Abstract
Background
The category B agent of bioterrorism, Entamoeba histolytica has a two-stage life cycle: an infective cyst stage, and an invasive trophozoite stage. Due to our inability to effectively induce encystation in vitro, our knowledge about the cyst form remains limited. This also hampers our ability to develop cyst-specific diagnostic tools.
Aims
Three main aims were (i) to identify E. histolytica proteins in cyst samples, (ii) to enrich our knowledge about the cyst stage, and (iii) to identify candidate proteins to develop cyst-specific diagnostic tools.
Methods
Cysts were purified from the stool of infected individuals using Percoll (gradient) purification. A highly sensitive LC-MS/MS mass spectrometer (Orbitrap) was used to identify cyst proteins.
Results
A total of 417 non-redundant E. histolytica proteins were identified including 195 proteins that were never detected in trophozoite-derived proteomes or expressed sequence tag (EST) datasets, consistent with cyst specificity. Cyst-wall specific glycoproteins Jacob, Jessie and chitinase were positively identified. Antibodies produced against Jacob identified cysts in fecal specimens and have potential utility as a diagnostic reagent. Several protein kinases, small GTPase signaling molecules, DNA repair proteins, epigenetic regulators, and surface associated proteins were also identified. Proteins we identified are likely to be among the most abundant in excreted cysts, and therefore show promise as diagnostic targets.
Major Conclusions
The proteome data generated here are a first for naturally-occurring E. histolytica cysts, and they provide important insights into the infectious cyst form. Additionally, numerous unique candidate proteins were identified which will aid the development of new diagnostic tools for identification of E. histolytica cysts.
Author Summary
We used tandem mass spectrometry to identify E. histolytica cyst proteins in 5 cyst positive stool samples. We report the identification of 417 non-redundant E. histolytica proteins including 195 proteins that were not identified in existing trophozoite derived proteome or EST datasets, consistent with cyst specificity. Because the cysts were derived directly from patient samples with incomplete purification, a limited number of proteins were identified (N = 417) that probably represent only a partial proteome. Nevertheless, the study succeeded in identifying proteins that are likely to be abundant in the cyst stage of the parasite. Several of these proteins may play roles in E. histolytica stage conversion or cyst function. Proteins identified in this study may be useful markers for diagnostic detection of E. histolytica cysts. Overall, the data generated in this study promises to aid the understanding of the cyst stage of the parasite which is vital for disease transmission and pathogenesis in E. histolytica.
Introduction
The parasitic protozoan Entamoeba histolytica is the causative agent of amebic colitis and amebic liver abscesses in humans [1], [2]. The World Health Organization estimates up to 50 million invasive infections world-wide annually [3]. E. histolytica has a simple, two-stage life cycle, consisting of the infective cyst and colon-invasive trophozoite forms. E. histolytica infections occur when cysts are ingested through contaminated food or water. In the lower intestine trophozoites emerge from cysts (a process known as excystation). As a result of unknown stimuli in the intestine, trophozoites again can differentiate into cysts (a process known as encystation), which may be excreted in feces to infect other humans. Although the cyst is the only form to transmit infections, most studies on E. histolytica have focused on the trophozoite form, which is the only form that can be readily cultured. The inability to encyst trophozoites in vitro has severely impaired our knowledge on the infectious stage of E. histolytica.
There is an increasing recognition of the burden of infection due to this protozoan parasite. Carefully conducted serologic studies in Mexico, where amebiasis is endemic, demonstrated antibody to E. histolytica in 8.4% of the population [4]. In the urban slum of Fortaleza, Brazil, 25% of the people tested carried antibody to E. histolytica; the prevalence of anti-amebic antibodies in children aged six to fourteen years was 40% [5]. A prospective study of preschool children in a slum of Dhaka, Bangladesh demonstrated new E. histolytica infection in 39% of children over a one year period of observation, with 10% of the children having an E. histolytica infection associated with diarrhea and 3% with dysentery [6].
The diagnosis of E. histolytica infection in endemic areas still relies on microscopy, which is neither sensitive nor specific [7]. PCR-based diagnostic methods have not replaced microscopy in endemic areas, as they require skilled people and sophisticated laboratory settings which are absent in these areas. Although there are simple (ELISA-based) diagnostic tools available to detect the trophozoite form of E. histolytica, none are designed to detect the cyst form of parasite. There are two reasons to produce tests that detect the cyst form of E. histolytica. First, the cyst is the infectious form of the parasite, and therefore of greatest importance for detection in potential food- or water-borne outbreaks, both natural and man-made. Second, cyst antigens are expected to be more abundant, stable, and readily detectable in stool samples, including formalin-preserved samples. Lack of such stability is the major limitation of the current E. histolytica antigen-detection test by TechLab [8]. However our understanding of E. histolytica cyst proteins remains the major factor limiting our ability to develop cyst specific diagnostic reagents.
Relatively more is known about the cyst stage of the reptilian parasite Entamoeba invadens [9]. E. invadens can be induced to encyst in vitro [10]–[12], a phenomenon that has made it a model system to study encystation. Studies have identified osmotic shock [13], low glucose [12] or carbon source [14], and levels of serum and mucin [15] as potential triggers of encystation in E. invadens in vitro. Additionally, cyst- and cyst-wall specific genes have been identified including chitin synthetase [16], chitinase [17], chitosan [18], and the lectins Jessie and Jacobs [19]. A 70-kDa heat shock protein [20], and proteins involved in proteasome function [21] were also found to be linked to encystation in E. invadens.
E. histolytica strains can undergo spontaneous encystation, although very inefficiently, when grown in presence of bacteria [22]. A pioneering microarray analysis of this process identified about 15% of all genes in the genome as developmentally regulated based on their mRNA transcript levels (>3-fold change, p-value<0.01) including 672 genes referred to as cyst-specific and 767 genes referred to as trophozoite-specific. The cyst-specific genes included cysteine proteases, putative DNA-binding or transcription factor-related proteins (such as Myb domain proteins) and signal transduction-related transmembrane protein kinases. The promoter motif for one of the Myb domain proteins was later characterized for and this motif appeared to function in the regulation of a subset of cyst-specific genes [23].
In contrast to transcriptomic data, no proteomic data are currently available for E. histolytica cysts. Proteomic analysis of cysts and trophozoites of E. invadens using high resolution 2D PAGE and digitized video image analysis of silver stained gels identified a total of 155 proteins unique to trophozoites and a total of 72 proteins unique to cysts [24]. Lack of knowledge of the E. histolytica cyst proteome hampers our ability to develop cyst-targeted diagnostic tools and to investigate stage conversion in this parasite. In this study, we applied mass spectrometry based whole genome shotgun sequencing approaches to 5 purified cyst samples in order to identify proteins expressed in E. histolytica cysts in vivo. Here we present for the first time the identification of 417 proteins (representing ∼5.1% of all 8201 predicted proteins) in the purified cyst samples. Out of the 417 proteins identified, 195 have never been detected in the E. histolytica trophozoite specific proteome or EST databases. This is the first proteomic analysis of naturally-occurring E. histolytica cysts.
Methods
Ethics statement
The collection of clinical samples used in this study has been approved by the Ethical Review Committee (ERC) of ICDDR,B. Since the study participants were minors (aged 2–6 years), parents or legal guardians provided written informed consent on behalf of all child participants. All clinical investigation has been conducted according to the principles expressed in the Declaration of Helsinki.
Screening of E. histolytica cysts
Fecal specimens were collected from children enrolled in an ongoing field study of human immunity to amebiasis in an urban slum community at Mirpur, Dhaka, Bangladesh. They were screened for the presence of E. histolytica cysts in their stools. Initially, fresh stool samples were checked by microscopy for cysts of E. histolytica/E. dispar/E. moshkovskii. Positive stools were then subject to E. histolytica-specific antigen detection by E. histolytica-II ELISA (TechLab, Blackburg, VA) as described previously [25]. Since this ELISA was incapable of distinguishing between a mixed infection of E. histolytica/E. dispar and a single infection of E. histolytica, ELISA positive stool samples were further subject to DNA purification and diagnostic PCR for E. histolytica and E. dispar. PCR was also performed for E. moshkovskii-specific DNA. Cysts were only purified from stool samples that were PCR positive solely for E. histolytica.
Cyst purification
Cysts were purified from stool samples as described previously with some modifications [26]–[28]. In brief, about 4–5 g of stool was dissolved in PBS, filtered through two layers of gauges, centrifuged at 3220×g for 20 minutes, and the supernatant discarded. After washing 3 times with PBS at 3220×g for 20 minutes, the pellet was suspended in 5 mL of ethyl acetate (to separate the cyst and the fecal debris) and centrifuged at 3220×g for 20 minutes. The supernatant was discarded and the pellet was washed 3 more times with PBS (at 3220×g for 20 minutes), and the pellet was resuspended in 2 mL of PBS. This was then carefully layered using a Pasteur pipette on top of a previously prepared 10–80% Percoll gradient in 17×120 mm, 15 mL high-clarity polypropylene conical tubes by layering 2 mLs each of 80% Percoll (in PBS), followed by 50%, 40%, 30%, 20% and 10% Percoll solutions. This was centrifuged at 3220×g for 20 minutes. The content between 80% and 40% of the Percoll gradient was transferred to a new tube, washed 3 times with PBS and checked by microscopy for purified cysts.
Production of polyclonal antibodies against Jacob
The rabbit sera developed against one member of E. histolytica Jacob (EHI_044500), which shows 83.1% identity and 88.1% similarity with the E. dispar Jacob at amino acid level, were purchased from the Cocalico Biologicals, Inc. (project number 2010-0158). This protocol has been approved by the Animal Care and Use Committee of Cocalico Biologicals, Inc. who follows the USDA and NIH guidelines. The Office of the Laboratory of Animal Welfare, Division of Assurance, National Institute of Health has approved the Animal Welfare Assurance (#A3669-01) of Cocalico Biologicals, Inc.
Fluorescence microscopy with the purified cysts
About 50 µl of the E. histolytica cysts purified using the Percoll gradient centrifugation were fixed on a microscopic slide by adding 4% formaldehyde solution for 1 hour at 37°C. After washing with 1× PBS containing 0.1% Triton X-100, cysts were treated with either 1∶200 diluted post-immune or pre-immune antisera against E. histolytica cyst-wall specific Jacob protein raised in rabbits. They were then stained with 1∶200 diluted Fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit secondary antibody and examined by immunofluorescence microscopy.
