Abstract
Apolipoprotein E (apoE) is an endogenous negative regulator of airway hyperreactivity (AHR) and mucous cell metaplasia in experimental models of house dust mite (HDM)-induced airway disease. The gene encoding human apoE is polymorphic, with three common alleles (ε2, ε3, and ε4) reflecting single amino acid substitutions at amino acids 112 and 158. The objective of this study was to assess whether the human apoE alleles modify airway responses to repeated nasal HDM challenges. Mice expressing the human apoE ε2 (huApoE2), ε3 (huApoE3), or ε4 (huApoE4) alleles received nasal HDM challenges, and airway responses were compared with mice expressing the endogenous murine apoE gene (muApoE). huApoE3 mice displayed significant reductions in AHR, mucous cell metaplasia, and airway inflammation compared with muApoE mice. The attenuated severity of airway inflammation in huApoE3 mice was associated with reductions in lung mRNA levels of Th2 and Th17 cytokines, as well as chemokines (CCL7, CCL11, CCL24). huApoE4 mice had an intermediate phenotype, with attenuated AHR and IgE production, compared with muApoE mice, whereas airway inflammation and mucous cell metaplasia were not reduced. In contrast, HDM-induced airway responses were not modified in mice expressing the huApoE2 allele. We conclude that the polymorphic huApoE alleles differentially modulate HDM-induced airway disease, which can be stratified, in rank order of increasing disease severity, ε3 < ε4 < ε2. These results raise the possibility that the polymorphic apoE alleles may modify disease severity in human asthma.
Keywords: asthma
asthma is a common disorder that affects 300 million individuals worldwide and results in 250,000 deaths each year (38a). The pathogenesis of asthma is complex and is manifested by airway inflammation, airway hyperreactivity (AHR), and airway remodeling responses, such as mucous cell metaplasia, epithelial desquamation, subepithelial fibrosis, mucosal edema, and airway smooth muscle hypertrophy and hyperplasia (2, 5, 12, 13). Key mediators of asthma pathogenesis include cytokines produced by Th2 and Th17 cells, chemokines, lipid mediators, and reactive oxygen species (3). Recently, an apolipoprotein E (apoE)-low-density lipoprotein receptor (LDLR) pathway has been found to play an unexpected role in modifying the pathogenesis of experimental house dust mite (HDM)-induced airway disease by functioning as an endogenous negative regulator of AHR and mucous cell metaplasia (39). Genome-wide profiling of the murine lung transcriptome identified apoE as a gene with persistently upregulated expression despite corticosteroid treatment following HDM challenge. apoE−/− mice were found to have augmented AHR and mucous cell metaplasia, which could be rescued by administration of an apoE mimetic peptide corresponding to the LDLR binding domain of apoE. ApoE, which is expressed by CD68+ alveolar macrophages, was shown to induce its effects through the LDLR, which is expressed by ciliated bronchial epithelial cells. Consistent with this, LDLR−/− mice also displayed an asthma phenotype of enhanced AHR and mucous cell metaplasia but were resistant to rescue by the apoE mimetic peptide.
The gene encoding human apoE (huApoE) is polymorphic, with three common alleles that have substitutions at amino acids 112 and 158 (24, 42). ApoE ε3, the most common allele, encodes for a cysteine at amino acid 112 and an arginine at amino acid 158. In contrast, apoE ε2 encodes for cysteines at both sites, whereas apoE ε4 encodes for arginines at both sites. The apoE alleles alter the conformation of the apoE protein and thereby modify its interaction with the LDLR. ApoE2 has markedly reduced binding to the LDLR, with less than 2% LDLR binding compared with apoE3, whereas apoE3 and apoE4 both avidly bind the LDLR (24, 42).
Because the apoE2 protein is selectively deficient in binding to the LDLR, we hypothesized that carriers of the apoE ε2 allele would have more severe manifestations of asthma compared with carriers of the apoE ε3 and ε4 alleles, which avidly bind the LDLR (24). To assess whether the polymorphic apoE alleles modify the severity and pathogenesis of asthma, we administered repeated nasal HDM challenges to knockin mice that have been genetically engineered to express the huApoE ε2, ε3, or ε4 alleles, which were compared with wild-type mice expressing the murine apoE (muApoE) allele (19, 32, 33). Here, we show that the polymorphic huApoE alleles differentially modulate airway responses to repeated nasal HDM challenges, which can be stratified, in rank order of increasing disease severity, ε3 < ε4 < ε2. These results suggest that the huApoE alleles might modify disease severity in asthma.
MATERIALS AND METHODS
Nasal HDM challenge model.
Female wild-type C57BL/6 mice, as well as homozygous female humanized apoE ε2, ε3 and ε 4 knockin mice [B6.129P2-ApoEtm1 (APOE*2)Mae N9, B6.129P2-ApoEtm2 (APOE*3)Mae N8, B6.129P2-ApoEtm3 (APOE*4)Mae N8], which had been backcrossed at least eight times onto a C57BL/6 background, were obtained from Taconic (Hudson, NY). The humanized apoE ε2, ε3, and ε4 knockin mice were created by replacing exons 2–4 of the muApoE gene with the corresponding exons of the huApoE ε2, ε3, and ε4 alleles to generate a chimeric locus that is regulated by murine regulatory elements and murine exon 1 but encodes huApoE proteins. Thus the expression of the humanized apoE isoforms remains under the control of the endogenous murine promoter (19, 32, 33). Airway disease was induced by nasal inhalation of Dermatophagoides pteronyssinus extract (Greer, Lenoir, NC), 25 μg of protein in 10 μl of saline, for 5 days each week, for 5 consecutive weeks, as previously described (16). The HDM extract contained 0.05 U per μl of endotoxin. Control mice received nasal inhalation of 10 μl of saline as a comparator. Experimental protocols were approved by the Animal Care and Use Committee of the National Heart, Lung and Blood Institute. Two independent experiments were performed with ten mice per group.
