Abstract
Summary: Progress in high-throughput genomic technologies has led to the development of a variety of resources that link genes to functional information contained in the biomedical literature. However, tools attempting to link small molecules to normal and diseased physiology and published data relevant to biologists and clinical investigators, are still lacking. With metabolomics rapidly emerging as a new omics field, the task of annotating small molecule metabolites becomes highly relevant. Our tool Metab2MeSH uses a statistical approach to reliably and automatically annotate compounds with concepts defined in Medical Subject Headings, and the National Library of Medicine's controlled vocabulary for biomedical concepts. These annotations provide links from compounds to biomedical literature and complement existing resources such as PubChem and the Human Metabolome Database.
Availability: http://metab2mesh.ncibi.org
Contact: akarnovs@umich.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
In recent years, many tools have been developed for automated annotation of genes, transcripts and proteins and linking them to published biomedical literature. The resulting information has proved valuable for a range of bioinformatics tools and research related to disease gene prioritization, gene set enrichment testing, and pathway visualization tools (Bresell et al., 2006; Gaulton et al., 2007; Leach et al., 2009; Sartor et al., 2010; Soldatos et al., 2010). Vast amounts of data describing biological roles of metabolites, drugs and other small molecules have been published. Several public databases have been developed to annotate the metabolome of human and other organisms (Jewison et al., 2012; Mochamad Afendi et al., 2012; Wishart et al., 2009). However, to date there are still few tools that attempt to automatically develop, search and present literature-derived compound annotations to the biomedical research community. PubChem is a public resource, used by individual researchers and as input data for other tools, that provides access to 30 million compounds and 85 million substances, and includes biomedical, structure, safety and additional information, including links to Pubmed abstracts via synonym mapping (Wang et al., 2009). ToxNet is another tool, developed by National Library of Medicine (NLM), that allows searches for compounds, and provides links to relevant literature through multiple sources (Wexler, 2001). Several tools geared towards chemists such as Compounds In Literature (CIL) and WENDI, allow users to search compounds using names, smiles strings or compound structures and identify relevant publications (Gruning et al., 2011; Zhu et al., 2010).
Rather than linking compounds to all available literature, we decided to utilize Medical Subject Headings (MeSH) to annotate compounds with the most commonly associated medical/biological terms and vice versa. We believe that this approach helps to provide more focused results and aids in finding relevant relationships and literature. MeSH is the NLM controlled vocabulary used to manually index articles for MEDLINE/PubMed. MeSH covers a broad range of biological topics and medical terms, with such general headings as Disease, Anatomy, Chemicals and Drugs, and Phenomena and Processes. There are currently 26 142 MeSH descriptors that are organized into a hierarchical structure. In addition, there are 83 MeSH qualifiers that can be used to narrow a descriptor to a more specific topic.
NLM provides a list of chemical substances that contain a wide range of synonyms used in biomedical publications that can be linked to the compound synonyms used in PubChem. Using the compounds and their occurrences in PubMed literature, we performed statistical tests to estimate the significance of the associations between compound—MeSH descriptor pairs. The resulting associations, supporting data and literature can be accessed through our web-based tool, Metab2MeSH. Web services are also available (http://ws.ncibi.org/m2m.html) and allow for batch queries. Users have the ability to identify either the MeSH descriptors significantly associated with a given compound or the compounds associated with a selected MeSH descriptor. Examples of both use cases are provided below.
