Abstract
Plants contain RNA-dependent RNA polymerase (RdRP) activities that synthesize short cRNAs by using cellular or viral RNAs as templates. During studies of salicylic acid (SA)-induced resistance to viral pathogens, we recently found that the activity of a tobacco RdRP was increased in virus-infected or SA-treated plants. Biologically active SA analogs capable of activating plant defense response also induced the RdRP activity, whereas biologically inactive analogs did not. A tobacco RdRP gene, NtRDRP1, was isolated and found to be induced both by virus infection and by treatment with SA or its biologically active analogs. Tobacco lines deficient in the inducible RDRP activity were obtained by expressing antisense RNA for the NtRDRP1 gene in transgenic plants. When infected by tobacco mosaic virus, these transgenic plants accumulated significantly higher levels of viral RNA and developed more severe disease symptoms than wild-type plants. After infection by a strain of potato virus X that does not spread in wild-type tobacco plants, the transgenic NtRDRP1 antisense plants accumulated virus and developed symptoms not only locally in inoculated leaves but also systemically in upper uninoculated leaves. These results strongly suggest that inducible RdRP activity plays an important role in plant antiviral defense.
RNA-dependent RNA polymerase (RdRP) activities have been detected in a number of plants (1–10). Recently, a tomato RdRP has been purified and its corresponding gene cloned (11–13). The purified tomato RdRP catalyzes in vitro the transcription of single-stranded RNA and DNA molecules into short cRNAs (11). Although similar RdRP activities have not been reported in other types of eukaryotic organisms, sequences homologous to plant RdRPs have been found in Schizosaccharomyces pombe (13), Neurospora crassa (14), and Caenorthaditis elegans (15).
Plant RdRPs have been implicated in posttranscriptional gene silencing (PTGS) (13), which is often manifested as an increased turnover of specific RNAs after introduction of homologous sequences in the plant genome. Consistent with an important role in PTGS, mutations in the genes encoding proteins similar to plant RdRPs abolish gene quelling in N. crass and RNA interference in C. elegans (14, 15). More recently, a RdRP gene (SDE1 or SGS2) has also been shown to be required for PTGS in Arabidopsis (16, 17). The current models indicate that PTGS involves double-stranded RNA (dsRNA) corresponding to a sense and antisense sequence of an endogenous mRNA (18). The dsRNA may be recognized and cleaved by a specific RNase to generate small dsRNAs that may subsequently serve as templates for sequence-specific cleavage of the corresponding mRNA (18). RdRPs may play an important role in the amplification of the dsRNA signal (18).
PTGS has been thought to be a major component in plant antiviral response (19). In a number of reported studies, viruses have been shown to induce an RNA-mediated defense similar to PTGS in plants that contain no homologous sequences in the nuclear genome (20, 21). Plants exhibiting the induced silencing state usually contain low levels of viral RNA, lack symptoms, and are immune to reinfection by viruses containing sequence homology to the silenced virus (20). Consistent with the role of PTGS in antiviral defense, several studies have reported mechanisms by which viruses counter this defense through suppression of PTGS (22–25). Because of the close connection between PTGS and antiviral defense, Arabidopsis sde1/sgs2 mutants were tested for virus resistance but, surprisingly, they exhibited enhanced susceptibility to only one of the five viruses tested (16, 17). These results raised a question of whether RdRPs have a general role in plant antiviral response (17).
In the present study, we report that infection by tobacco mosaic virus (TMV) or treatment with SA increased activity of a tobacco RdRP and induced expression of the corresponding gene. Transgenic antisense tobacco plants deficient in the inducible RdRP activity were more susceptible to both TMV and potato virus X (PVX). These results indicate that this inducible RdRP and/or other closely related RdRPs play an important role in plant antiviral defense.
Materials and Methods
Materials.
Tobacco (Nicotiana tabacum cv. Xanthi, nn genotype) plants used throughout the experiments were grown at 24°C in a 14-h light cycle. TMV (U1 strain) and PVX (a mild strain collected from a potato field in Idaho) were mechanically inoculated on leaf discs or whole leaves of 5-week-old tobacco plants by rubbing the virus (10 μg/ml solution in 5 mM sodium phosphate, pH 7.5) with carborundum. Mock inoculation was performed with the phosphate buffer only. Because mechanical inoculation caused wounding, TMV-induced RdRP activity and gene expression were analyzed 15 days after inoculation on the leaves directly above the inoculated ones (usually the fifth ones above).
Isolation and Assays of RdRP Activity.
Leaf tissues (≈250 mg) of 5-week-old tobacco plants were homogenized in 400 μl of buffer A (50 mM Tris-acetate, pH 7.4/10 mM potassium acetate/1 mM EDTA/10 mM β-mercaptoethanol/0.5 mM phenylmethylsulfonyl fluoride). The homogenates were centrifuged at 1,000 × g for 12 min, and the supernatants were collected and centrifuged again at 14,000 × g for 10 min after adding glycerol to a final concentration of 20%. An equal volume of 4 M (NH4)2SO4 was added to the supernatants, and precipitated proteins were collected by centrifugation at 10,000 × g for 20 min and resuspended in 80–100 μl of buffer B (25 mM Tris-acetate, pH 8.2/1 mM EDTA/20% glycerol/3 mM β-mercaptoethanol). The proteins were dialyzed overnight against buffer B. All of the preparation steps were carried out at 4°C.
