Abstract
Liver failure due to chronic hepatitis C virus infection is a major cause for liver transplantation worldwide. Recurrent infection of the graft is universal in HCV patients following transplant and results in rapid progression to severe fibrosis and end-stage liver disease in one-third of all patients. No single clinical variable, or combination thereof, has so far proven accurate in identifying patients at risk of hepatic decompensation in the transplant setting. A combination of longitudinal, dimensionality reduction, and categorical analysis of the transcriptome from 111 liver biopsy specimens taken from 57 HCV-infected patients over time identified a molecular signature of gene expression of patients at risk of developing severe fibrosis. Significantly, alterations in gene expression occur prior to histologic evidence of liver disease progression, suggesting that events which occur during the acute phase of infection influence patient outcome. Additionally, a common precursor state for different severe clinical outcomes was identified. Hence, based on this patient cohort, incidence of severe liver disease is a process initiated early during HCV infection of the donor organ. The probable cellular network at the basis of the initial transition to severe liver disease was identified and characterized.
Keywords: viral hepatitis, microarray, hepatic fibrosis
Liver failure due to chronic hepatitis C virus infection is the leading cause for orthotopic liver transplantation (OLT) in North America. Recurrent infection of the graft is universal in HCV patients following transplant, and in a subset of patients, the time of progression to severe fibrosis, eventual cirrhosis, and end-stage liver disease is greatly accelerated (1). Currently the only available recourse to patients with decompensated cirrhosis is retransplantation, which is both difficult for the patient and further depletes the limited supply of available donor organs. HCV patients undergoing retransplantation due to decompensated cirrhosis also have a lower graft survival rate than patients undergoing retransplantation for other indications (2).
The present standard for monitoring HCV recurrence and fibrosis progression relies on histopathological examination of core needle liver biopsies. This procedure is associated with significant morbidity, and frequently results in misdiagnoses of fibrosis progression due to the small size of the biopsy relative to the liver and the subjective nature of interpretation. Attempts to develop less invasive means of diagnosing hepatic fibrosis have not proved reliably accurate thus far, although such a method is highly desirable.
Previous studies demonstrated that distinct patterns of host gene expression are associated with different clinical outcomes in HCV transplant patients (3–5). However, these studies examined differential gene expression using standard analysis methodology. We applied mathematical modeling techniques to assess transcriptional dynamics contributing to severe liver disease temporally during HCV recurrence. We utilized a combination of longitudinal topographic profiling and singular value decomposition-initiated multidimensional scaling (SVD-MDS) to identify genes involved in progression to advanced hepatic fibrosis.
Results
Clinical data and sample grouping
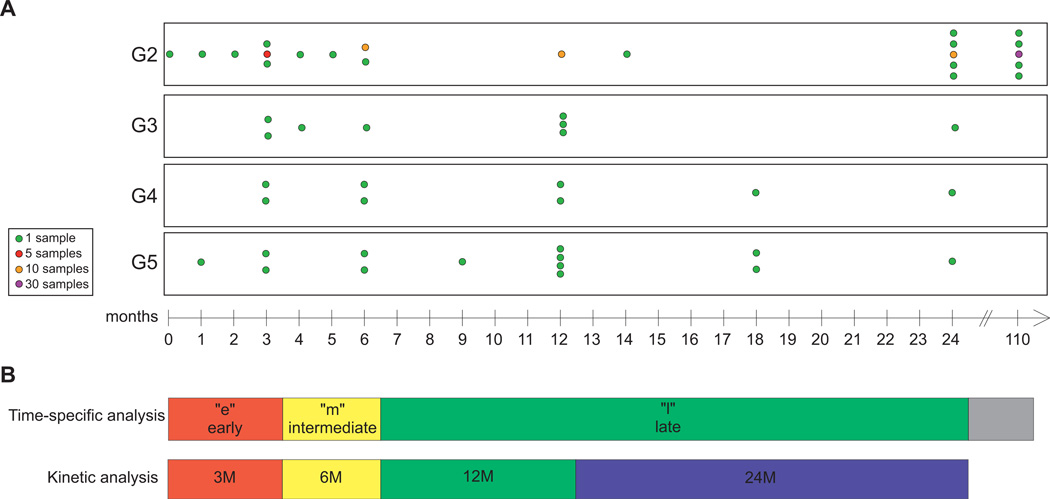
We identified 57 chronic HCV patients undergoing orthotopic liver transplantation (OLT) at the University of Washington Medical Center, and obtained core needle biopsies from various time points post-OLT (Fig. 1). We grouped patients by post-OLT clinical outcome. Of 57 patients, 14 (25%) developed an adverse clinical outcome post-OLT (Table 1). After classifying our control, uninfected normal pool (UNP) of liver tissue as group 1 (G1), we designated 43 HCV patients with no adverse clinical outcome as group 2 (G2). Three adverse clinical outcomes were defined for patient grouping. We first determined the patients’ most recent Batts-Ludwig stage of hepatic fibrosis by having one pathologist stage the most recent biopsy prior to June 1, 2009, when we stopped collecting clinical information on the cohort for this study. We identified four patients with most recent biopsies at stage 3–4 and designated them as group 3 (G3). We also determined whether patients presented clinical symptoms of cirrhosis (portal hypertension, encephalopathy, ascites, and bleeding esophageal varices) and identified three patients, designated group 4 (G4). Finally, we identified seven HCV patients who died or underwent retransplantation due to graft failure, designated group 5 (G5). All patients in G4 and G5 also developed stage 3–4 fibrosis prior to clinical cirrhosis or death/retransplantation. We confirmed that no patients demonstrated evidence of stage 3–4 fibrosis or symptoms of cirrhosis at the time the samples were collected. Therefore, gene expression changes determined by our analysis to be significantly associated with severe liver injury were identified from samples taken before clinical or histological evidence of disease progression. We also divided the 111 liver biopsy specimens based on time post-OLT sampling (Figure 1A).
Figure 1. Schematic representation of the transcriptome analysis strategy.
(A). Time-line illustrating sample distribution in the different groups (G2: HCV-infected patient biopsies with no adverse clinical outcome; G3: HCV-infected patient biopsies with advanced fibrosis; G4: HCV-infected patient biopsies with advanced fibrosis and clinical cirrhosis; G5: HCV-infected patient biopsies with advanced fibrosis, clinical cirrhosis, and death or retransplant). (B). Grouping of samples into time categories for the time-specific and the longitudinal analysis.
Table 1.
