Abstract
Bioremediation is one of the commonly applied remediation strategies at sites contaminated with polycyclic aromatic hydrocarbons (PAHs). However, remediation goals are typically based on removal of the target contaminants rather than on broader measures related to health risks. We investigated changes in the toxicity and genotoxicity of PAH-contaminated soil from a former manufactured-gas plant site before and after two simulated bioremediation processes: a sequencing batch bioreactor system and a continuous-flow column system. Toxicity and genotoxicity of the residues from solvent extracts of the soil were determined by the chicken DT40 B-lymphocyte isogenic cell line and its DNA-repair-deficient mutants. Although both bioremediation processes significantly removed PAHs from the contaminated soil (bioreactor 69% removal; column 84% removal), bioreactor treatment resulted in an increase in toxicity and genotoxicity over the course of a treatment cycle, whereas long-term column treatment resulted in a decrease in toxicity and genotoxicity. However, when screening with a battery of DT40 mutants for genotoxicity profiling, we found that column treatment induced DNA damage types that were not observed in untreated soil. Toxicity and genotoxicity bioassays can supplement chemical analysis-based risk assessment for contaminated soil when evaluating the efficacy of bioremediation.
Keywords: bioremediation, toxicity, genotoxicity, soil, PAHs, DT40
INTRODUCTION
Polycyclic aromatic hydrocarbons (PAHs) are of human health concern due to their known or suspected genotoxic, mutagenic or carcinogenic effects1, 2, and they are a major pollutant class at thousands of contaminated sites in the U.S.A.3. Bioremediation is an established technology for cleanup of PAH-contaminated soils and sediments,4 but like most remedial technologies it is typically evaluated based on the removal of target pollutants. U.S. Environmental Protection Agency (USEPA) guidelines for risk assessments of PAH-contaminated soil generally focus only on 16 priority-pollutant PAHs.5 However, in most cases it remains unknown whether the removal of the regulated PAHs during bioremediation corresponds to a reduction in health risk.6 Significant amounts of other carcinogenic polyaromatic compounds, such as dibenzo[a,l]pyrene, are also found in PAH-contaminated soils,6-8 and whether all hazardous compounds degrade concomitantly with the 16 priority PAHs monitored at contaminated sites is unknown7. Incomplete metabolism of PAHs in contaminated soil can also yield by-products, such as oxy-PAHs, during bioremediation which can exhibit greater toxicity than the parent PAHs.6-9 Although the parent compounds and their metabolites all contribute to the total risk of contaminated sites, it is not practical to monitor hundreds of these compounds throughout the bioremediation process. More importantly, the identities of many hazardous compounds in PAH-contaminated sites are rarely known. Another limitation of risk assessment based solely on chemical analysis is that the toxicity of a mixture is assumed to be simply the sum of the expected effects from each component10, and it does not account for the possible synergistic or antagonistic interactions between mixture components1, 11.
Toxicity and genotoxicity bioassays such as the Ames test6, Mutatox™ assay12, SOS Chromotest13, micronucleus test14, and Comet assay15 have been used to assess the potential hazard and risk of contaminated soil before and after bioremediation. However, all these bioassays have their limitations. The Ames test, Mutatox™ assay and SOS Chromotest are all bacterial-based genotoxicity bioassays. Whether a bacterial test is a suitable model for eukaryotic systems is still questionable.16 The micronucleus test and Comet assay can be applied to eukaryotes, but they are limited to a small range of detectable DNA injuries17, 18.
The DT40 genotoxicity bioassay is a novel reverse genetic approach to determine genotoxicity of chemicals and permits characterization of modes of action.19 Recently, DT40 cells have been applied to measure genotoxicity in environmental samples.20 The DT40 bioassay uses the chicken DT40 B-lymphocyte isogenic cell line and its DNA-repair-deficient mutants, which are ideal for reverse genetic studies19, 21, 22. Their strong phenotypic resemblance to murine cells in DNA repair genes makes it relatively easy to translate DT40 assay results to human exposures to genotoxins.23 The DT40 bioassay can detect not only whether test materials induce DNA damage but also determine the DNA repair or cell-cycle checkpoint genes required for cell survival after DNA damage. Since each individual repair pathway processes a distinct set of DNA lesion types, differential cytotoxicity as a function of which DNA repair pathway has been knocked out provides insight into the profile of genotoxicity induced.16, 19
The objective of this study was to investigate effects of bioremediation on toxicity and genotoxicity of PAH-contaminated soil from a former MGP site. Two representative biological treatment processes were evaluated in the laboratory, including a sequencing batch bioreactor system (simulating ex situ treatment) and a continuous-flow column system (simulating in situ treatment). The DT40 parent cell line and fifteen DNA-repair-deficient mutants were employed to understand the genotoxicity potential and profile of the contaminated soil before and after biological treatment.