2 METHODS
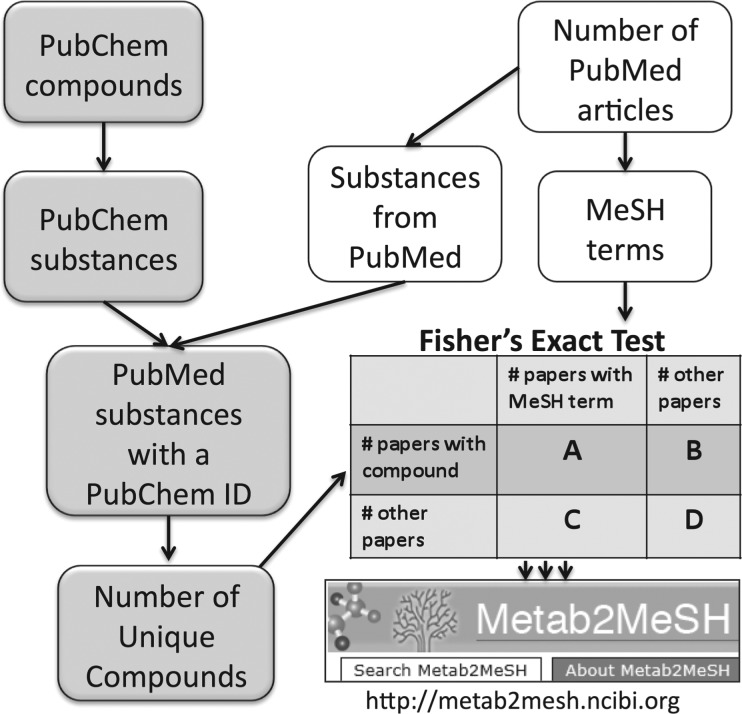
The PubChem Compound and Substance databases and NLM PubMed database were downloaded, parsed and loaded into a relational database. PubMed substances were extracted from the substance index for each PubMed abstract. The process used to create Metab2Mesh is outlined in Figure 1. The PubChem Substance database contains a list of user-supplied synonyms for each compound. We used those to identify matching synonyms in our list of PubMed Substances. This provided the links between PubChem compounds and literature citations. A total of 81 028 PubChem compounds were linked to literature citations. Links between MeSH descriptors and literature citations are provided directly by PubMed. To avoid irrelevant information in our Metab2MeSH tool, we filtered the MeSH descriptors to those under the following main headings: Diseases, Anatomy, Chemicals and Drugs, Phenomena and Processes, Organisms, Psychiatry and Psychology, Anthropology, Education, Sociology and Social Phenomena, Technology, Industry, and Agriculture. This resulted in a total of 21 818 unique MeSH descriptors. Next, we determined the total number of PubMed articles annotated with at least one of these MeSH descriptors and at least one PubMed substance that maps to a PubChem synonym (6 million), which served as the complete background set of articles for our tests. We also determined the PubMed articles annotated with each MeSH descriptor and each PubMed substance, and the total number of articles for each compound.
Fig. 1.
Flowchart describing the process used to build Metab2MeSH. All significant associations between compounds and MeSH terms (p<0.0001) are accessible in the web application.
Using these values and the numbers of PubMed articles co-annotated with each compound—MeSH term pair, we tested the significance of association between each compound—MeSH term pair using two-sided Fisher's exact tests, and p-values were calculated. This resulted in a total of 42 million tests, with 4 646 000 significant associations (p-value < 0.0001). Significant associations are presented in Metab2MeSH sorted by p-value. The False Discovery Rate method was used to adjust p-values for multiple comparisons, and the resulting q-values are provided (Benjamini and Hochberg, 1995). (In certain instances, the association is so significant that the precise p-value cannot be calculated. In these cases, the p-value is set to 0 and the χ2 statistic from the equivalent χ2-test is used to sort the associations by decreasing significance.) As an alternative measure of strength of association, we provide the Fold Change, defined for a specific compound and MeSH descriptor as follows:
where p is the number of articles that have the compound and MeSH descriptor pair, c is the total number of articles that contain the compound, d is the total number of articles that contain the MESH descriptor and t is the total number of articles considered. We combined the above significant results with links to the literature and PubChem compounds in a web interface to create our tool, Metab2MeSH (http://metab2mesh.ncibi.org). The data are updated twice a year. Supplementary Materials and Supplementary Figure S1 contain further details about the web interface.
The Metab2MeSH data set can also be accessed through our web services (http://ws.ncibi.org/m2m.html) which allows users to query the Metab2MeSH database programmatically, and perform batch queries if desired. The resulting computer-parsable XML data include compound-MeSH term pairs with significance values and a list of associated PMIDs (see Supplementary Material for an example web services bulk query).
3 RESULTS
To illustrate the uses and features of Metab2MeSH, we present two applications. Suppose we would like to determine whether cyclic AMP (cAMP) is commonly associated with any certain type of cancer. We can query the compound ‘cyclic AMP’ in Metab2MeSH using an exact match search and filter the MeSH headings by Disease to see that 29 diseases are found to be significantly associated with cAMP among the top 1000 displayed. To see the complete list of results (>1000), we can download the tab-delimited text file to view in a spreadsheet. Supplementary Figure S2 shows the Metab2MeSH results for the top diseases. The two diseases most strongly associated with cAMP levels are pseudohypoparathyroidism and neuroblastoma. There are several other types of cancers listed, but neuroblastomas are by far the most strongly associated MeSH Disease term by p-value (p=3.7e–262). ‘Leydig cell tumor’, although less studied in the literature, had the highest fold enrichment (fold = 12.5), with other cancers including Glioma, Osteosarcoma and Adrenal Gland Neoplasms. Clicking the literature link for Neuroblastoma and limiting to Review articles immediately brings our attention to ‘Defects in cAMP-pathway may initiate carcinogenesis in dividing nerve cells’ (Prasad et al., 2003).