The enzymatic assays were performed in a final volume of 50 μl containing 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 1 mM DTT, 0.1 mM phenylmethylsulfonyl fluoride, 1 mM each of ATP, GTP, and UTP, 2 μM CTP, 5μCi [32P]-CTP, 400 μg/ml bentonite, 4 μg total RNA extracted from TMV-infected tobacco leaves, and 20–25 μg protein extracts. The reaction was allowed to proceed for 1 h at 30°C and terminated by adding 50 μl phenol/chloroform/isoamylalcohol (25:24:1) mixture. The aqueous phase from the extraction was precipitated with isopropanol, resuspended in 20 μl of water containing 0.2% SDS, and resolved on an 8% polyacrylamide gel containing 8 M urea and 1% SDS in 1× TBE buffer (98 mM Tris-borate/2 mM EDTA).
Isolation and Characterization of NtRdRP1.
A tobacco cDNA library (in ZAP Express λ vector) of 106 phages was screened by using a 32P-labeled NtRdRP gene fragment amplified by PCR by using two NtRdRP1-specific primers (5′-CAGAGGGTTTTGAGGCAAAG-3′ and 5′-ACACTGCAAACTTCTTGTCTGA-3′). The hybridization was performed as described (26).
Northern Blotting.
Total RNA was extracted from frozen plants by using the TRIZOL reagent (BRL Life Technologies) according to the manufacturer's instructions. RNA was separated on agarose (1.2%)-formaldehyde gels and blotted onto nylon membrane. Hybridization was performed by using random-primed 32P-labeled DNA probes or strand-specific riboprobes, as described (26).
Western Blotting.
Preparation of soluble proteins, electrophoresis, and blotting were performed as described (27). Immunoblot analysis was performed with a 1:1,000 dilution of a rabbit PVX-specific antibody conjugated with radish peroxidase (Agdia, Elkhart, IN), as described (27).
Construction and Characterization of Transgenic NtRdRP1 Antisense Plants.
A 3.8-kb NtRdRP1 cDNA fragment was subcloned behind the cauliflower mosaic virus 35S promoter in antisense orientation in plant transformation vector pCK8 (28). Tobacco transformation was performed with the Agrobacterium-mediated procedure (29). Transformants were screened by directly assaying SA-inducible RdRP activity twice to ensure reproducibility.
Results
SA Induces a RdRP Activity in Tobacco.
We made our initial observation of induction of a tobacco RdRP activity from experiments designed to test whether TMV replicase was inhibited in SA-treated tobacco plants. In these experiments, tobacco plants were first inoculated with TMV and subsequently treated with SA. Crude membrane extracts were prepared from these plants and analyzed for TMV replicase activity. In these assays, we detected synthesis of not only the high molecular weight TMV RNAs by TMV replicase but also smaller RNAs by an unknown RNA polymerase. Synthesis of smaller RNAs has been previously observed in similar assays and attributed to the activity of a plant RdRP (5, 8–10). Interestingly, in our assays, synthesis of these smaller RNAs was drastically increased in SA-treated tobacco plants (data not shown).
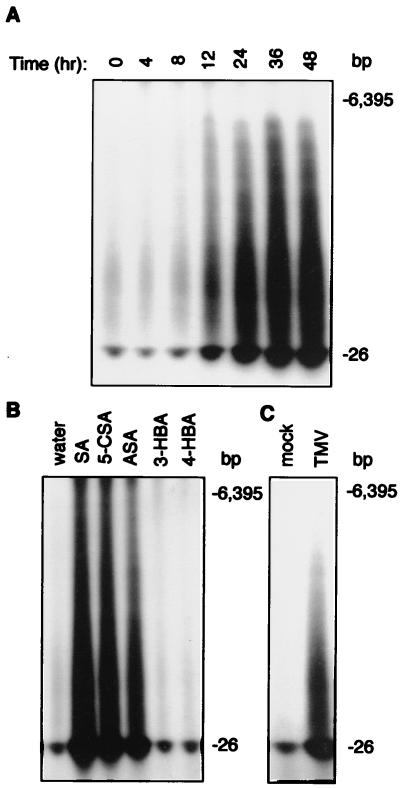
To confirm that increased synthesis of these small RNAs in TMV-infected SA-treated tobacco plants was because of an induced plant RdRP, we repeated the experiments in uninfected plants. As shown in Fig. 1A, this RdRP activity was very low in untreated tobacco leaves. In SA-treated leaves, the activity started to increase between 8 and 12 h and continued to rise up to 36 h after treatment (Fig. 1A). The RdRP activity in SA-treated leaves was observed to be still substantially elevated 72 h after the treatment (data not shown). This result confirmed the plant origin of the induced RdRP activity in TMV-infected SA-treated plants. To determine the biological significance of induced RdRP activity, we examined several SA analogs. AcetylSA and 5-chloroSA, two biologically active SA analogs capable of inducing plant defense response (27), were very effective in inducing the RdRP activity (Fig. 1B). By contrast, 3- and 4-hydroxybenzoic acid, two biologically inactive analogs of SA, failed to induce RdRP activity (Fig. 1B). Furthermore, TMV infection also induced RdRP activity (Fig. 1C). In all these assays, the labeled products of the RdRP activity separated on the polyacrylamide gels contained a strong band of small products of ≈26 nt and a smear of larger molecules with molecular weights up to 6–7 kb. The larger molecules appeared to be hybrids of small labeled products with unlabeled RNA templates of various sizes, because they could be converted to the small intensive band by treating with RNase A that degrades single-stranded RNA (data not shown).