Airway hyperreactivity.
Airway resistance was measured in anesthetized mice using an Elan RC Fine Pointe system (Buxco, Wilmington, North Carolina). Following cannulation of the trachea with a 19-gauge beveled metal catheter, mice received mechanical ventilation with a constant inspiratory flow. Mice then received increasing doses of nebulized methacholine or PBS. Airway resistance was recorded at 10-s intervals for 3 min, and average values are presented as cm H2O/ml per second.
Bronchoalveolar lavage fluid cells.
Bronchoalveolar lavage was performed three times with 0.5 ml of PBS. Red blood cells were lysed with ACK buffer for 2 min at 4°C, and cells were resuspended in 0.3 ml RPMI-1640 containing 10% FBS. Total cells were counted using a hemocytometer. Differential cell counts were performed on Diff-Quik-stained cytospin slides (Siemens, Deerfield, IL).
Lung histopathological examination.
Lungs were inflated to a pressure of 25 cm H20 before fixation in 10% formalin for 24 h, dehydrated through gradient ethanol, and embedded in paraffin before cutting of sagittal sections at a thickness of 5 μm. Sections were stained with hematoxylin and eosin or periodic acid-Schiff (PAS). Quantification of mucous cell metaplasia was performed as previously described (39). The number of airways containing PAS-positive cells in all the airways present [large (conducting), medium (central), and small (distal)] within representative lung sections was counted. Mucous cell metaplasia is presented as the percentage of airways containing PAS-positive cells. The number of airways inspected in each animal is also presented.
Quantitative RT-PCR.
Lungs were minced into 1-mm pieces and stored in RNAlater (Ambion, Austin, TX) at −80°C. Total RNA was subsequently isolated using the mirVana kit (Ambion), and contaminating DNA was removed by treatment with 10 U of DNase I per 20 μg of RNA. RNA was then reverse transcribed into cDNA using the high-capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA). cDNA was amplified using TaqMan Universal PCR Master Mix, FAM dye-labeled Taqman MGB probes, and a 7500 Real-Time PCR System running Sequence Detector version 2.1 software. apoE mRNA levels in muApoE mice were determined using primers that recognize muApoE, whereas apoE mRNA levels in huApoE mice were determined using primers that recognize huApoE. Gene expression was quantified relative to expression of 18S rRNA using one of the saline-challenged muApoE mice as the calibrator for all other groups to calculate the difference in Ct values (ΔΔCt). Data are presented as relative mRNA expression.
Measurement of serum IgE.
Total serum IgE was measured with an OptEIA kit (BD Biosciences Pharmingen, San Diego, CA).
Western blotting.
Mouse lungs were lysed in Tissue Protein Extraction Reagent with Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific, Waltham, MA), and protein concentrations were determined using a BCA kit (Thermo Scientific). Lung proteins (24 μg) were separated by SDS-PAGE using 10% Bis-Tris Nupage gels (Invitrogen), electroblotted onto nitrocellulose membranes, and reacted with primary antibodies for 2 h at 4°C. The goat polyclonal antibody that reacts with huApoE was from Millipore (Billerica, MA), whereas the mouse monoclonal anti-β-actin antibody was from Sigma-Aldrich (St. Louis, MO). Blots were washed five times with PBS plus 0.2% Tween 20 and incubated with appropriate secondary antibodies for 1 h. The donkey anti-rabbit and sheep anti-mouse horseradish peroxidase-conjugated antibodies were from GE Healthcare Lifesciences (Piscataway, NJ). Following repeat washing, signals were detected using a chemiluminescent substrate (Super Signal; Pierce, Rockford, IL). Densitometry was performed using ImageJ software (NIH, Bethesda, MD).
Statistics.
Results are presented as means ± SE. A one-way ANOVA with a Bonferroni's multiple-comparison test (to correct for 3 comparisons to the muApoE group) was utilized for all analyses with the exception of airway hyperreactivity experiments, in which case a two-way ANOVA with a Bonferroni posttest test was used for repeated measures of airway resistance in each animal to increasing doses of methacholine. A P value >0.05 was considered significant. Statistical analyses were performed with GraphPad Prism version 5.0a.
RESULTS
Humanized apoE ε3 and ε4 mice are protected from HDM-induced increases in airway hyperreactivity.
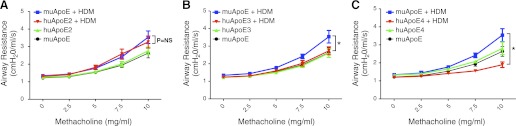
Humanized apoE knockin mice that express huApoE alleles ε2 (huApoE2), ε3 (huApoE3), and ε4 (huApoE4) were challenged with HDM or saline (Control) to assess whether allelic differences modify AHR responses compared with wild-type C57BL/6 mice that express the muApoE gene. As shown in Fig. 1, there was no difference in basal levels of AHR in saline-challenged wild-type mice expressing the muApoE allele and mice expressing the huApoE2, huApoE3, and huApoE4 alleles. Airway resistance to increasing doses of methacholine was similar between muApoE and huApoE2 mice. In contrast, huApoE3 and huApoE4 mice did not develop HDM-induced increases in AHR. Furthermore, airway resistance in HDM-challenged huApoE4 mice was significantly reduced compared with saline-challenged huApoE4 mice. These data suggest that the huApoE3 and E4 alleles confer a protective phenotype against HDM-induced AHR, whereas the huApoE2 allele does not.
