that miR-20a and -20b target CDKN1A/p21, and miR-194 regulates the expression of SOCS2. Functional studies in OCI-Ly8 cells support the TargetScan predictions that miR-20a and miR-20b regulate the expression of CDKN1A and that miR-194 regulates the expression of SOCS2. CDKN1A/p21 is a critical cell cycle inhibitor enforcing the restriction on G1/S transition of the cell cycle in response to DNA damage or other environmental insults.36,37 Others have also demonstrated that miR-20a targets CDKN1A/p21 in lymphoma cell lines.38 It is possible that decreased expression of the cell cycle inhibitor CDKN1A/p21 and SOCS2, which was observed in vivo in FL patients’ samples, may be a consequence of elevated expression of miR-20a, miR-20b, and miR-194. Taken together, altered expression of miR-20a, -20b, and -194 may contribute to cell proliferation and lymphomagenesis in FL through regulation of protein expression of CDKN1A/p21 and SOCS2 transcripts. It is interesting that patients with increased levels of miR-20a, -20b and -194 (group 2 miRNAs) showed improved response to chemotherapy. It is plausible to postulate that decreased expression of the cell cycle inhibitor, CDKN1A/p21, may lead to loss of the G1 cell cycle restriction point in FL tumor cells, conferring increased sensitivity to traditional chemotherapy agents.
MicroRNAs regulate gene expression by binding to the 3′ untranslated region of mRNAs, leading to either mRNA degradation or translational repression.12–14 In this study, we examined two miRNA targets (CDKN1A and SOCS2) whose expression differed between FL and FH. Additional potential miRNA targets remain to be investigated to determine whether target mRNAs are degraded or repressed by microRNAs during protein translation processes. The gene transcripts examined in this study are involved in the regulation of major signal transduction pathways (cell cycle control, apoptosis, NF-κB, PI-3K/AKT, JAK/STAT, and DNA damage signaling/repair pathways), which play important roles in biology and tumorigenesis. We found that the expression of AKT1, BAX, BCL2, BCL6, CUX1, DROSHA, EP300, IKBKG, IL4R, MAPK1, NUCB1, PAX5, PRKCE, and SYK was significantly increased, while the expression of BACH1, BRCA1, CDKN1A, CHEK1, EPHA2, GADD45B, IL1B, IL6, IL8, JUN, MKI67, PRDM1/BLIMP1, RAD51, SOCS2, STAT4, TNF and VEGFA was significantly decreased in FL tumor cells. We previously reported that IL-4 protein is increased, and MAPK1 is activated in FL, as determined by multiplex cytokine ELISA arrays and reverse-phase protein arrays.11 One recent study identified the source of increased IL-4 expression in FL as tumor-infiltrating T helper cells, with the consequence of over-expression of IL-4-regulated genes in FL tumor cells via activation of STAT6.10 In our current study, we found increased expression of IL4R, MAPK1 and AKT transcripts in enriched sorted FL tumor cells, further indicating that IL4/IL4R, MAPK and PI-3K/AKT pathways potentially play crucial roles in FL tumor proliferation and survival.
We observed that the expression of cell cycle inhibitor CDKN1A transcripts was decreased and the expression of cell cycle activator MAPK1/ERK transcripts was increased in FL compared to in FH. Consistent with these findings, in our previous study we also noted that ERK1/2 protein was basally or constitutively phosphorylated in FL, which was not seen in FH.11 These findings are interesting and somewhat unexpected because histological sections of clinical samples of FL and FH stained by immunohistochemistry with the proliferation marker Ki-67 consistently show greater staining for Ki-67 in FH than in grade 1 and 2 FL, indicating that the proliferation rate is higher in FH than in FL. Indeed, this immunohistochemical finding is used diagnostically by hematopathologists to differentiate FL from FH. It is unclear what factors may play a role in the reduced proliferation rate in FL as compared to FH. BCL2 is typically constitutively over-expressed in FL as a consequence of the t(14;18) chromosomal translocation. Interestingly, BCL2 has been shown not only to inhibit apoptosis and promote cell survival, but also to facilitate cell cycle arrest at the G0 phase and to delay G0 to S phase transition depending on cellular lineage and context.39 Constitutive activation of the MAPK signaling pathway may also contribute to the decreased proliferation rate in FL, since negative feedback loops within the MAPK signaling pathway have been reported, and non-physiological hyperactivation of RAS-ERK signaling has been shown to induce the expression of cyclin-dependent kinase inhibitors and trigger cell cycle arrest.40 Furthermore, FH typically occurs in the setting of inflammation with increased production of a variety of cytokines, which leads to a rapid expansion of lymphoid follicles and thus a high proliferation rate in FH.11 In the FL tumor environment, the expression of cytokines, with the exception of IL-4, is comparatively low.11 Increased expression and phosphorylation of MAPK1/ERK in FL cells may be a consequence of altered expression and constitutive activation of cell proliferation and survival signals related to oncoproteins, miRNAs, components of the NF-κB pathway, or other factors in the tumor microenvironment. Despite increased expression and phosphorylation of MAPK1/ERK in FL, the proliferation rate of FL is lower than that of FH and many other B-cell lymphomas including DLBCL, suggesting that other components of the cell proliferation pathway might be altered; or other cell cycle checkpoints or negative feedback mechanisms may be partially intact in FL. Clearly, when FL transforms into DLBCL, the proliferation rate and aggressiveness of the tumor increase.