Clinical characterization of patient cohort used in this study
| Stage 0–2 G2 (n=43) |
Stage 3–4 G345 (n=14) |
|
|---|---|---|
| Patient age (range) | 59 (45–78) | 62 (54–75) |
| % male patients | 86.1% | 64.3% |
| Donor age (range) | 31 (10–59) | 41 (13–60) |
| % male donors | 78.1% | 64.3% |
| Months post-OLT since 6/1/2009 | 79.6 ± 25.1 | 54.9 ± 18 |
| MELD score at time of transplant | 16.9 ± 8.2 | 14.6 ± 4.1 |
| % history of ethanol abuse | 72.1% | 21.4% |
| Incidence of acute cellular rejection | 18.6% | 57.1% |
Decomposition of transcriptome dynamics into functional stages
The relative heterogeneity of both timing of post-transplant biopsies from patients of this cohort and pathological phenotypes displayed by the different patients in the cohort (Figure 1A) required particular attention during data analysis. Clinical annotation defined disease categories (G2–G5), and samples were further subdivided into time categories (early, intermediate, late). These intervals were based on HCV reinfection kinetics and spreading in the donor organ and homogeneity of sample distribution. Non-progressors (G2) also encompassed samples beyond the two-year follow up period of the severe liver disease groups (Figure 1B).
We devised three analysis protocols for the transcriptomic data using different methodologies. First, we compared combined patient groups G3–G5 versus the entire G2 dataset for the early, intermediate, and late time categories separately and combined using the recently developed SVD-MDS method (6) to assess the prognostic value of the gene signatures generated with the two strategies and decompose these signatures into individual gene contributions. We also performed this comparison using time-matched G2 samples. Second, we performed longitudinal topographic profiling using a previously employed (7) self-organizing maps-based classifier to investigate transcriptional dynamics within each of the three severe disease patient groups G3–G5 and to also establish averaged gene expression profiles for the combined G3–G5 patient groups (Figure 1B). Finally, we used modified k-means clustering to identify a common precursor molecular signature distinguishing progression to severe fibrosis, and this transition occurred at early to intermediate timepoints post-OLT.
Early transcriptome dynamics determine severe liver disease following transplantation in HCV infected patients
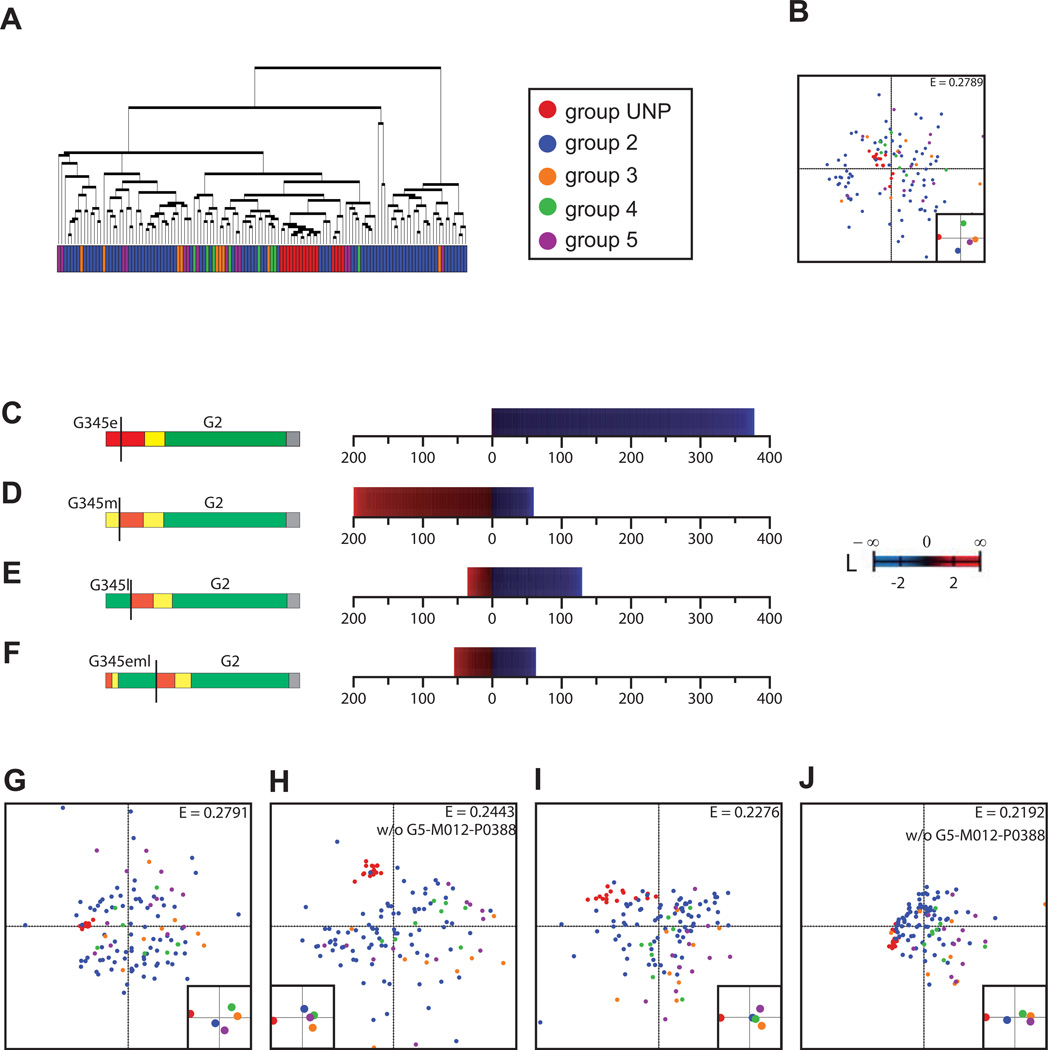
Single linkage hierarchical clustering based on Euclidean distances averaged over the entire microarray data set did not reveal an apparent structure of the entire set of samples (Figure 2A). Despite the variety of clinical phenotypes from asymptomatic to death, the overall profiles were not indicative of outcome. Time-specific profiling of the combined G345 patient groups using the early time category (G345e) as compared to the entire G2 dataset, however, identified almost 400 statistically significant differentially expressed genes (DEG; p<0.01, Figure 2C, Table S1). The vast majority of these genes were down-regulated compared to G2 expression.
Figure 2. Categorical time-specific profiling of genes differentially expressed when comparing severe forms of liver disease with all asymptomatic patient biopsies.