Another important feature of Metab2MeSH is the ability to search for compounds associated with a MeSH descriptor, e.g. a disease. Suppose one would like to identify compounds associated with phenylketonuria, a metabolic disease caused by a defect in the enzyme phenylalanine hydroxylase. This enzyme is required to convert phenylalanine to tyrosine. Supplementary Figure S3 shows the top 25 compounds associated with Phenylketonurias. Among these are phenylalanine, endophenyl, tetrahydrobiopterin, sapropterin, pterin and several other compounds that are known to be associated with this disease. This example illustrates that Metab2MeSH prioritizes known relationships. If users are unsure of the MeSH term they require, they can browse the MeSH tree via our provided link. Metab2MeSH also provides a quick way to retrieve the abstracts related to both disease and the compound. No other tool currently has this capability. Additional examples of use are provided in the Supplementary Material.
In conclusion, Metab2MeSH provides a quick and convenient method for identifying compounds commonly associated with a biological topic or medical term, or for identifying biological topics or medical terms commonly associated with a compound of interest. Although Metab2MeSH is not meant to predict novel associations (due to its dependence on what is in the literature), as the examples illustrate, it is very useful for identifying sets of associations that would otherwise be difficult to find. Metab2MeSH creates a valuable data set that can be accessed via our website or API, and will likely prove useful in other bioinformatics tools.
Funding: National Institutes of Health (U54DA21519); Michigan Diabetes Research and Training Center Pilot and Feasibility Grant (to A.K.); Michigan Nutrition Obesity Research Center (5P30DK089503 - A.K.) and 5R01DK079084 (M.A.S. and B.A.).
Conflict of Interest: none declared.
Supplementary Material
Footnotes
†The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
REFERENCES
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- Bresell A., et al. Ontology annotation treebrowser: an interactive tool where the complementarity of medical subject headings and gene ontology improves the interpretation of gene lists. Appl. Bioinform. 2006;5:225–236. doi: 10.2165/00822942-200605040-00005. [DOI] [PubMed] [Google Scholar]
- Gaulton K.J., et al. A computational system to select candidate genes for complex human traits. Bioinformatics. 2007;23:1132–1140. doi: 10.1093/bioinformatics/btm001. [DOI] [PubMed] [Google Scholar]
- Gruning B.A., et al. Compounds In Literature (CIL): screening for compounds and relatives in PubMed. Bioinformatics. 2011;27:1341–1342. doi: 10.1093/bioinformatics/btr130. [DOI] [PubMed] [Google Scholar]
- Jewison T., et al. YMDB: the Yeast Metabolome Database. Nucleic Acids Res. 2012;40:D815–D820. doi: 10.1093/nar/gkr916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach S.M., et al. Biomedical discovery acceleration, with applications to craniofacial development. PLoS Comput. Biol. 2009;5:e1000215. doi: 10.1371/journal.pcbi.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochamad Afendi,F., et al. KNApSAcK Family Databases: integrated metabolite-plant species databases for multifaceted plant researches. Plant Cell Physiol. 2012;53:e1. doi: 10.1093/pcp/pcr165. [DOI] [PubMed] [Google Scholar]
- Prasad K.N., et al. Defects in cAMP-pathway may initiate carcinogenesis in dividing nerve cells: a review. Apoptosis. 2003;8:579–586. doi: 10.1023/A:1026179324295. [DOI] [PubMed] [Google Scholar]
- Sartor M.A., et al. ConceptGen: a gene set enrichment and gene set relation mapping tool. Bioinformatics. 2010;26:456–463. doi: 10.1093/bioinformatics/btp683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldatos T.G., et al. Martini: using literature keywords to compare gene sets. Nucleic Acids Res. 2010;38:26–38. doi: 10.1093/nar/gkp876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., et al. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–W633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler P. TOXNET: an evolving web resource for toxicology and environmental health information. Toxicology. 2001;157:3–10. doi: 10.1016/s0300-483x(00)00337-1. [DOI] [PubMed] [Google Scholar]
- Wishart D.S., et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., et al. WENDI: a tool for finding non-obvious relationships between compounds and biological properties, genes, diseases and scholarly publications. J. Cheminform. 2010;2:6. doi: 10.1186/1758-2946-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.