BLIMP1 (PRDM1) and BCL6 are critical regulators of germinal center B-cell differentiation.41–43 BLIMP1 and BCL6 are expressed in a mutually exclusive pattern and evidence suggests that they repress each other in germinal center B cells.43,44 Consistent with this model, a marked decrease of BLIMP1 and an increase of BCL6 were observed in FL tumor cells in our study. BLIMP1 mutations are frequently found in activated B cell (ABC)-like DLBCL,45,46 and it has been demonstrated that BLIMP1 functions as a tumor suppressor.46,47 Let-7a, miR-9, miR-125b have been shown to regulate BLIMP1 expression,17,48,49 and miR-9 was reportedly increased in FL tissue samples and in germinal center B cells.17,20 A similar pattern was observed in our study; we found that let-7a and miR-9 were over-expressed in FL tumor cells with down-regulation of BLIMP1. The expression of several key DNA damage signaling and repair proteins, BRCA1, BACH1, CHEK1 and RAD51,36,50–52 was found to be decreased in FL in this study. These findings imply impairment of DNA repair in FL, which may contribute to the accumulation of genetic mutations during the course of the disease. In summary, the findings of this study indicate a role for aberrant microRNA expression in the biology of FL and provide insights for future studies on FL lymphomagenesis and tumor evolution.
Supplementary Material
Acknowledgments
We would like to thank Dr. Mark Minden of the Ontario Cancer Institute for generously providing the OCI-Ly8 cell line; Kaska Wloka for assistance with tonsil specimens, William Kopp for assistance with FL samples, Ryan Plyler for laboratory assistance, and Jamie Hahn for assistance with clinical records.
Footnotes
Funding: this work was supported by the Intramural Research Program of the NIH Clinical Center and the Center for Cancer Research, NCI.
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Vitolo U, Ferreri AJ, Montoto S. Follicular lymphomas. Crit Rev Oncol Hematol. 2008;66(3):248–61. doi: 10.1016/j.critrevonc.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 2.de Jong D. Molecular pathogenesis of follicular lymphoma: a cross talk of genetic and immunologic factors. J Clin Oncol. 2005;23(26):6358–63. doi: 10.1200/JCO.2005.26.856. [DOI] [PubMed] [Google Scholar]
- 3.Tan D, Horning SJ. Follicular lymphoma: clinical features and treatment. Hematol Oncol Clin North Am. 2008;22(5):863–82. viii. doi: 10.1016/j.hoc.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 4.Schuler F, Hirt C, Dolken G. Chromosomal translocation t(14;18) in healthy individuals. Semin Cancer Biol. 2003;13(3):203–9. doi: 10.1016/s1044-579x(03)00016-6. [DOI] [PubMed] [Google Scholar]
- 5.Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351(21):2159–69. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- 6.Janikova A, Tichy B, Supikova J, Stano-Kozubik K, Pospisilova S, Kren L, et al. Gene expression profiling in follicular lymphoma and its implication for clinical practice. Leuk Lymphoma. 2011;52(1):59–68. doi: 10.3109/10428194.2010.531412. [DOI] [PubMed] [Google Scholar]
- 7.Andreasson U, Eden P, Peterson C, Hogerkorp CM, Jerkeman M, Andersen N, et al. Identification of uniquely expressed transcription factors in highly purified B-cell lymphoma samples. Am J Hematol. 2010;85(6):418–25. doi: 10.1002/ajh.21701. [DOI] [PubMed] [Google Scholar]
- 8.Leich E, Salaverria I, Bea S, Zettl A, Wright G, Moreno V, et al. Follicular lymphomas with and without translocation t(14;18) differ in gene expression profiles and genetic alterations. Blood. 2009;114(4):826–34. doi: 10.1182/blood-2009-01-198580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Husson H, Carideo EG, Neuberg D, Schultze J, Munoz O, Marks PW, et al. Gene expression profiling of follicular lymphoma and normal germinal center B cells using cDNA arrays. Blood. 2002;99(1):282–9. doi: 10.1182/blood.v99.1.282. [DOI] [PubMed] [Google Scholar]
