Abstract
Extracellular invertase mediates phloem unloading via an apoplastic pathway. The gene encoding isoenzyme Nin88 from tobacco was cloned and shown to be characterized by a specific spatial and temporal expression pattern. Tissue-specific antisense repression of Nin88 under control of the corresponding promoter in tobacco results in a block during early stages of pollen development, thus, causing male sterility. This result demonstrates a critical role of extracellular invertase in pollen development and strongly supports the essential function of extracellular sucrose cleavage for supplying carbohydrates to sink tissues via the apoplast. The specific interference with phloem unloading, the sugar status, and metabolic signaling during pollen formation will be a potentially valuable approach to induce male sterility in various crop species for hybrid seed production.
In flowering plants the male gametophyte is formed within the anther, a complex organ composed of several tissues and cell types (1–3). The tapetum is a cell layer that plays a critical role in the process of pollen formation. It surrounds the pollen sac in early developmental stages, degenerates later during pollen development, and is degraded in mature anthers. It contributes to the development of the symplastically isolated pollen by secreting numerous substances into the loculus, such as carbohydrates and proteins, that are either important for pollen growth and development or become components of the outer pollen cell wall (4, 5).
Pollen development is a complex developmental process that is sensitive to mutations. Many cytoplasmic and nuclear mutations leading to male sterility have been shown to interfere with tapetal development and function (6, 7), supporting a critical function of this tissue for the production of functional pollen grains. Male sterile mutants are of agricultural importance for the production of hybrids to improve crop yield. Because of the heterosis effect, hybrids are characterized by increased resistance to diseases and enhanced performance in different environments compared with the parental lines (8–10). Hybrid production is limited to those plants in which sterility can be restored or in which the mechanical removal of anthers from flowers is feasible. Different strategies have been developed in the past to engineer male sterility by interfering with the development and the metabolism of the tapetum (11–14) or pollen in transgenic plants (15–19). However, any of the available systems has drawbacks such as interference with general development and metabolism or restriction to specific species. Therefore, the availability of a universal and dominant male sterility system with subtle although efficient effect on pollen growth offering the possibility to efficiently restore fertility would greatly facilitate the production of hybrid plants.
Carbohydrates were shown to play a critical role in anther and pollen development. They are nutrients used to sustain growth as well as signals to influence development in vivo and in vitro (20–22). Accordingly, different male sterile lines were shown to be characterized by perturbed carbohydrate metabolism (6, 20–23). Assimilates are produced in photosynthetically active source tissues and transported to photosynthetically less active or inactive sink tissues. An unloading pathway via the apoplasmic space is mandatory for symplastically isolated cells, such as developing pollen, and also can contribute prominently in other actively growing tissues. Sucrose is released from the sieve elements of the phloem into the apoplast via a sucrose transporter. An extracellular invertase ionically bound to the cell wall irreversibly hydrolyses the transport sugar sucrose. The hexose monomers are taken up into the sink cell by high-affinity hexose transporters. These key reactions create a localized concentration gradient, thus promoting phloem unloading via an apoplastic pathway and increasing the sink strength of the corresponding sink tissue. The importance of extracellular invertases for assimilate partitioning and source sink regulation has been suggested in recent years in a number of studies (24, 25), and the functional coupling with hexose transporters is supported by a coordinated regulation (26). It has been shown that extracellular invertases are encoded by small gene families that show a highly differential sink tissue-specific expression pattern (27). The identification of extracellular invertase isoenzymes from tomato (27), potato (28), and tobacco (this work) that are expressed in anther tissues support a link between extracellular sucrose cleavage and anther and pollen development.
Here, we report cloning of a gene encoding an extracellular invertase isoenzyme from tobacco that shows a specific spatial and temporal expression in anthers. Transgenic tobacco plants transformed with an antisense construct of extracellular invertase Nin88 under control of its own promoter are blocked in early pollen development, thus causing male sterility. These results prove the vital importance of extracellular invertase for pollen development by contributing to the supply of carbohydrates via an apoplastic pathway and metabolic signaling and offer the possibility to engineer male sterility for hybrid seed production.
Methods
Plant Material.
Nicotiana tabacum (cv. Samsun) and Lycopersicon peruvianum plants were grown under greenhouse conditions at 25°C with 16 h of light and 8 h of darkness. For DNA and RNA isolation, plant tissues were harvested, frozen in liquid nitrogen, and stored at −80°C. The material was ground with nitrogen-cooled mortar and pestle. Buds and flowers were harvested at different developmental stages from the plants. Pollen for in vitro germination assays was taken from dehisced anthers.
Cloning of the Nin88 cDNA and Promoter.
A 750-bp cDNA fragment of an extracellular invertase, designated Nin88, was cloned by reverse transcription–PCR (RT-PCR) by using RNA from tobacco anthers and the degenerate primers OIN3 and OIN4 (36). The Nin88 gene was cloned by screening of a tobacco genomic library in lambda gt10 with the Nin88 cDNA.
