Abstract
Comparative genomic studies have led to the recent identification of several novel globin types in the Metazoa. They have revealed a surprising evolutionary diversity of functions beyond the familiar O2 supply roles of hemoglobin and myoglobin. Here we report the discovery of a hitherto unrecognized family of proteins with a unique modular architecture, possessing an N-terminal calpain-like domain, an internal, circular permuted globin domain, and an IQ calmodulin-binding motif. Putative orthologs are present in the genomes of many metazoan taxa, including vertebrates. The calpain-like region is homologous to the catalytic domain II of the large subunit of human calpain-7. The globin domain satisfies the criteria of a myoglobin-like fold but is rearranged and split into two parts. The recombinantly expressed human globin domain exhibits an absorption spectrum characteristic of hexacoordination of the heme iron atom. Molecular evolutionary analyses indicate that this chimeric globin family is phylogenetically ancient and originated in the common ancestor to animals and choanoflagellates. In humans and mice, the gene is predominantly expressed in testis tissue, and we propose the name “androglobin” (Adgb). Expression is associated with postmeiotic stages of spermatogenesis and is insensitive to experimental hypoxia. Evidence exists for increased gene expression in fertile compared with infertile males.
Keywords: gene family, hexacoordination, hypoxia, protein domain, spermatogenesis
Introduction
Globins are small respiratory proteins found in all domains of life and mainly perform cellular functions involving the storage, transport, sensing, and enzymatic detoxification of gaseous ligands (Vinogradov and Moens 2008). They have been a prime model system for studying gene, protein, and species evolution. Over the last decade, comparative genomic surveys have transformed our views about the phylogenetic distribution and adaptive functional diversification of the globin gene superfamily. Complementing our extensive knowledge on hemoglobin (Hb) and myoglobin (Mb), the discovery of novel globin types like neuroglobin (Ngb) and cytoglobin (Cygb), which perform yet-to-be-illuminated functions in nerve cells and other cell types, has greatly enriched our appreciation of the structural and functional diversity of vertebrate globins (Burmester et al. 2000, 2002; Hankeln et al. 2005). The identification of additional globin types (GbX, GbE, and GbY) with unknown physiological functions and more restricted phyletic distributions has added further layers of complexity to our understanding of globin gene family evolution (Kugelstadt et al. 2004; Roesner et al. 2005; Fuchs et al. 2006). Phylogenetic analyses of these vertebrate globins revealed that erythroid-specific globins have independently evolved O2-transport functions in different lineages (Hoffmann et al. 2010). It also became clear that vertebrate, plant, and other metazoan Hbs with a classical 3/3 α-helical fold are likely to share a common ancestor with one of three bacterial globin types, the bacterial F (flavohemoglobin) globin family (Vinogradov et al. 2005, 2006, 2011). These F-type globins function in alleviating oxidative and nitrosative stress and, thus, may also reflect the ancestral role of vertebrate globins (Vinogradov and Moens 2008).
Motivated by recent discoveries, which suggest that the globin fold has served as a highly versatile functional module in the evolution of O2-binding and sensing proteins (Vinogradov and Moens 2008; Hoffmann et al. 2010), we conducted extensive in silico searches for previously unannotated globins in deuterostome sequence data. These searches led to the discovery of a novel family of large, chimeric proteins that contain putative calpain-like and globin-like domains. These chimeric sequences were documented in a phylogenetically diverse array of metazoan taxa, including humans, and in choanoflagellates. We provide experimental evidence for their preferential expression in vertebrate testis tissues and thus propose the name androglobin (Adgb) for this newly discovered globin type.
Materials and Methods
Identification of Adgb Proteins
Initially, Adgb sequences were discovered in the sea urchin and amphioxus genomes by BLAST and HMMR 3.0 searches (http://hmmer.org). Putative homologs were then identified via BLASTP and PSIBLAST searches (Altschul et al. 1997), by screening SUPERFAMILY (Gough et al. 2001), and by searching dedicated genome databases at the NCBI and the JGI via TBLASTN. All the protein sequences were subjected to domain identification and functional annotation via CDD (Marchler-Bauer et al. 2011), FUGUE (Shi et al. 2001), COILS (Lupas et al. 1991), and PSORT II (Yu et al. 2010). Orthology was inferred by bi-directional BLAST hits and by matching domain architecture.