Fig. 1.
Humanized apolipoprotein E (apoE) ε3 and ε4 mice are protected from house dust mite (HDM)-induced airway hyperreactivity. Airway hyperreactivity in wild-type C57BL/6 mice (murine apoE, muApoE) was compared with mice expressing human apoE2 (huApoE2) (A), apoE3 (huApoE3) (B), or apoE4 (huApoE4) (C) proteins, following daily nasal challenge with either saline (control) or HDM for 5 days per week for 5 wk. Airway resistance (cm H20/ml per s) was measured following nebulization of increasing doses of methacholine (n = 8–10 mice, *P < 0.001 at the 10 mg/ml dose of methacholine for huApoE + HDM vs. muApoE + HDM). Results are representative of 2 independent experiments.
HDM-induced mucous cell metaplasia is attenuated in humanized apoE ε3 mice.
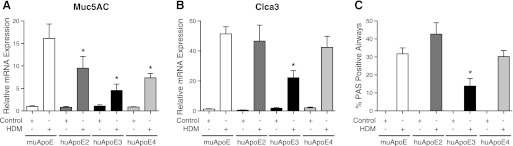
In addition to its role as a negative regulator of AHR, apoE also attenuates mucous cell metaplasia (39). Therefore, experiments were conducted to assess whether allelic differences in huApoE modify HDM-induced mucous cell metaplasia. As shown in Fig. 2, mRNA levels of the Muc5AC mucin gene and Clca3 (gob5), a calcium-activated chloride channel that is associated with mucous cell metaplasia, were significantly reduced in HDM-challenged huApoE3 mice compared with muApoE mice (26). In contrast, huApoE2 and huApoE4 mice showed reductions in mRNA levels for Muc5AC, but not for Clca3. Next, the effect of huApoE alleles on mucous cell metaplasia was assessed. As shown in Figs. 2C and 3, mucous cell metaplasia was reduced in HDM-challenged huApoE3 mice compared with muApoE mice, whereas the level of mucous cell metaplasia was not different between huApoE2 or huApoE4 mice compared with muApoE mice. Taken together, these results demonstrate that mice expressing the huApoE3 allele have a reduced burden of mucous cell metaplasia compared with mice expressing the muApoE allele.
Fig. 2.
HDM-induced mucous cell metaplasia is reduced in humanized apoE ε3 mice. A and B: quantification of lung mRNA levels for Muc5AC and Clca3 by qRT-PCR presented as relative mRNA expression (n = 6 mice, *P < 0.05, huApoE + HDM vs. muApoE + HDM). C: mucous cell metaplasia presented as the percentage of airways containing PAS-positive cells (n = 10 mice, * P < 0.001, huApoE + HDM vs. muApoE + HDM ). On average, 40 ± 1 airways were inspected in each mouse. Results are representative of 2 independent experiments. PAS, periodic acid-Schiff.
Fig. 3.
Pathological manifestations of HDM-induced airway inflammation and mucous cell metaplasia are attenuated in humanized apoE ε3 mice. Histological sections of lung were stained with hematoxylin and eosin (H & E) or PAS stains and images obtained at ×200 or ×1,000. Calibration bars indicate 100 μm for the ×200 images and 20 μm for the ×1,000 images. Results are representative of 2 independent experiments.
HDM-induced airway inflammation is reduced in mice expressing the huApoE3 allele.
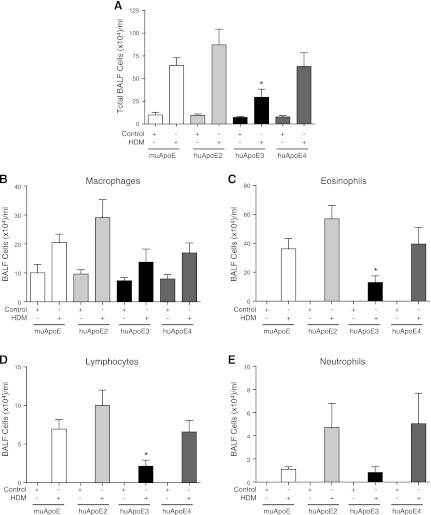
We have previously shown that, although HDM-induced airway inflammation is not altered in apoE−/− mice, administration of an apoE mimetic peptide corresponding to the LDLR binding domain of apoE can prevent the induction of airway inflammation (39). This suggests that apoE might play a role in the regulation of HDM-induced airway inflammation. Therefore, we assessed the role of huApoE alleles in modifying the inflammatory response to HDM challenge. As shown in Fig. 4, the total number of bronchoalveolar lavage fluid (BALF) cells, as well as the number of eosinophils and lymphocytes were decreased in HDM-challenged huApoE3 mice, compared with muApoE mice. Similarly, lung histology revealed a marked decrease in peribronchial inflammation in huApoE3 mice compared with muApoE mice (Fig. 3). In contrast, there was no difference in the total number of BALF inflammatory cells or inflammatory cell subtypes in either huApoE2 or huApoE4 mice compared with muApoE mice. Lung histology at baseline did not appear different between the groups. Taken together, these data demonstrate that mice expressing the huApoE3 allele have lower levels of HDM-induced airway inflammation compared with mice expressing the muApoE allele.
Fig. 4.
HDM-induced airway inflammation is attenuated in humanized apoE ε3 mice. Numbers of total cells (A) and inflammatory cell types (B–E) present in bronchoalveolar lavage fluid (BALF) (n = 10 mice, *P < 0.05, huApoE + HDM vs. muApoE + HDM). Results are representative of 2 independent experiments.
Effect of huApoE alleles on serum IgE levels following HDM challenge.