RNA Extraction and Northern Blot Analysis.
Total RNA was isolated from ground plant material essentially according to the method of Chomczynski and Sacchi (29). Northern blot analysis was performed as described (27) by using a radioactive labeled Nin88-specific probe. The cloned cDNA fragment of Nin88 was labeled by using a random primer DNA-labeling kit (MBI Fermentas, St. Leon-Rot, Germany).
RNA in Situ Hybridization and Nin88 Immunolocalization.
Samples were fixed by using 4% paraformaldehyde and 1% glutaraldehyde in PBS for 4 h at 4°C. Fifteen minutes of vacuum infiltration was applied at the beginning of the fixation. After fixation, samples were washed with PBS and left in PBS until sectioning or were embedded in paraffin.
For in situ hybridization, paraffin-embedded sections (7 μm) were treated as already described (30) with some modifications. The overnight hybridization was done at 42°C with a RNA digoxigenin-labeled probe (500 ng/ml) in 5× SSC buffer (0.15 M sodium chloride/0.015 M sodium citrate, pH 7) containing 5% dextran sulfate, 5× Denhardt's solution (0.02% polyvinylpyrrolidone/0.02% Ficoll/0.02% BSA), 0.5% SDS, and 50% formamide. After several washes, sections were treated with 20 mg/ml RNase A for 30 min and then incubated with antidigoxigenin antibody coupled to alkaline phosphatase (Boehringer Mannheim) diluted to 1 unit/ml. Revelation of alkaline phosphatase activity was performed by an overnight incubation with 5-bromo-4-chloro-3-indolyl phosphate (BCIP; Bio-Rad) and nitroblue tetrazolium (NBT; Bio-Rad). Hybridization with the sense probe was used as control.
For immunolocalization, fixed material was sectioned (60 μm) with a vibratome and washed twice in PBS. Sections were incubated in a blocking buffer (3 × 10 min) and then treated with 0.07% saponin for 20 min. Sections were incubated overnight at 4°C with the Nin88 antibody diluted at 1:1,500. Upon rinsing with PBS, sections were incubated with the secondary antibody (alkaline-phosphatase-linked sheep anti-rabbit IgG; Boehringer Mannheim). After three washes in PBS, sections were transferred for 5 min in an alkaline Tris buffer and then immunoreaction was revealed for 10 min by using BCIP and NBT as substrates. Sections incubated without the primary antibody were used as controls.
Plant Transformation.
The Nin88-GUS and the Nin88-Nin88–antisense constructs were transferred to tobacco (N. tabacum cv. Samsun) by using standard Agrobacterium transformation procedures (31). Transformation of L. peruvianum was done by following the protocol of Fillatti et al. (32).
In Vitro Germination Assay.
Isolated pollen was incubated in germination medium (16) for 5–12 h at 25°C and viewed under the microscope afterward. Determination of germination efficiencies was done by using a hemacytometer. Viability of pollen grains was examined by using Trypan blue solution (Sigma).
Electron Microscopy (EM) Pictures.
Pollen grains were isolated from different mutant lines and used for scanning EM (SEM) and transmission EM.
For SEM, pollen grains were air-dried, put on a sample plate, and coated with gold (layer thickness, 20 nm) in a Hummer VII-Sputter facility (Analtech). Samples were examined under a scanning electron microscope (Hitachi S450) at an operating voltage of 20 kV. Pictures were taken with a camera (RB67; Mamiya, Tokyo, Japan) that is connected to the scanning electron microscope.
Transmission EM was performed as described before (33) with the following modifications. Pollen was rinsed three times with EM buffer (50 mM KH2PO4/Na2HPO4, pH 7.0) and then fixed with 2.5% (wt/vol) glutaraldehyde in EM buffer for 1 h, washed three times with EM buffer, and subsequently treated with 2% (wt/vol) OsO4 in distilled water for 1 h. After dehydration, embedding, and cutting, samples were placed onto 400-mesh nickel grids. Finally, samples were counterstained and examined with a Hitachi H500 electron microscope at 75 kV as described (33).
Pollen Extracts and Invertase Activity Assay.
Pollen was isolated from anthers by stirring the anthers in I-Homogenization buffer (27) with a magnetic bar at maximum speed, until the mixture of microspores/anther debris and medium became milky. The buffer with the microspores was collected and washed three times with fresh buffer. In doing so, the anther debris was removed. The invertase activity assay was performed as described (27).
RT-PCR.