(A). Hierarchical clustering of the overall dataset (127 samples) computed over the entire set of genes. (B). SVD-MDS representation of the overall dataset computed over the entire set of genes. Inlet shows the centers of gravity (average weighted-averages) for the different groups. Coloring: see legend in (A). The E-value is the remaining Kruskal stress after dimensionality reduction and is directly proportional to the amount of lost information. (C). Visual representation of logarithmic fold change of the statistically significantly differentially expressed genes when comparing "early" post-transplant samples of the severe liver disease patients (G3–G5) to the entire set of recordings of the asymptomatic patients (G2). (D). Idem as (C) for "intermediate" post-transplant samples of the severe liver disease patients (G3–G5). (E). Idem as (C) for "late" post-transplant samples. (F). Idem as (C) for the combined "early", "intermediate", and "late" post-transplant samples. (G). Idem as (C) for the G345e vs. G2 statistically significantly differentially expressed genes (DEG). (H). Idem as (B) for the G345m vs. G2 DEG (without sample G5-M012-P038). (I). Idem as (B) for the G345l vs. G2 DEG. (J). Idem as (B) for the G345eml vs. G2 DEG (without sample G5-M012-P038). The distances in (B) and (J–G) signify the average correspondence value of any two data points. They are by convention scaled to unity as absolute normalization between datasets is not possible. Furthermore, the center of gravity of the geometric object (all data points together) is placed at the origin. This the same for the main and the inlet plots, which are intended to help the reader faster identify where different groups are located in the more detailed main plots.
Using Ingenuity Pathway Analysis (IPA), we performed functional analysis of these early DEG associated with progression to severe fibrosis. We found that 130 of these genes were associated with inflammatory disorders and infectious disease, including numerous human leukocyte antigen (HLA) genes (HLA-DMB, HLA-DPA1, HLA-DPB1, HLA-DQB1, HLA-DRA, HLA-DRB5, HLA-E, and HLA-G). Repression of antigen presentation is expected in a post-OLT cohort, as this is the goal of the immunosuppression regimens intended to prevent graft rejection. However, these were more repressed in G345 patients compared to G2 patients, as were other key immune and inflammatory genes such as immunoglobulins, Fc receptors, complement components, key signal transducers and transcriptional regulators, interferon stimulated genes (ISGs), protein modifiers such as ubiquitin, small ubiquitin-like modifier 2 (SUMO2), and interferon stimulated gene 15 (ISG15), proteasomal subunits, chemokines, cathepsins, and serine proteases.
Additionally, we observed 126 molecules functionally associated with cancer were strongly repressed in G345 patient samples compared to G2, including mediators of cell cycle arrest and DNA damage checkpoint control and apoptosis, indicating repressed cell cycle control and inhibition of apoptosis. This is consistent with studies indicating that cell cycle arrest occurs to facilitate viral replication in infected hepatocytes, and that apoptotic processes are disrupted by HCV infection (8, 9). Finally, we observed 70 molecules associated with metabolic functions, including lipid, vitamin and mineral, and cholesterol metabolism. Genes involved in multiple lipid biosynthetic processes, as well as those functioning in synthesis and transport of membrane phospholipids and cholesterol and fatty acid biosynthesis were also repressed in G345e biopsies. Together, these data suggest that during the first three months of HCV recurrence post-OLT, patients who eventually develop progressive HCV-induced liver disease experience more profound hepatic immunosuppression than G2 patients, while undergoing dramatic reprogramming of mitotic and metabolic functions characterized by repression of checkpoint regulators, cell cycle progression, and lipid biosynthesis and transport.
This initial repression was followed by general activation of gene expression during the intermediate stages post-transplantation as revealed by the G345m versus G2 comparison (Figure 2D), including many DEG related to cell cycle, cell death, and cancer. This contrasts with the G345l versus G2 comparison which revealed an increasingly restricted pattern of gene regulation (<200 DEG, Figure 2E), again primarily composed of reduced expression. As these different effects partially cancel themselves out, in the combined G345eml vs. G2 profile a limited set of DEG equally distributed between induced and repressed phenotypes was observed (Figure 2F). These further revealed distinct phases of transcriptome dynamics in severe liver disease patients compared to patients without evidence of progressive disease. Early down-regulation of a many genes related to inflammation, cell cycle regulation, and lipid metabolism was followed by an intermediate activation of another subset, and finally down-modulation of the overall transcriptional response but increased expression of fibrogenic genes such as type 1 collagens (COL1A1, COL1A2) and markers of hepatic stellate cell (HSC) activation such as secreted phosphoprotein 1/osteopontin (SPP1) and galectin 3 (LGALS3). Activated HSCs are the primary cellular mediators of collagen and extracellular matrix (ECM) deposition in HCV-induced fibrogenesis (10, 11). The temporal decrease in the number of DEG indicates increasing heterogeneity of gene expression patterns, leading to fewer statistically significant changes in gene expression. Heterogeneity in the molecular profiles is consistent with the increased heterogeneity of the phenotypes in the individual patients.
In order to assess the prognostic value of the different DEG identified here, we employed SVD-MDS, a method of non-linear dimensionality reduction for visualizing large datasets with many features such as microarray data. SVD-MDS reduces data to a matrix of Euclidean distances between features and generates a lower-dimensional representation, while maximally preserving inter-feature distances. Generally, non-linear dimensionality reduction methods such as SVD-MDS depict an additional 3–4 dimensions in a visualization. Therefore, while the hierarchical clustering shown in Fig. 2A only shows the first dimension of the biological condition space, representations 2B and 2G–2J visually represent approximately the first 5 dimensions, more faithfully addressing the structure of the data. This method allows data comparison between patients with different outcomes, as well as defining amongst statistically significant DEG those contributing most to distinguishing G345 progressors from G2 non-progressors. Generally, the more distant the groups and the closer the patient samples are within each group, the better the prognostic value of any given signature.
Hierarchical clustering of the entire set of genes did not clearly separate the samples into patient groups (Figures 2A, 2B). However, the DEG G345e vs. G2 (Figure 2G), G345m vs. G2 (Figure 2H), and G345l vs. G2 (Figure 2I) improved separation of the liver transplant patients from the UNP G1 control group and concomitantly provide fewer distinctions between G2 and G345. This behavior is concordant with the time-specific analysis discussed above and echoed by the G345eml vs. G2 DEG (Figure 2J). Therefore, DEG associated with severe disease were harder to detect over time, indicating that early events play a decisive role in the development of severe liver disease, and lead to a variety of observable phenotypes at later stages. Importantly, the SVD-MDS analysis also revealed that both G2 and G345 patient groups increasingly differentiated from the G1 UNP controls, which represent pooled normal liver gene expression profiles. This indicates a slow evolution to more heterogeneous gene expression regardless of clinical outcome. While the nature of this evolution is somewhat unclear, this poses important questions regarding the stochasticity of liver disease progression kinetics and suggests that decisive early transcriptional repression of select inflammatory mediators, cell cycle regulators, and genes involved in both lipid biogenesis and catabolism predict disease progression.