Increased IgE production is a hallmark of allergic asthma. As shown in Fig. 5, there was no difference in serum IgE levels between HDM-challenged muApoE and huApoE2 mice, whereas serum IgE levels were lower in huApoE4 mice. Although serum IgE levels appeared to be decreased in HDM-challenged huApoE3 mice compared with HDM-challenged muApoE mice, these reductions did not reach statistical significance. When serum IgE levels were compared amongst the HDM-challenged huApoE mice, the differences between the groups were statistically significant (n = 19–20, P < 0.02, 1-way ANOVA), with the highest levels in huApoE2 mice and the lowest levels in huApoE4 mice. This demonstrates that IgE production is reduced in HDM-challenged mice expressing the huApoE4 allele, whereas HDM-challenged mice expressing the huApoE3 allele have an intermediate phenotype.
Fig. 5.
Effect of huApoE alleles on serum IgE levels in HDM-challenged mice. Quantification of serum IgE levels. (n = 19–20, *P < 0.001, huApoE + HDM vs. muApoE + HDM). Results are pooled from 2 experiments.
Expression of Th2 and Th17 cytokines is attenuated in humanized apoE ε3 mice following HDM challenge.
We next assessed whether huApoE alleles modify the expression of Th2 and Th17 cytokines and chemokines as a mechanism by which the severity of airway inflammation may be attenuated in huApoE3 mice. As shown in Fig. 6, HDM-challenged huApoE3 mice had significant reductions in mRNA levels of the Th2 cytokines, IL-4 and IL-13, as well as the Th17 cytokine, IL-17A, compared with muApoE mice. IL-10 mRNA levels were also reduced in huApoE3 mice. huApoE4 mice also had reductions in mRNA levels of IL-4 and IL-13 compared with muApoE mice. In contrast, there was no difference in lung cytokine expression between huApoE2 and muApoE mice. These results show that, compared with mice expressing the muApoE allele, those expressing the huApoE3 or huApoE4 alleles have reduced expression of Th2 cytokines, whereas only mice expressing the human ε3 allele have reduced expression of Th17 cytokines.
Fig. 6.
Expression of lung Th2 and Th17 cytokines are decreased in HDM-challenged humanized apoE ε3 mice. Quantification of lung mRNA levels for IL-4, IL-13, IL-17A, and IL-10 by qRT-PCR presented as relative mRNA expression (n = 6 mice, *P < 0.05, huApoE + HDM vs. muApoE + HDM). Results are representative of 2 independent experiments.
Lung chemokine expression is modified in HDM-challenged mice expressing huApoE alleles.
Because chemokines participate in mediating the recruitment of lymphocytes and eosinophils to the airway, lung mRNA levels were quantified to assess whether the huApoE2, 3, and 4 alleles modify chemokine expression as an additional mechanism by which airway inflammation may be modulated. Compared with muApoE mice, those expressing the huApoE3 or 4 alleles had significant reductions in lung mRNA levels of CCL7 (MCP-3), CCL11 (eotaxin-1), and CCL24 (eotaxin-2), all of which induce the chemotaxis of T cells and eosinophils (Fig. 7) (2, 36, 40). In contrast, compared with muApoE mice, huApoE2 mice demonstrated increased expression of lung mRNA levels of CCL17 (TARC), a chemokine product of dendritic cells and airway epithelial cells that induces Th2 T cell chemotaxis to the lung via CCR4 during allergic airway inflammation (2, 29).
Fig. 7.
HDM-challenged humanized apoE ε3 mice have attenuated expression of lung chemokines. Quantification of lung mRNA levels for CCL11, CCL24, CCL7, and CCL17 by qRT-PCR presented as relative mRNA expression (n = 6 mice, *P < 0.05, huApoE + HDM vs. muApoE + HDM). Results are representative of 2 independent experiments.
Effect of huApoE alleles on lung apoE protein expression.
Lastly, we assessed whether the effects of the huApoE alleles in HDM-challenged mice reflected differences in protein expression. Western blots of lung proteins were performed, and apoE isoforms were detected using an antibody that recognizes huApoE. As shown in Fig. 8, lung apoE protein levels in huApoE2 mice were significantly elevated compared with those of huApoE3 or huApoE4 mice. Total lung apoE protein levels were not increased in response to HDM challenge in huApoE2, huApoE3, or huApoE4 mice. These data are consistent with the conclusion that reduced disease severity in huApoE3 and huApoE4 mice is not a consequence of increased expression of the apoE protein.
Fig. 8.
Expression of huApoE in the lungs of humanized apoE ε2, ε3, and ε4 mice. Lung protein expression of huApoE by humanized apoE ε2, ε3, and ε4 mice was assessed by Western blotting using an antibody directed against huApoE. Equivalency of protein loading was assessed using an antibody directed against β-actin. (n = 3 mice, *P < 0.001, huApoE2 + saline vs. huApoE3 + saline or huApoE4 + saline and huApoE2 + HDM vs. huApoE3 + HDM or huApoE4 + HDM). Duplicate samples are shown.
DISCUSSION
ApoE plays a key role in cholesterol metabolism and influences the risk of coronary artery disease (4). The primary function of apoE is to clear chylomicron, chylomicron remnants, and very-low-density lipoproteins from plasma via binding to LDLR in the liver, with resultant receptor-mediated endocytosis (10, 15, 22–24). Consistent with this, apoE knockout mice have defective clearance of remnant lipoproteins with resultant accelerated atherosclerosis (23, 30, 41). ApoE also plays an important role in the central nervous system, where it mediates cholesterol transport into neuronal cells and participates in synaptic repair and plasticity (6).