MATERIALS AND METHODS
Chemicals
PAH standards (EPA 610 PAH Mixture), benzo[a]pyrene diolepoxide (BPDE), methyl methanesulfonate (MMS), hydrogen peroxide (H2O2), dimethylsulfoxide (DMSO) and phosphate buffer solution (PBS) were obtained from Sigma-Aldrich (St. Louis, MO, U.S.A.). All solvents were high-performance liquid chromatography (HPLC) grade and were obtained from Fisher Scientific (Pittsburgh, PA, U.S.A.).
Soil, bioremediation processes, and sampling
Source soil used in this study was collected from a former MGP site in Salisbury, North Carolina, U.S.A., in the vicinity of the former tar well, 1.2 m below the surface. The soil was transferred by shovel to sample buckets and immediately transported to the laboratory, where it was blended and processed through a 10 mm sieve and stored at 4 °C prior to use. The sieved soil contained 66% sand, 28% silt, and 6% clay, with total organic matter of 16.6%. The total concentration of target PAHs (14 of the 16 priority PAHs, excluding acenaphthylene and indeno[1,2,3-cd]pyrene) was 556 ± 50 ng/mg (dry mass basis, wt/wt; individual PAH concentrations are shown in Supporting Information, Table S1).
Two bioremediation processes were employed to treat the source soil. One process involved treatment by a continuously stirred, semi-continuous, laboratory-scale aerobic bioreactor.24, 25 The bioreactor had a working volume of approximately 2 L, a solids concentration of 20% (wt/wt) and solids retention time of 35 d. Every week, 20% of the treated slurry was replaced with untreated source soil in a pH 7.5 buffer containing 5 mM phosphate and 5 mM ammonium nitrate. The other process was the 2.5-year-treatment by two continuous-flow columns (control column and biostimulated column), which were 110 cm long and 10.2 cm in diameter.26 Prior to column treatment, the source soil was mixed with sterile 40/50 grade silica sand (Unimin Corporation, Le Sueur, MN, U.S.A.) at a 50:50 ratio (dry weight) to maintain low-pressure flow during long-term column operation. The control column received simulated groundwater saturated with air. The biostimulated column received simulated groundwater saturated with pure oxygen and amended with ammonium nitrate and phosphate to yield final nitrogen and phosphorus concentrations of 1.0 mg/L and 0.3 mg/L, respectively. Detailed column design and operation are described elsewhere.26 Individual PAH concentrations of untreated bioreactor feed soil, bioreactor-treated soil, untreated column packing soil and column-treated soil are shown in Table S1.
To evaluate the temporal change in toxicity and genotoxicity in the bioreactor system, slurry from the bioreactor was sampled at five time intervals during each cycle: immediately after feeding (0 h), 8h, 1 d, 3d and 7 d after feeding. Soil from each column was sampled at the surface of the soil bed and at three sampling ports at 25-cm intervals along the column length (Ports A, B and C, respectively, in the direction of flow) after 2.5 years of continuous operation.
Sample extraction, PAH analysis and residue preparation
Soil samples were centrifuged at 3,500 rpm for 15 min, after which the supernatant was discarded. Triplicate aliquots of 3 g (wet weight) centrifuged soil were each extracted overnight twice, each time with a mixture of 10 mL acetone and 10 mL dichloromethane as described elsewhere27. Each extract was filtered through a 0.2 μm pore-size nylon filter (Millipore, Burlington, MA, U.S.A.) and was brought to a volume of 50 mL with acetonitrile. An aliquot of 1 mL of each extract was removed and analyzed by HPLC for PAH quantification27. An aliquot of 10 mL of each triplicate extract from the same soil sample was combined in a pre-weighed vial (total 30 mL) and evaporated to dryness with a mild flow of nitrogen. The mass of dry residue was determined gravimetrically. The residue was then re-dissolved with DMSO to 10,000 μg/mL and stored in liquid nitrogen before use.
DT40 DNA damage response analysis
DNA damage was determined by 24-well plate-based DNA damage response analysis using a DT40 isogenic cell line and its mutants knocked out in specific DNA repair and cell cycle pathways as described elsewhere19. Cells were exposed to the residue re-dissolved in DMSO that was serially diluted with PBS. The concentration of DMSO was adjusted so that the final concentration for all cell exposures was 0.3%. BPDE, MMS and H2O2 were used as positive controls (Table S2); while a vehicle blank (DMSO diluted in PBS) was used as negative control. Fifteen DT40 mutants were tested in this study, including base excision repair (BER)-deficient mutants (Polβ−/−, Fen1−/−), DNA damage sensor-deficient mutants (Rad9−/−, Rad17−/−), a nucleotide excision repair (NER)-deficient mutant (Xpa−/−), a mis-match repair (MMR)-deficient mutant (Msh2−/−), a nonhomologous end-joining (NHEJ)-deficient mutant (Ku70−/−), homologous recombination (HR)-deficient mutants (Rad54−/−, FancD2−/−), and tans-lesion synthesis (TLS)-deficient mutants (Rad18−/−, Rev1−/−, Rev3−/−, Polκ−/−, Polη−/−, Polθ−/−).
The DT40 system has not been tested previously for its ability to activate compounds that require metabolic activation before exerting a genotoxic effect. Therefore, we conducted a preliminary evaluation of the response of DT40 and the mutant Rev3−/− to exposure to benzo[a]pyrene (BaP). Details are provided in Supporting Information.