Pollen was isolated as before, and total RNA was isolated by using the E.Z.N.A. Plant RNA Kit (Peqlab, Erlangen, Germany) according to the manufacturer's instruction. RT was done with M-MLV Reverse Transcriptase (Promega) by following the manufacturer's instruction using RTN2 (5′-ACA TAT TTT CCG TTG TAC GAT GC-3′) as a specific primer for the RT reaction. Three microliters of the cDNA was used in the following standard PCRs by using RTN1 (5′-GGA GTC TAT CAT CTA TTC TAC-3′) and RTN2 as primers.
4′,6-Diamidino-2-phenylindole Staining.
Staining the nuclei with the fluorescent dye 4′,6-diamidino-2-phenylindole was done according to Vergne et al. (34). The stained pollen was observed under a fluorescence microscope (Zeiss).
In Vitro Pollen Maturation.
The experiments on the in vitro pollen maturation were done according to Touraev and Heberle-Bors (35). T1 medium was used with 250 mM or 500 mM of either glucose or sucrose or 250 mM sucrose and 250 mM glucose.
Results
Specific Temporal and Spatial Expression Pattern of Nin88.
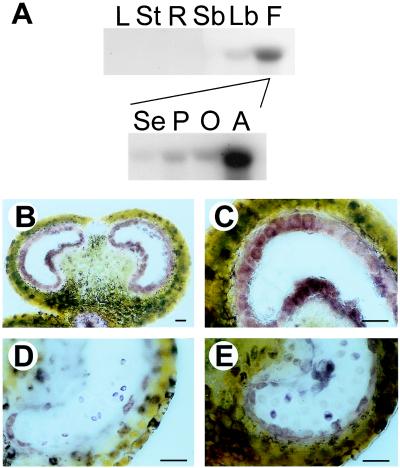
The Nin88 cDNA was cloned from tobacco anthers and shown to encode an extracellular invertase based on the high degree of sequence homology to other cell wall invertases and the presence of a conserved amino acid motif (37). Northern blot analysis of RNA from different tobacco tissues using this cDNA fragment as a probe revealed an exclusive expression of the Nin88 gene in flowers from all tissues analyzed. Further analysis of the different floral organs showed that the strong expression is restricted to anthers (Fig. 1A). RNA in situ hybridization studies demonstrated that the Nin88 mRNA is present in the tapetal cell layer and in developing microspores (data not shown). The tissue-specific localization of Nin88 is supported further by immunolocalization studies by using a polyclonal antiserum directed against a fusion protein of Nin88 with the maltose-binding protein (Fig. 1 B– E). At early stages of development, when the intact tapetum surrounds the pollen mother cells, the Nin88 protein is present only in the tapetal cell layer (Fig. 1 B and C). At the time when the tapetum starts getting degraded, the Nin88 protein can be detected in the tetrads (Fig. 1D), and when the tapetum is completely degraded, Nin88 is found in the developing microspores (Fig. 1E). These findings reveal a complex and specific spatial and temporal expression pattern of extracellular invertase Nin88 during anther development and pollen maturation.
Figure 1.
Tissue-specific expression pattern of Nin88. (A) RNA blot analysis of Nin88 expression in different tobacco tissues: L, leaves; St, stem; R, roots; Sb, small buds; Lb, large buds; F, flowers; Se, sepals; P, petals; O, ovary; A, anther. (B– E) Immunolocalization of Nin88 with cross-sections of anthers at different developmental stages. (B) Tapetum surrounding the pollen mother cells. (Bar = 50 μm.) (C) Tapetum surrounding the pollen mother cells. (Bar = 50 μm.) (D) Microspores in tetrad stage. (Bar = 50 μm.) (E) Tapetum shrunken, pollen grains separated. (Bar = 50 μm.)
Cloning of the Nin88 Promoter.
The complete structural gene of Nin88 and 4.3 kb of 5′ sequence were cloned from a tobacco genomic library. To show that 5′ sequences control the Nin88 gene specifically, the Escherichia coli GUS gene (38) was fused with a 3.3-kb Nin88 gene upstream fragment (nucleotides −3,304 to +12), including the Nin88 start codon, and then tobacco plants were transformed with the chimeric Nin88-GUS gene. Nineteen independent transformants containing the Nin88-GUS gene were generated as confirmed by PCR. Expression of the Nin88-GUS gene was analyzed by histochemical staining with X-gluc as substrate (38) and showed different levels of expression, suggesting different numbers of inserted copies. The GUS activity is restricted to anther tissue and pollen grains (Fig. 2) whereas no GUS activity could be detected in any other part of the flowers or tobacco plants or in the wild-type control plants. This finding demonstrates that the chimeric Nin88-GUS gene was regulated correctly and that the 3.3-kb Nin88 gene 5′ fragment contains all primary regulatory elements to program the specific expression pattern.
Figure 2.
GUS staining of Nin88-GUS tobacco plants and wild-type plants. (A) Anther from wild-type control plant (Upper) and anthers from Nin88-GUS plants (Lower) after incubation with X-Glc. (Bar = 1 mm.) (B) Pollen from Nin88-GUS plants. (Bar = 20 μm.) (C) Pollen from wild-type control plants. (Bar = 20 μm.)