Preparation of E. histolytica proteins from the purified cysts
Crude proteins from purified cyst samples were isolated in either of two ways. In the first protocol, cysts were subject to 5 freeze-and-thaw cycles to facilitate the breakage of cyst wall, followed by 20–25 minutes of sonication (30-second pulse followed by 30-second rest) in presence of protease inhibitor (Sigma-Aldrich). This procedure was followed for only one sample, AM951. In the second protocol, cysts were subject to 20–25 minutes of sonication (30-second pulse followed by 30-second rest) in presence of protease inhibitor (Sigma-Aldrich) eliminating the first freeze-and-thaw cycles as above. This procedure was followed for all 5 samples, including AM951. Protein concentration was measured using the Bio-Rad Protein Assay (Bio-Rad, USA) according to the manufacturer's instructions.
Mass spectrometry
All the cyst samples (after extraction of crude proteins) were divided into two parts (except for one, sample 4268, which had too little protein to divide into two parts) – supernatant (which should contain soluble proteins) and pellet (which should contain insoluble proteins).
The sample supernatant was removed, dried, and suspended in SDS-PAGE loading buffer. The remaining pellet was soaked in SDS-PAGE loading buffer. For sample 4268, the whole sample (containing both the supernatant and the beads) was dried, and suspended in SDS-PAGE loading buffer. They were loaded on a gel and run until the dye front was ∼1 cm into the gel. This 1 cm×1 cm area of gel was cut for processing (2 gel pieces for each sample, except sample 4268). For sample 4268, the entire sample (containing both the supernatant and the beads) was dried, suspended in SDS-PAGE loading buffer, and processed as above as single gel piece.
The gel piece for each sample was cubed and transferred to a siliconized tube and washed and destained in 200 µL 50% methanol for 4 h. The gel pieces were dehydrated in acetonitrile, rehydrated in 30 µL of 10 mM dithiolthreitol in 0.1 M ammonium bicarbonate and reduced at room temperature for 0.5 h. The DTT solution was removed and the sample alkylated in 30 µL 50 mM iodoacetamide in 0.1 M ammonium bicarbonate at room temperature for 0.5 h. The reagent was removed and the gel pieces dehydrated in 100 µL acetonitrile. The acetonitrile was removed and the gel pieces rehydrated in 100 µL 0.1 M ammonium bicarbonate. The pieces were dehydrated in 100 µL acetonitrile, the acetonitrile removed and the pieces completely dried by vacuum centrifugation. The gel pieces were rehydrated in 20 ng/µL trypsin in 50 mM ammonium bicarbonate on ice for 10 min. Any excess enzyme solution was removed and 20 µL 50 mM ammonium bicarbonate added. The sample was digested overnight at 37°C and the peptides formed extracted from the polyacrylamide in two 50 µL aliquots of 50% acetonitrile/5% formic acid. These extracts were combined and evaporated to 15 µL for MS analysis.
The LC-MS/MS system consisted of a Thermo Electron Orbitrap Velos ETD mass spectrometer system with a Protana nanospray ion source interfaced to a self-packed 8 cm×75 um id Phenomenex Jupiter 10 um C18 reversed-phase capillary column. Seven microliters of the extract was injected and the peptides eluted from the column by an acetonitrile/0.1 M acetic acid gradient at a flow rate of 0.5 µL/min over 1 hr. The nanospray ion source was operated at 2.5 kV. The digest was analyzed using the double play capability of the instrument acquiring a full scan mass spectrum (MS – Orbitrap 60K resolution) to determine peptide molecular weights and 20 product ion spectra (MS/MS – Ion trap) to determine amino acid sequence over the gradient elution.
Mass spectrometry data analysis
The mass spectrometry data were analyzed by database searching using the Sequest search algorithm (Bioworks 3.3.1) against E. histolytica (downloaded from NCBI Oct 2010), IPI Human (Ver 3.78), and NCBI NR (downloaded Jan 2011). The data were loaded in Scaffold v3 and minimal filters were set (Peptide score >60%, Protein score >90%, Xcorr >1.8 (+1), 2.2 (+2), 2.5(+3) and 3.5 (+4 or greater)). The p-values were determined using the two-tailed Fisher's exact test.
Results
Five E. histolytica cyst positive samples were selected for proteomic study
In order to identify E. histolytica cyst positive samples, we performed microscopical examination of stool specimens from children enrolled in an ongoing study on amebiasis. However, since the cyst of E. histolytica is morphologically indistinguishable from that of E. dispar and E. moshkovskii, microscopy-positive samples were then subject to species-specific E. histolytica II ELISA (TechLab, Blacksburg, VA). One limitation of E. histolytica II ELISA is that it cannot differentiate between a co-infection of E. histolytica with E. dispar or E. moshkovskii and a single infection of E. histolytica. Therefore, microscopy/ELISA positive samples were then subject to E. histolytica-, E. dispar- and E. moshkovskii-specific PCRs. Using these methods, we identified 5 stool specimens that were positive only for E. histolytica (data not shown). Microscopy revealed that 4 of these samples had only the cyst form of the parasite, while the 5th sample (4268) had both cyst and trophozoite forms in the original stool sample, although cysts were the predominant form. Five samples were derived from children aged 2–6 years, three of them were from males (4268, AM951, and AM797), and two of them were from females (8076 and CMS33-7132).
Verification of cyst structures after Percoll purification
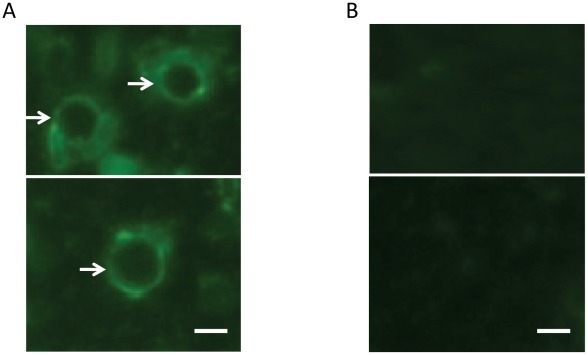
Following a positive microscopical identification, Percoll purification was used to significantly enrich the cyst form of the parasite (data not shown). In order to verify that the cyst-like structures in the cyst sample after Percoll purification were indeed E. histolytica cysts, we performed immunofluorescence assay using a polyclonal antibody developed in rabbits against a highly abundant cyst protein Jacob (EHI_044500). While the rabbit preimmune sera could not recognize cyst structures, the immune sera clearly stained the round-shaped cyst wall of E. histolytica in the Percoll-purified sample as expected (Figure 1).
Figure 1. Detection of Entamoeba histolytica cysts in Percoll purified cyst samples by immunofluorescence to the Jacob protein.
E. histolytica cysts were purified using the Percoll density gradient centrifugation [26]–[28], and about 50 µL of the purified cysts were fixed in microscopic slides by adding 4% formaldehyde solution for 1 hour at 37°C. After washing with 1× PBS containing 0.1% Triton X-100, cells were treated with either 1∶200 diluted post-immune or pre-immune antisera against E. histolytica cyst-wall specific Jacob protein raised in rabbits. They were then labeled with 1∶200 diluted FITC-conjugated goat anti-rabbit secondary antibody and examined by immunofluorescence microscopy. (a) Anti-Jacob antisera stained the cyst wall (shown by white arrows), (b) while pre-immune antisera failed to stain cyst wall. White bar is approximately 10 microns.
In order to investigate whether the Percoll gradient cyst purification protocol may have also co-purified the trophozoite form of the parasite, we added 0, 100, and 1000 E. histolytica trophozoites into 1 g each of three E. histolytica-negative stool samples (by microscopy, culture, ELISA and PCR), and they were subject to identical Percoll gradient cyst purification as performed with the five test samples. We then performed E. histolytica II ELISA using the contents from the 40–80% fractions of Percoll gradient to detect lectin antigen (which expresses most abundantly in the trophozoite form), and found that all three samples were negative for E. histolytica lectin (data not shown). This suggested that the Percoll gradient purification of stool specimens selectively enriched cysts but not trophozoites. Since only 1 out of 5 cyst samples (ID: 4268) had microscopically positive trophozoite in the initial stool examination, and the number of trophozoites in this sample was much smaller than that of cysts (data not shown), we conclude that our proteomic data was comprised predominantly (if not entirely) of proteins expressed in cysts.
In order to determine which method of lysate preparation worked best for mass spectrometry analysis, we applied two methods on 1 of the 5 cyst samples (ID: AM951). The first method was sonication only, while the second method employed freeze-thaw followed by sonication. Mass spectrometry results suggested that a simple sonication-only approach was able to identify more proteins than an alternative approach involving a prior freeze-thaw step. Samples processed by sonication yielded about 20% more identifiable E. histolytica proteins (total 101) than samples processed by the freeze-thaw-sonication approach, which yielded only 81 identifiable E. histolytica proteins. Fifty-two proteins overlapped between the two approaches (Tables S1 and S2). We investigated if there were any qualitative differences in proteins identified by the two different methods of lysate preparation. Two calcium binding or signal related proteins, and two cyst wall specific proteins were detected in both methods of lysate preparations (Table S2). In contrary, out of 3 DNA repair proteins 2 were identified in the sonication only method and the third one was identified by freeze-thaw-sonication method only. The only surface associated protein (EHI_065330) was isolated by the sonication only method. Proportionately more proteins with putative enzymatic function were identified by freeze-thaw-sonication (19/81 or 23.5%) compared to sonication only (18/101 or 17.8%). In contrast, slightly more actin-related proteins were identified in sonication only method (7/101 or 6.9%) compared to freeze-thaw-sonication method (3/81 or 3.7%). However, these differences were not statistically significant. So, we conclude that there was no particular trend that could be detected in differentiating protein categories identified by sonication only or freeze-thaw-sonication methods.
Mass spectrometry was carried out for 4 out of 5 cyst samples in such a way as to detect both soluble and insoluble proteins (for details, see the materials and methods section). For the 5th sample (4268), there was not enough cyst material to separate soluble and insoluble proteins and the two fractions were analyzed together. In most cases more proteins were identified in insoluble fraction than soluble fraction (59/48 for AM951, 99/42 for AM797, and 57/35 for CMS33-7132; Figure S1).