- 10.Pangault C, Ame-Thomas P, Ruminy P, Rossille D, Caron G, Baia M, et al. Follicular lymphoma cell niche: identification of a preeminent IL-4-dependent T(FH)-B cell axis. Leukemia. 2010;24(12):2080–9. doi: 10.1038/leu.2010.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calvo KR, Dabir B, Kovach A, Devor C, Bandle R, Bond A, et al. IL-4 protein expression and basal activation of Erk in vivo in follicular lymphoma. Blood. 2008;112(9):3818–26. doi: 10.1182/blood-2008-02-138933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 14.Di Leva G, Croce CM. Roles of small RNAs in tumor formation. Trends Mol Med. 2010;16(6):257–67. doi: 10.1016/j.molmed.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 16.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Jima DD, Jacobs C, Fischer R, Gottwein E, Huang G, et al. Patterns of microRNA expression characterize stages of human B-cell differentiation. Blood. 2009;113(19):4586–94. doi: 10.1182/blood-2008-09-178186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353(17):1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 19.Lawrie CH, Soneji S, Marafioti T, Cooper CD, Palazzo S, Paterson JC, et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int J Cancer. 2007;121(5):1156–61. doi: 10.1002/ijc.22800. [DOI] [PubMed] [Google Scholar]
- 20.Roehle A, Hoefig KP, Repsilber D, Thorns C, Ziepert M, Wesche KO, et al. MicroRNA signatures characterize diffuse large B-cell lymphomas and follicular lymphomas. Br J Haematol. 2008;142(5):732–44. doi: 10.1111/j.1365-2141.2008.07237.x. [DOI] [PubMed] [Google Scholar]
- 21.Lawrie CH, Chi J, Taylor S, Tramonti D, Ballabio E, Palazzo S, et al. Expression of microRNAs in diffuse large B cell lymphoma is associated with immunophenotype, survival and transformation from follicular lymphoma. J Cell Mol Med. 2009;13(7):1248–60. doi: 10.1111/j.1582-4934.2008.00628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C, Kim SW, Rai D, Bolla AR, Adhvaryu S, Kinney MC, et al. Copy number abnormalities, MYC activity, and the genetic finger-print of normal B cells mechanistically define the microRNA profile of diffuse large B-cell lymphoma. Blood. 2009;113(26):6681–90. doi: 10.1182/blood-2009-01-202028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jima DD, Zhang J, Jacobs C, Richards KL, Dunphy CH, Choi WW, et al. Deep sequencing of the small RNA transcriptome of normal and malignant human B cells identifies hundreds of novel microRNAs. Blood. 2010;116(23):e118–27. doi: 10.1182/blood-2010-05-285403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y, Chen L, Barlogie B, Stephens O, Wu X, Williams DR, et al. High-risk myeloma is associated with global elevation of miRNAs and overexpression of EIF2C2/AGO2. Proc Natl Acad Sci USA. 2010;107(17):7904–9. doi: 10.1073/pnas.0908441107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bendandi M, Gocke CD, Kobrin CB, Benko FA, Sternas LA, Pennington R, et al. Complete molecular remissions induced by patient-specific vaccination plus granulocyte-monocyte colony-stimulating factor against lymphoma. Nat Med. 1999;5(10):1171–7. doi: 10.1038/13928. [DOI] [PubMed] [Google Scholar]
- 26.Neelapu SS, Gause BL, Harvey L, Lee ST, Frye AR, Horton J, et al. A novel proteoliposomal vaccine induces antitumor immunity against follicular lymphoma. Blood. 2007;109(12):5160–3. doi: 10.1182/blood-2006-12-063594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang H, Blondal JA, Benchimol S, Minden MD, Messner HA. p53 mutations, c-myc and bcl-2 rearrangements in human non-Hodgkin’s lymphoma cell lines. Leuk Lymphoma. 1995;19(1–2):165–71. doi: 10.3109/10428199509059672. [DOI] [PubMed] [Google Scholar]
- 28.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26(3):317–25. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 29.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 30.Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31–q32 amplification in malignant lymphoma. Cancer Res. 2004;64(9):3087–95. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 31.Tagawa H, Seto M. A microRNA cluster as a target of genomic amplification in malignant lymphoma. Leukemia. 2005;19(11):2013–6. doi: 10.1038/sj.leu.2403942. [DOI] [PubMed] [Google Scholar]
- 32.Knuutila S, Bjorkqvist AM, Autio K, Tarkkanen M, Wolf M, Monni O, et al. DNA copy number amplifications in human neoplasms: review of comparative genomic hybridization studies. Am J Pathol. 1998;152(5):1107–23. [PMC free article] [PubMed] [Google Scholar]