Sequence Alignment and Molecular Phylogeny
Multiple sequence alignments were obtained using COBALT (Papadopoulos and Agarwala 2007), MAFFT 6.833 (Katoh et al. 2005), CLUSTALW 2.0.3 (Larkin et al. 2007), and MUSCLE 3.7 (Edgar 2004). The globin domain alignments were checked manually for preservation of the Mb-fold, the pattern of predominantly hydrophobic residues at 36 conserved positions (Lesk and Chothia 1980; Bashford et al. 1987) and the invariant His at F8. Our criteria for a satisfactory globin required in addition a FUGUE Z score >6 (99% probability). For reconstruction of the globin phylogeny, we aligned the same protein data with different alignment algorithms and then concatenated the slightly different individual alignments before tree building to avoid a potentially subjective selection of a particular alignment. With regard to the ADGB affiliation in the tree, three out of the four individual alignments gave the same topology as the concatenated dataset. Alignment files are available from the authors upon request.
Poorly aligned positions and divergent regions of the alignments of the complete Adgb sequences were eliminated by Gblocks 0.91b (Castresana 2000), choosing the “less stringent selection” parameter set (http://www.phylogeny.fr). As suggested by ProtTest 1.2.7 (Abascal et al. 2005), we used the WAG (globin domain alignment) and JTT (complete Adgb alignment) models of amino acid evolution (Jones et al. 1992; Whelan and Goldman 2001), assuming a Γ distribution of evolution rates. Bayesian inference trees were obtained employing MrBayes 3.1.2 (Ronquist and Huelsenbeck 2003). Metropolis-coupled Markov chain Monte Carlo sampling was performed in two parallel runs with six separate chains for 15 × 106 generations, sampling every 2 500 generations, and using default priors. Once convergence was verified by tracking the split frequency, support for the nodes and parameter estimates were derived from a majority rule consensus of the last 1 000 trees. Maximum likelihood–based phylogenetic trees were obtained using RAxML 7.2.3 (Stamatakis et al. 2008). The resulting trees were tested by bootstraps with 100 replicates. We calculated dN/dS ratios to assess the level of functional constraint (Steinway et al. 2010).
Adgb mRNA Expression Analyses
Mouse RNA samples were kindly provided by Drs. H. Marti (University of Heidelberg) and D. Katschinski (University of Göttingen). Reverse transcriptase quantitative PCR (RT-qPCR) was performed using internal calibration standards and a SybrGreen qPCR reagent kit (Sigma) on the MX3000P light cycler (Stratagene). Primer sequences were as follows: mouse Adgb-F 5′-GTGACTCACCATGCAACACC-3′; mouse ADGB-R 5′-AACCTCCTCACTCTCCAGCA-3′; mouse S12-F 5′-GAAGCTGCCAAAGCCTTAGA-3′; mouse S12-R 5′-AACTGCAACCAACCACCTTC-3′; mouse beta-actin-F 5′-GAGCGTGGCTACAGCTTCAC-3′; mouse beta-actin-R 5′-GGCATAGAGGTCTTTACGGATG-3′; human ADGB-F 5′-GCATTACCTTAGCGGGTTCA-3′; human ADGB-R 5′-ATTGCCACTTCTTCCACACC-3′; human L28-F 5′-GCAATTCCTTCCGCTACAAC-3′; human L28-R 5′-TGTTCTTGCGGATCATGTGT-3′; human PHD2-F 5′-GAAAGCCATGGTTGCTTGTT-3′; human PHD2-R 5′-TTGCCTTCTGGAAAAATTCG-3′. PCR was performed by initial denaturation at 95 °C for 10 min, followed by 40 cycles of 30 s at 95 °C, 60 s at 60 °C, and 60 s at 72 °C. Amplicon specificity was checked by melting curve analysis. In silico analyses of ADGB mRNA levels in human tissues were performed using the NCBI UniGene (http://www.ncbi.nlm.nih.gov/unigene), ONCOMINE (Rhodes et al. 2004), and R2 (http://r2.amc.nl) databases. mRNA in situ-hybridization to 15 μm cryo-sections of mouse testes was performed using Digoxigenin™-labeled (Roche) in vitro-transcribed antisense RNA probes followed by alkaline phosphatase–conjugated antibody detection and NBT/BZIP staining as described (Reuss et al. 2002).