Data analysis
Statistical analyses were conducted with SPSS® (v16.0, SPSS Inc.). Student’s t-test and one-way analysis of variance (ANOVA) with Tukey’s test were employed to test for statistically significant differences between two groups and among multiple groups, respectively. Spearman test and partial correlation analysis with Spearman test were applied to investigate relation between LD50 and total PAH concentrations (CtPAHs) or total organic residue concentration (Cresidue). LD50 was calculated based on the dose-response relation and converted from residue dose to equivalent soil dose as described in Supporting Information.
RESULTS
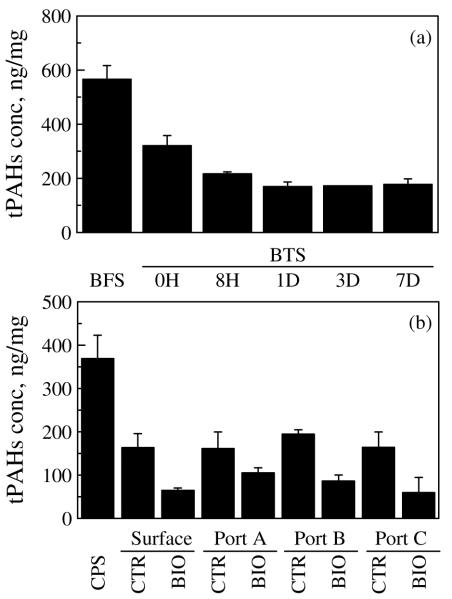
Both the bioreactor system and the column system significantly removed PAHs from the contaminated soil (Figure 1). For the bioreactor system, during each cycle (7 d), total PAH concentration of the treated soil decreased with time and approached a minimum 24 h after feeding (Figure 1a). For the column system, the PAH concentration of both control-column and biostimulated-column treated soil was significantly lower than that of the untreated column packing soil (Figure 1b). Overall, the bioreactor system had total PAH removal of 69% and the biostimulated column had total PAH removal of 84%.
Figure 1.
Total PAH concentration of soil before and after bioremediation. (a) Soils from five consecutive sampling times during 7-d cycle in the bioreactor treatment. (b) Soils from both the control column and biostimulated column at four sampling points along each column after 2.5-year column treatment. Values are mean ± SD of triplicates. BFS: untreated bioreactor feed soil; BTS: bioreactor treated soil; CPS: untreated column packing soil; CTR: control-column treated soil; BIO: biostimulated-column treated soil.
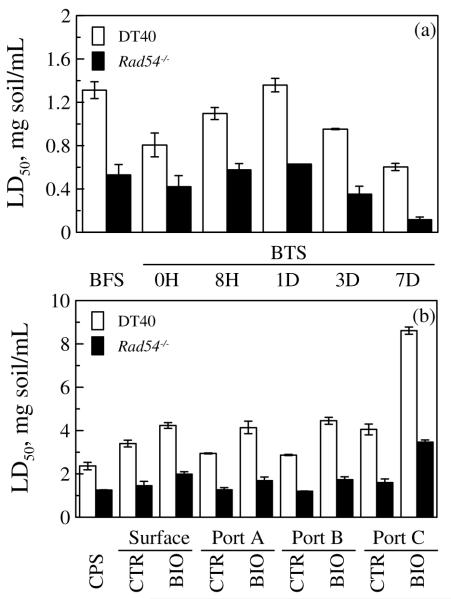
Based on its sensitivity to a broad range of DNA damage and its application in measuring genotoxicity in crude oil-contaminated sediments20, the Rad54−/− mutant was selected for detailed analysis of the effects of the two bioremediation processes on genotoxicity of the soil. For the bioreactor system, the LD50 of the bioreactor-treated soil for DT40 and the Rad54−/−mutant increased through Day 1, then decreased (Figure 2a). The LD50 of the bioreactor-treated soil for DT40 was significantly lower than that of the untreated bioreactor soil, except at Day 1; the LD50 of the bioreactor-treated soil for the Rad54−/− mutant was not significantly different from that of the untreated bioreactor soil through Day 1, but was significantly lower on Day 3 and Day 7 (Figure 2a). For the column system, the LD50 of both control-column and biostimulated-column treated soils for DT40 was significantly higher than that of the untreated column soil; the LD50 of the control-column treated soil for Rad54−/− was not significantly different from that of the untreated column soil, while the LD50 of the biostimulated-column treated soil for Rad54−/− was significantly higher than that of the untreated column soil (Figure 2b).
Figure 2.
LD of soil before and after bioremediation for parental DT40 cell line and its Rad54−/− mutant. (a) Soils from five consecutive sampling times during 7-d cycle in the bioreactor treatment. (b) Soils from both control column and biostimulated column at four sampling points along each column after 2.5-year column treatment. Values are mean ± SD of three separate experiments. Abbreviations are as defined in Figure 1.
