Abstract
Purpose
Renal cell carcinoma is the eighth most common cancer in the United States and clear cell renal carcinoma is the most common type. Many signaling pathways are implicated in clear cell renal carcinoma development, including the inflammation pathway. However, less is known about how gene expression variation in this pathway influences clear cell renal carcinoma development and clinical outcomes.
Materials and Methods
Gene expression in tumor and adjacent normal tissues from 93 patients was detected using a genome-wide expression array. A panel of 661 inflammation related genes was then analyzed. Differential expression patterns between tumor and normal tissues were identified. Association with recurrence or survival was evaluated with genes showing significant association tested further in a validation set of 258 tumors using an independent platform (quantitative real-time polymerase chain reaction).
Results
We identified 151 genes with at least a two-fold change in gene expression between adjacent normal tissue and tumor, of which most were up-regulated in tumors. A total of 20 genes significantly associated with recurrence and/or overall survival were selected for further validation. In the replication data set high expression of GADD45G was significantly associated with a 2.09-fold (95% CI 1.08 – 6.14, p = 0.034) increased risk of recurrence while high CARD9, NCF2 and CIITA expression was significantly associated with a 2.52-fold (95% CI 1.24 –5.12, p = 0.010), 2.26-fold (95% CI 1.12– 4.58, p = 0.023) and 2.11-fold (95% CI 1.05– 4.27, p = 0.037) increased risk of death, respectively.
Conclusions
Results suggest that inflammation gene expression may be significant prognostic biomarkers for the risk of recurrence (GADD45G) and death (CARD9, CIITA and NCF2) in patients with clear cell renal carcinoma.
Keywords: kidney, carcinoma, renal cell, inflammation, gene expression, neoplasm recurrence, local
It was estimated that 57,760 new cases of cancer of the kidney and renal pelvis were diagnosed and 12,980 deaths occurred in 2009, of which most represented RCC.1 RCC is a heterogeneous group of malignant tumors consisting of various subtypes that are classified based on morphological and genetic features. Of these subtypes the clear cell subtype or ccRCC is most common, accounting for approximately 80% of all renal neoplasms.2 Large epidemiological studies identified that several factors are associated with the risk of RCC, including cigarette smoking, chemical agents, obesity, hypertension, antihypertensive medication and end stage renal disease.3,4
Another known RCC risk factor is chronic inflammation. This has long been known to be associated with RCC development and progression, as shown by epidemiological and clinical studies.5,6 However, the detailed transition mechanism from inflammation to cancer is still unclear. Previously the inflammatory response was thought to help the host against the developing tumor but recent studies showed that the inflammatory response may contribute to cancer growth and metastasis.5,6 Cancer and inflammation are complicated processes under the control of many signal pathways rather than a single one.7,8 In RCC cases the tumor expression of several inflammation related proteins was noted, such as cyclooxygenase-2, interleukin-6 and 8, tumor necrosis factor-α and nuclear factor-κB.9–11
However, to our knowledge a systematic, comprehensive assessment of the expression of inflammation related genes has not been performed. Thus, we used a microarray based screen to assess the expression patterns of inflammation related genes in ccRCC cases to determine whether they may have a role in ccRCC development, recurrence and/or overall survival. We then validated these findings in an independent set of tissues. These results identify potential biomarkers for ccRCC risk and clinical outcomes.
MATERIALS AND METHODS
Patient Population and Data Collection
For this study patients with ccRCC were recruited from our institution. There were no age, gender, ethnicity or cancer stage restrictions on recruitment. All patients provided written informed consent and the study protocol was approved by the M. D. Anderson Cancer Center institutional review board. Patient demographic variables, tobacco and alcohol use history, family history of cancer, weight and height to calculate BMI and medical history were obtained by interview in person. Clinical information was abstracted from the medical records, including clinical stage, grade, comorbidities, tumor size, pathological stage, histology, treatment, recurrence, survival and progression.