Recombinant Expression and Characterization of the Human ADGB Globin Domain
The globin domain of ADGB was RT-PCR amplified from human testis cDNA and cloned and expressed in the pET3a/E. coli BL21(DE3)pLys vector/host system as described (Dewilde et al. 2001). Samples were purified by low-spin centrifugation (10 min, 10 000 g) and ion exchange chromatography (DEAE-Sepharose Fast Flow). Molecular modeling of the globin domain was conducted using the I-TASSER software (Zhang 2008) and PyMOL (http://www.pymol.org).
Results
Identification of a Novel Chimeric Globin Protein Family in Animals
BLASTP, PSI-BLAST, and FUGUE database searches using human NGB (NP_067080) and the polymeric globin from Strongylocentrotus (XP_001197673) identified in the sea urchin and in the amphioxus genome a cognate chimeric protein of 1 724 and 1 676 amino acid residues (XP_001186068; JGI:scaffold146:751504-811458), respectively, each containing a putative rearranged globin domain. A group of related chimeric globins was detected by further BLAST searches in the genomes of over 30 metazoan taxa by employing the sea urchin and amphioxus sequences as query. This set of taxa included humans and 22 other vertebrates, uro-, cephalo-, and hemi-chordates, lophotrochozoa, ecdysozoa, coelenterates, placozoa, and the choanoflagellate Monosiga brevicollis, which may represent the closest relative to metazoans (fig. 1: supplementary table S1, Supplementary Material online). Surprisingly, we found no trace of putative orthologs in the genomes of fungi, Drosophila spp., Daphnia pulex, or nematodes (Caenorhabditis spp., Brugia malayi, Pristionchus pacificus, Trichinella spiralis, Loa loa, Wuchereria bancrofti, and Onchocerca volvulus).
FIG. 1.
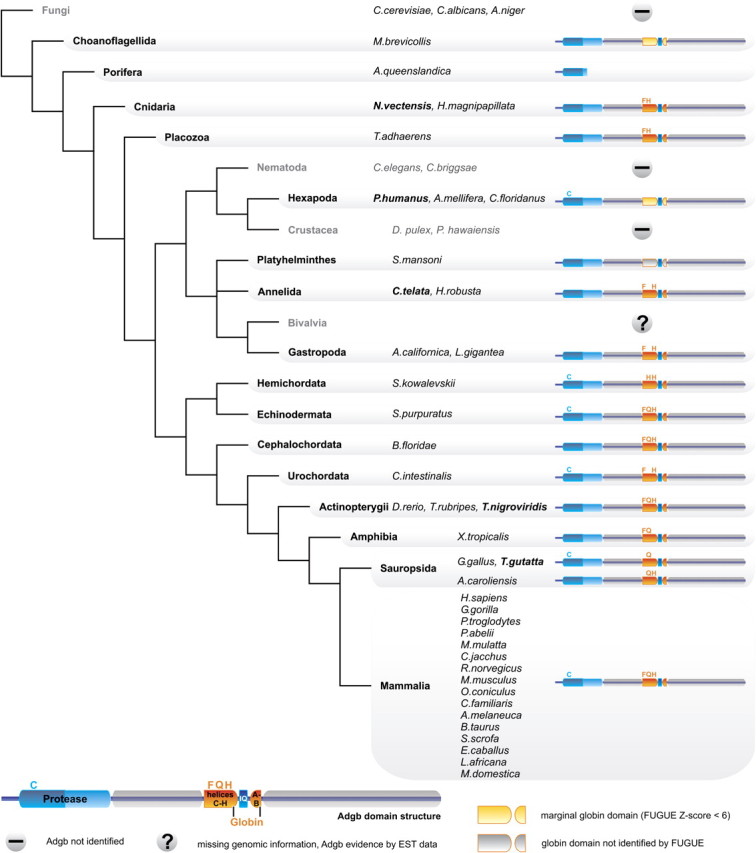
Diagrammatic representation of the phylogenetic distribution of Adgb orthologs. The Adgb domain structure, the conserved Cys (C) residues of the calpain-like domains and essential residues of the globin domain (CD1, E7, F8) are indicated by the amino acid one-letter code (for details see fig. 2). If a taxon contains different Adgb variants, the Adgb structure is shown for the species displayed in bold letters.