Identification of E. histolytica proteins in cyst samples
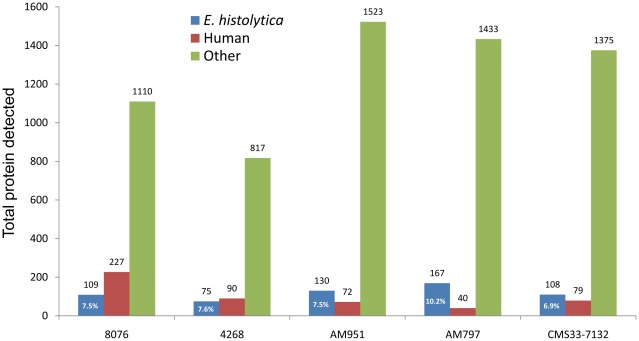
A total of 417 unique E. histolytica proteins were identified in the 5 cyst preparations. Human, bacterial, and rice-related proteins comprised about 90% of all proteins identified in the purified cyst samples (Figure 2). Human and rice-related proteins were abundant because of the source of cyst material (human feces) and nutritional habit of people in that region of the world (Dhaka, Bangladesh). Bacteria attached to the surfaces of cysts or inside the cysts might be the predominant source of bacterial proteins in the proteomic data. The proportions of human or rice related proteins may be reduced by using more stringent washing steps (from large amount of initial stool volume). However, cyst protein yields would be reduced and it would be very difficult, if not impossible, to get rid of bacteria that are attached to or ingested by the cysts. The 5 cyst samples came from 5 children aged 2–6 years, as a result, the amount of original stools that could be collected was very low, which was a limitation of this work. Nevertheless, E. histolytica proteins were identified in each of the 5 cyst samples as expected. The percentage of total proteins identified that were E. histolytica proteins in various cyst samples ranged from 6.9 to 10.2% (Figure 2).
Figure 2. Total number of E. histolytica, human and other proteins detected in 5 cyst samples.
In 5 cyst samples, the total number of proteins detected by MS/MS analysis ranged from 982 (in sample 4268) to 1725 (in sample AM951). The maximum number of E. histolytica proteins detected in a sample was 167 (sample AM797), while the least number of E. histolytica protein was 75 (sample 4268). The percentages of total proteins belong to E. histolytica are shown inside the bar graphs for each sample, which ranged from 6.9% (for sample CMS33-7133 to 10.2% (for sample AM797).
Comparison of proteomic data with other E. histolytica data
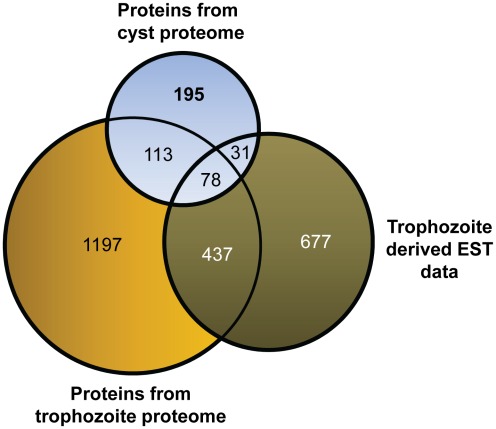
Since the total number of proteins identified in the naturally-occurring cyst samples was small, we asked how unique these proteins are in comparison with the publicly available protein datasets in AmoebaDB (http://amoebadb.org/amoeba/). Two LC-MS/MS proteomic datasets were available for the trophozoite stage of E. histolytica comprising a total of 1825 proteins (as of November 30, 2011) [29], [30]. There was also available an EST database for E. histolytica trophozoite comprising a total of 1223 proteins (as of October 1, 2011; from a total of 20,812 public entries at http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html). A three-way comparison of protein overlaps between the three datasets (i.e. cyst proteome, trophozoite proteome, and trophozoite EST) was done using the freely available software in the internet (http://www.cmbi.ru.nl/cdd/biovenn/index.php) [31]. The Venn diagram shows that a total of 191 proteins overlapped between the cyst proteomic and trophozoite proteomic (LC-MS/MS) datasets, 109 proteins overlapped between the cyst proteomic and trophozoite EST datasets, and 78 proteins overlapped between all three datasets (Figure 3). It was surprising to see that only 515 proteins overlapped between the trophozoite proteome (515/1825 or 28.2%) and the EST datasets (515/1223 or 42.1%). This disparity is probably because only limited numbers of trophozoite-derived proteome data are available for E. histolytica. The two proteome works available at the AmoebaDB public database when this manuscript was being written (November 2011) were highly specific in nature - one attempted to identify trophozoite proteins from phagosome [29], while the other attempted to identify Concavalin A-enriched glycoproteins from the E. histolytica trophozoite [30]. So, we think many of the trophozoite-specific proteins are not represented in them, resulted in showing poor overlap with the trophozoite-derive EST datasets from E. histolytica. Nevertheless, a unique set of 195 proteins in the cyst proteomic dataset did not overlap with the other two datasets. This set might include E. histolytica cyst-specific proteins.
Figure 3. Venn diagram for 3-way comparison between the proteins of this study, proteins from the trophozoite proteome and the EST data on trophozoite.
Overlaps between the E. histolytica cyst proteomic data (N = 417), trophozoite proteomic data (N = 1825), and the trophozoite specific EST data (N = 1223) were checked. A total of 191 proteins overlapped between the cyst proteomic and trophozoite proteomic datasets. A total of 109 proteins overlapped between the cyst proteomic and the trophozoite EST datasets. Seventy eight proteins overlapped between all three datasets. A total of 195 proteins of cyst proteomic study did not show any overlaps with other two datasets derived from trophozoites of E. histolytica, suggesting these are E. histolytica cyst-specific proteins.
The cyst proteomic data was also compared with the only available mRNA transcriptome data for encysting cultures grown in vitro [22]. The encysting transcriptome was performed on recently isolated E. histolytica strains that can spontaneously encyst in vitro when grown in complex diphasic Robinson's medium. Six hundred and seventy two genes were identified to be up-regulated in E. histolytica cysts (based on ≥3-fold expression change and p-value of <0.05). Out of 417 cyst proteins detected in this study, 23 had no mRNA expression data available. Forty-seven of the remaining 394 proteins identified here overlapped with the 672 cyst-specific genes identified by transcriptomics (p-value 0.0058) (Figure S2). We then asked whether the unique set of 195 proteins that did not overlap with the trophozoite proteomic and the EST datasets exhibited a stronger overlap with the 672 cyst-specific genes. Out of 195 unique cyst proteins, 10 had no mRNA expression data available. However, 28 of the remaining 185 proteins identified by cyst proteomic analysis overlapped with the 672 cyst-specific genes identified by transcriptomic analysis. As expected, this overlap between cyst-specific transcripts and unique cyst proteins was stronger (p-value 0.0014) than the overlap between cyst-specific transcripts and cyst proteins in general (p-value 0.0058) (Figure S3). Despite the fact that there was a statistically significant overlap between the cyst-specific transcripts [22] and cyst proteins of this study, we did notice a large discrepancy between the two datasets. Several factors could explain this discrepancy: (i) our cyst proteomic data was based on naturally occurring cyst samples, while the Ehrenkaufer and colleague's cyst transcriptome data was based on cyst-like cultures obtained in vitro; (ii) the number of proteins identified in the cyst proteome was very low, and they represent only ∼5.1% (417/8201) of all proteins; (iii) cyst-specific genes were categorized based on >3-fold higher mRNA levels compared to that of trophozoites, as a result, there will be some proteins that are expressed in lower (>3-fold or more) levels in cysts compared to trophozoites, but will still be regarded as cyst-specific by Ehrenkaufer et al; and (iv) some mRNA transcript levels may not correspond to protein levels due to post-transcriptional regulation or protein degradation.
Comparison of proteomic data with the Entamoeba invadens data
The cyst proteomic data was also compared with available data from developmental studies on the reptilian species E. invadens, whose encystation can be induced easily in vitro. Two E. histolytica chitinases, EHI_109890 and EHI_152170, show 79% and 47% identities with the E. invadens chitinases, EiChit1 and EiChit4, respectively [32]. In E. invadens, the mRNA expression of 4 chitinases was studied by Makioka and colleagues [32] during in vitro encystation and excystation. In the early phase of encystation, mRNA expression of all 4 chitinases increased although the greatest increase was seen for EiChit1 and EiChit4. However, following 5 hours of excystation, mRNA levels of these two chitinases dropped sharply compared to pre-induction stage, suggesting that these are highly cyst-specific. Both of these chitinase homologues (EHI_109890 and EHI_152170) were detected in our cyst samples as expected. Three actin depolymerizing factor (ADF) family proteins have been identified in E. invadens [33]. These proteins are thought to be important in actin cytoskeleton reorganization during development of E. invadens, and can be detected in both trophozoite and cyst stages of this parasite. The single member of ADF family protein in E. histolytica (actophorin/EHI_197480), which shows virtually no mRNA differences between trophozoite and cyst stages (Table S3; [22]) was identified in our cyst proteome, consistent with a similar role in the development of E. histolytica. An E. histolytica glycolytic enzyme enolase (EHI_130700) shows 85% identity with the E. invadens enolase (EIN_093390). This protein was present in the cytoplasmic vesicles as well as in the cyst wall of E. invadens as revealed by immunofluorescence microscopy [34]. However, this protein was present only in the cytoplasmic vesicles of E. histolytica trophozoites recovered from amebic liver lesions of experimental animals [34]. Additionally, enolase was detected in the mature cyst wall of E. histolytica in samples derived from human infections. Consistent with this finding, we could also detect enolase in 3 out of 5 cyst samples in our proteomic study. An earlier study has shown that heat shock treatment of E. invadens can result in strong induction of cyst-specific chitinase and Jacob mRNAs, and moderate induction of heat shock protein mRNAs, including a 70-kDa heat shock protein known as BiP (AF252299) [20]. Although heat shock alone cannot produce matured, chitin-walled cysts, these authors suggest that amebic heat shock proteins are involved in degradation of cytoskeletal proteins during encystation. In our cyst proteome study, 5 putative heat shock proteins were detected (Table S3), and one of these (EHI_199590) is a homoologue of E. invadens BiP (with 88% identity), suggesting a similar role of heat shock proteins in E. histolytica encystation.
However, other proteins that appeared to be important in E. invadens development were not detected in our study. For example, the E. histolytica homologue of one of the cysteine proteases EiCP-B9 [35], or profilins (such as EiPFN1 and EiPFN4; [36]) that are expressed in the cyst stage of E. invadens, were not detected in our proteome study. These discrepancies could be due to the fact that we were only able to identify a fraction of all cyst-specific proteins. Alternatively, they might reflect differences between parasite species and/or between cysts generated in hosts and in vitro.