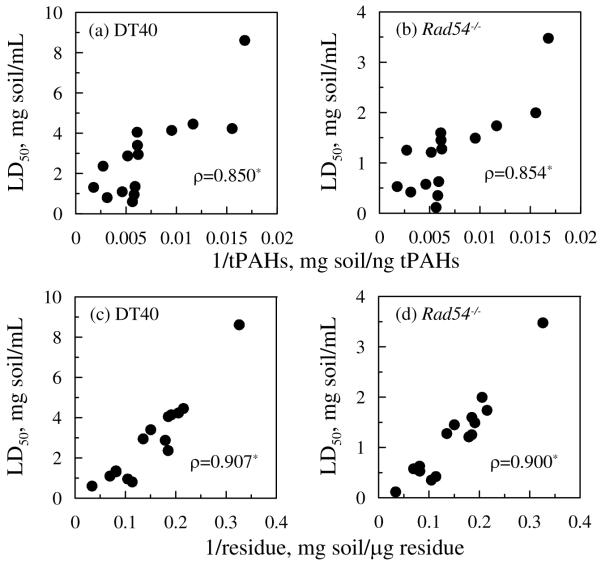
Inverse correlations between LD50 and CtPAHs or Cresidue were both highly positive and statistically significant (Figure 3). However, when 1/Cresidue was controlled, partial correlations between LD50 and 1/CtPAHs were not statistically significant; conversely, when 1/CCtPAHs was controlled, partial correlations between LD50 and 1/Cresidue were highly positive and statistically significant (Table S3).
Figure 3.
Inverse correlations between LD50 and concentrations of tPAH for parental DT40 cell line (a) and its Rad54−/− mutant (b), and between LD50 and concentrations of total residue for parental DT40 cell line (c) and its Rad54−/− mutant (d). Each data point represents the mean for each soil sample (total 15 samples) including untreated column packing soil, all sampling points along each column, untreated bioreactor feed soil, and all sampling events for bioreactor-treated soil during the 7-d. Asterisks indicate the correlation is statistically significant (p< 0.05).
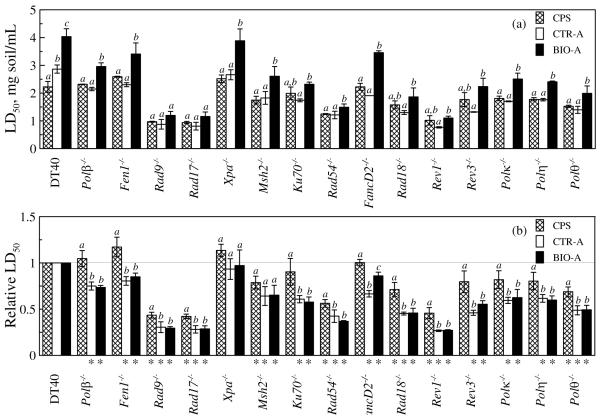
The column system soils were also screened with a battery of DT40 cell lines for genotoxicity profiling (Figure 4a). There were no significant differences in LD50 between control-column treated soil and untreated column packing soil, except for the parent DT40 cells. In general, the LD50 of biostimulated-column treated soil was significantly higher than the corresponding LD50 of untreated column packing soil, except for Rad9−/−, Rad17−/−, Ku70−/−, Rad18−/−, Rev1−/− and Rev3−/−. The LD50 of biostimulated-column treated soil was significantly higher than that of the control-column treated soil, except for Rad9−/− and Rad17−/−.
Figure 4.
LD50 (a) and relative LD50 (b) of soil before and after 2.5 year column treatment in the test with a battery of DT40 cell lines. Values are mean ± SD of three separate experiments. Different letters are assigned to conditions for which there was a significant difference (p<0.05). Asterisks indicate values significantly less than 1 (p< 0.05). CPS: untreated column packing soil; CTR-A: control-column treated soil at Port A; BIO-A: biostimulated-column treated soil at Port A.
For a quantitative comparison, we also calculated the relative LD50 of column-system soils (Figure 4b) as described by Ji et al.28, where the relative LD50 of the parental DT40 cell was defined as 1. If the relative LD50 of a mutant to a sample is significantly less than 1, that mutant is defined as sensitive to that sample. Seven mutants were sensitive to both untreated and treated columns soils, including Rad9−/−, Rad17−/−, Msh2−/−, Rad54−/−, Rad18−/−, Rev1−/− and Polθ−/−. Seven mutants were sensitive only to column-treated soils but not to untreated column packing soil, including Polβ−/−, Fen1−/−, Ku70−/−, FancD2−/−, Rev3−/−, Polκ−/− and Polη−/−. Xpa−/− was not sensitive to either untreated column packing soil or column-treated soils.