ApoE is a 34-kDa protein that is comprised of 299 amino acids and is encoded by a gene located on chromosome 19 (24). When bound to lipid, the apoE protein has a circular horseshoe configuration. Its amino-terminus is a four-helix amphipathic helical bundle that contains the LDLR-binding region, corresponding to amino acids 134–150, whereas the lipid-binding domain is located in the carboxy terminus. Three common polymorphic alleles of the huApoE gene have been identified (24, 42). The apoE3 allele is present in ∼65–70% of humans, whereas the ε2 and ε4 alleles are present in ∼5–10% and 15–20% of general populations, respectively (6, 22, 24). The three apoE alleles result in six corresponding genotypes (ε3/ε3, ε3/ε4, ε2/ε3, ε4/ε4, ε2/ε4, and ε2/ε2, listed in descending order of frequency) that modify plasma lipid levels and the risk of coronary heart disease (6, 37). Carriers of the ε4 allele have higher plasma levels of low-density lipoprotein cholesterol (LDL-C), as well as a higher risk of coronary artery disease than those carrying the ε3 allele (4, 9). In contrast, carriers of the ε2 allele have lower levels of LDL-C and lower cardiovascular risk, except in 5–10% of ε2/ε2 individuals who develop type III familial hyperlipoproteinemia and accelerated atherosclerosis (4, 19, 33). ApoE genotypes also modify longevity in Caucasian populations, with survival to advanced age being most likely in carriers of the ε2 allele and least likely in carriers of the ε4 allele (9). The ε4 allele is a major genetic risk factor for Alzheimer's disease and may be associated with accelerated neurodegeneration in Parkinson's disease and multiple sclerosis (6). In HIV-infected individuals, the apoE ε4 allele enhances cellular entry and accelerates the progression to development of AIDS and death (7, 24). Lastly, although several genome-wide associations have been performed on asthmatic populations, an association has not been reported between the huApoE gene and asthma (14, 21, 25, 31).
Work from our laboratory has shown a role for an apoE-LDLR pathway in the lung as an endogenous negative regulator of AHR and mucous cell metaplasia in an experimental murine model of HDM-induced airway disease (39). Therefore, we hypothesized that the polymorphic huApoE alleles, which differ in their binding affinities for the LDLR, might differentially modify the pathogenic manifestations of repeated nasal HDM challenges. To address this question, we utilized humanized knockin mice that express the huApoE alleles in lieu of endogenous muApoE (19, 32, 33). Here, we show that the polymorphic huApoE alleles differentially modify disease severity in response to repeated nasal HDM challenges. Increases in airway inflammation, mucous cell metaplasia, and AHR were not modified in HDM-challenged mice expressing the huApoE2 allele compared with mice expressing the muApoE allele. In contrast, mice expressing the huApoE3 allele, and to a lesser extent the huApoE4 allele, displayed a phenotype with attenuated manifestations of HDM-induced airway disease compared with mice expressing the muApoE allele, which was utilized as a comparator (summarized in Fig. 9). In particular, mice expressing the huApoE3 allele displayed significant reductions in AHR, mucous cell metaplasia, and airway inflammation compared with mice expressing the muApoE gene. Expression of Th2 and Th17 cytokines, as well as chemokines that participate in mediating the recruitment of eosinophils and T cells (CCL7, CCL11, and CCL24), was also reduced in huApoE3 mice. Although serum IgE levels appeared decreased in huApoE3 mice, the reduction did not reach statistical significance compared with muApoE mice. This shows that the human ε3 allele has a milder effect on serum IgE levels than the human ε4 allele.
Fig. 9.
Summary of airway responses to repeated HDM challenges in humanized apoE knockin mice.
Mice expressing the huApoE4 allele displayed an intermediate asthma phenotype with attenuated AHR and serum IgE production compared with mice expressing the muApoE allele, whereas airway inflammation was not decreased despite a reduction in mRNA levels of Th2 cytokines and C-C chemokines. Similarly, neither Clca3 mRNA levels nor mucous cell metaplasia were reduced in huApoE4 mice despite a decrease in Muc5AC mRNA levels. It is not clear, however, why the reduction in mRNA levels of Th2 cytokines and C-C chemokines in HDM-challenged huApoE4 mice did not result in a reduction in airway inflammation or mucous cell metaplasia. This suggests that huApoE4 did not suppress additional proinflammatory mediators or pathways that are sufficient to induce airway inflammation and mucous cell metaplasia. We also found that huApoE4 mice had reduced AHR in response to HDM challenge compared with saline-challenged huApoE4 mice. The reason for the reduced AHR in HDM-challenged huApoE4 mice is not clear, but a similar finding has been observed in another study of Der p1-challenged C57BL/6 mice, which also showed a reduction in AHR in response to HDM challenge, compared with saline challenge, despite the induction of airway inflammation and mucous cell metaplasia (17). Taken together, these findings suggest that huApoE4 may modulate distinct pathways or sets or genes that regulate AHR independently of airway inflammation and mucous cell metaplasia in C57BL/6 mice.