Figure 1.
Induction of tobacco RdRP activity by SA and TMV infection. (A) Tobacco leaves were treated by floating on 1 mM SA and harvested at indicated times before preparation and assays of RdRP activity. (B) Tobacco leaves were treated with 1 mM SA, 5-chloroSA (5-CSA), acetylSA (ASA), 3-hydroxybenzoic acid (3HBA), or 4-hydroxybenzoic acid (4-HBA) for 48 h before preparation and assays of RdRP activity. (C) Tobacco plants were inoculated on a low leaf with TMV. Leaf tissues were harvested 15 days after inoculation from the upper systemically infected leaves for preparation and assays for RdRP activity. Migration positions of two RNA size markers (generated by in vitro transcription of the TMV genome and a 26-bp DNA fragment cloned in a pBluscript vector) are indicated.
Cloning and Expression of NtRdRP1.
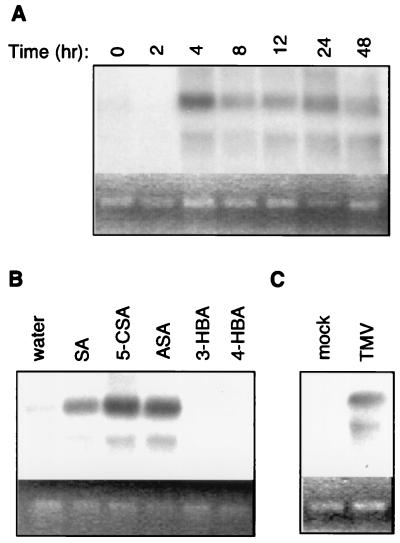
Previously, a RDRP protein from tomato has been purified and its corresponding cDNA clone isolated (11–13). The sequence of a tobacco cDNA clone encoding a RDRP (NtRdRP1) highly homologous to the tomato RdRP was also available in the GenBank database (accession no. AJ011576). A full-length tobacco cDNA clone for the RdRP gene was isolated by using a PCR-amplified NtRdRP1 fragment as a probe, and the expression of the gene was analyzed with Northern blotting. As shown in Fig. 2A, little transcript for the RdRP gene was detected in untreated tobacco plants. Between 2 and 4 h after SA treatment, two transcripts of 3.8 and 2.5 kb were induced, and the elevated transcript levels were steadily maintained throughout the 48 h of experiments. The 3.8-kb transcript likely corresponds to the full-length mRNA of NtRdRP1, whereas the smaller transcript may be a product of alternative splicing or degradation of NtRdRP1 or mRNA of a closely related gene. Reprobing of the same blot with tobacco PR1 (27) and tWRKY3 (26) showed no significant degradation of total RNA used in the blotting (data not shown), suggesting that if the smaller transcript resulted from mRNA turnover, it was specific to NtRdRP1. Interestingly, when the total RNA isolated from SA-treated Arabidopsis leaves was probed with a similar Arabidopsis RdRP gene located on BAC F10B6 (17), two similar transcripts induced by SA were also detected (Z.X. and Z.C., unpublished results). Sequence search of the Arabidopsis genome found no highly homologous RdRP gene that would produce a transcript of this size. Thus, the two transcripts for the RdRP genes probably resulted from posttranscriptional modification. As with the RdRP activity, induced expression of NtRdRP1 occurred in plants treated with SA or its biologically active analogs but not in plants treated with a biologically inactive analogue (Fig. 2B). Furthermore, induction of the RdRP gene was also observed in TMV-infected tobacco plants (Fig. 2C). Thus, there was a correlation between the increase of RdRP activity and the induction of NtRdRP1 in SA-treated or TMV-infected tobacco plants.
Figure 2.
Northern blot analysis of the NtRdRP1 gene expression. Total RNA was prepared from tobacco plants and probed with an 860-bp HindIII fragment of the NtRdRP1 cDNA. (A) Expression of NtRdRP1 in tobacco leaves treated with 1 mM SA for indicated times. (B) Expression of NtRdRP1 in tobacco leaves treated for 24 h with 1 mM SA, 5-chloroSA (5-CSA), acetylSA (ASA), 3-hydroxybenzoic acid (3HBA), or 4-hydroxybenzoic acid (4-HBA). (C) Expression of NtRdRP1 in mock- or TMV-inoculated tobacco plants. Chemical treatments and TMV infection were performed as described in Fig. 1. The ethidium bromide stain of rRNA is shown for each lane to allow assessment of equal loading.