DISCUSSION
Effects of bioremediation on toxicity and genotoxicity
Bioremediation is an established technology to remove PAHs from contaminated soil and sediment.4 However, some researchers have advised caution about bioremediation, since the removal of the monitored PAHs during bioremediation of contaminated soil or sediment might not correspond to a reduction in health risk.6, 9 In some studies toxicity decreased as treatment progressed,12-14, 29, 30 while in other studies there was either no reduction or even a substantial increase in toxicity following bioremediation.15, 31-33 Increases in toxicity might be caused by formation of toxic metabolites or increased bioavailability of native toxins over the course of bioremediation.31
Our study confirmed that bioremediation reduced PAH levels in the contaminated soil (Figure 1), but the effect of bioremediation on toxicity is complicated. Generally, we observed increased toxicity (decreased LD50) in the bioreactor system but decreased toxicity (increased LD50) in the column system after bioremediation (Figure 2). Remediation methods and the specific ways they are implemented can substantially influence the community of PAH-degrading microorganisms in contaminated soil, thus influencing the collective balance between complete and incomplete metabolism of PAHs by these organisms and, therefore, potential variation in toxicity and genotoxicity. Longer periods of bioremediation, such as that used in the column systems, may be required to significantly reduce the genotoxic hazard of a contaminated soil.6 Hughes et al.32 also found variations of genotoxicity changes in creosote-contaminated soil before and after four bioremediation processes. However, they could not determine whether observed increases in genotoxicity were due to the processes themselves or to the amendments added to the soil.32
Temporal change in toxicity and genotoxicity in the bioreactor system
A temporal change in toxicity was observed in the bioreactor system following a feeding event (Figure 2a). Toxicity to both the DT40 parent cell line and its Rad54−/− mutant initially decreased (increased LD50), then increased (decreased LD50) during the feeding cycle. Other researchers have also observed temporal changes in the genotoxicity of PAH-contaminated soils undergoing bioremediation.6, 34 The somewhat cyclical nature of toxicity and genotoxicity may suggest the formation, and subsequent degradation, of toxic compounds,6 although if that were the case with our bioreactor system then we would have observed a temporal trend opposite to that shown in Figure 2a. Sampling of the column system was not designed to evaluate temporal trends in toxicity and genotoxicity, so only the long-term treatment effects were observed.
The source of toxicity
Compounds responsible for toxicity and genotoxicity of PAH-contaminated soil other than the USEPA 16 priority PAHs might not degrade concomitantly with PAHs during bioremediation.6-8 Moreover, in a complex system such as contaminated soil, some transformations that do not lead to complete metabolism of the parent compound are inevitable. Although the correlation between LD50 and total PAH concentration was significant (Figure 3a and 3b), the partial correlation between LD50 and total PAH concentration was poor and insignificant, when controlling for the effects of total organic residue. We conclude that the total organic compounds present in soil extracts are responsible for the toxicity and genotoxicity of PAH-contaminated soil undergoing bioremediation. Further research is needed to identify the toxic and genotoxic compounds.
Genotoxicity profiling
In order to understand the effects of bioremediation on the genotoxicity potential of PAH-contaminated soil in the column system, we screened 15 DNA-repair-deficient DT40 mutants. When compared to the untreated soil, the control column did not reduce toxicity except for the parental DT40 cell line; in contrast, the biostimulated column significantly reduced toxicity for both the parental DT40 cell line and most of the mutants (Figure 4a). We also observed that the genotoxicity profiles (relative LD50) of control-column treated soil and biostimulated-column treated soil were similar but both were different from that of the untreated soil (Figure 4b). Several mutants were sensitive to treated soil but not untreated soil, including Polβ−/−, Fen1−/−, Ku70−/−, FancD2−/−, Rev3−/−, Polκ−/− and Polη−/−, indicating that more types of DNA damage were induced by remediation. This finding suggests that genotoxic compounds were generated during bioremediation, although their concentrations must have been low enough not to lead to an overall increase in genotoxicity per unit soil mass.
RAD9 and RAD17 are intra-S-phase DNA damage checkpoint control proteins and are in the cellular response to stalled DNA replication.35 Both Rad9−/− and Rad17−/− were sensitive to treated and untreated soil, strongly suggesting that bioremediation could not eliminate genotoxic compounds in PAH-contaminated soil that can induce DNA replication block. RAD54 is a DNA repair and HR protein.36 Rad54−/− was sensitive to both treated and untreated soil, indicating that the soil both before and after bioremediation could induce DNA double-strand breaks or DNA damage leading to replication blockage36. NER mediated by the Xpa gene is thought to be involved in the elimination of bulky DNA adducts37. However, Xpa−/− was not sensitive to either treated soil or untreated soil, indicating that the potential for formation of bulky DNA adducts may be negligible before and after bioremediation. Metabolic activation of PAHs may lead to bulky DNA adducts2, but the capacity of DT40 cells for metabolic activation has not been reported before. Our preliminary results indicate that Rev3−/− was sensitive to BaP (Figure S1), indicating that DT40 cells may have a metabolic activation system for PAHs. Regardless, this study was not intended to elucidate the genotoxicity of PAHs per se, but to evaluate the changes in genotoxicity of the combination of soil contaminants as a result of bioremediation.