Tissue-Specific Antisense Repression of Nin88 Causes Male Sterility.
To specifically reduce extracellular invertase activity in anthers to interfere with carbohydrate supply during pollen development, an 820-bp fragment of the third exon of Nin88 was fused in antisense orientation to the 3.3-kb Nin88 promoter fragment. The construct was introduced into tobacco plants, and 42 independent transformants were regenerated. The transfer of the antisense construct was confirmed by PCR analysis. The Nin88-antisense plants were identical to untransformed control plants with respect to growth rate, height, morphology of vegetative and floral organs and tissues, time of flowering, and flower coloration pattern. However, 26% (11 of 42) of the Nin88-antisense plants failed to produce seed capsules, and all other plants showed reduced seed number in the capsules. The flowers senesced and fell off or only small capsules with few seeds were formed. This inability or reduced ability to produce seeds correspond to the germination efficiency of pollen isolated from mature anthers in an in vitro germination assay. The plants that were unable to produce seeds showed a germination efficiency of less than 2%, although more than 90% of the corresponding pollen grains were vital as revealed by staining with trypan blue. Pollen grains from other antisense lines germinated with efficiencies ranging from 10% to 50% (Table 1). Staining for starch with KI/I2 solution (39) showed that pollen from plants with germination efficiencies less than 10% did not stain, whereas the pollen from plants germinating with 10–50% efficiency was only slightly reduced in staining for starch (Table 1).
Table 1.
Germination efficiencies and starch accumulation of Nin88-antisense plants
| Line | Germination efficiency | Starch staining |
|---|---|---|
| NT23-6 | 0.46% (SD 0.65%) | − |
| NT23-81 | 0.60% (SD 0.73%) | − |
| NT23-59 | 1.80% (SD 0.83%) | − |
| NT23-55 | 10.66% (SD 1.68%) | + |
| NT23-17 | 26.97% (SD 3.71%) | ++ |
| NT23-4 | 28.56% (SD 6.86%) | ++ |
| NT23-8 | 34.74% (SD 4.30%) | ++ |
| NT23-14 | 51.40% (SD 6.25%) | ++ |
| Wild type | 73.90% (SD 1.06%) | +++ |
Germination efficiency is given as percentage of pollen with a pollen tube of total viable pollen. The intensity of starch staining with KI/I2-solution is given in relative values. −, no staining of pollen grains; +, more than 50% staining of pollen grains; ++, more than 85% staining of pollen grains; +++, more than 95% staining of pollen grains.
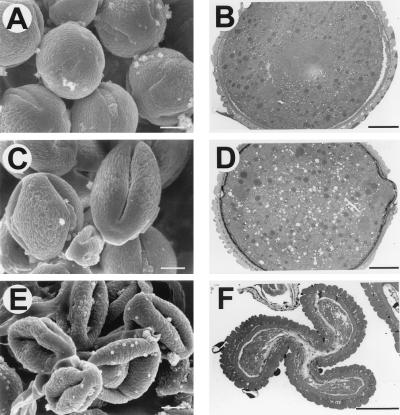
Pollen from the Nin88-antisense plants is also characterized by a different morphology. Analysis with a light microscope revealed that pollen grains from wild-type plants are of uniform size and filled with starch granules. In contrast, pollen grains from the antisense lines are a heterogeneous population with respect to pollen grains of different size and shape. The proportion of wild-type-like pollen of the different antisense lines corresponds to the germination efficiencies. The differences in morphology between wild-type pollen and pollen from antisense lines, in particular, are evident by the analysis with electron microscopes (Fig. 3). Analysis with the scanning electron microscope reveals that pollen from antisense lines is less turgescent, resulting in a distorted and invaginated morphology, which indicates a reduced amount of intracellular material or turgor. Distinct differences are also evident in cross-sections analyzed under the transmission electron microscope. Pollen from antisense lines with an intermediate germination efficiency has an increased number of vacuoles compared with wild-type pollen whereas the amount of intracellular material is highly reduced in the strong antisense lines, resulting in compression of the outer layers.
Figure 3.
Electron microscopic pictures of pollen from wild-type (A and B) and transgenic plants (C–F). (A) SEM picture of wild-type pollen. (Bar = 5 μm.) (B) Transmission EM (TEM) picture of wild-type pollen. (Bar = 5 μm.) (C) SEM picture of pollen from NT23–4. (Bar = 5 μm.) (D) TEM picture of pollen from NT23–4. (Bar = 5 μm.) (E) SEM picture of pollen from NT23–6. (Bar = 5 μm.) (F) TEM picture of pollen from NT23–6. (Bar = 5 μm.) NT23–4 represents one of the plants whose pollen shows germination efficiencies between 25% and 40%; NT23–6 is one of the plants with germination efficiency <2%.