Transgenic NtRdRP1 Antisense Tobacco.
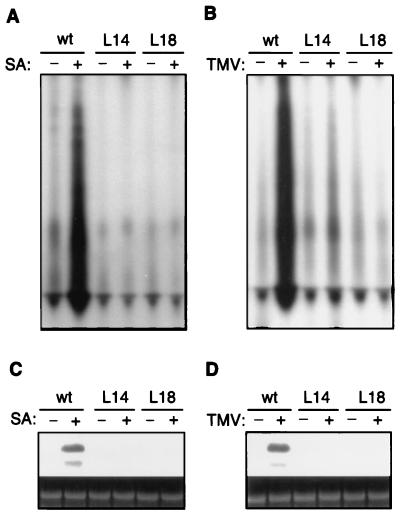
To study its functions, we constructed transgenic tobacco plants that expressed antisense RNA for NtRdRP1. Approximately 20 independent transgenic lines were obtained, and about 70% of them had reduced RdRP activity after SA treatment. Among them, lines 14 and 18 had the most depressed RdRP activity and contained no detectable NtRdRP1 transcripts after SA treatment or TMV infection (Fig. 3). The basal RdRP activity found in healthy untreated control plants was still detected in the transgenic antisense lines (Fig. 3 A and B), suggesting that the antisense plants were deficient specifically in inducible RdRP activity. NtRdRP1-deficient transgenic tobacco plants grew and developed normally.
Figure 3.
Suppression of inducible RdRP activity in transgenic tobacco NtRdRP1 antisense plants. (A) RdRP activity from vector-transformed wild-type plants (wt) or transgenic NtRdRP1 antisense lines 14 (L14) and 18 (L18) after treatment with water (−) or 1 mM SA (+) for 48 h. (B) RdRP activity from wild-type plants (wt) or transgenic antisense lines in upper uninoculated leaves 15 days after mock (−) or TMV (+) infection on lower leaves. (C) Transcript levels of NtRdRP1 in wild-type or antisense lines after treatment with water (−) or 1 mM SA (+) for 48 h. (D) Transcript levels of NtRdRP1 in wild-type or antisense lines in upper systemically infected leaves 15 days after mock (−) or TMV (+) infection on lower leaves. Northern blots were hybridized with an antisense-strand RNA probe transcribed from an 860-bp HindIII fragment of the NtRdRP1 cDNA clone. The ethidium bromide stain of rRNA is shown for each lane.
Response to Viral Pathogens.
SA inhibits accumulation of TMV viral RNA in tobacco plants (30). If inducible RdRP plays an important role in SA-induced antiviral defense, transgenic NtRdRP1 antisense plants would be compromised in SA-induced inhibition of the viral RNA accumulation. However, SA inhibited TMV RNA accumulation equally well in wild-type and antisense plants (data not shown). Thus, additional factors appear responsible for inhibiting viral RNA accumulation in SA-treated tobacco plants.
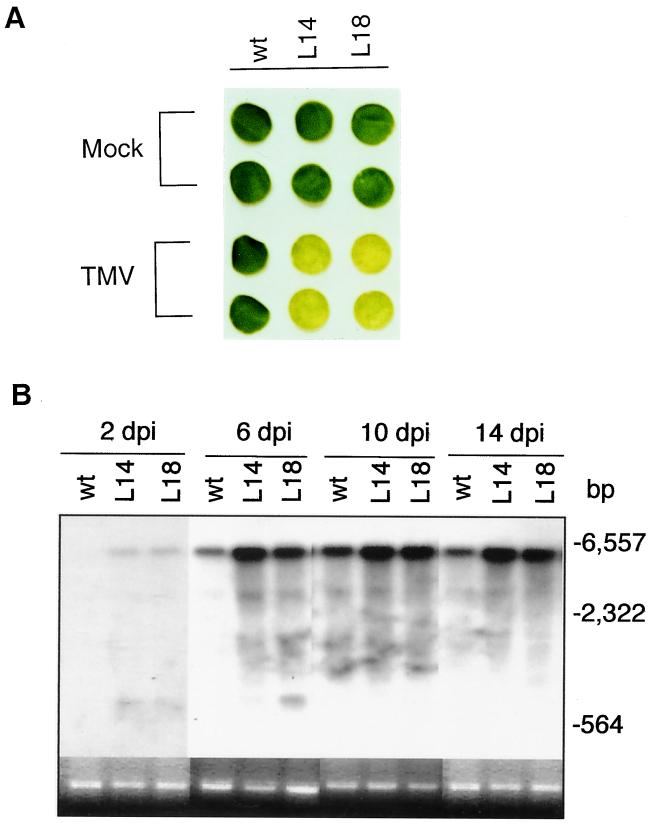
The importance of RdRP became apparent when we examined untreated plants for their response to viral infection. In these studies, leaf discs were taken from fully expanded leaves of both control and antisense plants and were inoculated with TMV. As shown in Fig. 4A, leaf discs from control plants showed no visible symptoms 2 weeks after inoculation. By contrast, those from antisense plants started to develop chlorotic symptoms 10 days after inoculation and became extensively chlorotic a few days later (Fig. 4A). Northern blot analysis indicated that viral RNA levels in antisense leaves were significantly higher than in leaves from control plants (Fig. 4B). Assays were also conducted with the same results in progeny of these transgenic lines, indicating that these altered phenotypes were reproducible from generation to generation.