Functional categories of proteins identified in the E. histolytica cyst preparations
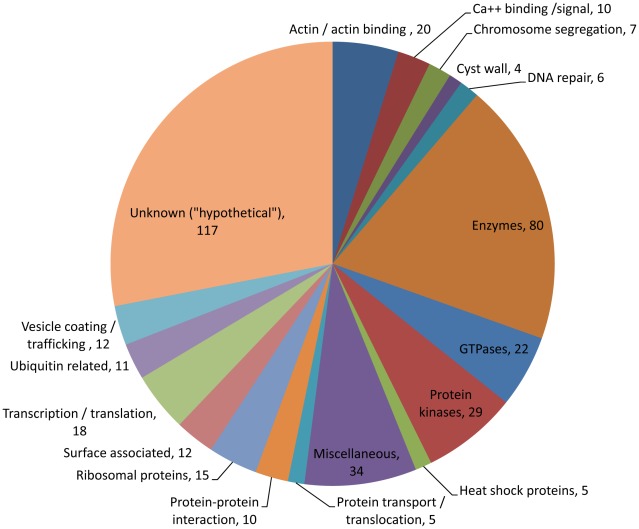
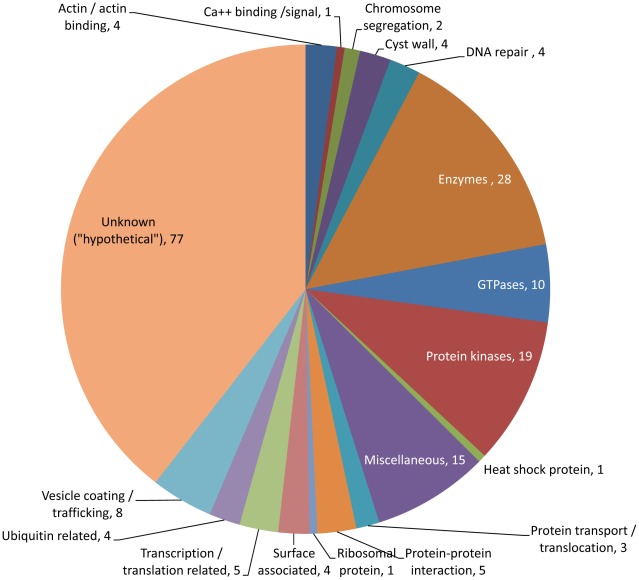
The 417 E. histolytica proteins (Table S3) fell into several broad functional groups. About 28% (117 proteins) did not have a known function and are referred to as ‘hypothetical proteins’ (Figure 4). About 12.9% (54 proteins) contained putative transmembrane (TM) domains (Table S3). Perhaps the most important were the categories of 195 unique proteins that showed no overlaps with the trophozoite specific proteome or EST datasets (Figure 5). About 39.5% of these proteins (77 proteins) did not have a known function.
Figure 4. Categories of all 417 proteins detected in 5 cyst samples.
E. histolytica proteins that were present in 1 or more cyst preparations fell into functional categories based on their annotations in the database. Out of 417 proteins, 117 did not have known function and are designated as hypothetical proteins. Four E. histolytica cyst-wall specific proteins (such as Jacob, two chitinases and chitin synthetase) were present in 1 or more cyst preparations. A total of 133 various enzymes (mostly protein kinases and GTPases) were also detected. Among other proteins, 18 transcription or translation related proteins (important for understanding of encystation biology), 12 surface associated proteins, 15 ribosomal proteins, 10 protein-protein interacting proteins, 12 vesicle coating or trafficking protein, 10 calcium signal related proteins, and 6 DNA repair proteins are notable.
Figure 5. Categories of 195 unique cyst specific proteins.
E. histolytica proteins that were present in 1 or more cyst preparations and those that were not found in trophozoite proteome or trophozoite EST datasets were categorized in various functional categories based on their annotations in the database. Out of a total 195 proteins, 77 did not have any known function, and they are designated as hypothetical proteins. Four E. histolytica cyst-wall specific proteins (such as Jacob, 2 chitinases and chitin synthetase) were present in 1 or more cyst preparations. A total of 60 various enzymes (including 19 protein kinases, 10 GTPases, 2 chitinase and a chitin synthetase) were also detected. Among other proteins, 8 vesicle coating or trafficking protein, 4 DNA repair proteins, 5 transcription or translation related proteins, 4 surface associated proteins, are notable.
The E. invadens cyst wall is composed of chitin (a homopolymer of ß-1,4-linked GlcNAc), and chitin-binding Jacob and Jessie lectins and chitinases [37]–[39]. The prior transcriptome data from encysting cultures in vitro supported that these proteins were components of the E. histolytica cyst [22]. In this study, among the 195 unique cyst proteins, we identified one member of the Jacob gene family (EHI_028930), two chitinases (EHI_109890 and EHI_152170), and one chitin synthetase 2 (EHI_044840) supporting prior suppositions as to the composition of the cyst wall [22], [39]. Besides cyst wall proteins, 4 putative surface associated proteins were identified in the 195 cyst proteins. Three of these belonged to the BspA-like leucine rich repeat (LRR) protein family. An important aspect of BspA family proteins is to mediate protein-protein interactions [40]. In E. histolytica, more than 70 proteins belong to BspA-like LRR protein family, although only 4 of these were previously identified as being cyst-specific based on mRNA transcriptome data [22]. One BspA family member (EAL42510/849.m00008) was previously reported to be primarily located on the plasma membrane of E. histolytica trophozoite [41]. Consistent with the trophozoite-specificity of this protein, we could not identify this protein in the present cyst proteome. Another BspA family protein has been implicated in trifluoromethionine resistance [42]. Immunofluorescence studies with the episomal overexpression of this protein suggest that it is expressed in the cytoplasm of the E. histolytica trophozoites. Again, this protein was not identified in our cyst proteome, consistent with trophozoite-specificity. The other potential surface protein identified was a putative member of lectin family protein (EHI_110830).
Actin is one of the most conserved and ubiquitous proteins in eukaryotes. During encystation, morphological changes occur in the trophozoite structure of E. histolytica, and elongated trophozoites become spherical cysts. We anticipate a reorganization of the actin cytoskeleton during this process. One study shows that jasplakinolide, an actin-polymerizing and filament-stabilizing drug, inhibits both growth and encystation of E. histolytica and E. invadens through perturbation of the actin cytoskeleton [43]. Four actin or actin-related proteins were identified in the cyst proteome that might be involved in these morphological and structural changes, and deserve further studies.
Transmembrane domain containing protein kinases (TMKs) are involved in signal transduction in higher eukaryotes. Over 100 TMKs have been identified in the E. histolytica genome [44]. Since these proteins have extracellular domains coupled to cytoplasmic kinase domains, they have the potential to sense environmental cues. Fourteen of these TMKs showed significantly differential mRNA expression in encystating E. histolytica cultures compared to that of trophozoites [22]. Additionally, a number of cytoplasmic protein kinases have also been identified to be cyst-specific [22]. The E. histolytica genome encodes for about 343 protein kinases (including >100 TMKs), and 19 of these were identified in the 195 unique cyst proteins. Protein kinases regulate cellular pathways, especially those involved in signal transduction. More than half of these protein kinases (11/19) were predicted to have transmembrane domains, suggesting their potential roles in sensing outside clues for stage conversion (Table S3) [44].
GTPases are a large family of GTP-binding hydrolase enzymes involved in various biological functions including signal transduction, protein biosynthesis, cell division, translocation of proteins through membranes and transport of vesicles within the cell. The GTP-binding proteins are classified into five families – Ras, Rho, Rab, Arf and Ran [45]. Recent data suggest that the E. histolytica genome codes for 17, 22, 73, 12 and 1 member(s) of Ras, Rho, Rab, Arf and Ran proteins, respectively [45]. Additionally there are 41 unclassified small G proteins in the E. histolytica genome. The Ras proteins are mainly involved in cell proliferation. The Rho proteins are implicated in cytoskeleton regulation. Both Rab and Arf proteins are involved in membrane trafficking in the cytosol. The Ran proteins are implicated in nuclear-cytosol transport. It is anticipated that a dramatic change of cellular components occurs in a rapid, yet highly regulated fashion during encystation, involving de novo synthesis of new proteins and degradation of unwanted proteins. In order to achieve this, the amoeba is expected to have a developmental–specific system of membrane trafficking. The transcriptomic changes of Rab genes have been studied during the encystation of E. invadens, and 23 cyst-specific, 36-trophozoite-specific and 31 constitutively expressed Rab genes were identified [46]. In the 195 unique cyst proteins, we identified 10 GTPases (including members of Rab or related proteins) that may have stage-specific functions in E. histolytica (Figure 5; Table S3).
Four DNA repair proteins, including two DNA double-strand break repair proteins (EHI_053200 and EHI_125910), one DNA mismatch repair protein (EHI_126120) and a type A flavoproteins (EHI_129890) were identified in the 195 cyst proteins. We also found 8 vesicle coating (or trafficking) related proteins including 3 putative clathrin proteins which are generally involved in shaping rounded vesicles in the cytoplasm for intracellular trafficking [47]. In addition, a SET domain containing histone lysine methyltransferase (EHI_0319060) that adds methylmarks on histone tails, and two chromodomain-containing proteins (EHI_000780 and EHI_031370) that bind methylmarks on histone tails and facilitate the recruitment of transcriptional effector molecules were also identified, suggesting a role for epigenetic machineries in the development of E. histolytica.
E. histolytica cyst proteins with diagnostic potential
Among various species of Entamoeba that can infect humans, only E. histolytica can cause intestinal and extraintestinal diseases in humans. At least two other human-infecting Entamoeba species, E. dispar and E. moshkovskii, are morphologically identical to E. histolytica. These commensal species create diagnostic challenges. Although E. dispar has never been documented to cause diseases, some emerging data suggest E. moshkovskii may cause diseases in humans [48], [49]. It is recommended that individuals with E. histolytica infection regardless of clinical status be treated due to the risk of the development of invasive amebiasis even in asymptomatic individuals. However, no simple E. histolytica cyst-specific diagnostic test is currently available to detect carriers. Here, we developed an immunofluorescence assay (IFA) using rabbit anti-Jacob antiserum that could detect E. histolytica cysts (Figure 1); however this polyclonal antibody might not be specific to E. histolytica. We conclude that proteins identified in this study are likely to be at least partially specific to E. histolytica, and therefore warrant further investigation as to their diagnostic potential.
In order to identify candidate diagnostic targets, we focused initially on the list of 195 unique proteins that were not detected in any trophozoite specific proteome or EST datasets. Twenty-five of these proteins showed 80% or less identity on the amino acid level compared to homologues in E. dispar (Table 1). These proteins displayed even lower identities with other proteins in the database including E. moshkovskii and human proteins. Over half (13/25) of the 25 proteins have no known functions and are annotated as hypothetical proteins. Five of these had previously shown 141- to 1012-fold higher mRNA transcript levels in cyst-like cultures compared to cultured trophozoite of E. histolytica [22]. Three out of 25 proteins in the list have no homologues in E. dispar including two that are putative reverse transcriptases (416.m00035 and 453.m00043) and one hypothetical protein (112.m00115).