We also directly compared time-matched G2 and G345 samples. Consistent with the first analysis, clustering analysis showed that gene expression alone was insufficient to segregate patients according to clinical outcome (Figure S1). These DEG were similarly repressed and functionally consistent with significant DEG identified in the first analysis. These results thus confirm that early events post-OLT are detrimental to liver physiology. Note that we refrained from providing direct G2 versus G3 or G4 or G5 comparisons, as the amount of available biopsies in this cohort is too small to provide for robust insights. Such direct comparisons will become feasible in the analysis of a larger independent cohort, or in meta-analysis across different cohorts.
Gene expression profiles associated with liver disease progression over time
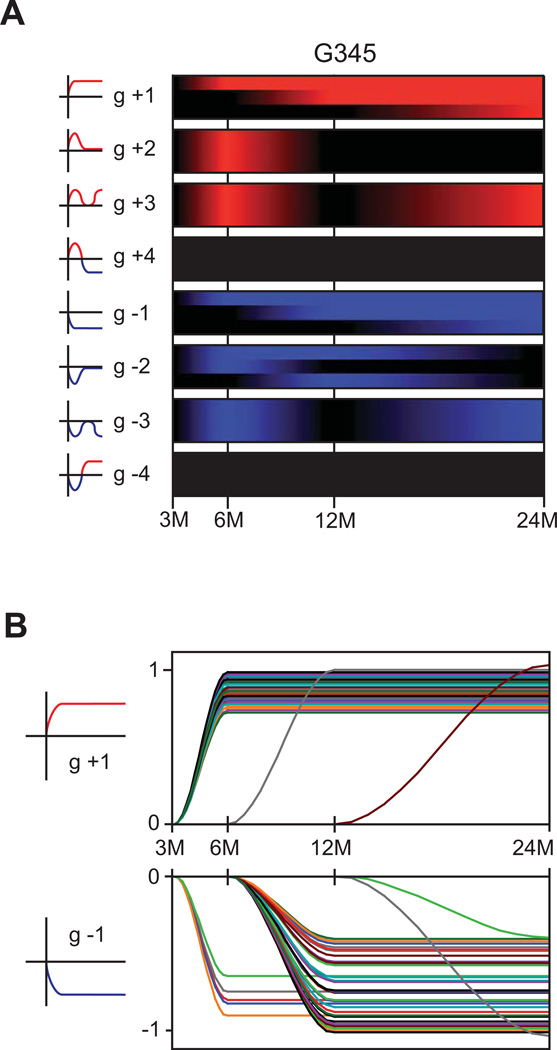
To better understand how early events lead to severe liver disease and compare differences in gene expression patterns over time within each individual disease group, we performed a longitudinal kinetic analysis. We used a recently utilized classifier (7) derived from the analysis of many different longitudinal publicly available and in-house datasets using a Kohonen Maps approach, which fits gene expression to topographic maps representing distinct regulatory patterns. Our classifier comprised relevant topographic groups: g1– 6 for initial positive regulation, a neutral g0 group for genes expressed but unchanged, and the mirror g-1 to g-6 groups for negative regulation (Figure 3A). The classifier tests for statistical significance (fold change-based Z test) of association with individual topographic groups by testing the statistical significance of the logarithmic fold change difference in expression of every individual gene at every time point against its estimated baseline, with absolute expression change rescaled to unity. Thus, gene expression was analyzed for its characteristic, statistically significant “shape” over time, rather than magnitude of change. We further sub-divided the time categories to generate a fourth category (Figure 1B). As we were mainly interested in identifying genes involved in severe liver disease development, we focused on genes which permanently change expression over time (Figure 3B; Table S2).
Figure 3. Longitudinal analysis of the transcriptome dynamics over time in the different patient groups.
(A). Heatmaps in form of the topographic profiles of the longitudinal analysis of combined patient groups G3–G5. A schematic representation of the topographic profiles is given to the left. Time categories as defined in Figure 1. Red coloring indicates upregulation, blue, downregulation. Time-line is in months (M) post-transplantation. (B). Gene-level kinetics for genes according to the topographic profiles g+1 and g-1 for the combined G3–G5 patient groups. Note that all statistically significant changes from the base-line estimate were scaled to unity and splines were estimated for the graphical representation
Using IPA, we categorized 48 genes related to inflammatory responses and immune cell trafficking, particularly phagocyte and lymphocyte recruitment and chemotaxis, including many C-X-C and C-C chemokines and chemokine receptors. Also we observed molecules bridging innate and adaptive immune functions, including signal transduction and activation of immune and inflammatory transcriptional responses, proinflammatory cytokines, Fc receptors, complement components, ISGs, HLA alleles, and lymphocyte activation. We also identified increases in genes associated with HSC activation and collagen deposition, including TIMP metalloproteinase inhibitor (TIMP1), LGALS3, and multiple collagen transcripts. Finally, 59 genes associated with cancer also gradually increased, including many associated with metastasis, cell proliferation, and cell death, indicating that dysregulation of normal cell division and apoptotic mechanisms underlie hepatic inflammation and collagen deposition.