Tissue Samples
Tumor and adjacent normal tissues for the discovery stage were obtained from 93 newly diagnosed patients with histologically confirmed ccRCC who underwent partial or radical nephrectomy. Only those with tumor and normal tissue available were included in analysis. A set of additional 258 ccRCC tumor samples was used for replication. All tissue samples were snap frozen in liquid nitrogen immediately after excision and stored at −80C.
RNA Isolation and Microarray Processing
Approximately 20 mg frozen tissue were placed in 100 μl RNAlater®-ICE Frozen Tissue Transition Solution (Ambion®) at −30C for more than 16 hours to minimize RNA degradation. Total RNA was isolated using the mirVana™ RNA Isolation Kit according to standard protocol. Labeled cRNA was synthesized, amplified and purified from 200 ng total RNA using the Total Prep™ RNA Amplification Kit according to manufacturer recommendations. Each sample was then hybridized to Human-6 v2 Expression BeadChips and read using the BeadStation 500 scanner (Illumina®). Array data were quantile normalized and log2 transformed.
Inflammation Related Gene Selection
We compiled the genes involved in inflammatory response regulation based on the WKINFLAM panel12 and the T1Dbase database of inflammation genes (http://www.t1dbase.org). Predefined canonical pathways and custom built pathways were generated using the Ingenuity® Pathway Analysis tool. We selected 23 pathways, for example nuclear factor-κB, cytokine signaling and tumor necrosis factor-α, and 1,546 genes. After excluding genes from further analysis if less than 20% of the 93 samples showed less than a 1.5-fold difference from the median, 661 inflammatory related genes of the total of 1,546 had suitable expression above background and sufficient variation among samples.
Quantitative Real-Time PCR
Genes were selected for validation using quantitative real-time PCR if they were most significantly associated with recurrence and/or survival in the discovery population. Total RNA (10 ng) was reverse transcribed using Super-Script™ III and random hexamer primers (Invitrogen™) according to manufacturer instructions. Real-time quantitative PCR was done using TaqMan® assays with a 384-well optic plate on the ABI Prism® 7900HT Sequence Detection System. GAPDH served as an endogenous control to normalize the amount of RNA input and reverse transcription efficiency. Thermal cycling conditions consisted of 1 cycle for 2 minutes at 50C and 10 minutes at 95C, followed by 40 cycles for 15 seconds at 95C and 1 minute at 60C. The PCR reaction for each sample was done in duplicate. Data were analyzed by the δ-δ method (2−ΔΔCt) using the equation, ΔΔCt = (Ct gene – Ct gene average)/(Ct GAPDH = Ct GAPDH average), where Ct represents threshold cycle.
Statistical Analysis
Comparison of patient characteristics between the discovery and validation populations was analyzed using the Student t, Mann-Whitney or Fisher exact test, as appropriate. For smoking history patients who had never smoked or had smoked fewer than 100 cigarettes in a lifetime were considered never smokers while those who had smoked at least 100 cigarettes in a lifetime were considered ever smokers. Differences in gene expression between tumor and adjacent normal tissues were compared by Student’s t test. Time to recurrence was defined as the interval between the date of diagnosis and recurrence or last followup. Recurrence in patients with metastatic stage IV disease was defined as a secondary metastatic recurrence after treatment. Overall survival duration was defined as the interval from diagnosis to patient death or last followup. The HR and the corresponding 95% CI for recurrence and survival end points were estimated by applying the Cox proportional hazards regression model while adjusting for patient age at diagnosis, gender, BMI, family history of cancer, race, hypertension, smoking status and stage.
To assess the dose-response trend in the discovery and validation populations we applied a spline modeling procedure implemented in R, version 2.5 to identify potential cutoffs at which optimal thresholds to assess dose-response pattern were identified. The association between individual genes and survival time was estimated by Ka-plan-Meier analysis and assessed for significance by the log rank test. Kaplan-Meier survival curves were constructed comparing the survival of patients with ccRCC who had high vs low gene expression based on the expression level assessed by the spline model. Statistical analysis was done with Stata®, version 10. All statistical tests were 2-sided with p <0.05 considered statistically significant.