Table 1. E. histolytica cyst proteins that show 80% or less identity with E. dispar proteins.
| E. histolytica Locus ID | *Detected in samples | Annotation | Transcriptome data** | E. dispar homolog | |||
| p-value | fold change: cyst/troph | HM-1 expr | Locus ID | % identity | |||
| 416.m00035 | 2 | reverse transcriptase, putative | NF | NF | NF | None | N/A |
| 453.m00043 | 2 | reverse transcriptase, putative | NF | NF | NF | None | N/A |
| 112.m00115 | 2, 4 | hypothetical protein | NF | NF | NF | None | N/A |
| EHI_117680 | 3 | protein kinase, putative | 0.1170 | 2.9 | 0.05 | EDI_160040 | 34 |
| EHI_087690 | 3 | hypothetical protein | 0.0282 | 140.8 | 0.05 | EDI_102140 | 44 |
| EHI_075310 | 1, 2 | hypothetical protein | 0.2720 | 382.5 | 0.04 | EDI_204760 | 47 |
| EHI_182030 | 2 | EhRabF2 | 0.1280 | −1.7 | 1.96 | EDI_154730 | 48 |
| EHI_133790 | 1, 2 | hypothetical protein | 0.0161 | 84.9 | 0.04 | EDI_063010 | 49 |
| EHI_066720 | 4 | MAEBL, putative | 0.3900 | −1.6 | 2.27 | EDI_281920 | 54 |
| EHI_019630 | 1, 4, 5 | hypothetical protein | 0.0001 | 515.8 | 0.08 | EDI_199570 | 54 |
| EHI_174540 | 5 | Tyrosyl-DNA phosphodiesterase, putative | 0.5250 | 1.6 | 0.04 | EDI_259130 | 59 |
| EHI_113200 | 1 | hypothetical protein | 0.3480 | 2.5 | 0.04 | EDI_078590 | 59 |
| EHI_047820 | 2 | BspA-like leucine rich repeat protein, putative | 0.2670 | 1.8 | 0.05 | EDI_238280 | 63 |
| EHI_104230 | 1 | hypothetical protein | 0.0001 | 1012.0 | 0.05 | EDI_083080 | 64 |
| EHI_146120 | 1, 2, 4 | hypothetical protein | 0.0001 | 59.1 | 0.04 | EDI_105270 | 65 |
| EHI_133780 | 1 | hypothetical protein | 0.0001 | 1012.0 | 0.05 | EDI_083080 | 66 |
| EHI_158620 | 1 | Opioid growth factor receptor (OGFr) conserved region | 0.3260 | 1.7 | 0.26 | EDI_036800 | 69 |
| EHI_059040 | 3 | protein kinase domain containing protein | NF | NF | NF | EDI_142140 | 70 |
| EHI_126120 | 1, 2 | DNA mismatch repair protein mutL | 0.4520 | 1.7 | 0.06 | EDI_012590 | 72 |
| EHI_034210 | 2 | Protein kinase, putative | 0.0450 | −1.6 | 0.05 | EDI_142140 | 72 |
| EHI_174190 | 1, 2 | hypothetical protein | 0.1110 | −3.4 | 0.54 | EDI_018190 | 72 |
| EHI_175920 | 4 | protein kinase, putative | NF | NF | NF | EDI_280160 | 74 |
| EHI_092210 | 3 | hypothetical protein | 0.1160 | 2.9 | 0.04 | EDI_154030 | 75 |
| EHI_163520 | 4 | hypothetical protein 256.t00008 | 0.0563 | −2.5 | 1.60 | EDI_222270 | 80 |
| EHI_138010 | 1 | hypothetical protein | 0.3790 | 1.9 | 0.20 | EDI_134120 | 80 |
*: Detected in samples: 1 = 8076; 2 = 4268; 3 = AM951; 4 = AM797; and 5 = CMS33-7132;
**: From [22], 2007; NF = not found; troph = trophozoite; expr = expression; N/A = Not applicable. NB. During the revision of this manuscript, two new trophozoite-specific proteome datasets became available at the AmoebaDB on January 25, 2012. However, none of the proteins listed in Table 1 could be detected in any of the 4 trophozoite-specific proteome datasets available at the AmoebaDB or EST datasets (as of February, 2012).
For diagnostic purposes, proteins that were identified in a majority of the cyst samples are hypothesized to be consistently expressed in excreted cysts at detectable levels, and therefore potentially interesting as candidate diagnostic targets. Eight proteins were identified (from the list of 195 unique proteins) in 3 or more (out of 5) cyst samples (Table 2). Four of the 8 proteins displayed 90% or less amino acid level homologies with proteins from E. dispar. Two of these were putative cyst-wall specific proteins: chitinase (EHI_109890, which showed 87-fold higher mRNA transcript in encysting culture) and chitinase Jessie 3 (EHI_152170, which showed 44-fold higher mRNA transcript in encysting culture).
Table 2. E. histolytica cyst proteins detected in 3 or more cyst samples.
| E. histolytica Locus ID | *Detected in samples | Annotation | Transcriptome data** | E. dispar homolog | |||
| p-value | fold change: cyst/troph | HM-1 expr | Locus ID | % identity | |||
| EHI_012500 | 3, 4, 5 | coatomer gamma subunit, putative | 0.0881 | −2.7 | 1.90 | EDI_129270 | 92 |
| EHI_038630 | 2, 3, 4, 5 | TBC domain containing protein, putative | 0.3190 | 1.7 | 0.06 | EDI_259490 | 91 |
| EHI_019630 | 1, 4, 5 | hypothetical protein | 0.0001 | 515.8 | 0.08 | EDI_199570 | 54 |
| 200.m00090 | 1, 4, 5 | clathrin heavy chain, putative | 0.7530 | −1.0 | 32.75 | EDI_237980 | 92 |
| EHI_109890 | 1, 3, 4, 5 | chitinase, putative | 0.0001 | 87.4 | 0.20 | EDI_120190 | 82 |
| EHI_152170 | 1, 3, 4, 5 | chitinase Jessie 3, putative | 0.0001 | 43.5 | 0.67 | EDI_038370 | 90 |
| EHI_146120 | 1, 2, 4 | hypothetical protein | 0.0001 | 59.1 | 0.04 | EDI_105270 | 65 |
| EHI_196570 | 1, 2, 3, 4, 5 | hypothetical protein | 0.0001 | 37.2 | 0.55 | EDI_039770 | 93 |
*: Detected in samples: 1 = 8076; 2 = 4268; 3 = AM951; 4 = AM797; and 5 = CMS33-7132;
**: From [22]; NF = not found; troph = trophozoite; expr = expression; N/A = Not applicable; shown in bold fonts are those that show 90% or less identity with E. dispar proteins; ideal candidates for E. histolytica specific diagnostic tool development. NB. None of the proteins listed in Table 2 could be detected in any of the 4 trophozoite-specific proteome datasets available at the AmoebaDB or EST datasets (as of February, 2012).
Proteins detected in our study have variable number of spectrum counts as detected by the LC-MS/MS experiments. For diagnostic purposes, proteins with greater numbers of spectrum counts may be better candidates for diagnostic assay development, as they may be more stable and more abundant in samples. Seven proteins were identified (from the list of 195 unique proteins) with more than 10 spectrum counts (Table 3). Four of the 7 proteins displayed 90% or less amino acid level identities with proteins from E. dispar. Two of these are chitinase (EHI_109890) and chitinase Jessie 3 (EHI_152170) with 52 and 24 spectrums detected, respectively, while the remaining two are hypothetical proteins (EHI_146120, 36 spectrums; and EHI_019630, 15 spectrums). Overall, Tables 1, 2, and 3 describe a total of 32 unique (non-redundant) proteins with diagnostic potential.
Table 3. E. histolytica cyst proteins with more than 10 spectrum counts detected in MS/MS experiments.
| E. histolytica Locus ID | *Detected in samples | Annotation | Transcriptome data** | E. dispar homolog | No. of spectrum counts in LC-MS/MS | |||
| p-value | fold change: cyst/troph | HM-1 expr | Locus ID | % identity | Total | |||
| 10.m00319 | 4, 5 | actin | 0.2720 | 1.1 | 127.75 | EDI_251490 | 100 | 193 |
| EHI_109890 | 1, 3, 4, 5 | chitinase, putative | 0.0001 | 87.4 | 0.20 | EDI_120190 | 82 | 52 |
| EHI_146120 | 1, 2, 4 | hypothetical protein | 0.0001 | 59.1 | 0.04 | EDI_105270 | 65 | 36 |
| EHI_196570 | 1, 2, 3, 4, 5 | hypothetical protein | 0.0001 | 37.2 | 0.55 | EDI_039770 | 93 | 36 |
| EHI_152170 | 1, 3, 4, 5 | chitinase Jessie 3, putative | 0.0001 | 43.5 | 0.67 | EDI_038370 | 90 | 24 |
| EHI_019630 | 1, 4, 5 | hypothetical protein | 0.0001 | 515.8 | 0.08 | EDI_199570 | 54 | 15 |
| 200.m00090 | 1, 4, 5 | clathrin heavy chain, putative | 0.7530 | −1.0 | 32.75 | EDI_237980 | 92 | 11 |
*: Detected in samples: 1 = 8076; 2 = 4268; 3 = AM951; 4 = AM797; and 5 = CMS33-7132;
**: From [22]; NF = not found; troph = trophozoite; expr = expression; N/A = Not applicable; shown in bold fonts are those that show 90% or less identity with E. dispar proteins; ideal candidates for E. histolytica specific diagnostic tool development.
NB. Only a single spectrum was detected for a high proportion of 195 cyst proteins (137 or 70%) compared to that detected in 914/1825 (or 50%) of trophozoite MS/MS proteins deposited into the AmoebaDB database (http://amoebadb.org/amoeba/). In fact, other proteins other than E. histolytica in the cyst samples hampered an efficient identification of cyst specific proteins. The sources of these other proteins include (1) numerous bacteria that were attached to the surface of the cyst, or ingested by the cyst, and (2) the human feces, which was the original source of cyst material contributed the human proteins and diet-related (mainly rice) proteins. In contrast, the trophozoite MS/MS data in the AmoebaDB was coming from two studies that used axenically grown E. histolytica trophozoites, which should be free from all other organisms including bacteria [29], [30]. Also, note that none of the proteins listed in Table 3 could be detected in any of the 4 trophozoite-specific proteome datasets available at the AmoebaDB or EST datasets (as of February, 2012).