We also evaluated the functional significance of DEG downregulated over time following OLT. We identified 12 genes associated with lipid, drug, vitamin and mineral, and carbohydrate metabolism. These are involved in lipid biosynthesis, fatty acid oxidation, and amino acid and glucose metabolism. G345 patients therefore demonstrated reduced hepatic metabolic function, consistent with reductions in metabolic activity previously observed at late timepoints in HCV-infected hepatic cells in vitro (12). Additionally, we identified 11 genes associated with cancer, specifically those involved in cell cycle control such as retinoblastoma-like 2 (RBL2) and cyclin-dependent kinase inhibitor 3 (CDKN3), and regulators of cellular differentiation. As cancer-related genes associated with cellular proliferation steadily increased, those associated with cell cycle checkpoint control and cell type specification were downregulated. This indicates that patients with progressive liver disease experience a loss of differentiation and checkpoint cell cycle arrest, consistent with the concordant gradual increase in proliferative capacity. This also suggests a mechanism by which chronic HCV infection contributes to tumorigenesis of hepatocellular carcinoma
Gene signature characterizing a precursor state for severe liver disease
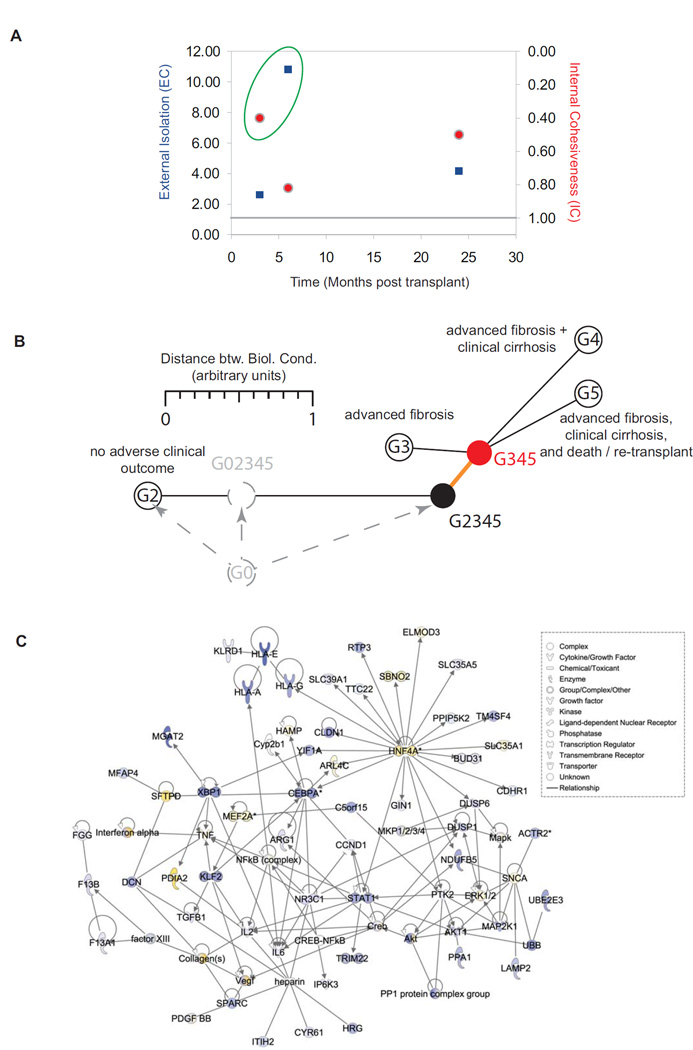
The SVD-MDS method used in the analysis presented in figures 2G–J and figure S1G–J allows the computation of two additional parameters aside from the Kruskal stress (information-loss during dimensionality reduction): external isolation (the arithmetic average inter-group distance) and internal cohesiveness (the intra-group distance). Both parameters determined for the analyses peak 3–6 months post-OLT (Figure 4A), indicating that the signatures derived from these time-categories generate the relative maximal resolution. Hence, the early stages of HCV reinfection best characterize overall clinical outcome. We then used the time-specific analysis to define a gene expression pattern-based distance measure between any of the individual groups and with combined G2345 and G345, as well as G45 longitudinal analysis. To investigate severe liver disease progression according to time and patient outcome, these measures were then subjected to k-means clustering (13) using inter-group distances as additional constraints. This analysis indicates the existence of a common precursor state G345 for all progressor groups (Figure 4B, red), from which all three adverse outcomes split individually. This precursor state is comprised of 35 DEG (Table 2), which distinguish the transformation to a progressive disease outcome long before histological or clinical evidence of severe disease.
Figure 4. The cellular network of HCV-induced progression to severe liver disease.
(A) The two parameters characterizing the predictive performance of the different gene expression signatures plotted against time. The External Isolation (blue) parameter is derived from the analyses presented in Figure 2, the Internal Cohesiveness (red) is computed from the analyses in Figure 3. Both parameters are expressed as fractions of their values over the entire transcrioptomes for which they were arbitrary set to unity. The peak-values with respect to performance are highlighted by the green circle. (B) A common precursor state to clinical outcomes G3–G5 can be identified by distance-based k-means clustering under constraint using the joint longitudinal profiles G2345 and G345 but not G45 as seeds. Distance between biologic conditions is used as additional constraint in the distance to seed based clustering. In absence of non-infected liver biopsies over time (provisionally: "G0"), the order between G2 and G2345 cannot be established. (C) The cellular network differentially expressed between joint G2345 and joint G345 responsible for the G2345 to G345 transition (orange edge), merged with the G345e time-specific analysis gene sets, and built based on direct interactions between molecules. Data overlaid is the averaged G345e/G2 log 2 ratio expression data. Upregulated genes are shaded in yellow, and downregulated genes are shaded in blue.
Table 2.
Differentially expressed genes associated with transition to progressive disease
| Accession Number |
Symbol | Entrez Gene Name | Location | Molecule Type |
|---|---|---|---|---|
| NM_018849 | ABCB4 | ATP-binding cassette, sub-family B (MDR/TAP), member 4 | PM | transporter |
| NM_005891 | ACAT2 | acetyl-CoA acetyltransferase 2 | Cytoplasm | enzyme |
| ACTR2 | ARP2 actin-related protein 2 homolog (yeast) | PM | other | |
| NM_001005386 | ||||
| NM_000045 | ARG1 | arginase, liver | Cytoplasm | enzyme |
| NM_018840 | C20orf24 | chromosome 20 open reading frame 24 | Cytoplasm | other |
| NM_020199 | C5orf15 | chromosome 5 open reading frame 15 | unknown | other |
| NM_173060 | CAST | calpastatin | Cytoplasm | peptidase |
| NM_053056 | CCND1 | cyclin D1 | Nucleus | other |
| NM_021101 | CLDN1 | claudin 1 | PM | other |
| NM_001554 | CYR61 | cysteine-rich, angiogenic inducer, 61 | ECS | other |
| NM_001946 | DUSP6 | dual specificity phosphatase 6 | Cytoplasm | phosphatase |
| NM_001994 | F13B | coagulation factor XIII, B polypeptide | Cytoplasm | enzyme |
| NM_013402 | FADS1 | fatty acid desaturase 1 | PM | enzyme |
| NM_014923 | FNDC3A | fibronectin type III domain containing 3A | Cytoplasm | other |
| NM_002116 | HLA-A | major histocompatibility complex, class I, A | PM | TM receptor |
| U88244 | HLA-G | major histocompatibility complex, class I, G | PM | TM receptor |
| NM_000412 | HRG | histidine-rich glycoprotein | ECS | other |
| NM_015525 | IBTK | inhibitor of Bruton agammaglobulinemia tyrosine kinase | Cytoplasm | other |
| NM_004221 | IL32 | interleukin 32 | ECS | other |
| NM_002216 | ITIH2 | inter-alpha (globulin) inhibitor H2 | ECS | other |
| AB067508 | KLHL29 | kelch-like 29 (Drosophila) | unknown | other |
| NM_002294 | LAMP2 | lysosomal-associated membrane protein 2 | PM | enzyme |
| NM_002404 | MFAP4 | microfibrillar-associated protein 4 | ECS | other |
| NM_002408 | MGAT2 | mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase | Cytoplasm | enzyme |
| NM_000277 | PAH | phenylalanine hydroxylase | Cytoplasm | enzyme |
| NM_021129 | PPA1 | pyrophosphatase (inorganic) 1 | Cytoplasm | enzyme |
| NM_015216 | PPIP5K2 | diphosphoinositol pentakisphosphate kinase 2 | Cytoplasm | other |
| NM_020532 | RTN4 | reticulon 4 | Cytoplasm | other |
| NM_014624 | S100A6 | S100 calcium binding protein A6 | Cytoplasm | transporter |
| NM_006918 | SC5DL | sterol-C5-desaturase (ERG3 delta-5-desaturase homolog, S. cerevisiae)-like | Cytoplasm | enzyme |
| NM_019844 | SLCO1B3 | solute carrier organic anion transporter family, member 1B3 | PM | transporter |
| NM_001060 | TBXA2R | thromboxane A2 receptor | PM | GPCR |
| NM_004617 | TM4SF4 | transmembrane 4 L six family member 4 | PM | other |
| NM_006074 | TRIM22 | tripartite motif containing 22 | Cytoplasm | transcription regulator |
| NM_006357 | UBE2E3 | ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) | Nucleus | enzyme |
In the absence of time-resolved samples from healthy, non-HCV patients, we were not able to determine whether a common G2345 (Figure 4B, black) state exists or how this hypothetical intermediate state would relate to G2 and G345. More importantly, the predicted common G345 precursor state confirmed our observation that eventual severe liver disease is programmed early post-OLT, and in combination with the time-specific analyses described above, identified DEG distinguishing progressors and non-progressors within 6 months of transplantation.