- 33.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17–92 expression in lymphocytes. Nat Immunol. 2008;9(4):405–14. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, et al. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16(6):720–8. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408(6811):433–9. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 37.Sherr CJ. The Pezcoller lecture: cancer cell cycles revisited. Cancer Res. 2000;60(14):3689–95. [PubMed] [Google Scholar]
- 38.Inomata M, Tagawa H, Guo YM, Kameoka Y, Takahashi N, Sawada K. MicroRNA-17-92 down-regulates expression of distinct targets in different B-cell lymphoma subtypes. Blood. 2009;113(2):396–402. doi: 10.1182/blood-2008-07-163907. [DOI] [PubMed] [Google Scholar]
- 39.Zinkel S, Gross A, Yang E. BCL2 family in DNA damage and cell cycle control. Cell Death Differ. 2006;13(8):1351–9. doi: 10.1038/sj.cdd.4401987. [DOI] [PubMed] [Google Scholar]
- 40.Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci. 2011;36(6):320–8. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Angelin-Duclos C, Cattoretti G, Lin KI, Calame K. Commitment of B lymphocytes to a plasma cell fate is associated with Blimp-1 expression in vivo. J Immunol. 2000;165(10):5462–71. doi: 10.4049/jimmunol.165.10.5462. [DOI] [PubMed] [Google Scholar]
- 42.Kuo TC, Shaffer AL, Haddad J, Jr, Choi YS, Staudt LM, Calame K. Repression of BCL-6 is required for the formation of human memory B cells in vitro. J Exp Med. 2007;204(4):819–30. doi: 10.1084/jem.20062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martins G, Calame K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol. 2008;26:133–69. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
- 44.Tunyaplin C, Shaffer AL, Angelin-Duclos CD, Yu X, Staudt LM, Calame KL. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J Immunol. 2004;173(2):1158–65. doi: 10.4049/jimmunol.173.2.1158. [DOI] [PubMed] [Google Scholar]
- 45.Pasqualucci L, Compagno M, Houldsworth J, Monti S, Grunn A, Nandula SV, et al. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J Exp Med. 2006;203(2):311–7. doi: 10.1084/jem.20052204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mandelbaum J, Bhagat G, Tang H, Mo T, Brahmachary M, Shen Q, et al. BLIMP1 is a tumor suppressor gene frequently disrupted in activated B cell-like diffuse large B cell lymphoma. Cancer Cell. 2010;18(6):568–79. doi: 10.1016/j.ccr.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calado DP, Zhang B, Srinivasan L, Sasaki Y, Seagal J, Unitt C, et al. Constitutive canonical NF-kappaB activation cooperates with disruption of BLIMP1 in the pathogenesis of activated B cell-like diffuse large cell lymphoma. Cancer Cell. 2010;18(6):580–9. doi: 10.1016/j.ccr.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nie K, Gomez M, Landgraf P, Garcia JF, Liu Y, Tan LH, et al. MicroRNA-mediated down-regulation of PRDM1/Blimp-1 in Hodgkin/Reed-Sternberg cells: a potential pathogenetic lesion in Hodgkin lymphomas. Am J Pathol. 2008;173(1):242–52. doi: 10.2353/ajpath.2008.080009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malumbres R, Sarosiek KA, Cubedo E, Ruiz JW, Jiang X, Gascoyne RD, et al. Differentiation stage-specific expression of microRNAs in B lymphocytes and diffuse large B-cell lymphomas. Blood. 2009;113(16):3754–64. doi: 10.1182/blood-2008-10-184077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu X, Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol Cell Biol. 2004;24(21):9478–86. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumaraswamy E, Shiekhattar R. Activation of BRCA1/BRCA2-associated helicase BACH1 is required for timely progression through S phase. Mol Cell Biol. 2007;27(19):6733–41. doi: 10.1128/MCB.00961-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gupta R, Sharma S, Sommers JA, Kenny MK, Cantor SB, Brosh RM., Jr FANCJ (BACH1) helicase forms DNA damage inducible foci with replication protein A and interacts physically and functionally with the single-stranded DNA-binding protein. Blood. 2007;110(7):2390–8. doi: 10.1182/blood-2006-11-057273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.