In addition to candidate proteins for cyst specific diagnosis, we also identified proteins in the remaining 222 proteins out of the 417 (that were also present in the trophozoite specific proteome and EST datasets). These may also be useful to develop diagnostic tools to detect both cysts and trophozoites of E. histolytica. These proteins are listed in Tables S4, S5, S6.
Host (human) proteins and bacterial species identified in at least 3 out of 5 cyst samples
Thirty-one human proteins were identified in at least 3 out of 5 cyst samples including 7 that were identified across all 5 samples, and 10 that were identified in 4 samples (Table S7). Multiple lectin- or sugar-binding proteins such as galectin-3 (IPI00023673), galectin-4 (IPI00009750), intelectin-1 (IPI00291737), intelectin-2 (IPI00103436), glycoprotein 2 (IPI01014468), and proteoglycan 3 (IPI00005778) were detected. The possible interaction of some of these proteins with the cyst or trophozoite surface and their potential roles in stage-conversion may warrant further investigation. Additional proteins detected in the cyst samples derived from bacterial origins (Table S8).
Annotation of the genome of E. histolytica
The E. histolytica gene annotation is limited at present, although it is improving through the combined efforts of biostatisticians and researchers [50]. More than half of the genes (53.8%; 4413 genes out of a total predicted 8201 genes) are designated as “hypothetical” proteins of unknown function [50]. This is mainly because the E. histolytica genes are highly diverse in sequence compared to other organisms, which makes prediction of gene function difficult. One hundred and seventeen (or 28%) of 417 proteins identified in this study were predicted “hypothetical” proteins including 40 that were previously identified in trophozoite derived proteome or EST datasets. For the remaining 77 cyst specific “hypothetical” genes we now have evidence of protein level expression. We also found that 9 out of 417 protein genes identified in this study were missing in the most recent genome annotation (http://amoebadb.org/amoeba/). Three of these are putative clathrin heavy chain containing proteins (gi|103484580, found in sample AM951; 200.m00090, found in samples 8076, AM797, CMS33-7132; and 141.m00078, found in sample AM797) that are involved in vesicle trafficking in other organisms. Additional non-annotated genes encode 2 putative reverse transcriptases (416.m00035 and 453.m00043, both found in sample 4268), a putative Sec61 alpha subunit (gi|52352493, found in sample AM951), an actin (10.m00319, found in samples AM797 and CMS33-7132), and two hypothetical proteins (112.m00115, found in samples 4268 and AM797; and 270.m00054, found in sample CMS33-7132). We now have evidence that these genes are expressed as proteins in E. histolytica cysts.
Discussion
Previous studies characterized gene expression in E. histolytica and E. invadens cells that have encysted under laboratory conditions. The goal of the present study was to obtain the proteomic profiles of E. histolytica cells that have encysted under natural conditions. To achieve this we employed a highly sensitive, whole genome shotgun sequencing approach using mass spectrometric methods. We successfully identified 417 proteins representing 5.1% of all predicted proteins in E. histolytica. Of these 417 proteins 191 overlapped with the proteins previously identified in trophozoite proteome. Similarly, 109 proteins overlapped with the previously identified trophozoite specific EST datasets (Figure 3). The remaining195 proteins have not been seen in trophozoite proteomes and are likely to be specific to cyst stage of the parasite.
Analysis of each of the cyst preparations identified 195 unique proteins, many of which warrant further investigation as potential diagnostic targets. The 417 identified proteins comprise approximately 5.1% of all predicted proteins in E. histolytica, suggesting that the proteomic data constitutes a partial representation of all proteins present in cysts. However, we cannot know the true percentage of representation since the total number of proteins upregulated during encystation is not known. Part of the difficulty in identifying cyst-specific proteins lies in part to the low proportion of E. histolytica proteins in the Percoll-purified cyst samples, with ≥90% of the proteins identified of human, rice, bacteria or other origin. An improved method of separation of E. histolytica cysts from other materials present in stool samples should be considered in future proteomic work. Although this study may not represent a comprehensive proteome of E. histolytica cysts, it succeeded in identifying some of the most abundant proteins in the naturally occurring cysts. These proteins will likely be useful targets for developing improved diagnostic tests for E. histolytica infection, as demonstrated by the ability to identify cysts in fecal specimens using antibodies directed to the cyst-specific Jacob protein.
There were several sample-based and technical limitations in this work. First, 5 asymptomatically infected 2–6 year-old children provided all the E. histolytica cyst positive stool samples. As a result, the amount of stool was small in each case. Efforts to collect additional samples from these patients were not successful as the number of cysts usually decreased in successive stools. Second, due to the nature of source material (human stool), unwanted host proteins, diet related (e.g., rice) proteins, and bacterial proteins co-purified with cysts despite our efforts to reduce these by using vigorous washing steps. Bacterial proteins may have been derived from bacteria attached to the surface of cysts; these would be extremely difficult (if not impossible) to exclude. Overall, the presence of contaminating proteins compromised the identification of E. histolytica cyst proteins. An alternate approach to Percoll gradient purification (such as CsCl gradient) should be considered in future cyst purification. However, despite these technical limitations, 195 novel E. histolytica proteins were still identified marking a significant advancement in our knowledge of the cyst proteome.
The E. histolytica cyst is a biodefense threat to water and food supplies due to its resistance to chlorination and low infectious dose (<10 cysts). It is also a public health threat, especially as a cause of diarrhea in children in Africa, Asia, and Latin America. However, the presence of morphologically indistinguishable cysts of non-pathogenic species E. dispar and E. moshkovskii severely complicates current microscopy based diagnosis which is neither sensitive nor specific. E. histolytica infections lead to intestinal and extra-intestinal amebiasis. Each year 40 to 50 million cases of colitis and liver abscess due to the pathogenic E. histolytica occur, causing up to 100,000 deaths. So, the development of sensitive molecular based techniques to detect E. histolytica cyst is a priority. In this study, we have been successful in identifying several target proteins that may lead to develop improved diagnostic tools to detect cysts in asymptomatic cyst passers (Tables 1, 2, 3). It is worth mentioning here that two new trophozoite-derived proteome works became available at the AmoebaDB public database (on January 25, 2012) while this manuscript was under revision [51], [52]. However, none of the 32 (non-redundant) cyst-specific candidate proteins listed in Tables 1, 2, and 3 could be detected in the new trophozoite-derived proteomic datasets. So, we are confident that some of these proteins would be highly useful as cyst-specific diagnostic targets. In addition, some of the proteins identified in this study will help to develop diagnostic tools capable of detecting both cyst and trophozoite forms of the parasite (Tables S4, S5, S6). A recent study shows that sera of amebic liver abscess patients can recognize a 110 kDa protein (annotated as pyruvate phosphate dikinase, EHI_009530) [53]. This protein was identified in all 5 cyst samples in our study (see Table 2 or Table S3), supporting the notion that proteins identified in this study have promise as candidate diagnostic targets.
Our knowledge about the cyst stage of E. histolytica is very limited due in part to our inability to induce encystation in vitro. We do not know if the involvement of host proteins is a necessary factor. It appears likely that bacterial involvement is essential for encystation to take place as encystation cannot be induced in axenic (bacteria-free) culture. However, we do not know the mechanism of bacterial association in encystation. For example, we do not know if certain bacterial species are essential or if certain bacterial proteins can efficiently induce encystation. Carefully designed functional studies using some of the human and bacterial proteins detected in this study (Tables S7, S8) may provide clues as to their involvement in encystation of E. histolytica.
This study provides evidence of protein level expression of several genes that are missing in the current gene annotation. Likewise, this study also provides evidence of expression at the protein level for over a hundred genes that were annotated as “hypothetical” in the current gene annotation. Therefore these data will help improve the present gene annotation. These data will be made publicly available through the AmoebaDB.
The data generated may aid the understanding of biochemistry and physiology of cysts and the developmental switch between the trophozoite and cyst stage in E. histolytica, a process that is vital for disease transmission and pathogenesis in this parasite. Among the cyst specific 195 proteins, it is intriguing to find numerous transmembrane domain containing protein kinases, vesicle coating or trafficking related proteins, DNA repair proteins, and GTPase-based signal molecules in the present study. Further functional work on these cyst proteins may provide insight into the signaling pathways that trigger encystation, as well as mechanisms of stage conversion, which will aid in developing better therapeutic agents and preventive measures to control amebiasis.
List of genes/proteins discussed in this paper
E. histolytica : EHI_044500, EHI_065330, EHI_109890, EHI_152170, EHI_197480, EHI_130700, EHI_028930, EHI_044840, EAL42510, EHI_110830, EHI_199590, EHI_053200, EHI_125910, EHI_126120, EHI_129890, 416.m00035, 453.m00043, 112.m00115, EHI_146120, EHI_019630, gi|103484580, 200.m00090, 141.m00078, gi|52352493, 10.m00319, 270.m00054, EHI_0319060, EHI_000780 and EHI_031370. E. invadens : EiChit1, EiChit4, AF252299/BiP, EIN_093390, EiCP-B9, EiPFN1, and EiPFN4. Human: galectin-3/IPI00023673, galectine-4/IPI00009750, intelectin-1/IPI00291737, intelectin-2/IPI00103436, glycoprotein 2/IPI01014468, and proteoglycan 3/IPI00005778.
Supporting Information
Total number of E. histolytica proteins detected in soluble, insoluble or both fractions of mass spectrometry experiments for 4 cyst samples. The mass spectrometry was carried out for 4 out of 5 cyst samples in such a way that it could detect both soluble and insoluble proteins (for details, see the Methods section). For the 5th sample (4268), there was not enough cyst material to proceed by this method, and proteins identified in this sample represented both soluble and insoluble proteins. Except for the sample 8076 (which has a protein distribution such as 48 in soluble fraction, 43 in insoluble fraction, and 18 in both fractions), there was a general trend that relatively more proteins were identified in the insoluble fraction compared with the soluble fraction (59/48 for AM951, 99/42 for AM797, and 57/35 for CMS33-7132).
(TIF)
Overlap between the 417 proteins (from this study) and the cyst-specific mRNA transcripts (from [22] ). The overlap between 672 cyst-specific transcripts (p-value<0.05 and fold-change ≥3) and 394 proteins out of all 417 proteins from cyst proteomic study (except for the remaining 23 proteins, that were not found in microarray data) was tested using the Venn diagram. The overlap between the cyst protein data and the cyst-specific mRNA transcript data was statistically significant (p-value 0.0058). The p-value was determined using the two-tailed Fisher's exact test using the GraphPad software freely available in the internet at http://www.graphpad.com/quickcalcs/contingency1.cfm.