BER plays an essential role in protecting cells from DNA damage caused by hydrolysis, oxidative agents and alkylating agents.38 We observed that BER-deficient mutants (Polβ−/−, Fen1−/−) were sensitive to treated soils but not to untreated soil, indicating that bioremediation generated genotoxic compounds that could induce oxidative stress, unstable depurinating DNA adducts or alkylation DNA damage39, 40. Certain TLS-deficient mutants (Rad18−/−, Rev1−/− and Polθ−/−) were sensitive to untreated soil, indicating that unstable depurinating DNA adducts and alkylated DNA bases could also be generated by exposure to untreated soil. While oxidative DNA damage is thought to be repaired by BER, it has been proposed that DNA lesions caused by oxidative stress could also be repaired by NHEJ involving protein KU7041. Ku70−/− was sensitive to treated soil but not to untreated soil, further indicating the likelihood that bioremediation generated genotoxic compounds causing oxidative stress, which might be attributed to the formation of oxy-PAHs during incomplete biodegradation42.
Value of genotoxicity testing
Although bioremediation is an effective tool to remove PAHs from contaminated soil, its effects on toxicity and genotoxicity of PAH-contaminated soil need thorough study if the ultimate goal of remediation is to reduce human health risk. This study demonstrated that different bioremediation strategies could lead to different outcomes of toxicity and genotoxicity for PAH-contaminated soil. We also observed enhanced oxidative DNA damage caused by the soil after bioremediation in the column system. Overall, toxicity and genotoxicity bioassays can be an effective supplement to chemical analysis-based risk assessment for contaminated soil. Further research is still needed to isolate, characterize, and quantify the toxic and genotoxic compounds in the contaminated soil as remediation progresses.
Supplementary Material
ACKNOWLEDGEMENT
We thank Hiroshi Arakawa, Jean-Marie Buerstedde, and Shunichi Takeda for providing various DT40 mutants. We also thank Wei Sun for assistance in statistical analysis. This work was supported by the U.S. National Institute of Environmental Health Sciences’ Superfund Research Program (grant 5 P42ES005948).
Footnotes
SUPPORTING INFORMATION AVAILABLE
Table of concentrations of individual PAHs in the soil before and after bioremediation; Table of LD50 for BPDE, MMS and H2O2 as positive control; LD50 calculation method; Table of partial correlation coefficients and corresponding p-values among LD50, 1/CtPAHs and 1/Cresidue; Test of BaP metabolic activation by DT40 cell lines. This information is available free of charge via the Internet at http://pubs.acs.org/.
REFERENCES
- (1).White PA. The genotoxicity of priority polycyclic aromatic hydrocarbons in complex mixtures. Mutat. Res. 2002;515(1-2):85–98. doi: 10.1016/s1383-5718(02)00017-7. [DOI] [PubMed] [Google Scholar]
- (2).Xue WL, Warshawsky D. Metabolic activation of polycyclic and heterocyclic aromatic hydrocarbons and DNA damage: A review. Toxicol. Appl. Pharmacol. 2005;206(1):73–93. doi: 10.1016/j.taap.2004.11.006. [DOI] [PubMed] [Google Scholar]
- (3).USEPA . Cleaning up the nation’s waste sites: Markets and technology trends. 2004 Edition. United States Environmental Protection Agency; Washington, DC: 2004. EPA/542/R/04/015. [Google Scholar]
- (4).USEPA . Treatment technologies for site cleanup: Annual status report. United States Environmental Protection Agency; Washington, DC: 2007. EPA/542/R/07/012. [Google Scholar]
- (5).USEPA . Supplementary guidance for conducting health risk assessment of chemical mixtures. United States Environmental Protection Agency; Washington, DC: 2000. EPA/630/R/00/002. [Google Scholar]
- (6).Lemieux CL, Lynes KD, White PA, Lundstedt S, Oberg L, Lambert IB. Mutagenicity of an aged gasworks soil during bioslurry treatment. Environ. Mol. Mutagen. 2009;50(5):404–412. doi: 10.1002/em.20473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Lundstedt S, Haglund P, Oberg L. Degradation and formation of polycyclic aromatic compounds during bioslurry treatment of an aged gasworks soil. Environ. Toxicol. Chem. 2003;22(7):1413–1420. [PubMed] [Google Scholar]
- (8).Lemieux CL, Lambert AB, Lundstedt S, Tysklind M, White PA. Mutagenic hazards of complex polycyclic aromatic hydrocarbon mixtures in contaminated soil. Environ. Toxicol. Chem. 2008;27(4):978–990. doi: 10.1897/07-157.1. [DOI] [PubMed] [Google Scholar]