The plasma lipoprotein profiles of mice expressing huApoE alleles reflect the relative LDLR binding affinities of the huApoE proteins (18, 28). Furthermore, these mice have previously been demonstrated to have phenotypes consistent with the function of the corresponding huApoE alleles. For example, mice expressing the huApoE2 allele fed an atherogenic diet recapitulate a type III hyperlipoproteinemia phenotype, whereas mice expressing the apoE ε4 allele had increased plasma cholesterol compared with mice expressing the huApoE3 allele (19, 33). Consequently, this model system has been utilized to assess the biological effects of huApoE polymorphisms on the pathogenesis of complex human diseases, such as atherosclerosis (28). Our results suggest that altered binding affinities of the huApoE2 protein to the LDLR may in part contribute to the differential effects of the polymorphic apoE alleles in modulating the pathogenesis of airway responses to repeated HDM challenges. Mice expressing the huApoE2 allele, which has significantly reduced binding to the LDLR, did not modify the manifestations of HDM-induced airway disease. In contrast, mice expressing the huApoE3 allele, which binds avidly to the LDLR, significantly attenuated the key pathogenic manifestations of HDM-induced airway disease. The huApoE4 allele, which has similar LDLR binding but altered structural properties compared with the huApoE3 allele, had an intermediary effect on disease severity (24). The differential effects of the huApoE3 and huApoE4 mice on HDM-induced airway disease, however, suggest that additional factors, such as structural differences that modify molecular interactions, might in part account for the contrasting ability of apoE3 and apoE4 to attenuate airway inflammation and mucous cell metaplasia despite similar binding affinities for the LDLR. For example, the huApoE3 and apoE4 proteins have different side chain orientations and salt bridge rearrangements that modify how their amino- and carboxy-termini interact with lipoproteins (24). These structural differences result in preferential binding of huApoE4 to large, triglyceride-rich very-low-density lipoproteins, whereas huApoE3 and apoE2 preferentially bind to small, phospholipid-rich high-density lipoproteins.
Prior studies have suggested several potential mechanisms by which apoE may attenuate the pathogenesis of HDM-induced airway disease. Recently, it has been shown that apoE may mediate its effects via binding to the SET protein, which functions as a physiological inhibitor of protein phosphatase 2A (PP2A) (8, 34). Following internalization, apoE binds to SET, resulting in an increase in PP2A activity, which attenuates signaling by NF-κB, MAPK, and Akt pathways. HuApoE3 and apoE4 have also been shown to suppress proliferative and delayed type hypersensitivity responses by human T cells (20). ApoE can bind and facilitate the presentation of lipid antigens by antigen presenting cells via a mechanism that involves receptor-mediated endocytosis (35). Similarly, B cells have been shown to present lipid antigens to NKT cells via a pathway that involves both apoE and the LDLR (1). Consistent with this being a LDLR-mediated event, B cells can present lipid antigens and activate NKT cells utilizing either huApoE3 or apoE4, but not apoE2, which is defective in LDLR binding. Facilitation of antigen presentation by apoE, however, would be expected to augment immune responses, rather than attenuate airway inflammation. In addition, our laboratory is actively investigating additional mechanisms by which the apoE-LDLR pathway attenuates disease severity in HDM-induced airway disease.
We also found that levels of apoE in the lungs of mice expressing the huApoE2 allele were significantly greater than mice expressing either the huApoE3 or E4 alleles. This demonstrates that the greater ability of huApoE3 and huApoE4 to attenuate HDM-induce airway disease did not simply reflect an increase in protein levels. Of note, plasma apoE protein levels have previously been shown to be increased in huApoE2 mice compared with huApoE3 or apoE4 mice (19). Furthermore, we were surprised that HDM-challenge did not increase apoE protein levels in lung homogenates from mice expressing either the huApoE2, 3, or 4 alleles, as we had previously demonstrated that HDM-challenge increases apoE mRNA expression in the lung (39). This suggests that constitutive levels of apoE protein in the lung modulate the severity of HDM-induced airway disease.
In summary, we have shown that huApoE alleles differentially modify key pathogenic responses to nasal repeated HDM challenges, which can be stratified, in rank order of increasing disease severity, ε3 < ε4 < ε2. Compared with muApoE, the huApoE3 allele confers a protective phenotype with reduced airway hyperreactivity, mucous cell metaplasia, and airway inflammation, whereas the huApoE4 allele has an intermediate asthma phenotype with selectively attenuated AHR and serum IgE production. Lastly, the huApoE2 allele did not modify HDM-induced airway disease. These results suggest that the polymorphic apoE alleles might modify asthma severity in human subjects. Further studies, however, would be required to establish whether the results from this experimental murine model system are applicable to human disease. If applicable, our results suggest that asthmatic carriers of the ε2 allele might have increased disease severity compared with carriers of the ε3 or ε4 alleles. This would be most relevant for individuals with the ε2/ε2 genotype, who comprise less than 1% of the population (11, 38). In contrast, ∼59–66% of individuals have the ε3/ε3 genotype, whereas ∼1–2% have the ε4/ε4 genotype. Additional studies would also be required to assess the effect of heterozygous huApoE genotypes on asthma severity.
GRANTS
This work was funded by the Division of Intramural Research, NHLBI, NIH and the Department of Laboratory Medicine, Clinical Center, NIH.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
ACKNOWLEDGMENTS
We are extremely appreciative of the staff of the NHLBI Laboratory of Animal Medicine and Surgery, whose commitment, professional advice, and excellent technical support made this study possible. We are also most appreciative of Drs. Joel Moss and Martha Vaughan for helpful discussions.