RESULTS
Patient Characteristics
Table 1 lists the demographic and clinical characteristics of the 93 patients in the discovery population and the 258 in the validation population. No significant differences were noted between the 2 populations in age at diagnosis, BMI, family history of cancer, grade, recurrence and survival status. The validation population included more men than the discovery population (p = 0.028). A significant difference in stage distribution between the 2 populations was also evident. The proportion of patients with stage I and IV tumors was similar but there was an excess of those with stage III disease and fewer with stage II in the validation population (p = 0.0021).
Table 1.
Patient characteristics
| Discovery* | Validation* | p Value | |
|---|---|---|---|
| No. pts | 93 | 258 | |
| Mean ± SD age | 59.1 ± 10.3 | 59.8 ± 10.6 | 0.59 |
| No. gender (%): | 0.028 | ||
| F | 39 (41.94) | 75 (29.07) | |
| M | 54 (58.06) | 183 (70.93) | |
| Mean ± SD BMI (kg/m2) | 30.7 ± 6.5 | 30.4 ± 6.5 | 0.74 |
| No. smoking (%): | 0.28 | ||
| Never | 49 (52.69) | 118 (46.46) | |
| Former | 28 (30.11) | 100 (39.37) | |
| Current | 16 (17.20) | 36 (14.17) | |
| No. hypertension history (%): | 0.81 | ||
| Yes | 52 (55.91) | 139 (53.88) | |
| No | 41 (44.09) | 119 (46.12) | |
| No. Ca family history (%): | 0.057 | ||
| Yes | 53 (56.99) | 176 (68.22) | |
| No | 40 (43.01) | 82 (31.78) | |
| No. stage (%): | 0.0021 | ||
| I | 45 (48.39) | 104 (40.63) | |
| II | 18 (19.35) | 20 (7.81) | |
| III | 16 (17.20) | 84 (32.81) | |
| IV | 14 (15.05) | 48 (18.75) | |
| No. grade (%): | 0.96 | ||
| 1 | 24 (25.81) | 69 (26.96) | |
| 2 | 53 (56.99) | 140 (54.69) | |
| 3 | 16 (17.20) | 47 (18.36) | |
| No. recurrence (%): | 0.65 | ||
| Yes | 21 (29.17) | 61 (26.29) | |
| No | 51 (70.83) | 171 (73.71) | |
| No. death (%): | 0.89 | ||
| Yes | 21 (22.83) | 57 (22.09) | |
| No | 71 (77.17) | 201 (77.91) |
Some categories do not total 100% due to missing values.
ccRCC Differential Gene Expression
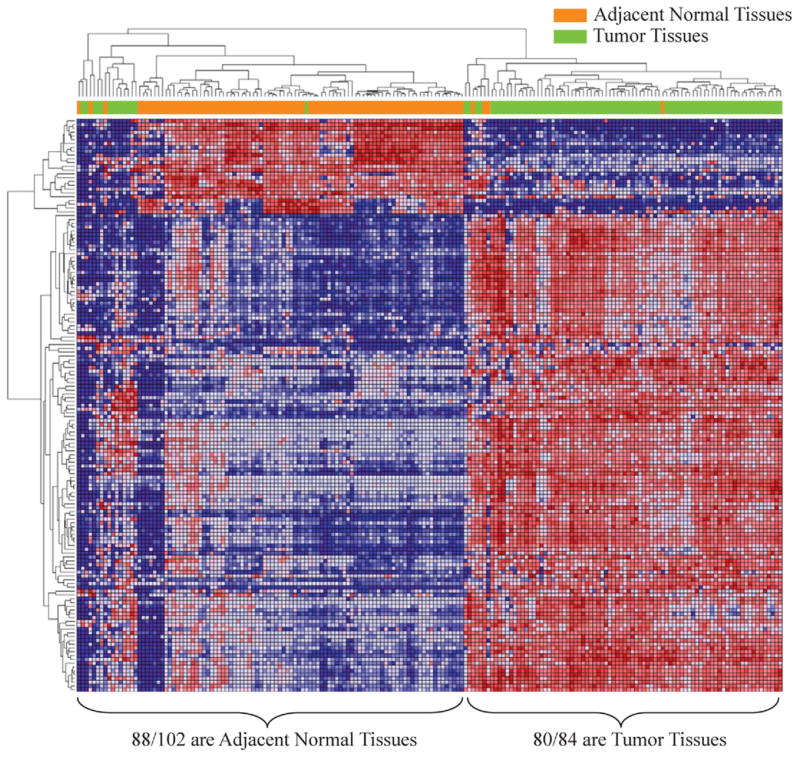
To identify which genes were differentially expressed in tumor vs adjacent normal tissue we analyzed 93 pairs of ccRCC tissues in the discovery population using genome-wide expression microarrays. Of the 661 inflammatory related genes 464 showed significant differences in expression between tumor and normal samples. A total of 151 genes had at least a two-fold change in gene expression between normal and tumor samples (fig. 1). Clustering these significant genes revealed 2 clusters, including 24 down-regulated and 127 up-regulated genes with respect to adjacent normal tissue. These genes differentiated tumor and normal samples except for the misclassification of 5 normal and 14 tumor tissues.