(TIF)
Overlap between the 195 cyst –specific proteins (from this study) and the cyst-specific mRNA transcripts (from [22] ). The overlap between 672 cyst-specific transcripts (p-value<0.05 and fold-change ≥3) and the 185 proteins out of 195 proteins from the cyst proteomic study that were not identified in trophozoite-specific proteome or EST datasets (except for the remaining 10 proteins, that were not found in the microarray data) was tested using the Venn diagram. The overlap was statistically significant (p-value 0.0014). The p-value for the overlap of this comparison is better than the previous comparison shown in Figure S2 (0.0014 versus 0.0058, respectively) as expected. The p-values were determined using the two-tailed Fisher's exact test using the GraphPad software freely available in the internet at http://www.graphpad.com/quickcalcs/contingency1.cfm.
(TIF)
Proteins detected in sample AM951 using two different processing methods prior to LC-MS/MS experiments.
(XLSX)
Categories of proteins identified in sonication only and freeze-thaw-sonication, or both approaches for sample AM951.
(XLSX)
All the E. histolytica proteins detected in 5 E. histolytica cyst samples (N = 417).
(XLSX)
E. histolytica proteins detected in cyst samples that show 80% or less identity with E. dispar proteins. These are candidate proteins for development of diagnostic tool for both trophozoites and cysts of E. histolytica.
(XLSX)
E. histolytica proteins detected in 3 or more of 5 cyst samples. These are candidate proteins for development of diagnostic tool for both trophozoites and cysts of E. histolytica.
(XLSX)
E. histolytica proteins with maximum number of spectrum counts detected in cyst samples by LC-MS/MS experiments. These are candidate proteins for development of diagnostic tool for both trophozoites and cysts of E. histolytica.
(XLSX)
Host (human) proteins detected in cyst samples. Human proteins that were identified in at least 3 out of 5 E. histolytica cyst samples are shown in this Table.
(XLSX)
Bacterial species detected in 3 or more E. histolytica cyst samples. Many bacterial peptides were identified in the E. histolytica cyst samples by LC-MS/MS experiments. From the database search using the peptide sequences, the bacterial species were identified. At least 19 bacterial species were detected in 3 or more E. histolytica cyst samples.
(XLSX)
Acknowledgments
We thank all the children and/or their legal guardians for providing the samples. We also thank Maegan Ashworth Dirac for her assistance with human subjects' aspects of the study.
Footnotes
The authors have declared that no competing interests exist.
This research was supported by National Institutes of Health (NIH) grants 5R01AI043596 (to WAP) and U01 AI82186 (to GAC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Haque R, Huston CD, Hughes M, Houpt E, Petri WA., Jr Amebiasis. N Engl J Med. 2003;348:1565–1573. doi: 10.1056/NEJMra022710. [DOI] [PubMed] [Google Scholar]
- 2.Stanley SL., Jr Amoebiasis. Lancet. 2003;361:1025–1034. doi: 10.1016/S0140-6736(03)12830-9. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous WHO/PAHO/UNESCO report. A consultation with experts on amoebiasis. Mexico City, Mexico 28–29 January, 1997. Epidemiol Bull. 1997;18:13–14. [PubMed] [Google Scholar]
- 4.Caballero-Salcedo A, Viveros-Rogel M, Salvatierra B, Tapia-Conyer R, Sepulveda-Amor J, et al. Seroepidemiology of amebiasis in Mexico. Am J Trop Med Hyg. 1994;50:412–419. doi: 10.4269/ajtmh.1994.50.412. [DOI] [PubMed] [Google Scholar]
- 5.Braga LL, Gomes ML, Da Silva MW, Facanha FE, Jr, Fiuza L, et al. Household epidemiology of Entamoeba histolytica infection in an urban community in northeastern Brazil. Am J Trop Med Hyg. 2001;65:268–271. doi: 10.4269/ajtmh.2001.65.268. [DOI] [PubMed] [Google Scholar]
- 6.Haque R, Mondal D, Karim A, Molla IH, Rahim A, et al. Prospective case-control study of the association between common enteric protozoal parasites and diarrhea in Bangladesh. Clin Infect Dis. 2009;48:1191–1197. doi: 10.1086/597580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petri WA, Jr, Haque R, Lyerly D, Vines RR. Estimating the impact of amebiasis on health. Parasitol Today. 2000;16:320–321. doi: 10.1016/s0169-4758(00)01730-0. [DOI] [PubMed] [Google Scholar]
- 8.Tanyuksel M, Petri WA., Jr Laboratory diagnosis of amebiasis. Clin Microbiol Rev. 2003;16:713–729. doi: 10.1128/CMR.16.4.713-729.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McConnachie EW. The morphology, formation and development of cysts of Entamoeba. Parasitology. 1969;59:41–53. doi: 10.1017/s003118200006981x. [DOI] [PubMed] [Google Scholar]
- 10.Rengpien S, Bailey GB. Differentiation of Entamoeba: a new medium and optimal conditions for axenic encystation of E. invadens. J Parasitol. 1975;61:24–30. [PubMed] [Google Scholar]
- 11.Thepsuparungsikul V, Seng L, Bailey GB. Differentiation of Entamoeba: encystation of E. invadens in monoxenic and axenic cultures. J Parasitol. 1971;57:1288–1292. [PubMed] [Google Scholar]
- 12.Vazquezdelara-Cisneros LG, Arroyo-Begovich A. Induction of encystation of Entamoeba invadens by removal of glucose from the culture medium. J Parasitol. 1984;70:629–633. [PubMed] [Google Scholar]
- 13.Bailey GB, Rengypian S. Osmotic stress as a factor controlling encystation of Entamoeba invadens. Arch Invest Med (Mex) 1980;11:11–16. [PubMed] [Google Scholar]
- 14.Avron B, Stolarsky T, Chayen A, Mirelman D. Encystation of Entamoeba invadens IP-1 is induced by lowering the osmotic pressure and depletion of nutrients from the medium. J Protozool. 1986;33:522–525. doi: 10.1111/j.1550-7408.1986.tb05655.x. [DOI] [PubMed] [Google Scholar]
- 15.Eichinger D. Encystation in parasitic protozoa. Curr Opin Microbiol. 2001;4:421–426. doi: 10.1016/s1369-5274(00)00229-0. [DOI] [PubMed] [Google Scholar]
- 16.Das S, Gillin FD. Chitin synthase in encysting Entamoeba invadens. Biochem J. 1991;280(Pt 3):641–647. doi: 10.1042/bj2800641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villagomez-Castro JC, Calvo-Mendez C, Lopez-Romero E. Chitinase activity in encysting Entamoeba invadens and its inhibition by allosamidin. Mol Biochem Parasitol. 1992;52:53–62. doi: 10.1016/0166-6851(92)90035-i. [DOI] [PubMed] [Google Scholar]
- 18.Das S, Van Dellen K, Bulik D, Magnelli P, Cui J, et al. The cyst wall of Entamoeba invadens contains chitosan (deacetylated chitin). Mol Biochem Parasitol. 2006;148:86–92. doi: 10.1016/j.molbiopara.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 19.Van Dellen K, Ghosh SK, Robbins PW, Loftus B, Samuelson J. Entamoeba histolytica lectins contain unique 6-Cys or 8-Cys chitin-binding domains. Infect Immun. 2002;70:3259–3263. doi: 10.1128/IAI.70.6.3259-3263.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Field J, Van Dellen K, Ghosh SK, Samuelson J. Responses of Entamoeba invadens to heat shock and encystation are related. J Eukaryot Microbiol. 2000;47:511–514. doi: 10.1111/j.1550-7408.2000.tb00083.x. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez J, Bai G, Frevert U, Corey EJ, Eichinger D. Proteasome-dependent cyst formation and stage-specific ubiquitin mRNA accumulation in Entamoeba invadens. Eur J Biochem. 1999;264:897–904. doi: 10.1046/j.1432-1327.1999.00682.x. [DOI] [PubMed] [Google Scholar]
- 22.Ehrenkaufer GM, Haque R, Hackney JA, Eichinger DJ, Singh U. Identification of developmentally regulated genes in Entamoeba histolytica: insights into mechanisms of stage conversion in a protozoan parasite. Cell Microbiol. 2007;9:1426–1444. doi: 10.1111/j.1462-5822.2006.00882.x. [DOI] [PubMed] [Google Scholar]
- 23.Ehrenkaufer GM, Hackney JA, Singh U. A developmentally regulated Myb domain protein regulates expression of a subset of stage-specific genes in Entamoeba histolytica. Cell Microbiol. 2009;11:898–910. doi: 10.1111/j.1462-5822.2009.01300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bond VC, Beanan MJ, Bailey GB. High resolution two-dimensional protein gel evidence of differential gene expression during Entamoeba encystation. Arch Med Res. 1992;23:11–13. [PubMed] [Google Scholar]
- 25.Haque R, Faruque AS, Hahn P, Lyerly DM, Petri WA., Jr Entamoeba histolytica and Entamoeba dispar infection in children in Bangladesh. J Infect Dis. 1997;175:734–736. doi: 10.1093/infdis/175.3.734. [DOI] [PubMed] [Google Scholar]
- 26.Avron B, Bracha R, Deutsch MR, Mirelman D. Entamoeba invadens and E. histolytica: separation and purification of precysts and cysts by centrifugation on discontinuous density gradients of Percoll. Exp Parasitol. 1983;55:265–269. doi: 10.1016/0014-4894(83)90022-x. [DOI] [PubMed] [Google Scholar]
- 27.Segovia-Gamboa NC, Chavez-Munguia B, Medina-Flores Y, Cazares-Raga FE, Hernandez-Ramirez VI, et al. Entamoeba invadens, encystation process and enolase. Exp Parasitol. 2010;125:63–69. doi: 10.1016/j.exppara.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 28.Weber R, Bryan RT, Juranek DD. Improved stool concentration procedure for detection of Cryptosporidium oocysts in fecal specimens. J Clin Microbiol. 1992;30:2869–2873. doi: 10.1128/jcm.30.11.2869-2873.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boettner DR, Huston CD, Linford AS, Buss SN, Houpt E, et al. Entamoeba histolytica phagocytosis of human erythrocytes involves PATMK, a member of the transmembrane kinase family. PLoS Pathog. 2008;4:e8. doi: 10.1371/journal.ppat.0040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carpentieri A, Ratner DM, Ghosh SK, Banerjee S, Bushkin GG, et al. The antiretroviral lectin cyanovirin-N targets well-known and novel targets on the surface of Entamoeba histolytica trophozoites. Eukaryot Cell. 2010;9:1661–1668. doi: 10.1128/EC.00166-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makioka A, Kumagai M, Hiranuka K, Kobayashi S, Takeuchi T. Different structure and mRNA expression of Entamoeba invadens chitinases in the encystation and excystation. Parasitol Res. 2011;109:417–423. doi: 10.1007/s00436-011-2270-2. [DOI] [PubMed] [Google Scholar]