Figure 4.
TMV symptom development and viral RNA accumulation in detached leaf discs. (A) Leaf discs from wild-type or antisense lines 14 (L14) and 18 (L18) were inoculated with TMV. The inoculated leaves were incubated in Petri dishes for 14 days before photographing. The enhanced chlorotic symptoms typically began in the antisense lines 10 days after inoculation. (B) Total RNA was isolated from TMV-infected leaf discs at indicated days postinoculation (dpi), separated on an agarose (1.2%)-formaldehyde gel, and probed with a DNA fragment corresponding to the TMV coat protein subgenomic RNA. The blot for the 2-dpi time point was exposed twice longer to detect low levels of TMV RNAs. The ethidium bromide stain of rRNA is shown for each lane. Migration positions of size markers (generated from HindIII-digested phage λDNA fragments) are indicated.
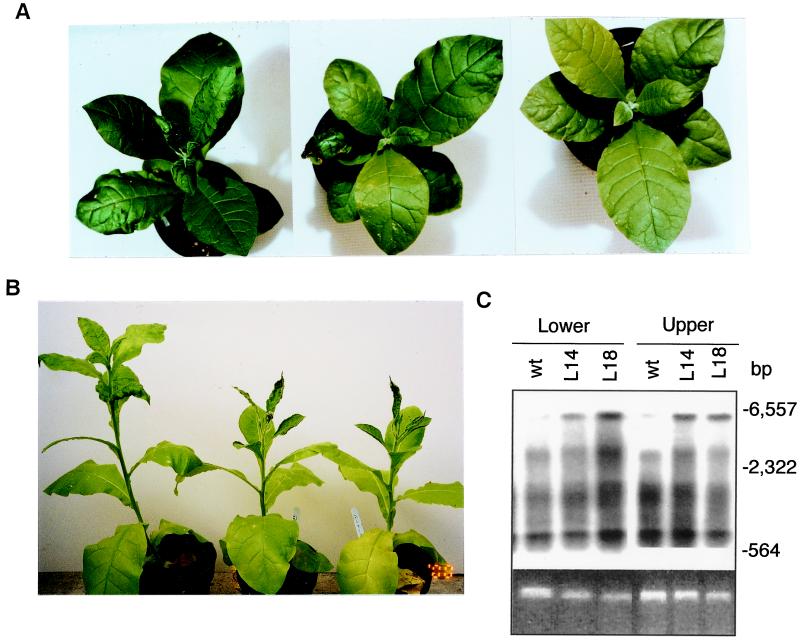
With more antisense plants in the second generation, we also evaluated their response to TMV on whole plants. Control plants developed typical mosaic symptoms on upper expanding leaves approximately 1–2 weeks after TMV inoculation (Fig. 5A). When the antisense plants were inoculated, development of mosaic symptoms in upper uninoculated leaves was delayed, apparently because of the absence of the “green islands” found in TMV-infected control plants (Fig. 5A). Interestingly, the difference in symptoms between control and antisense plants became greater a few weeks after inoculation. Although the control plants continued to produce leaves of nearly normal sizes and shapes with typical mosaic symptoms, the newly emerging leaves from the antisense plants become narrower or even needle-like as a result of severely stunted expansion of leaf lamina (Fig. 5B). RNA gel blotting revealed that the NtRdRP1 antisense plants accumulated higher levels of full-length viral RNAs in both lower inoculated and upper systemically infected leaves, although the accumulation of three subgenomic RNAs was not significantly changed (Fig. 5C).
Figure 5.
TMV symptom development and viral RNA accumulation on whole plants. (A) TMV-susceptible wild-type (Left) or antisense lines 14 (Center) and 18 (Right) were inoculated on one of the lower leaves with TMV. The plants were photographed 15 days after inoculation. (B) The difference in symptom development between the wild-type (Left) and antisense lines 14 (Center) and 18 (Right) 35 days after TMV inoculation. (C) Total RNA was isolated 15 days after TMV inoculation from the inoculated leaves and the fifth leaves directly above the inoculated ones of the wild-type (wt), antisense lines 14 (L14) and 18 (L18) and probed with a DNA fragment corresponding to the TMV coat protein subgenomic RNA. The ethidium bromide stain of rRNA is shown for each lane. Migration positions of size markers are indicated. No significant hybridization signal was detected in uninoculated plants.