Using IPA, we generated a network of directly interacting molecules based on the network analysis of the transitional signature and the G345e time-specific gene sets (Figure 4C, Figure S2). We confirmed that repression of genes involved in cell cycle regulation and stress responses (cyclin D1, CCND1, and X-box binding protein, XBP1), innate immunity (signal transducer and activator of transcription 1, STAT1), and antigen presentation (HLA-A, HLA-G, and HLA-E) characterize transition to a progressive phenotype. Additionally, collagen upregulation was detected within several months of transplantation, months or years before fibrosis is histologically detectable. Coupled with our finding that the statistically significant upregulation of collagen expression correlates with disease progression over time, this indicates that collagen transcription is both critical to the mechanism of fibrogenesis and potentially useful as a predictive marker to identify patients at risk of HCV-induced liver disease prior to extensive collagen deposition and associated liver damage.
Discussion
Investigating the influence of transcriptional profiles on clinical outcome in patients following transplantation could lead to more refined prognostic models. This study represents the first in which SVD-MDS analysis has been used to identify contributions of significant DEG associated with HCV-induced liver disease progression. The SVD-MDS method reduced dimensionality by removing dimensions with little information (high biological noise) and emphasizing the main contributing dimensions. This is a significant advantage over clustering techniques, which here failed to provide meaningful biological insight. In this context, SVD-MDS demonstrates that pertinent information contained in the entire set of measured transcript abundances is enriched during the statistical analysis. The unique molecular profiles that distinguish patients who develop severe liver disease provide insight into the biological mechanism of disease progression, both prior to the advent of disease and over time. Furthermore, they provide a basis for larger validation studies or meta-analysis across additional different cohorts of HCV patients in future efforts to establish definite molecular correlates.
Our transitional signature suggests that the key regulators of a precursor state leading to progression play the most critical role at early to intermediate time points post-OLT. Patients who eventually develop the most severe liver disease may be most clearly distinguished by DEG within three months post-OLT compared to patients who do not progress. Specifically we observed a broad repression of genes related to antigen presentation, immune responses, and cell cycle regulation in patients who progress. This suggests that long-term clinical outcome is determined by early reprogramming of the donor liver during recurrence, and specifically by blunting responses that prevent unchecked inflammation and cell division. These processes are directly connected to hallmarks of HCV-induced hepatic disease such as chronic inflammatory hepatitis, cirrhosis, and hepatocellular carcinoma.
While downregulation of immune transcripts is expected in patients taking immunosuppressant drugs to prevent allograft rejection, in patients who developed severe disease we observed profound repression of genes related to antiviral responses. This distinguishes this signature with regard to long-term outcome compared to the clinical situation early post-OLT. Acute cellular rejection (ACR) is difficult to distinguish from HCV recurrence based on analysis of patient liver biopsies, due to common histological features. Previous studies comparing HCV patients with and without ACR demonstrate that many of the repressed genes are significantly upregulated during ACR in HCV patients (3, 14). Additionally, repression of innate and inflammatory genes was characteristic of HCV recurrence rather than ACR in HCV transplant patients (15). This indicates that while short-term clinical factors such as ACR may confound long-term efforts to develop molecular signatures of liver disease pathogenesis, repression of these innate immune genes is more widespread and of greater magnitude.
Immune repression early in infection may contribute to increased hepatocyte infection during HCV recurrence, and thus create a more favorable environment for progression to severe disease. Although antigen presentation has been associated with HCV pathogenesis (16–18), these pathways are normally suppressed by allograft rejection drugs, causing impaired T cell responses in HCV transplant patients. However, it is difficult to determine the effect of specific immunosuppressive regimens as patients are routinely treated with different drugs and dosing regimens. Generally, the immunosuppressive regimens used are less likely to repress innate immune responses that could attenuate the severity of HCV recurrence. Innate immune antagonism by HCV infection may result in the virus eliciting a transcriptional program that eventually results in fibrosis and disease progression, which is partially reflected by the increase in inflammatory genes over time caused by infiltrating leukocytes.
HCV facilitates its replication by antagonizing induction of antiviral interferons, ISGs, and antiviral cytokines through the action of the viral NS3/4 protease and NS5A non-structural protein (19–22). Clinicians have not routinely treated HCV patients with post-OLT ribavirin and PEG-interferon, primarily because the high expense and harsh side effects of this treatment regimen do not justify its use in patients recovering from organ transplantation. However, a recent study demonstrated that post-OLT treatment resulted in stable or improved fibrosis scores, even in some patients who did not demonstrate sustained virologic response (SVR) (23). Our data indicating that repressed antiviral gene expression early in infection determines transition to severe disease and suggests that patients may benefit from early therapeutic intervention during HCV recurrence to boost innate immune genes not impacted by immunosuppressant drugs during the first 3 months post-OLT.