Figure 1.
Expression profile of 151 highly significant genes with at least 2-fold change between 93 pairs of tumor and adjacent normal tissues.
Recurrence and Overall Survival
Association in discovery population
Using expression microarray data 18 inflammation related genes in the discovery population were significantly associated with recurrence. These genes showed a dose recurrence trend (p <0.05). Another 7 genes associated with overall survival showed a dose-death trend (p <0.05, table 2). Two of these genes, ADORA3 and CARD9, were significantly associated with each clinical outcome.
Table 2.
Association of inflammation related genes with overall survival in discovery population using expression microarrays
| Gene Expression | No. Death (%)
|
HR (95% CI)* | p Value | |
|---|---|---|---|---|
| No | Yes | |||
| ADORA3:† | 0.00036 | |||
| Low | 55 (77.46) | 14 (66.67) | 1 (referent) | |
| High | 16 (22.54) | 7 (33.33) | 21.34 (3.97–114.61) | |
| CARD9:† | 0.0026 | |||
| Low | 51 (71.83) | 10 (47.62) | 1 (referent) | |
| High | 20 (28.17) | 11 (52.38) | 7.11 (1.98–25.49) | |
| CASP2 + RIPK1 domain containing adaptor with death domain: | 0.0050 | |||
| Low | 34 (47.89) | 14 (66.67) | 1 (referent) | |
| High | 37 (52.11) | 7 (33.33) | 0.14 (0.036–0.56) | |
| NCF2: | 0.0051 | |||
| Low | 41 (57.75) | 7 (33.33) | 1 (referent) | |
| High | 30 (42.25) | 14 (66.67) | 5.87 (1.70–20.26) | |
| Suppressor of cytokine signaling 1: | 0.0075 | |||
| Low | 56 (78.87) | 13 (61.90) | 1 (referent) | |
| High | 15 (21.13) | 8 (38.10) | 4.98 (1.54–16.13) | |
| Epstein-Barr virus induced 3: | 0.017 | |||
| Low | 38 (53.52) | 8 (38.10) | 1 (referent) | |
| High | 33 (46.48) | 13 (61.90) | 4.40 (1.30–14.87) | |
| CIITA: | 0.021 | |||
| Low | 62 (87.32) | 15 (71.43) | 1 (referent) | |
| High | 9 (12.68) | 6 (28.57) | 4.19 (1.24–14.14) | |
Adjusted for age at diagnosis, gender, BMI, family history of cancer, race, hypertension, smoking status and stage.
Also significant for recurrence.