REFERENCES
- 1.Allan LL, Hoefl K, Zheng DJ, Chung BK, Kozak FK, Tan R, van den Elzen P. Apolipoprotein-mediated lipid antigen presentation in B cells provides a pathway for innate help by NKT cells. Blood 114: 2411–2416, 2009 [DOI] [PubMed] [Google Scholar]
- 2.Barnes PJ. The cytokine network in asthma and chronic obstructive pulmonary disease. J Clin Invest 118: 3546–3556, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes PJ. Immunology of asthma and chronic obstructive pulmonary disease. Nat Rev Immunol 8: 183–192, 2008 [DOI] [PubMed] [Google Scholar]
- 4.Bennet AM, Di Angelantonio E, Ye Z, Wensley F, Dahlin A, Ahlbom A, Keavney B, Collins R, Wiman B, de Faire U, Danesh J. Association of apolipoprotein E genotypes with lipid levels and coronary risk. JAMA 298: 1300–1311, 2007 [DOI] [PubMed] [Google Scholar]
- 5.Bergeron C, Al-Ramli W, Hamid Q. Remodeling in asthma. Proc Am Thorac Soc 6: 301–305, 2009 [DOI] [PubMed] [Google Scholar]
- 6.Bu G. Apolipoprotein E and its receptors in Alzheimer's disease: pathways, pathogenesis and therapy. Nat Rev Neurosci 10: 333–344, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burt TD, Agan BK, Marconi VC, He W, Kulkarni H, Mold JE, Cavrois M, Huang Y, Mahley RW, Dolan MJ, McCune JM, Ahuja SK. Apolipoprotein (apo) E4 enhances HIV-1 cell entry in vitro, and the APOE epsilon4/epsilon4 genotype accelerates HIV disease progression. Proc Natl Acad Sci USA 105: 8718–8723, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen DJ, Ohkubo N, Oddo J, Van Kanegan MJ, Neil J, Li F, Colton CA, Vitek MP. Apolipoprotein E and peptide mimetics modulate inflammation by binding the SET protein and activating protein phosphatase 2A. J Immunol 186: 2535–2542, 2011 [DOI] [PubMed] [Google Scholar]
- 9.Drenos F, Kirkwood TB. Selection on alleles affecting human longevity and late-life disease: the example of apolipoprotein E. PLos One 5: e10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eichner JE, Dunn ST, Perveen G, Thompson DM, Stewart KE, Stroehla BC. Apolipoprotein E polymorphism and cardiovascular disease: a HuGE review. Am J Epidemiol 155: 487–495, 2002 [DOI] [PubMed] [Google Scholar]
- 11.Elosua R, Ordovas JM, Cupples LA, Fox CS, Polak JF, Wolf PA, D'Agostino RA, Sr, O'Donnell CJ. Association of APOE genotype with carotid atherosclerosis in men and women: the Framingham Heart Study. J Lipid Res 45: 1868–1875, 2004 [DOI] [PubMed] [Google Scholar]
- 12.Evans CM, Williams OW, Tuvim MJ, Nigam R, Mixides GP, Blackburn MR, DeMayo FJ, Burns AR, Smith C, Reynolds SD, Stripp BR, Dickey BF. Mucin is produced by clara cells in the proximal airways of antigen-challenged mice. Am J Respir Cell Mol Biol 31: 382–394, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fanta CH. Asthma. N Engl J Med 360: 1002–1014, 2009 [DOI] [PubMed] [Google Scholar]
- 14.Ferreira MA, Matheson MC, Duffy DL, Marks GB, Hui J, Le Souef P, Danoy P, Baltic S, Nyholt DR, Jenkins M, Hayden C, Willemsen G, Ang W, Kuokkanen M, Beilby J, Cheah F, de Geus EJ, Ramasamy A, Vedantam S, Salomaa V, Madden PA, Heath AC, Hopper JL, Visscher PM, Musk B, Leeder SR, Jarvelin MR, Pennell C, Boomsma DI, Hirschhorn JN, Walters H, Martin NG, James A, Jones G, Abramson MJ, Robertson CF, Dharmage SC, Brown MA, Montgomery GW, Thompson PJ. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet 378: 1006–1014, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldstein JL, Brown MS. The LDL receptor. Arterioscler Thromb Vasc Biol 29: 431–438, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson JR, Wiley RE, Fattouh R, Swirski FK, Gajewska BU, Coyle AJ, Gutierrez-Ramos JC, Ellis R, Inman MD, Jordana M. Continuous exposure to house dust mite elicits chronic airway inflammation and structural remodeling. Am J Respir Crit Care Med 169: 378–385, 2004 [DOI] [PubMed] [Google Scholar]
- 17.Kelada SN, Wilson MS, Tavarez U, Kubalanza K, Borate B, Whitehead G, Maruoka S, Roy MG, Olive M, Carpenter DE, Brass DM, Wynn TA, Cook DA, Evans CM, Schwartz DA, Collins FS. Strain-dependent genomic factors affect allergen-induced airway hyper-responsiveness in mice. Am J Respir Cell Mol Biol 45: 817–824, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knouff C, Briand O, Lestavel S, Clavey V, Altenburg M, Maeda N. Defective VLDL metabolism and severe atherosclerosis in mice expressing human apolipoprotein E isoforms but lacking the LDL receptor. Biochim Biophys Acta 1684: 8–17, 2004 [DOI] [PubMed] [Google Scholar]
- 19.Knouff C, Hinsdale ME, Mezdour H, Altenburg MK, Watanabe M, Quarfordt SH, Sullivan PM, Maeda N. Apo E structure determines VLDL clearance and atherosclerosis risk in mice. J Clin Invest 103: 1579–1586, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laskowitz DT, Lee DM, Schmechel D, Staats HF. Altered immune responses in apolipoprotein E-deficient mice. J Lipid Res 41: 613–620, 2000 [PubMed] [Google Scholar]
- 21.Li X, Howard TD, Zheng SL, Haselkorn T, Peters SP, Meyers DA, Bleecker ER. Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J Allergy Clin Immunol 125: 328–335; e311, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science 240: 622–630, 1988 [DOI] [PubMed] [Google Scholar]