Early repression of cell division mediators in patients who progress also indicates that these transcriptional profiles are altered. These genes are largely involved in controlling cell cycle progression and transcriptional activation of related pathways, and their repression suggests induction of cell cycle arrest. In vitro, HCV infection induces cell cycle arrest early in infection to facilitate viral replication (8, 24, 25). The repression of genes promoting cell cycle progression may represent a greater number of infected hepatocytes, suggesting that more severe cases of recurrence facilitate a hepatic environment which augments further cell cycle dysregulation. As our kinetic analysis indicates, cell cycle regulators decrease over time in patients who progress, while genes promoting cell division, such as growth factors, continue to increase. One of these genes, CDKN3, regulates specific cell cycle networks related to HCV-induced cirrhosis and hepatocellular carcinoma (26), supporting the notion that early cell cycle arrest occurring in infected hepatocytes can result in the loss of key regulatory functions over time and promote eventual tumorigenesis. Additional repression of genes such as BRCA1, which are critical mediators of DNA damage repair, may result in genetic lesions that also contribute to cell death and eventual oncogenesis, as is the case for the BRCA1-interacting gene BRE, which promotes HCC growth (27). Increasingly altered cell cycle regulation contributes to the altered mitotic state created by initial repression of cell cycle regulators early in infection, and ultimately leads to cell death, aberrant proliferation, and potentially cancer.
Our analysis demonstrates the dynamic transcriptional response elicited by HCV in the post-transplant setting. Early repression of innate immunity and cell cycle progression may establish a state in the donor organ facilitating viral replication and establishment of more widespread chronic infection. Also contributing to this is the increasing presence of collagens and other fibrogenic transcripts. After three months post-OLT, this hepatic reprogramming mediates transition to progressive disease characterized by gradual increases in DEG associated with inflammation, HSC activation, collagen deposition, cell proliferation, and cell death, and decreases in genes related to cell cycle control. Our study identifies a signature early during recurrence consistent with early cellular responses to HCV infection distinguishing progressors in the post-transplant setting. This yields insight into the earliest host responses to HCV recurrence and raises the exciting possibility of identifying and treating patients based on transcriptional profiling long before disease progression or significant damage to the donor organ.
Experimental Procedures
Additional detail regarding methods can be found in supplementary material.
Human Liver Tissue Samples. Core needle liver biopsies were obtained from liver transplant patients at the University of Washington Medical Center (UWMC). All patients provided informed consent according to protocols approved by the Human Subject Review Committee at the University of Washington. No donor organs were obtained from executed prisoners or other institutionalized persons.
Data processing and normalization for SVD-MDS analysis. Microarray raw data were extracted using the Bioconductor limma package (28), and median normalized. For inter-assay comparisons and longitudinal analysis the NeONORM method was used for normalization (29).
Identification of statistically significantly differentially expressed genes. Differentially expressed genes have been identified using a fold-change based z-test statistic (with a fold-change parameter of 1.2; p<0.01).
SVD-MDS dimensionality reduction and derived representations. SVD-MDS dimensionality reduction and subsequent 2D representations have been obtained using the SVD-MDS method (6). Kruskal Stress represents information loss due to dimensionality reduction/representation as a fraction of total information. The geometric objects (transcriptomic data for individual genes in different samples at different times) are non-linearly deformed (MDS), rotated into the principal non-linear dimensions (SVD), and then projected onto the plane. Therefore, the 2D representation captures features of the geometric objects which would otherwise only be visible in a space of higher dimension. As the non-linearity is not uniform, this space of higher dimension is not exactly defined but typically corresponds to a space of 2–4 dimensions higher than that of the visual representation. SVD-MDS performs better than hierarchical clustering in this setting because it accounts for several of the principal dimensions of the data.
Longitudinal time-series analysis. Longitudinal analysis was achieved using the same methodology as employed in (7). Briefly, a Kohonen Maps-based classifier tests association of any given gene with 13 topographic profiles. The profiles correspond to increasing regulatory complexity and subsequent profiles are modeled using increasing numbers of events.
Data accessibility. Data were warehoused in a Labkey system (Labkey, Inc., Seattle, WA). Primary data are available in accordance with proposed Minimum Information About a Microarray Experiment (MIAME) standards (http://viromics.washington.edu). Also, data are available from the MACE database (http://mace.ihes.fr) using accession number: 2491581318.
Supplementary Material
Acknowledgements
The authors thank James Perkins and Renuka Bhattacharya for clinical support.
Financial support: This work was funded by the National Institute on Drug Abuse grant 1P30DA01562501.
Abbreviations
- ACR
acute cellular rejection
- BRCA1
breast cancer 1, early onset
- BRE
brain and reproductive organ-expressed
- CCND1
cyclin D1
- CDKN3
cyclin-dependent kinase inhibitor 3
- COL
collagen
- DEG
differentially expressed genes
- ECM
extracellular matrix
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HLA
human leukocyte antigen
- HSC
hepatic stellate cell
- IPA
Ingenuity Pathways Analysis
- ISG
interferon-stimulated gene
- ISG15
interferon stimulated gene 15
- LGALS3
galectin 3
- MELD
model for end-stage liver disease
- MDS
multidimensional scaling
- OLT
orthotopic liver transplant
- RBL2
retinoblastoma-like 2
- SPP1
secreted phosphoprotein 1 (osteopontin)
- STAT1
signal transducer and activator of transcription 1
- SUMO2
small ubiquitin-like modifier 2
- SVD
singular value decomposition
- SVR
sustained virologic response
- TIMP1
TIMP metalloproteinase inhibitor 1
- UNP
universal normal pool
- XBP1
X-box binding protein 1
References
- 1.Berenguer M, Prieto M, Rayon JM, Mora J, Pastor M, Ortiz V, Carrasco D, et al. Natural history of clinically compensated hepatitis C virus-related graft cirrhosis after liver transplantation. Hepatology. 2000;32:852–858. doi: 10.1053/jhep.2000.17924. [DOI] [PubMed] [Google Scholar]
- 2.Rowe IA, Barber KM, Birch R, Curnow E, Neuberger JM. Retransplantation for graft failure in chronic hepatitis C infection: a good use of a scarce resource? World J Gastroenterol. 2010;16:5070–5076. doi: 10.3748/wjg.v16.i40.5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asaoka T, Kato T, Marubashi S, Dono K, Hama N, Takahashi H, Kobayashi S, et al. Differential transcriptome patterns for acute cellular rejection in recipients with recurrent hepatitis C after liver transplantation. Liver Transpl. 2009;15:1738–1749. doi: 10.1002/lt.21883. [DOI] [PubMed] [Google Scholar]