The protein family is defined by its typical modular domain structure, as identified by CD searches (fig. 2A). The chimeric proteins characteristically comprise four domains, an N-terminal ∼350 residue calpain-like cysteine protease domain, a region of ∼300 residues without known motifs/domains, followed by an ∼150 residue circularly permutated globin domain, and a second, uncharacterizable ∼750 residues C-terminal domain. Based on its predominant expression pattern in mammalian testes (see below), we have named this newly discovered chimeric protein “androglobin” (ADGB; registered by the HUGO Gene Nomenclature Committee).
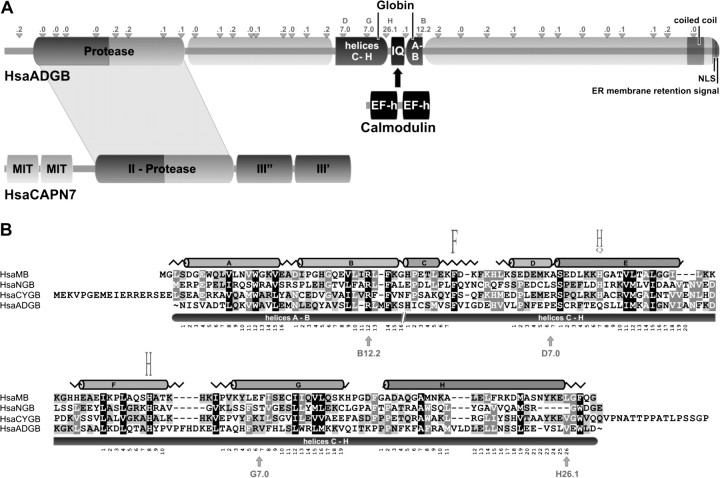
FIG. 2.
(A) The chimeric domain structure of human ADGB (HsaADGB). The calpain-like protease domain, the rearranged globin domain, the IQ motif, a C-terminal coiled-coil region, and candidate nuclear localisation (NLS) and ER membrane retention signals are indicated. The ADGB IQ motif may mediate binding to calmodulin, which reacts to Ca2+ levels via its EF hand (EFh) motifs. Small triangles indicate the position of introns in the human ADGB gene and intron phases are given (e.g., ‘.1’ indicates insertion of an intron in phase 1 between codon positions 1 and 2). For comparison, the structure of human calpain-7 (HsaCAPN7) is shown below (MIT, microtubule-interacting and trafficking domain; III’’ and III’, domains moderately similar to the Ca2+ binding domain of other calpains). (B) Amino acid sequence alignment of the concatenated human ADGB globin domain (helices A/B plus C-H) with human NGB, CYGB and Mb. The globin α-helical structure is drawn on top of the alignment. The functionally conserved Phe (F) in the C-D region and the proximal and distal His (H) residues at positions F8 and E7 are indicated. Note that ADGB contains a Gln (Q) instead of the distal His. Gray-scale shading indicates conservation of iso-functional amino acid residues. Intron positions within the globin domain of the ADGB gene are shown by arrows below the alignment.
The Adgb Globin Domain
We used NCBI Conserved Domain searches and alignments with Mb to demarcate the boundaries of the globin domain in the chimeric Adgb sequence. Interestingly, the globin domain, which normally consists of eight consecutive α-helices (named A-H), is circularly permutated and split into two parts within Adgb (fig. 2A). The part containing helices A and B has been shifted in the C-terminal direction and is separated from the main globin sequence (helices C–H) by a calmodulin-binding IQ motif (supplementary file S2, Supplementary Material online). The shifted A-B helix segment is unambiguously confirmed by the presence of a B12.2 intron position, which is conserved in most globin family members known to date.