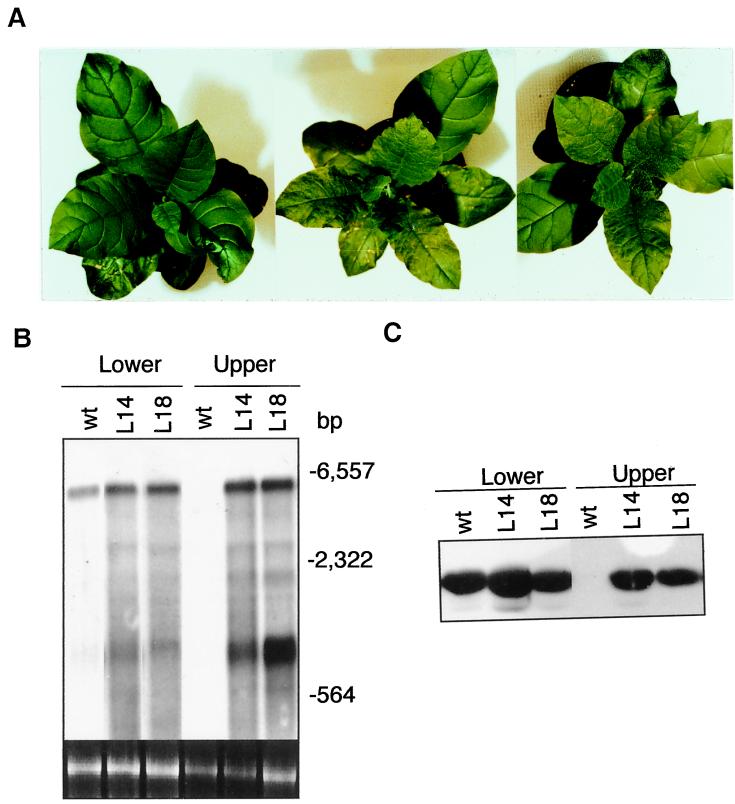
We also examined the transgenic plants for response to a mild PVX strain. Inoculation of lower leaves of control plants with the PVX strain did not lead to development of detectable symptoms in upper uninoculated leaves throughout the 3-week experiment (Fig. 6A). During the same period, all NtRdRP1 antisense plants developed symptoms not only in lower but also in upper uninoculated leaves (Fig. 6A). Both Northern and Western blot analyses indicated that, whereas high levels of viral RNA and proteins were detected only in lower inoculated leaves of control plants, they were present in both the lower and upper leaves of the antisense plants (Fig. 6 B and C). These results demonstrated that deficiency in the expression of the inducible RDRP gene promoted systemic spread of the virus in transgenic plants.
Figure 6.
PVX symptom development and virus accumulation in transgenic NtRdRP1 antisense plants. (A) The wild-type (Left) or antisense lines 14 (Center) and 18 (Right) were inoculated on one of the lower leaves with PVX. The plants were photographed 15 days after inoculation. (B) Total RNA was isolated 15 days after PVX inoculation from the inoculated leaves and the fifth leaves directly above the inoculated ones of the wild-type (wt) and antisense lines 14 (L14) and 18 (L18) and probed with a DNA fragment corresponding to the coding region for the PVX coat protein. The ethidium bromide stain of rRNA is shown for each lane. Migration positions of size markers are indicated. (C) Total soluble proteins were isolated 15 days after PVX inoculation from the lower inoculated and upper systemically infected leaves of the wild-type (wt) and antisense lines 14 (L14) and 18 (L18) and probed with a PVX-specific polyclonal antibody.
Discussion
Since their discovery in Chinese cabbage 3 decades ago (2), RdRPs have been found in different plant species (1). In several studies, elevated RdRP activity was found in virus-infected plants and has been speculated to be one of the host factors involved in virus replication (5, 8–10). In the present study, we have shown that RdRP activity is induced not only by virus infection but also by defense-inducing compounds (Fig. 1). Enhanced RdRP activity was correlated with increased expression of NtRdRP1 (Fig. 2). These observations suggest that induction of RdRP activity in virus-infected plants is part of activated plant antiviral defense, rather than a host mechanism exploited by invading viruses.
To determine the biological function of inducible RdRP, we have constructed transgenic antisense plants and obtained transgenic lines deficient in inducible RdRP activity (Fig. 3). These transgenic plants accumulated higher levels of viral RNAs and developed more severe symptoms after infection by TMV or PVX (Figs. 4–6). Thus, even though wild-type tobacco plants used in the study contain no resistance gene specific for either TMV or PVX, they are able to mount a generalized defense that restricts either the proliferation or the spread of the viruses and reduces symptom development.