- 4.Smith MW, Walters KA, Korth MJ, Fitzgibbon M, Proll S, Thompson JC, Yeh MM, et al. Gene expression patterns that correlate with hepatitis C and early progression to fibrosis in liver transplant recipients. Gastroenterology. 2006;130:179–187. doi: 10.1053/j.gastro.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 5.Mas V, Maluf D, Archer KJ, Potter A, Suh J, Gehrau R, Descalzi V, et al. Transcriptome at the time of HCV recurrence may predict the severity of fibrosis progression after liver transplantation. Liver Transpl. 2011 doi: 10.1002/lt.22309. [DOI] [PubMed] [Google Scholar]
- 6.Becavin C, Tchitchek N, Mintsa-Eya C, Lesne A, Benecke A. Improving the efficiency of multidimensional scaling in the analysis of high-dimensional data using singular value decomposition. Bioinformatics. 2011;27:1413–1421. doi: 10.1093/bioinformatics/btr143. [DOI] [PubMed] [Google Scholar]
- 7.Jacquelin B, Mayau V, Targat B, Liovat AS, Kunkel D, Petitjean G, Dillies MA, et al. Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J Clin Invest. 2009;119:3544–3555. doi: 10.1172/JCI40093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walters KA, Syder AJ, Lederer SL, Diamond DL, Paeper B, Rice CM, Katze MG. Genomic analysis reveals a potential role for cell cycle perturbation in HCV-mediated apoptosis of cultured hepatocytes. PLoS Pathog. 2009;5:e1000269. doi: 10.1371/journal.ppat.1000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blackham S, Baillie A, Al-Hababi F, Remlinger K, You S, Hamatake R, McGarvey MJ. Gene expression profiling indicates the roles of host oxidative stress, apoptosis, lipid metabolism, and intracellular transport genes in the replication of hepatitis C virus. J Virol. 2010;84:5404–5414. doi: 10.1128/JVI.02529-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramirez S, Perez-Del-Pulgar S, Forns X. Virology and pathogenesis of hepatitis C virus recurrence. Liver Transpl. 2008;14(Suppl 2):S27–S35. doi: 10.1002/lt.21644. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe N, Aizaki H, Matsuura T, Kojima S, Wakita T, Suzuki T. Hepatitis C virus RNA replication in human stellate cells regulates gene expression of extracellular matrix-related molecules. Biochem Biophys Res Commun. 2011;407:135–140. doi: 10.1016/j.bbrc.2011.02.125. [DOI] [PubMed] [Google Scholar]
- 12.Diamond DL, Syder AJ, Jacobs JM, Sorensen CM, Walters KA, Proll SC, McDermott JE, et al. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 2010;6:e1000719. doi: 10.1371/journal.ppat.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lloyd SP. Least-Squares Quantization in Pcm. Ieee Transactions on Information Theory. 1982;28:129–137. [Google Scholar]
- 14.Sreekumar R, Rasmussen DL, Wiesner RH, Charlton MR. Differential allograft gene expression in acute cellular rejection and recurrence of hepatitis C after liver transplantation. Liver Transpl. 2002;8:814–821. doi: 10.1053/jlts.2002.35173. [DOI] [PubMed] [Google Scholar]
- 15.Gehrau R, Maluf D, Archer K, Stravitz R, Suh J, Le N, Mas V. Molecular pathways differentiate hepatitis C virus (HCV) recurrence from acute cellular rejection in HCV liver recipients. Mol Med. 2011;17:824–833. doi: 10.2119/molmed.2011.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romero-Gomez M, Eslam M, Ruiz A, Maraver M. Genes and hepatitis C: susceptibility, fibrosis progression and response to treatment. Liver Int. 2011;31:443–460. doi: 10.1111/j.1478-3231.2011.02449.x. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt J, Neumann-Haefelin C, Altay T, Gostick E, Price DA, Lohmann V, Blum HE, et al. Immunodominance of HLA-A2-restricted hepatitis C virus-specific CD8+ T cell responses is linked to naive-precursor frequency. J Virol. 2011;85:5232–5236. doi: 10.1128/JVI.00093-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt J, Thimme R, Neumann-Haefelin C. Host genetics in immune-mediated hepatitis C virus clearance. Biomark Med. 2011;5:155–169. doi: 10.2217/bmm.11.19. [DOI] [PubMed] [Google Scholar]
- 19.Kriegs M, Burckstummer T, Himmelsbach K, Bruns M, Frelin L, Ahlen G, Sallberg M, et al. The hepatitis C virus non-structural NS5A protein impairs both the innate and adaptive hepatic immune response in vivo. J Biol Chem. 2009;284:28343–28351. doi: 10.1074/jbc.M109.038877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sene D, Levasseur F, Abel M, Lambert M, Camous X, Hernandez C, Pene V, et al. Hepatitis C virus (HCV) evades NKG2D-dependent NK cell responses through NS5A-mediated imbalance of inflammatory cytokines. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001184. e1001184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson CL, Owen DM, Gale M., Jr Functional and therapeutic analysis of hepatitis C virus NS3.4A protease control of antiviral immune defense. J Biol Chem. 2007;282:10792–10803. doi: 10.1074/jbc.M610361200. [DOI] [PubMed] [Google Scholar]
- 22.Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, et al. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A. 2005;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Testino G, Sumberaz A, Ansaldi F, Borro P, Leone S, Ancarani AO, Gentile R, et al. Treatment of recurrent hepatitis C (genotype 1) with pegylated interferon alfa-2b and ribavirin combination and maintenance therapy. Hepatogastroenterology. 2011;58:536–538. [PubMed] [Google Scholar]
- 24.Denard B, Seemann J, Chen Q, Gay A, Huang H, Chen Y, Ye J. The Membrane-Bound Transcription Factor CREB3L1 Is Activated in Response to Virus Infection to Inhibit Proliferation of Virus-Infected Cells. Cell Host Microbe. 2011;10:65–74. doi: 10.1016/j.chom.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kannan RP, Hensley LL, Evers LE, Lemon SM, McGivern DR. Hepatitis C virus infection causes cell cycle arrest at the level of initiation of mitosis. J Virol. 2011;85:7989–8001. doi: 10.1128/JVI.00280-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Sun L, Huang J, Jiang M. Cyclin-dependent kinase inhibitor 3 (CDKN3) novel cell cycle computational network between human non-malignancy associated hepatitis/cirrhosis and hepatocellular carcinoma (HCC) transformation. Cell Prolif. 2011;44:291–299. doi: 10.1111/j.1365-2184.2011.00752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chui YL, Ching AK, Chen S, Yip FP, Rowlands DK, James AE, Lee KK, et al. BRE over-expression promotes growth of hepatocellular carcinoma. Biochem Biophys Res Commun. 2010;391:1522–1525. doi: 10.1016/j.bbrc.2009.12.111. [DOI] [PubMed] [Google Scholar]
- 28.Smyth GK. Limma : Linear Models for Microarray Data. In: Gentleman VC R, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 29.Noth S, Brysbaert G, Benecke A. Normalization using weighted negative second order exponential error functions (NeONORM) provides robustness against asymmetries in comparative transcriptome profiles and avoids false calls. Genomics Proteomics Bioinformatics. 2006;4:90–109. doi: 10.1016/S1672-0229(06)60021-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.