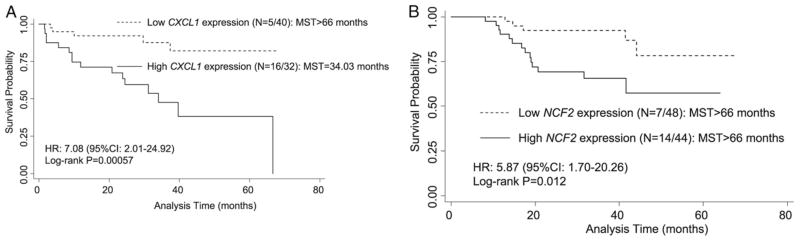
Six of the 18 genes suggested a protective effect for recurrence risk, including amyloid β precursor. High expression of this gene was associated with a significant decrease in the recurrence risk (HR 0.19, 95% CI 0.058–0.64, p = 0.0075). In contrast, the remaining 12 genes with higher expression resulted in an increased recurrence risk. We also noted that increased gene expression of B-cell linker; CXCL1; superoxide dismutase 2; mucin 1, cell surface associated; and CARD9 was significantly associated with the risk of recurrence and shorter recurrence-free survival while other genes were only associated with an increased risk of recurrence. For example, patients with high CXCL1 expression were at 7.08-fold increased risk (95% CI 2.01–24.92) than those with low expression (fig. 2, A). This resulted in a greater than 30-month survival disadvantage from more than 66 months in patients with low expression to only 34.03 months in those with high expression (log rank test p = 0.00057).
Figure 2.
Recurrence-free and overall survival by inflammation related gene expression in discovery population. A, CXCL1 effect on recurrence. B, NCF2 effect on overall survival. N, number of events/total number of patients. MST, median event-free survival.
For overall survival high expression of 6 of the 7 genes resulted in a significantly increased risk of death (4.19 to 21.34-fold). The most significant association was for ADORA3 expression, which resulted in an increased risk of death (HR 21.34, 95% CI 3.97–114.61). Of these genes associated with increased risk higher NCF2 expression was associated with a 5.87-time increased risk of death (95% CI 1.70–20.26), resulting in significantly worse survival (log rank test p = 0.012, fig. 2, B).
Validation in independent population
We selected 20 inflammatory related genes for further study, including 13 for recurrence (B-cell linker; interleukin-8; CXCL1; complement component 1, r subcomponent; matrix metallopeptidase 9; GADD45G; superoxide dismutase 2; mucin 1, cell surface associated; amyloid β precursor; nuclear receptor subfamily 2, group F, member 1; bone morphogenetic protein 4; kallikrein 1; and interleukin 10 receptor, α) and 5 for survival (CASP2 and RIPK1 domain containing adaptor with death domain, NCF2, suppressor of cytokine signaling 1, Epstein-Barr virus induced 3 and CIITA). ADORA3 and CARD9 were significant for recurrence and overall survival, and they were selected for validation.
We measured the expression of these candidate genes in an independent population of 258 cases. Of the 15 genes that showed a significant association with recurrence in the discovery population only GADD45G remained significant in the validation population. High GADD45G expression was associated with an increased risk of recurrence (HR 2.09, 95% CI 1.06–4.14, p = 0.034). In the context of overall survival CARD9, NCF2 and CIITA remained significant in the validation population. The effect of these 3 genes was even more dramatic in patients with higher gene expression than in those with low gene expression, resulting in a 2.52-fold (95% CI 1.24 –5.12, p = 0.010), 2.26-fold (95% CI 1.12– 4.58, p = 0.023) and 2.11-fold (95% CI 1.05– 4.27, p = 0.037) increased risk of death for CARD9, NCF2 and CIITA, respectively.
DISCUSSION
Studies have demonstrated that the expression patterns of inflammatory related genes are associated with ccRCC.13,14 However, most of these studies focused on only a few genes and on differential gene expression between normal and tumor tissue with far less published information on the effect of expression differences among inflammation related genes and clinical outcomes. In this study we used a comprehensive approach to investigate the differential expression of inflammation related genes between ccRCC tumors and adjacent normal as well as the association between expression and clinical outcome. Candidate genes for overall survival and recurrence were analyzed in a large independent population to validate findings in the discovery population. To our knowledge this is the first comprehensive analysis of the role of inflammation related gene expression in terms of ccRCC development and clinical outcome.