Antisense RNA-induced gene silencing can occur to highly homologous genes. This raises a question of whether the transgenic NtRdRP1 antisense plants were deficient in expression only of NtRdRP1 or of other RdRP genes as well. In transgenic NtRdRP1 antisense plants, basal RdRP activity was still observed, but inducible RdRP activity was undetectable (Fig. 3). In addition, because transgenic antisense plants deficient in inducible RdRP activity could easily be obtained, there must be sufficient levels of RdRP(s) required for antisense RNA-induced gene silencing, a process likely to be related to transgene-induced PTGS. These observations would argue against suppression of all RdRP genes in antisense plants. However, suppression of RdRP genes highly homologous to NtRdRP1 might be possible in the antisense plant, and the difference in the antiviral activity between the control and antisense plants may be attributed to NtRdRP1 and/or other closely related RdRPs.
On the basis of the biochemical activity of RdRPs (11, 13) and the enhanced accumulation of viral RNAs in transgenic RdRP antisense plants (Figs. 4–6), the induced RdRP could protect plants from viral infection by synthesizing small cRNAs by using viral RNAs as templates and target their degradation. This mechanism, analogous to virus-induced PTGS, is consistent with the similarity of the phenotypes of TMV or PVX-infected NtRdRP1 antisense plants and the synergistic diseases caused by mixed infection of TMV or PVX and a potyvirus that contains suppressors of PTGS (31). The cRNAs synthesized by inducible RdRP activity may also interfere with the replication and/or translation of viral RNAs, resulting in reduced synthesis of viral components important for virus proliferation and symptom development. The apparent block of systemic movement of a PVX strain in wild-type tobacco plants may result from interference of the functions of the viral movement protein, a known RNA-binding protein (32), by the cRNA synthesized by RdRP activity. Interestingly, a recent study has revealed that the PVX movement protein is able to prevent spread of the PTGS signal that is likely to be an RNA molecule (33). Thus, there might be a functional antagonism between components important for viral movement and cRNAs produced by RdRPs. In addition, the enhanced symptom development in virus-infected antisense plants may be attributed to dysregulation of host gene expression that normally develops in virus-infected control plants and is kept in check by the inducible RdRP activity. Targeting of both viral and host RNA molecules by inducible RdRP activity is consistent with the observations that the enzyme can use both viral and plant RNAs as templates for synthesis of cRNAs (Fig. 7, which is published as supplemental data on the PNAS web site, www.pnas.org).
In Arabidopsis, there are a number of genes encoding proteins similar to RdRPs (17). This raises the question of whether different RdRP proteins have distinct or overlapping biological functions. It has recently been shown that the Arabidopsis RdRP gene SDE1/SGS2 is required for transgene-induced PTGS but may not play a general role in plant antiviral response (16, 17). In the present study, we have demonstrated that transgenic antisense plants deficient in an inducible RdRP activity can be obtained easily, suggesting that RdRP is not required for antisense suppression of plant gene expression, a process that may bear mechanistic resemblance to PTGS. However, these transgenic plants exhibited markedly enhanced susceptibility to both TMV and PVX (Figs. 4–6). Thus, it appears that certain members of the RdRP gene family may have an important role in transgene-induced PTGS, whereas others could be more involved in antiviral defense. Interestingly, among the RdRP genes identified in Arabidopsis, the one (on BAC F10B6) most homologous to the tobacco NtRdRP1 gene is also induced by SA and its biologically active analogs. By contrast, the recently isolated SGS2/SDE1 important for PTGS shares only a limited sequence similarity to NtRdRP1. Thus, at least one member of the Arabidopsis RdRP gene family may play a role as an antiviral defense gene equivalent to that played by NtRdRP1 in tobacco.
Supplementary Material
Acknowledgments
We thank Allan Caplan for critically reading the manuscript, Phil Berger and Pat Shield (University of Idaho, Moscow, ID) for PVX and antibodies, and Daniel Klessig (Rutgers University, Piscataway, NJ) for the cDNA library. Z.X. was supported by a University of Idaho Plant Biotechnology Graduate Student Assistantship.