Selective Repression of Invertase Activity in the Tapetum and the Developing Pollen.
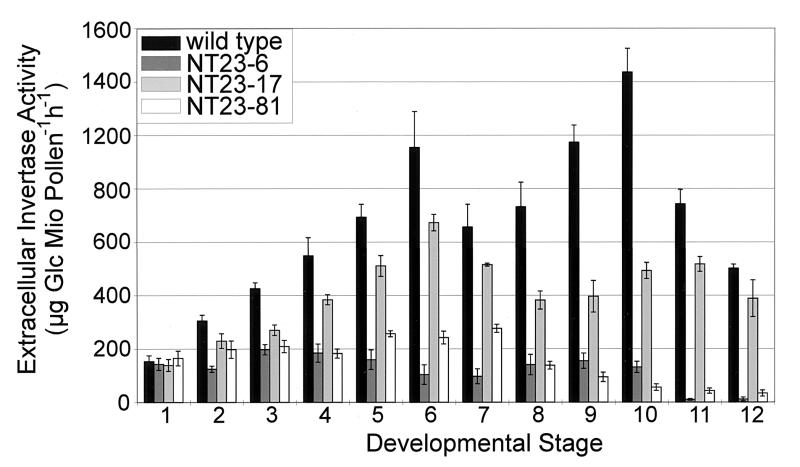
Determination of extracellular invertase activity in total anthers showed no differences between wild-type and Nin88 antisense plants, indicating a highly cell type-specific reduction of invertase activity (data not shown). Immunolocalization of Nin88 in anther cross-sections revealed that in the strong antisense lines NT23–6 and NT23–81, the amount of the Nin88 protein in the tapetum was either highly reduced or absent (data not shown). In further experiments, the effect of Nin88 antisense repression on the invertase activity in developing pollen was determined. The development of the tobacco flower can be divided into 12 stages (40), and pollen from these different developmental stages was isolated and tested for extracellular invertase activity. Fig. 4 shows that extracellular invertase activity in wild-type pollen is increasing during pollen maturation, with two activity peaks at stages 6 and 10. This pattern of extracellular invertase activity was reflected by the Nin88 mRNA level, as determined by RT–PCR analysis of selected developmental stages (data not shown). Nin88-antisense plants showing an intermediate phenotype, with 25–35% germination efficiencies such as NT23–17, showed a reduction in extracellular invertase activity up to 60%, whereas the pattern with the two activity peaks was similar to the wild type. Pollen extracts from those Nin88-antisense plants showing less than 2% germination efficiency (e.g., NT23–6 and NT23–81) have a drastically reduced extracellular invertase activity compared with the wild type, which, in particular, was evident at later stages of development, where the activity was reduced up to 99%. The two-peak pattern is also not evident any more. These data demonstrate that Nin88 antisense repression results in a specific repression of extracellular invertase in the tapetal cell layer and during pollen development and that the germination efficiency correlates with the reduction in enzyme activity.
Figure 4.
Extracellular invertase activity of pollen at different developmental stages.
The phenotypical characterization of pollen derived from antisense plants indicated that the inability to germinate is due to a block during development. Staining the DNA of pollen grains from dehisced anthers with the fluorescent dye 4′,6-diamidino-2-phenylindole showed that the wild-type pollen developed normally and has two stained nuclei, whereas in the transgenic pollen only one nucleus is visible (data not shown), which suggests a blockage in pollen development in the unicellular microspore stage, before microspore mitosis I, which takes place around stage 6 (40). At this developmental stage, the wild-type pollen shows the highest extracellular invertase activities and the reduction of invertase activity in pollen from the antisense lines is most pronounced (see Fig. 4).
In vitro maturation assays (35) were carried out to assess whether the block in development of pollen from antisense lines could be overcome by an exogenous supply of sugars. Pollen isolated from wild-type plants at the unicellular microspore stage developed normally and was able to germinate after maturation in the presence of either 250 mM glucose or sucrose. Cell division and further development with the typical starch accumulation also could be observed in pollen derived from antisense plants exogenously supplied with these sugars. This finding demonstrates that the developmentally arrested pollen is vital and is principally able to develop further. However, no complete maturation could be achieved, the proportion of developing pollen was about 60% lower when compared with wild-type pollen, and no significant difference was evident between glucose and sucrose, despite the reduction in the sucrose-cleaving enzyme activity. These results indicates that the block is not solely due to a perturbation of carbohydrate supply but that the function of extracellular invertase is also critical for metabolic signaling required for pollen development. Sugars are known to be important for gene regulation, and experimental evidence has been obtained that the sugar status and, in particular, the hexose/sucrose ratio are important for cell division and development (41).
Anther-Specific Expression of the Nin88 Promoter in Other Plants.