- 23.Mahley RW, Huang Y. Atherogenic remnant lipoproteins: role for proteoglycans in trapping, transferring, and internalizing. J Clin Invest 117: 94–98, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer's disease to AIDS. J Lipid Res 50 Suppl: S183–188, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, von Mutius E, Farrall M, Lathrop M, Cookson WO. A large-scale, consortium-based genome-wide association study of asthma. N Engl J Med 363: 1211–1221, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakanishi A, Morita S, Iwashita H, Sagiya Y, Ashida Y, Shirafuji H, Fujisawa Y, Nishimura O, Fujino M. Role of gob-5 in mucus overproduction and airway hyperresponsiveness in asthma. Proc Natl Acad Sci USA 98: 5175–5180, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pendse AA, Arbones-Mainar JM, Johnson LA, Altenburg MK, Maeda N. Apolipoprotein E knock-out and knock-in mice: atherosclerosis, metabolic syndrome, and beyond. J Lipid Res 50 Suppl: S178–S182, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pilette C, Francis JN, Till SJ, Durham SR. CCR4 ligands are upregulated in the airways of atopic asthmatics after segmental allergen challenge. Eur Respir J 23: 876–884, 2004 [DOI] [PubMed] [Google Scholar]
- 30.Plump AS, Smith JD, Hayek T, Aalto-Setala K, Walsh A, Verstuyft JG, Rubin EM, Breslow JL. Severe hypercholesterolemia and atherosclerosis in apolipoprotein E-deficient mice created by homologous recombination in ES cells. Cell 71: 343–353, 1992 [DOI] [PubMed] [Google Scholar]
- 31.Schauberger EM, Ewart SL, Arshad SH, Huebner M, Karmaus W, Holloway JW, Friderici KH, Ziegler JT, Zhang H, Rose-Zerilli MJ, Barton SJ, Holgate ST, Kilpatrick JR, Harley JB, Lajoie-Kadoch S, Harley IT, Hamid Q, Kurukulaaratchy RJ, Seibold MA, Avila PC, Rodriguez-Cintron W, Rodriguez-Santana JR, Hu D, Gignoux C, Romieu I, London SJ, Burchard EG, Langefeld CD, Wills-Karp M. Identification of ATPAF1 as a novel candidate gene for asthma in children. J Allergy Clin Immunol 128: 753–760, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sullivan PM, Mezdour H, Aratani Y, Knouff C, Najib J, Reddick RL, Quarfordt SH, Maeda N. Targeted replacement of the mouse apolipoprotein E gene with the common human APOE3 allele enhances diet-induced hypercholesterolemia and atherosclerosis. J Biol Chem 272: 17972–17980, 1997 [DOI] [PubMed] [Google Scholar]
- 33.Sullivan PM, Mezdour H, Quarfordt SH, Maeda N. Type III hyperlipoproteinemia and spontaneous atherosclerosis in mice resulting from gene replacement of mouse Apoe with human Apoe*2. J Clin Invest 102: 130–135, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Switzer CH, Cheng RY, Vitek TM, Christensen DJ, Wink DA, Vitek MP. Targeting SET/I (2)PP2A oncoprotein functions as a multi-pathway strategy for cancer therapy. Oncogene 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van den Elzen P, Garg S, Leon L, Brigl M, Leadbetter EA, Gumperz JE, Dascher CC, Cheng TY, Sacks FM, Illarionov PA, Besra GS, Kent SC, Moody DB, Brenner MB. Apolipoprotein-mediated pathways of lipid antigen presentation. Nature 437: 906–910, 2005 [DOI] [PubMed] [Google Scholar]
- 36.Walsh ER, Sahu N, Kearley J, Benjamin E, Kang BH, Humbles A, August A. Strain-specific requirement for eosinophils in the recruitment of T cells to the lung during the development of allergic asthma. J Exp Med 205: 1285–1292, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward H, Mitrou PN, Bowman R, Luben R, Wareham NJ, Khaw KT, Bingham S. APOE genotype, lipids, and coronary heart disease risk: a prospective population study. Arch Intern Med 169: 1424–1429, 2009 [DOI] [PubMed] [Google Scholar]
- 38.Ward H, Mitrou PN, Bowman R, Luben R, Wareham NJ, Khaw KT, Bingham S. APOE genotype, lipids, and coronary heart disease risk: a prospective population study. Arch Intern Med 169: 1424–1429, 2009 [DOI] [PubMed] [Google Scholar]
- 38a.World Health Organization Global Surveillance, Prevention and Control of Chronic Respiratory Diseases: A Comprehensive Approach. Geneva, Switzerland: World Health Organization, 2007 [Google Scholar]
- 39.Yao X, Fredriksson K, Yu ZX, Xu X, Raghavachari N, Keeran KJ, Zywicke GJ, Kwak M, Amar MJ, Remaley AT, Levine SJ. Apolipoprotein E negatively regulates house dust mite-induced asthma via a LDL receptor-mediated pathway. Am J Respir Crit Care Med 182: 1228–1238, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ying S, Robinson DS, Meng Q, Barata LT, McEuen AR, Buckley MG, Walls AF, Askenase PW, Kay AB. C-C chemokines in allergen-induced late-phase cutaneous responses in atopic subjects: association of eotaxin with early 6-h eosinophils, and of eotaxin-2 and monocyte chemoattractant protein-4 with the later 24-hour tissue eosinophilia, and relationship to basophils and other C-C chemokines (monocyte chemoattractant protein-3 and RANTES). J Immunol 163: 3976–3984, 1999 [PubMed] [Google Scholar]
- 41.Zhang SH, Reddick RL, Piedrahita JA, Maeda N. Spontaneous hypercholesterolemia and arterial lesions in mice lacking apolipoprotein E. Science 258: 468–471, 1992 [DOI] [PubMed] [Google Scholar]
- 42.Zhong N, Weisgraber KH. Understanding the association of apolipoprotein E4 with Alzheimer disease: clues from its structure. J Biol Chem 284: 6027–6031, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]