A total of 151 inflammation genes were differentially expressed between tumor and adjacent normal tissues. Several of these genes were previously noted to have altered expression in cancer, suggesting a role for them in ccRCC tumorigenesis. Pathway analysis revealed that down-regulated genes were mainly involved in complement and coagulation cascades (serpin peptidase inhibitor, clade A [α-1 anti-proteinase, antitrypsin], member 1; plasminogen activator, tissue; plasminogen; kininogen 1; and complement component 7), and in the calcium signaling pathway (calmodulin, phosphodiesterase 1, G-protein coupled receptor and protein tyrosinekinase). A previous group noted that plasminogen showed decreased expression in several ovarian tumor derived cell lines and primary tumor samples.15 Another down-regulated gene, cysteinyl-tRNA synthetase, encodes for a calcium sensing receptor, which was also strongly down-regulated in a proportion of parathyroid carcinomas with a high proliferation index.16 This result suggested that alteration in cysteinyl-tRNA synthetase expression might have a role in cell proliferation and possibly in carcinogenesis.
A subset of the 127 inflammatory related genes with increased expression in ccRCC tumor tissue is involved in antigen processing and presentation, the chemokine signaling pathway and other cancer related pathways. The human leukocyte antigen MHC II, DR α was highly differentially expressed. This protein functions in antigen presentation and immune activation. Our result was consistent with that in other reports of MHC II, DR α expression in colon cancer17 and hepatocellular carcinoma18 cases. Rasrelated C3 botulinum toxin substrate 2 also had higher expression in ccRCC tumors than in normal tissue. A similar change was found in prostate cancer.19 The consistency of the results of our studies and those of other cancer sites provides support that these genes are involved in ccRCC development and may provide information on the mechanisms responsible.
Of the 15 genes selected as the most significant candidates associated with recurrence only GADD45G expression was validated in an independent large population. GADD45G is a signal transducer involved in the regulation of many cellular functions, such as growth arrest, differentiation, cell survival and apoptosis. GADD45G is deficient in a number of cancers, for example hepatocellular20 and prostate21 carcinoma. However, to our knowledge no correlation has previously been reported between GADD45G expression and clinical outcome. In our study statistical analysis revealed that high GADD45G expression correlated significantly with an increased risk of recurrence. Since the accumulation of DNA damage leads to carcinogenesis, the increased GADD45G expression associated with functional defects or abnormal transcriptional regulation may also have a major influence on cell growth. This suggests that GADD45G might be a useful biomarker to predict ccRCC recurrence.
Of the 7 genes identified as promising candidates in our discovery data set the modulating effects of variations in CARD9, NCF2 and CIITA expression on survival were confirmed in the validation population. CARD9 is an apoptosis inducing gene involved in inflammation and immune activation.22 CARD9 over expression is presumably associated with the development or progression of gastric B-cell lymphoma.23 However, to our knowledge no group has previously evaluated the correlation between CARD9 expression and clinical outcome. We found that high CARD9 expression was associated with an increased risk of death. CARD9 over expression might activate immune responses, leading to catastrophic reactions.
The gene encoding NCF2 is actively transcribed after the promyelocyte stage of myelopoiesis and transcription continues until cell death.24 Previously groups evaluated the association of NCF2 polymorphisms and cancer risk.25,26 We found that increased NCF2 expression was associated with a significantly increased risk of death from cancer. NCF2 is known to be involved in reactive oxygen species metabolism. Thus, it is reasonable to believe that NCF2 over expression might lead to the dysregulation of reactive oxygen species metabolism.
CIITA is an important regulator of MHC II transcription in human T-cell lines and in patient tumor samples.27 The transcription of MHC II genes is tightly regulated by CIITA in healthy B cells with dysregulation of CIITA observed in tumors.18 Our results indicate that high CIITA expression is associated with an increased risk of death. CIITA over expression induces MHC II transcription in tumors involved in the immune response, resulting in an increased inflammatory state that contributes to cancer progression.
To our knowledge this is the first report showing a strong association of CARD9, NCF2 and CIITA expression with survival in patients with ccRCC. The specific roles of these genes in the context of ccRCC would be worth further study.