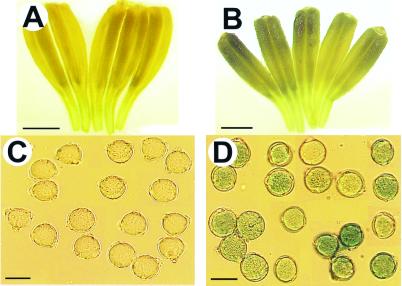
The Nin88-GUS reporter gene fusion was used for transformation of tomato plants to determine whether the Nin88 5′ regulatory region also could function in different plants. Analysis of 17 transgenic tomato plants proved that the Nin88 promoter sequence also confers its specific temporal and spatial expression pattern in plants other than tobacco. Histochemical detection of GUS activity in the transgenic tomato plants demonstrated that the expression of the reporter gene driven by the Nin88 promoter was restricted to anthers and in pollen (Fig. 5) whereas GUS activity could not be detected in any other tissue. This finding demonstrates that the 5′ upstream sequences of the Nin88 gene of tobacco are sufficient in other species to confer the same specific expression pattern as in tobacco.
Figure 5.
GUS staining of Nin88-GUS tomato plants and wild-type plants. (A) Anthers from wild-type control plant. (Bar = 1 mm.) (B) Anthers from Nin88-GUS plants. (Bar = 1 mm.) (C) Pollen from wild-type control plants. (Bar = 20 μm.) (D) Pollen from Nin88-GUS plants. (Bar = 20 μm.)
Discussion
Extracellular invertases of different species were shown to be encoded by small gene families with a differential regulation and expression pattern (25). Whereas most of the invertases characterized so far are not restricted to one specific plant organ, the extracellular invertase Nin88 of tobacco was shown to be characterized by a specific temporal and spatial expression pattern in anthers. At early stages of flower development, this invertase is present exclusively in the tapetal cell layer of anthers followed by a distinct expression pattern during pollen development. Reporter gene fusions demonstrated that the Nin88 upstream sequence is sufficient for the cell-specific expression and that the Nin88 gene is regulated primarily at the transcriptional level. The latter result is consistent with earlier publications that showed that most anther-specific genes are under transcriptional control (11).
The essential function of extracellular invertase Nin88 for pollen development could be proven by tissue-specific antisense repression under control of the corresponding promoter. The strong reduction of apoplastic invertase activity led to a developmental arrest of the symplastically isolated pollen at the unicellular microspore stage causing male sterility. This block could be partially overcome by exogenous supply of carbohydrates in an in vitro maturation assay, further supporting that the interference with the substrate availability for heterotrophic growth is responsible for the block in development. The finding that full maturation could not be achieved indicates that the antisense repression of invertase not only interferes with the carbohydrate supply but also with metabolic signals that are required for development, consistent with the increasing evidence for distinct signaling functions of hexoses and sucrose (41). The set of transgenic plants that have been generated for this study, with different degree of inhibition of cell wall-bound invertase activity, will be valuable tools to evaluate further the regulatory function of specific hexose/sucrose ratios determined by extracellular invertase.
The expression pattern indicates that the transport sugar sucrose initially is cleaved by Nin88 synthesized in the tapetal cell layer, whereas after degradation of the tapetum, the corresponding sucrose-cleaving activity is provided by Nin88 that is synthesized by the developing pollen. Thus, the apoplastic carbohydrate supply pathway for the pollen is controlled both by the surrounding tissues of the anther and by the developing pollen in a sequential manner (21). This study strongly supports the importance of apoplastic cleavage of sucrose by extracellular invertase for supplying carbohydrates to sink tissues and for normal development of plants, which has been suggested by an increasing number of studies in recent years (42–45). A number of studies have shown that cell wall invertases are sink tissue specifically expressed and up-regulated by stimuli that are associated with an increased demand for carbohydrates such as stress-related stimuli and growth-stimulating phytohormones (25). The specific involvement of invertases in anther development was suggested before for Lilium longiflorum (46, 20), tomato (27), and potato (28). It also has been shown that an arrest in pollen development in wheat because of a water deficit during meiosis correlated with alterations in carbohydrate metabolism and a drastic decrease in invertase activity (23). Antisense repression of an extracellular invertase in carrot under control of the constitutive 35S promoter strongly interferes with regeneration of the transformed plants and resulted in pleiotropic effects (43). This further supports the highly restricted expression pattern of the Nin88 promoter during anther development because we could not observe any effect during plant growth and development, and even anther formation was phenotypically normal. The high level of tissue specificity of the Nin88 promoter is supported further by the finding that antisense constructs of the extracellular invertases Cin1 from Chenopodium rubrum (36) and Ntbfruct1 from N. tabacum (47) under control of the Nin88 promoter also affected only pollen development (M.G. and T.R., unpublished observations), ruling out an interference of the corresponding antisense mRNA with invertases in other tissues.