- 33.Makioka A, Kumagai M, Hiranuka K, Kobayashi S, Takeuchi T. Entamoeba invadens: identification of ADF/cofilin and their expression analysis in relation to encystation and excystation. Exp Parasitol. 2011;127:195–201. doi: 10.1016/j.exppara.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 34.Segovia-Gamboa NC, Talamas-Rohana P, Angel-Martinez A, Cazares-Raga FE, Gonzalez-Robles A, et al. Differentiation of Entamoeba histolytica: a possible role for enolase. Exp Parasitol. 2011;129:65–71. doi: 10.1016/j.exppara.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 35.Ebert F, Bachmann A, Nakada-Tsukui K, Hennings I, Drescher B, et al. An Entamoeba cysteine peptidase specifically expressed during encystation. Parasitol Int. 2008;57:521–524. doi: 10.1016/j.parint.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Makioka A, Kumagai M, Hiranuka K, Kobayashi S, Takeuchi T. Expression analysis of Entamoeba invadens profilins in encystation and excystation. Parasitol Res. 2011 doi: 10.1007/s00436-011-2735-3. [DOI] [PubMed] [Google Scholar]
- 37.Arroyo-Begovich A, Carabez-Trejo A, Ruiz-Herrera J. Identification of the structural component in the cyst wall of Entamoeba invadens. J Parasitol. 1980;66:735–741. [PubMed] [Google Scholar]
- 38.Garcia-Zapien AG, Gonzalez-Robles A, Mora-Galindo J. Congo red effect on cyst viability and cell wall structure of encysting Entamoeba invadens. Arch Med Res. 1999;30:106–115. doi: 10.1016/s0188-0128(98)00029-3. [DOI] [PubMed] [Google Scholar]
- 39.Ghosh SK, Van Dellen KL, Chatterjee A, Dey T, Haque R, et al. The Jacob2 lectin of the Entamoeba histolytica cyst wall binds chitin and is polymorphic. PLoS Negl Trop Dis. 2010;4:e750. doi: 10.1371/journal.pntd.0000750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirt RP, Harriman N, Kajava AV, Embley TM. A novel potential surface protein in Trichomonas vaginalis contains a leucine-rich repeat shared by micro-organisms from all three domains of life. Mol Biochem Parasitol. 2002;125:195–199. doi: 10.1016/s0166-6851(02)00211-6. [DOI] [PubMed] [Google Scholar]
- 41.Davis PH, Zhang Z, Chen M, Zhang X, Chakraborty S, et al. Identification of a family of BspA like surface proteins of Entamoeba histolytica with novel leucine rich repeats. Mol Biochem Parasitol. 2006;145:111–116. doi: 10.1016/j.molbiopara.2005.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Penuliar GM, Furukawa A, Nakada-Tsukui K, Husain A, Sato D, et al. Transcriptional and functional analysis of trifluoromethionine resistance in Entamoeba histolytica. J Antimicrob Chemother. 2012;67:375–386. doi: 10.1093/jac/dkr484. [DOI] [PubMed] [Google Scholar]
- 43.Makioka A, Kumagai M, Ohtomo H, Kobayashi S, Takeuchi T. Effect of jasplakinolide on the growth, encystation, and actin cytoskeleton of Entamoeba histolytica and Entamoeba invadens. J Parasitol. 2001;87:399–405. doi: 10.1645/0022-3395(2001)087[0399:EOJOTG]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 44.Beck DL, Boettner DR, Dragulev B, Ready K, Nozaki T, et al. Identification and gene expression analysis of a large family of transmembrane kinases related to the Gal/GalNAc lectin in Entamoeba histolytica. Eukaryot Cell. 2005;4:722–732. doi: 10.1128/EC.4.4.722-732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chung Chau Hon KN-T, Nozaki T, Guillén G. Dissecting the Actin Cytoskeleton of Entamoeba histolytica from a Genomic Perspective. In: Clark CGraham, Johnson PatriciaJ, Adam RD., editors. Anaerobic Parasitic Protozoa: Genomics and Molecular Biology. Caister Academic Press; 2010. [Google Scholar]
- 46.Nozaki KN-TaT. Genomic and post-genomic approaches to understand the pathogenesis of the enteric protozoan parasite Entamoeba histolytica. In: Fratamico Pina, Kathariou Sophia, Liu Y., editors. Genomes of Food- and Water-borne Pathogens. Washington, D.C.: ASM Press; 2011. [Google Scholar]
- 47.Pearse BM. Clathrin: a unique protein associated with intracellular transfer of membrane by coated vesicles. Proc Natl Acad Sci U S A. 1976;73:1255–1259. doi: 10.1073/pnas.73.4.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yakoob J, Abbas Z, Beg MA, Naz S, Khan R, et al. Entamoeba species associated with chronic diarrhoea in Pakistan. Epidemiol Infect. 2011:1–6. doi: 10.1017/S0950268811000215. [DOI] [PubMed] [Google Scholar]
- 49.Shimokawa C, Kabir M, Taniuchi M, Mondal D, Kobayashi S, et al. Entamoeba moshkovskii is Associated with Diarrhea in Infants and Causes Diarrhea and Colitis in Mice. J Infect Dis. 2012 doi: 10.1093/infdis/jis414. [In press] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lorenzi HA, Puiu D, Miller JR, Brinkac LM, Amedeo P, et al. New assembly, reannotation and analysis of the Entamoeba histolytica genome reveal new genomic features and protein content information. PLoS Negl Trop Dis. 2010;4:e716. doi: 10.1371/journal.pntd.0000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marion S, Laurent C, Guillen N. Signalization and cytoskeleton activity through myosin IB during the early steps of phagocytosis in Entamoeba histolytica: a proteomic approach. Cell Microbiol. 2005;7:1504–1518. doi: 10.1111/j.1462-5822.2005.00573.x. [DOI] [PubMed] [Google Scholar]
- 52.Marquay Markiewicz J, Syan S, Hon CC, Weber C, Faust D, et al. A proteomic and cellular analysis of uropods in the pathogen Entamoeba histolytica. PLoS Negl Trop Dis. 2011;5:e1002. doi: 10.1371/journal.pntd.0001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong WK, Tan ZN, Othman N, Lim BH, Mohamed Z, et al. Analysis of Entamoeba histolytica Excretory-Secretory Antigen and Identification of a New Potential Diagnostic Marker. Clin Vaccine Immunol. 2011;18:1913–1917. doi: 10.1128/CVI.05356-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Total number of E. histolytica proteins detected in soluble, insoluble or both fractions of mass spectrometry experiments for 4 cyst samples. The mass spectrometry was carried out for 4 out of 5 cyst samples in such a way that it could detect both soluble and insoluble proteins (for details, see the Methods section). For the 5th sample (4268), there was not enough cyst material to proceed by this method, and proteins identified in this sample represented both soluble and insoluble proteins. Except for the sample 8076 (which has a protein distribution such as 48 in soluble fraction, 43 in insoluble fraction, and 18 in both fractions), there was a general trend that relatively more proteins were identified in the insoluble fraction compared with the soluble fraction (59/48 for AM951, 99/42 for AM797, and 57/35 for CMS33-7132).
(TIF)
Overlap between the 417 proteins (from this study) and the cyst-specific mRNA transcripts (from [22] ). The overlap between 672 cyst-specific transcripts (p-value<0.05 and fold-change ≥3) and 394 proteins out of all 417 proteins from cyst proteomic study (except for the remaining 23 proteins, that were not found in microarray data) was tested using the Venn diagram. The overlap between the cyst protein data and the cyst-specific mRNA transcript data was statistically significant (p-value 0.0058). The p-value was determined using the two-tailed Fisher's exact test using the GraphPad software freely available in the internet at http://www.graphpad.com/quickcalcs/contingency1.cfm.
(TIF)
Overlap between the 195 cyst –specific proteins (from this study) and the cyst-specific mRNA transcripts (from [22] ). The overlap between 672 cyst-specific transcripts (p-value<0.05 and fold-change ≥3) and the 185 proteins out of 195 proteins from the cyst proteomic study that were not identified in trophozoite-specific proteome or EST datasets (except for the remaining 10 proteins, that were not found in the microarray data) was tested using the Venn diagram. The overlap was statistically significant (p-value 0.0014). The p-value for the overlap of this comparison is better than the previous comparison shown in Figure S2 (0.0014 versus 0.0058, respectively) as expected. The p-values were determined using the two-tailed Fisher's exact test using the GraphPad software freely available in the internet at http://www.graphpad.com/quickcalcs/contingency1.cfm.
(TIF)
Proteins detected in sample AM951 using two different processing methods prior to LC-MS/MS experiments.
(XLSX)
Categories of proteins identified in sonication only and freeze-thaw-sonication, or both approaches for sample AM951.
(XLSX)
All the E. histolytica proteins detected in 5 E. histolytica cyst samples (N = 417).
(XLSX)
E. histolytica proteins detected in cyst samples that show 80% or less identity with E. dispar proteins. These are candidate proteins for development of diagnostic tool for both trophozoites and cysts of E. histolytica.
(XLSX)
E. histolytica proteins detected in 3 or more of 5 cyst samples. These are candidate proteins for development of diagnostic tool for both trophozoites and cysts of E. histolytica.
(XLSX)
E. histolytica proteins with maximum number of spectrum counts detected in cyst samples by LC-MS/MS experiments. These are candidate proteins for development of diagnostic tool for both trophozoites and cysts of E. histolytica.
(XLSX)
Host (human) proteins detected in cyst samples. Human proteins that were identified in at least 3 out of 5 E. histolytica cyst samples are shown in this Table.
(XLSX)
Bacterial species detected in 3 or more E. histolytica cyst samples. Many bacterial peptides were identified in the E. histolytica cyst samples by LC-MS/MS experiments. From the database search using the peptide sequences, the bacterial species were identified. At least 19 bacterial species were detected in 3 or more E. histolytica cyst samples.
(XLSX)