The expression patterns of several genes were significantly associated with the clinical outcome in the discovery population but they did not show the same effect in the validation population. Microarray based expression analysis is hindered by numerous false-positive findings due to the large number of genes that are simultaneously analyzed for expression. This characteristic of microarrays makes it essential to replicate any findings in a large, well matched, independent population using a different platform. A real strength of our study is the availability of such a patient population in which to replicate our microarray based findings from the discovery population using quantitative real-time PCR to exclude that the associations were due to chance. This difference in the platforms used to assess expression in the samples may also have contributed to the differences observed in the HRs between the discovery and validation populations. We further adjusted for several known factors associated with ccRCC risk and outcome to remove any potentially confounding effects from analysis, strengthening the likelihood that our findings of an association with overall survival are not artifacts.
CONCLUSIONS
We performed microarray expression analysis of 93 pairs of ccRCC tissues. We identified inflammation related genes associated with cancer development by comparing differences between tumor and adjacent normal tissues, and we also identified candidate genes for overall survival and recurrence risk. Validation of the clinical outcome analysis in a larger patient population implicated GADD45G as an indicator of recurrence, and CARD9, NCF2 and CIITA as modulators of overall survival. These 4 genes represent potential candidate biomarkers to predict the outcome in patients with ccRCC.
Supplementary Material
Acknowledgments
Supported by National Institutes of Health Grant R01 CA098897.
Abbreviations and Acronyms
- ADORA3
adenosine A3 receptor
- BMI
body mass index
- CARD9
caspase recruitment domain family, member 9
- ccRCC
clear cell RCC
- CIITA
major histocompatibility complex, class II transactivators
- CXCL1
chemokine C-X-C ligand 1
- GADD45G
growth arrest and DNA-damage-inducible, γ
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- MHC
major histocompatibility complex
- NCF2
neutrophil cytosolic factor 2
- PCR
polymerase chain reaction
- RCC
renal cell carcinoma
Footnotes
Study received M. D. Anderson Cancer Center institutional review board approval.
Supplementary material for this article can be obtained at http://epi.mdanderson.org/WuJUrol2011/.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Eble JN, Togashi K, Pisani P. Renal cell carcinoma. In: Eble JN, Sauter G, Epstein JI, et al., editors. Pathology and Genetics of Tumors of the Urinary System and Male Genital Organs. Lyon, France: IARC Press; 2004. [Google Scholar]
- 3.Ng CS, Wood CG, Silverman PM, et al. Renal cell carcinoma: diagnosis, staging, and surveillance. AJR Am J Roentgenol. 2008;191:1220. doi: 10.2214/AJR.07.3568. [DOI] [PubMed] [Google Scholar]
- 4.Yu MC, Mack TM, Hanisch R, et al. Cigarette smoking, obesity, diuretic use, and coffee consumption as risk factors for renal cell carcinoma. J Natl Cancer Inst. 1986;77:351. [PubMed] [Google Scholar]
- 5.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 8.Nathan C. Points of control in inflammation. Nature. 2002;420:846. doi: 10.1038/nature01320. [DOI] [PubMed] [Google Scholar]
- 9.Konig B, Steinbach F, Janocha B, et al. The differential expression of proinflammatory cyto-kines IL-6, IL-8 and TNF-alpha in renal cell carcinoma. Anticancer Res. 1999;19:1519. [PubMed] [Google Scholar]