The antisense expression of Nin88 leads to male sterility in the transgenic plants that are normal in all aspects of plant growth and development except for failure to produce functional pollen. Thus, the perturbation of the carbohydrate supply provides a subtle although highly efficient biotechnological method to engineer male sterility in plants for practical applications. The ability of inducing male sterility by metabolic engineering and the possibility to use this system in other agriculturally important plants provide a new strategy for the production of hybrid crop plants. Transferring this system to crop plants such as rape or maize should allow the production of hybrids without mechanical emasculation of the plants. In plants, where the fruits are not the harvested products (e.g., lettuce or carrot), male sterile plants can be crossed with any pollinator line to produce hybrid seeds. By contrast, in other plants in which the fruits are harvested (e.g., rice, corn, or tomato), a restorer line is needed to restore full male fertility. This restorer line could be plants that express a distantly related invertase, such as from bacteria or yeast that are not subject to the antisense repression through a plant invertase. Alternatively, introduction of a sucrose transporter could bypass the requirement for extracellular sucrose cleavage. This male sterility system also could be useful as a biological safety precaution for the increasing number of genetically engineered plants in field trials and agricultural production. Male sterility prevents an outcrossing of transgenes to wild-type species or to neighboring fields with the same species. Finally, the strong and specific temporal and spatial expression pattern of the Nin88 promoter provides the opportunity to introduce subtle and cell type-specific modifications to study regulatory signals such as phytohormones or the function of specific proteins at the cellular level.
Acknowledgments
We thank G. Stühler, P. Schittko, E. Herold, and A. Taffner for excellent technical assistance and G. Peissig for taking care of the plants. We are also grateful to W. Tanner for his continuing interest in our work and helpful discussions. We also thank U. Sonnewald for help with the plant transformation and A. Touraev for help with the in vitro maturation assay.
Abbreviations
- EM
electron microscopy
- SEM
scanning EM
- RT-PCR
reverse transcription–PCR
Footnotes
References
- 1.Bedinger P. Plant Cell. 1992;4:879–887. doi: 10.1105/tpc.4.8.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCormick S. Plant Cell. 1993;5:1265–1275. doi: 10.1105/tpc.5.10.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldberg R B, Beals T P, Sanders P M. Plant Cell. 1993;5:1217–1229. doi: 10.1105/tpc.5.10.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pacini E, Franchi G G, Hesse M. Plant Syst Evol. 1985;149:155–185. [Google Scholar]
- 5.Pacini E, Franchi G G. In: Microspores: Evolution and Ontogeny. Blackmore S, Barnes S H, editors. San Diego: Academic; 1991. pp. 213–237. [Google Scholar]
- 6.Kaul M L H. Monographs on Theoretical and Applied Genetics. Vol. 10. Berlin: Springer; 1988. [Google Scholar]
- 7.Chaudhury A M. Plant Cell. 1993;5:1277–1283. doi: 10.1105/tpc.5.10.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brewbaker J L. Agricultural Genetics. Englewood Cliffs, NJ: Prentice–Hall; 1964. [Google Scholar]
- 9.Feistritzer W R, Kelly A F. Hybrid Seed Production of Selected Cereal Oil and Vegetable Crops. Rome: Food and Agriculture Organization of the United Nations; 1987. [Google Scholar]
- 10.Mayo H. The Theory of Plant Breeding. Oxford: Clarendon; 1980. [Google Scholar]
- 11.Mariani C, DeBeuckeleer M, Truettner J, Leemans J, Goldberg R B. Nature (London) 1990;347:737–741. [Google Scholar]
- 12.Mariani C, Gossele V, DeBeuckeleer M, DeBlock M, Goldberg R B, DeGreef W, Leemans J. Nature (London) 1992;357:384–387. [Google Scholar]
- 13.Tsuchiya T, Toriyama K, Yoshikawa M, Ejiri S, Hinata K. Plant Cell Physiol. 1995;36:487–494. doi: 10.1093/oxfordjournals.pcp.a078784. [DOI] [PubMed] [Google Scholar]