- (9).Lundstedt S, White PA, Lemieux CL, Lynes KD, Lambert LB, Oberg L, Haglund P, Tysklind M. Sources, fate, and toxic hazards of oxygenated polycyclic aromatic hydrocarbons (PAHs) at PAH-contaminated sites. Ambio. 2007;36(6):475–485. doi: 10.1579/0044-7447(2007)36[475:sfatho]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- (10).USEPA . Risk assessment guidance for superfund volume I: Human health evaluation manual (Part E, Supplemental guidance for dermal risk assessment) United States Environmental Protection Agency; Washington, DC: 2004. EPA/540/R/99/005. [Google Scholar]
- (11).Park J, Ball LM, Richardson SD, Zhu HB, Aitken MD. Oxidative mutagenicity of polar fractions from polycyclic aromatic hydrocarbons-contaminated soils. Environ. Toxicol. Chem. 2008;27(11):2207–2215. doi: 10.1897/07-572.1. [DOI] [PubMed] [Google Scholar]
- (12).Mendonca E, Picado A. Ecotoxicological monitoring of remediation in a coke oven soil. Environ. Toxicol. 2002;17(1):74–79. doi: 10.1002/tox.10034. [DOI] [PubMed] [Google Scholar]
- (13).Haeseler F, Blanchet D, Druelle V, Werner P, Vandecasteele JP. Ecotoxicological assessment of soils of former manufactured gas plant sites: Bioremediation potential and pollutant mobility. Environ. Sci. Technol. 1999;33(24):4379–4384. [Google Scholar]
- (14).Baud-Grasset S, Baud-grasset F, Bifulco JM, Meier JR, Ma TH. Reduction of genotoxicity of a creosote-contaminated soil after fungal treatment determined by the Tradescantia-micronucleus test. Mutat. Res. 1993;303(2):77–82. doi: 10.1016/0165-7992(93)90098-g. [DOI] [PubMed] [Google Scholar]
- (15).Gandolfi I, Sicolo M, Franzetti A, Fontanarosa E, Santagostino A, Bestetti G. Influence of compost amendment on microbial community and ecotoxicity of hydrocarbon-contaminated soils. Bioresour. Technol. 2010;101(2):568–575. doi: 10.1016/j.biortech.2009.08.095. [DOI] [PubMed] [Google Scholar]
- (16).Evans TJ, Yamamoto KN, Hirota K, Takeda S. Mutant cells defective in DNA repair pathways provide a sensitive high-throughput assay for genotoxicity. DNA Repair. 2010;9(12):1292–1298. doi: 10.1016/j.dnarep.2010.09.017. [DOI] [PubMed] [Google Scholar]
- (17).Collins AR. The comet assay for DNA damage and repair: Principles, applications, and limitations. Mol. Biotechnol. 2004;26(3):249–261. doi: 10.1385/MB:26:3:249. [DOI] [PubMed] [Google Scholar]
- (18).Barile FA. Introduction to in vitro Cytotoxicology: Mechanisms and Methods. CRC Press; Boca Raton, FL: 1994. [Google Scholar]
- (19).Ridpath JR, Takeda S, Swenberg JA, Nakamura J. Convenient, multi-well plate-based DNA damage response analysis using DT40 mutants is applicable to a high-throughput genotoxicity assay with characterization of modes of action. Environ. Mol. Mutagen. 2011;52(2):153–160. doi: 10.1002/em.20595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Ji K, Seo J, Liu X, Lee S, Lee W, Park J, Khim JS, Hong S, Choi Y, Shim WJ, Takeda S, Giesy JP, Choi K. Genotoxicity and endocrine-disruption potentials of sediment near an oil spill site: Two years after the Hebei Spirit oil spill. Environ. Sci. Technol. 2011;45(17):7481–7488. doi: 10.1021/es200724x. [DOI] [PubMed] [Google Scholar]
- (21).Ulrich E, Boehmelt G, Bird A, Beug H. Immortalization of conditionally transformed chicken cells: Loss of normal p53 expression in an early step that is independent of cell transformation. Genes Dev. 1992;6(5):876–887. doi: 10.1101/gad.6.5.876. [DOI] [PubMed] [Google Scholar]
- (22).Yamazoe M, Sonoda E, Hochegger H, Takeda S. Reverse genetic studies of the DNA damage response in the chicken B lymphocyte line DT40. DNA Repair. 2004;3(8-9):1175–1185. doi: 10.1016/j.dnarep.2004.03.039. [DOI] [PubMed] [Google Scholar]
- (23).Sonoda E, Morrison C, Yamashita YM, Takata M, Takeda S. Reverse genetic studies of homologous DNA recombination using the chicken B-lymphocyte line, DT40. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2001;356(1405):111–117. doi: 10.1098/rstb.2000.0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Singleton DR, Powell SN, Sangaiah R, Gold A, Ball LM, Aitken MD. Stable-isotope probing of bacteria capable of degrading salicylate, naphthalene, or phenanthrene in a Bioreactor treating contaminated soil. Appl. Environ. Microbiol. 2005;71(3):1202–1209. doi: 10.1128/AEM.71.3.1202-1209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Singleton DR, Richardson SD, Aitken MD. Pyrosequence analysis of bacterial communities in aerobic bioreactors treating polycyclic aromatic hydrocarbon-contaminated soil. Biodegradation. 2011;22(6):1061–1073. doi: 10.1007/s10532-011-9463-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Richardson SD, Aitken MD. Desorption and bioavailability of PAHs in contaminated soil subjected to long-term in situ biostimulation. Environ. Toxicol. Chem. 2011;30(12):2674–2681. doi: 10.1002/etc.