- 10.Meteoglu I, Erdogdu IH, Meydan N, et al. NF-kappaB expression correlates with apoptosis and angiogenesis in clear cell renal cell carcinoma tissues. J Exp Clin Cancer Res. 2008;27:53. doi: 10.1186/1756-9966-27-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mungan MU, Gurel D, Canda AE, et al. Expression of COX-2 in normal and pyelonephritic kidney, renal intraepithelial neoplasia, and renal cell carcinoma. Eur Urol. 2006;50:92. doi: 10.1016/j.eururo.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 12.Loza MJ, McCall CE, Li L, et al. Assembly of inflammation-related genes for pathway-focused genetic analysis. PLoS One. 2007;2:e1035. doi: 10.1371/journal.pone.0001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cozar JM, Romero JM, Aptsiauri N, et al. High incidence of CTLA-4 AA (CT60) polymorphism in renal cell cancer. Hum Immunol. 2007;68:698. doi: 10.1016/j.humimm.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Krambeck AE, Thompson RH, Dong H, et al. B7-H4 expression in renal cell carcinoma and tumor vasculature: associations with cancer progression and survival. Proc Natl Acad Sci USA. 2006;103:10391. doi: 10.1073/pnas.0600937103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denison SR, Callahan G, Becker NA, et al. Characterization of FRA6E and its potential role in autosomal recessive juvenile parkinsonism and ovarian cancer. Genes Chromosomes Cancer. 2003;38:40. doi: 10.1002/gcc.10236. [DOI] [PubMed] [Google Scholar]
- 16.Haven CJ, van Puijenbroek M, Karperien M, et al. Differential expression of the calcium sensing receptor and combined loss of chromosomes 1q and 11q in parathyroid carcinoma. J Pathol. 2004;202:86. doi: 10.1002/path.1489. [DOI] [PubMed] [Google Scholar]
- 17.Schetter AJ, Nguyen GH, Bowman ED, et al. Association of inflammation-related and mi-croRNA gene expression with cancer-specific mortality of colon adenocarcinoma. Clin Cancer Res. 2009;15:5878. doi: 10.1158/1078-0432.CCR-09-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie XW, Mei MH, Liao WJ, et al. Expression of CIITA-related MHCII molecules in tumors linked to prognosis in hepatocellular carcinoma. Int J Oncol. 2009;34:681. doi: 10.3892/ijo_00000194. [DOI] [PubMed] [Google Scholar]
- 19.Engers R, Ziegler S, Mueller M, et al. Prognostic relevance of increased Rac GTPase expression in prostate carcinomas. Endocrinol Relat Cancer. 2007;14:245. doi: 10.1677/ERC-06-0036. [DOI] [PubMed] [Google Scholar]
- 20.Sun L, Gong R, Wan B, et al. GADD45gamma, down-regulated in 65% hepatocellular carcinoma (HCC) from 23 Chinese patients, inhibits cell growth and induces cell cycle G2/M arrest for hepatoma Hep-G2 cell lines. Mol Biol Rep. 2003;30:249. doi: 10.1023/a:1026370726763. [DOI] [PubMed] [Google Scholar]
- 21.Jiang F, Wang Z. Gadd45gamma is androgen-responsive and growth-inhibitory in prostate cancer cells. Mol Cell Endocrinol. 2004;213:121. doi: 10.1016/j.mce.2003.10.050. [DOI] [PubMed] [Google Scholar]
- 22.Hsu YM, Zhang Y, You Y, et al. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat Immunol. 2007;8:198. doi: 10.1038/ni1426. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura S, Matsumoto T, Yada S, et al. Over-expression of caspase recruitment domain (CARD) membrane-associated guanylate kinase 1 (CARMA1) and CARD9 in primary gastric B-cell lymphoma. Cancer. 2005;104:1885. doi: 10.1002/cncr.21421. [DOI] [PubMed] [Google Scholar]
- 24.Leto TL, Lomax KJ, Volpp BD, et al. Cloning of a 67-kD neutrophil oxidase factor with similarity to a noncatalytic region of p60c-src. Science. 1990;248:727. doi: 10.1126/science.1692159. [DOI] [PubMed] [Google Scholar]
- 25.Lee JY, Park AK, Lee KM, et al. Candidate gene approach evaluates association between innate immunity genes and breast cancer risk in Korean women. Carcinogenesis. 2009;30:1528. doi: 10.1093/carcin/bgp084. [DOI] [PubMed] [Google Scholar]
- 26.Rajaraman P, Brenner AV, Butler MA, et al. Common variation in genes related to innate immunity and risk of adult glioma. Cancer Epidemiol Biomarkers Prev. 2009;18:1651. doi: 10.1158/1055-9965.EPI-08-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Bruin EC, van de Velde CJ, van Krieken JH, et al. Epithelial human leukocyte antigen-DR expression predicts reduced recurrence rates and prolonged survival in rectal cancer patients. Clin Cancer Res. 2008;14:1073. doi: 10.1158/1078-0432.CCR-07-1597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.