- 14.van der Meer I M, Stam M E, van Tunen A J, Mol J N M, Stuitje A R. Plant Cell. 1992;4:253–262. doi: 10.1105/tpc.4.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmülling T, Schell J, Spena A. EMBO J. 1988;7:2621–2629. doi: 10.1002/j.1460-2075.1988.tb03114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmülling T, Röhrig H, Pilz S, Walden R, Schell J. Mol Gen Genet. 1993;237:385–394. doi: 10.1007/BF00279442. [DOI] [PubMed] [Google Scholar]
- 17.Worral D, Hird D L, Hodge R, Paul W, Draper J, Scott R. Plant Cell. 1992;4:759–771. doi: 10.1105/tpc.4.7.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hernould M, Suharsono, Zabaleta E, Carde J P, Litvak S, Araya A, Mouras A. Plant Mol Biol. 1998;36:499–508. doi: 10.1023/a:1005946104983. [DOI] [PubMed] [Google Scholar]
- 19.Napoli C A, Fahy D, Wang H Y, Taylor L P. Plant Phys. 1999;120:615–622. doi: 10.1104/pp.120.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clément C, Burrus M, Audran J C. Am J Bot. 1996;83:459–469. [Google Scholar]
- 21.Clément C, Audran J C. Protoplasma. 1995;187:172–181. [Google Scholar]
- 22.Clément C, Audran J C. In: Anther and Pollen. From Biology to Biotechnology. Clément C, Pacini E, Audran J C, editors. Heidelberg: Springer; 1999. pp. 69–90. [Google Scholar]
- 23.Dorion S, Lalonde S, Saini H S. Plant Physiol. 1996;111:137–145. doi: 10.1104/pp.111.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sturm A. Plant Physiol. 1999;121:1–7. doi: 10.1104/pp.121.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roitsch T, Ehness R, Goetz M, Hause B, Hofmann M, Sinha A K S. Aust J Plant Physiol. 2000;27:815–825. [Google Scholar]
- 26.Ehness R, Roitsch T. Plant J. 1997;11:539–548. doi: 10.1046/j.1365-313x.1997.11030539.x. [DOI] [PubMed] [Google Scholar]
- 27.Godt D E, Roitsch T. Plant Physiol. 1997;115:273–282. doi: 10.1104/pp.115.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maddison A L, Hedley P E, Meyer R C, Aziz N, Davidson D, Machray G C. Plant Mol Biol. 1999;41:741–751. doi: 10.1023/a:1006389013179. [DOI] [PubMed] [Google Scholar]
- 29.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 30.Guivarc'h A, Carneiro M, Vilaine F, Pautot V, Chriqui D. Plant Mol Biol. 1996;30:125–134. doi: 10.1007/BF00017807. [DOI] [PubMed] [Google Scholar]
- 31.Horsch R B, Fry J B, Hoffmann N L, Wallroth M, Eichholtz D, Rogers S G, Fraley R T. Science. 1985;227:1229–1231. [Google Scholar]
- 32.Fillatti J J, Kiser J, Rose R, Comai L. Biotechnology. 1987;5:726–730. [Google Scholar]
- 33.Engels A, Kahmann U, Ruppel H G, Pistorius E K. Biochim Biophys Acta. 1997;1340:33–44. doi: 10.1016/s0167-4838(97)00025-3. [DOI] [PubMed] [Google Scholar]
- 34.Vergne P, Delvallee I, Dumas C. Stain Technol. 1987;62:299–304. doi: 10.3109/10520298709108014. [DOI] [PubMed] [Google Scholar]
- 35.Touraev A, Heberle-Bors E. In: Methods in Molecular Biology 111. Hall R D, editor. Totowa, NJ: Humana; 1999. pp. 281–291. [DOI] [PubMed] [Google Scholar]
- 36.Roitsch T, Bittner M, Godt D E. Plant Physiol. 1995;108:285–294. doi: 10.1104/pp.108.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goetz M, Roitsch T. Plant J. 1999;20:707–711. doi: 10.1046/j.1365-313x.1999.00628.x. [DOI] [PubMed] [Google Scholar]
- 38.Jefferson R A. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- 39.Nelson O E. Genetics. 1968;60:507–524. doi: 10.1093/genetics/60.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koltunow A M, Truettner J, Cox K H, Wallroth M, Goldberg R B. Plant Cell. 1990;2:1201–1224. doi: 10.1105/tpc.2.12.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roitsch T. Curr Opin Plant Biol. 1999;2:198–206. doi: 10.1016/S1369-5266(99)80036-3. [DOI] [PubMed] [Google Scholar]
- 42.Miller E M, Chourey P S. Plant Cell. 1992;4:297–305. doi: 10.1105/tpc.4.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang G Q, Lüscher M, Sturm A. Plant Cell. 1999;11:177–189. doi: 10.1105/tpc.11.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.von Schaewen A, Stitt M, Schmidt R, Sonnewald U, Willmitzer L. EMBO J. 1990;9:3033–3043. doi: 10.1002/j.1460-2075.1990.tb07499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weber H, Borisjuk L, Heim U, Buchner P, Wobus U. Plant Cell. 1995;7:1835–1846. doi: 10.1105/tpc.7.11.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ranwala A P, Miller W B. Physiol Plant. 1998;103:541–550. [Google Scholar]
- 47.Greiner S, Weil M, Krausgrill S, Rausch T. Plant Physiol. 1995;108:825–826. doi: 10.1104/pp.108.2.825. [DOI] [PMC free article] [PubMed] [Google Scholar]