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Richardson SD, Lebron BL, Miller CT, Aitken MD. Recovery of phenanthrene-degrading bacteria after simulated in situ persulfate oxidation in contaminated soil. Environ. Sci. Technol. 2011;45(2):719–725. doi: 10.1021/es102420r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Ji K, Kogame T, Choi K, Wang X, Lee J, Taniguchi Y, Takeda S. A novel approach using DNA-repair-deficient chicken DT40 cell lines for screening and characterizing the genotoxicity of environmental contaminants. Environ. Health Perspect. 2009;117(11):1737–1744. doi: 10.1289/ehp.0900842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Sasek V, Bhatt M, Cajthaml T, Malachova K, Lednicka D. Compost-mediated removal of polycyclic aromatic hydrocarbons from contaminated soil. Arch. Environ. Contam. Toxicol. 2003;44(3):336–342. doi: 10.1007/s00244-002-2037-y. [DOI] [PubMed] [Google Scholar]
- (30).Sayles GD, Acheson CM, Kupferle MJ, Shan Y, Zhou Q, Meier JR, Chang L, Brenner RC. Land treatment of PAH contaminated soil: Performance measured by chemical and toxicity assays. Environ. Sci. Technol. 1999;33(23):4310–4317. [Google Scholar]
- (31).Andersson E, Rotander A, von Kronhelm T, Berggren A, Ivarsson P, Hollert H, Engwall M. AhR agonist and genotoxicant bioavailability in a PAH-contaminated soil undergoing biological treatment. Environ. Sci. Pollut. Res. Int. 2009;16(5):521–530. doi: 10.1007/s11356-009-0121-9. [DOI] [PubMed] [Google Scholar]
- (32).Hughes TJ, Claxton LD, Brooks L, Warren S, Brenner R, Kremer F. Genotoxicity of bioremediated soils from the Reilly Tar site, St. Louis Park, Minnesota. Environ. Health Perspect. 1998;106(6):1427–1433. doi: 10.1289/ehp.98106s61427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Gillespie AM, Wang S, McDonald T, Garcia SG, Cosgriff D, He L-Y, Huebner H, Donnelly KC. Genotoxicity assessment of wood-preserving waste-contaminated soil undergoing bioremediation. Biorem. J. 2007;11(4):171–182. [Google Scholar]
- (34).Belkin S, Stieber M, Tiehm A, Frimmel FH, Abeliovich A, Werner P, Ulitzur S. Toxicity and genotoxicity enhancement during polycyclic aromatic hydrocarbons biodegradation. Environ. Toxicol. Water Qual. 1994;9(4):303–309. [Google Scholar]
- (35).Kobayashi M, Hirano A, Kumano T, Xiang SL, Mihara K, Haseda Y, Matsui O, Shimizu H, Yamamoto K. Critical role for chicken Rad17 and Rad9 in the cellular response to DNA damage and stalled DNA replication. Genes Cells. 2004;9(4):291–303. doi: 10.1111/j.1356-9597.2004.00728.x. [DOI] [PubMed] [Google Scholar]
- (36).Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair. 2006;5(9-10):1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- (37).van Gent DC, Hoeijmakers JHJ, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2001;2(3):196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- (38).Yoshimura M, Kohzaki M, Nakamura J, Asagoshi K, Sonoda E, Hou E, Prasad R, Wilson SH, Tano K, Yasui A, Lan L, Seki M, Wood RD, Arakawa H, Buerstedde JM, Hochegger H, Okada T, Hiraoka M, Takeda S. Vertebrate POLQ and POL beta cooperate in base excision repair of oxidative DNA damage. Mol. Cell. 2006;24(1):115–125. doi: 10.1016/j.molcel.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Asagoshi K, Tano K, Chastain PD, Adachi N, Sonoda E, Kikuchi K, Koyama H, Nagata K, Kaufman DG, Takeda S, Wilson SH, Watanabe M, Swenberg JA, Nakamura J. FEN1 Functions in Long Patch Base Excision Repair Under Conditions of Oxidative Stress in Vertebrate Cells. Mol. Cancer Res. 2010;8(2):204–215. doi: 10.1158/1541-7786.MCR-09-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Tano K, Nakamura J, Asagoshi K, Arakawa H, Sonoda E, Braithwaite EK, Prasad R, Buerstedde JM, Takeda S, Watanabe M, Wilson SH. Interplay between DNA polymerases beta and lambda in repair of oxidation DNA damage in chicken DT40 cells. DNA Repair. 2007;6(6):869–875. doi: 10.1016/j.dnarep.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Narasimhaiah R, Tuchman A, Lin SL, Naegele JR. Oxidative damage and defective DNA repair is linked to apoptosis of migrating neurons and progenitors during cerebral cortex development in Ku70-deficient mice. Cereb. Cortex. 2005;15(6):696–707. doi: 10.1093/cercor/bhh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Zielinska-Park J, Nakamura J, Swenberg JA, Aitken MD. Aldehydic DNA lesions in calf thymus DNA and HeLa S3 cells produced by bacterial quinone metabolites of fluoranthene and pyrene. Carcinogenesis. 2004;25(9):1727–1733. doi: 10.1093/carcin/bgh174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.