Abbreviations
- RdRP
RNA-dependent RNA polymerase
- SA
salicylic acid
- PTGS
posttranscriptional gene silencing
- TMV
tobacco mosaic virus
- PVX
potato virus X
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Astier-Manifacier S, Cornuet P. C R Hebd Seances Acad Sci D. 1978;287:1043–1046. [PubMed] [Google Scholar]
- 2.Astier-Manifacier S, Cornuet P. Biochim Biophys Acta. 1971;232:484–493. doi: 10.1016/0005-2787(71)90602-2. [DOI] [PubMed] [Google Scholar]
- 3.Boege F. Biosci Rep. 1982;2:379–389. doi: 10.1007/BF01119300. [DOI] [PubMed] [Google Scholar]
- 4.Boege F, Rohde W, Sanger H L. Biosci Rep. 1982;2:185–194. doi: 10.1007/BF01116382. [DOI] [PubMed] [Google Scholar]
- 5.Dorssers L, Zabel P, van der Meer J, van Kammen A. Virology. 1982;116:236–249. doi: 10.1016/0042-6822(82)90416-0. [DOI] [PubMed] [Google Scholar]
- 6.Duda C T, Zaitlin M, Siegel A. Biochim Biophys Acta. 1973;319:62–71. doi: 10.1016/0005-2787(73)90041-5. [DOI] [PubMed] [Google Scholar]
- 7.Duda C T. Virology. 1979;92:180–189. doi: 10.1016/0042-6822(79)90223-x. [DOI] [PubMed] [Google Scholar]
- 8.Khan Z A, Hiriyanna K T, Chavez F, Fraenkel-Conrat H. Proc Natl Acad Sci USA. 1986;83:2383–2386. doi: 10.1073/pnas.83.8.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romaine C P, Zaitlin M. Virology. 1978;86:241–253. doi: 10.1016/0042-6822(78)90024-7. [DOI] [PubMed] [Google Scholar]
- 10.Takanami Y, Fraenkel-Conrat H. Biochemistry. 1982;21:3161–3167. doi: 10.1021/bi00256a020. [DOI] [PubMed] [Google Scholar]
- 11.Schiebel W, Haas B, Marinkovic S, Klanner A, Sanger H L. J Biol Chem. 1993;268:11858–11867. [PubMed] [Google Scholar]
- 12.Schiebel W, Haas B, Marinkovic S, Klanner A, Sanger H L. J Biol Chem. 1993;268:11851–11857. [PubMed] [Google Scholar]
- 13.Schiebel W, Pelissier T, Riedel L, Thalmeir S, Schiebel R, Kempe D, Lottspeich F, Sanger H L, Wassenegger M. Plant Cell. 1998;10:2087–2101. doi: 10.1105/tpc.10.12.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cogoni C, Macino G. Nature (London) 1999;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- 15.Smardon A, Spoerke J M, Stacey S C, Klein M E, Mackin N, Maine E M. Curr Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- 16.Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel J B, Jouette D, Lacombe A M, Nikic S, Picault N, et al. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 17.Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe D C. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 18.Bass B L. Cell. 2000;101:235–238. doi: 10.1016/s0092-8674(02)71133-1. [DOI] [PubMed] [Google Scholar]
- 19.Baulcombe D. Arch Virol Suppl. 1999;15:189–201. doi: 10.1007/978-3-7091-6425-9_14. [DOI] [PubMed] [Google Scholar]
- 20.Ratcliff F, Harrison B D, Baulcombe D C. Science. 1997;276:1558–1560. doi: 10.1126/science.276.5318.1558. [DOI] [PubMed] [Google Scholar]
- 21.Al-Kaff N S, Covey S N, Kreike M M, Page A M, Pinder R, Dale P J. Science. 1998;279:2113–2115. doi: 10.1126/science.279.5359.2113. [DOI] [PubMed] [Google Scholar]
- 22.Anandalakshmi R, Pruss G J, Ge X, Marathe R, Mallory A C, Smith T H, Vance V B. Proc Natl Acad Sci USA. 1998;95:13079–13084. doi: 10.1073/pnas.95.22.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brigneti G, Voinnet O, Li W X, Ji L H, Ding S W, Baulcombe D C. EMBO J. 1998;17:6739–6746. doi: 10.1093/emboj/17.22.6739. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Kasschau K D, Carrington J C. Cell. 1998;95:461–470. doi: 10.1016/s0092-8674(00)81614-1. [DOI] [PubMed] [Google Scholar]
- 25.Voinnet O, Pinto Y M, Baulcombe D C. Proc Natl Acad Sci USA. 1999;96:14147–14152. doi: 10.1073/pnas.96.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C, Chen Z. Plant Mol Biol. 2000;42:387–396. doi: 10.1023/a:1006399311615. [DOI] [PubMed] [Google Scholar]
- 27.Conrath U, Chen Z, Ricigliano J W, Klessig D F. Proc Natl Acad Sci USA. 1995;92:7143–7147. doi: 10.1073/pnas.92.16.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu D, Xie Z, Chen C, Fan B, Chen Z. Plant Mol Biol. 1999;139:477–488. doi: 10.1023/a:1006180708533. [DOI] [PubMed] [Google Scholar]
- 29.Horsch R, Fry J, Hoffmann N, Neidermeyer J, Rogers S G, Fraley R T. In: Plant Molecular Biology Manual. Gelvin S B, Schilperoort R A, editors. A5. Dordrecht, The Netherlands: Kluwer; 1988. pp. 1–9. [Google Scholar]
- 30.Chivasa S, Murphy A M, Naylor M, Carr J P. Plant Cell. 1997;9:547–557. doi: 10.1105/tpc.9.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pruss G, Ge X, Shi X M, Carrington J C, Bowman Vance V. Plant Cell. 1997;9:859–868. doi: 10.1105/tpc.9.6.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morozov S Y, Solovyev A G, Kalinina N O, Fedorkin O N, Samuilova O V, Schiemann J, Atabekov J G. Virology. 1999;260:55–63. doi: 10.1006/viro.1999.9788. [DOI] [PubMed] [Google Scholar]
- 33.Voinnet O, Lederer C, Baulcombe D C. Cell. 2000;103:157–167. doi: 10.1016/s0092-8674(00)00095-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.