Alignments of the split Adgb globin domain with mammalian Mb, Ngb, and Cygb sequences (fig. 2B; supplementary file S2, Supplementary Material online) revealed that Adgb—despite its rearrangement—conforms to the criteria of the “globin fold” tertiary structure. This was confirmed by molecular modelling of the human ADGB globin domain 3D structure, showing that the helix C–H segment alone is able to produce a bona fide globin fold (supplementary file S3, Supplementary Material online). Highly conserved, functionally important residues such as the Phe in interhelical region CD and the proximal His F8, which coordinates the heme iron, are present in most Adgbs. Adgb shares with several other known globins the substitution of the ligand-binding distal His E7 for a Gln. Amino acid identity/similarity values (PAM250 matrix) of the artificially re-ordered and concatenated human ADGB globin domain (helices A–H) are 16.6/37.3% to human NGB, 17.4/44.7% to MB, and 17.4/40.0% to CYGB.
To ascertain the functionally important heme coordination chemistry, helix region C–H of the human ADGB globin domain (together with 20 N-terminal amino acids of non-globin origin) was recombinantly expressed in E. coli. Interestingly, the partial C–H globin domain produced an absorption spectrum that is typical of complete globins (supplementary file S4, Supplementary Material online). The reduced deoxy form exhibits peaks at 427, 531, and 560 nm, characteristic of a hexacoordinated low-spin ferrous heme group (Kakar et al. 2010). Unlike other hexacoordinated Hbs, all Adgbs have an E7 Gln (fig. 2B); hence, the only possible hexacoordination scheme is [Gln]-Fe-[His], as also evidenced by the molecular modeling (supplementary file S3, Supplementary Material online).
Notably, not all Adgb homologs appear to contain the rearranged, but possibly functional, globin domain (fig. 1; supplementary table S1, Supplementary Material online). Globin domains are unrecognizable in Adgbs of the arthropod Camponotus floridanus, the annelid Helobdella robusta, and the platyhelminth Schistosoma mansoni. We detected weak globin matches in Adgbs of the choanoflagellate Monosiga brevicollis and the arthropods Pediculus humanus and Apis mellifera (FUGUE Z-scores <6 [99% confidence limit]). The functionally critical F8 His is lacking in the globin domains of Adgb from the zebra finch Taeniopygia guttata, the clawed frog Xenopus tropicalis, the cnidarian Nematostella vectensis, and the placozoan Trichoplax adhaerens.
The Calpain-Like Domain and Additional Conserved Motifs in Adgb
Calpains are calcium-regulated cytoplasmic cysteine proteases involved in the intracellular processing of proteins (Goll et al. 2003; Sorimachi et al. 2010). Mammalian calpains are heterodimers of identical 28 kDa regulatory subunits and 80 kDa subunits encoded by up to 14 genes. The crystal structures of the latter demonstrate that they consist of four domains, with the catalytic domain II comprising two subdomains sharing the three active site residues: Cys in subdomain IIa and His and Asn in subdomain IIb (Strobl et al. 2000). FUGUE searches showed that the Adgb N-terminal ∼350 residues are homologous to the catalytic domain II of human calpains. A phylogenetic analysis of the Adgb calpain-like domains with 14 human calpains demonstrated that they were most closely related to human calpain-7 (fig. 3A). Similarities of ADGB calpain subdomains IIa and IIb amounted to 47% and 43% compared with CAPN7 and 43% and 41% relative to the variant CAPN15, which is second next to ADGB in the tree. The calpain domain of Adgb has retained only one of three calpain-typical active site residues (the Cys in subdomain IIa), but it contains several His and Asn residues at non-standard positions (supplementary file S2, Supplementary Material online).
FIG. 3.
Phylograms describing relationships among (A) the calpain domain of Adgb and the repertoire of human calpains, (B) the complete Adgb sequences, and (C) the globin domain of Adgb and representatives from other metazoan, fungal, plant, and bacterial globin lineages. Bayesian posterior probabilities and bootstrap support values are provided next to the nodes. For species abbreviations, see figure 1.
In addition to the globin- and calpain-like domains, Adgbs contain several known motifs (fig. 2A), that is, the calmodulin-binding IQ motif interrupting the two globin segments (supplementary file S2, Supplementary Material online), a coiled-coil motif, possibly involved in protein dimerization, a putative nuclear localization signal (prediction value 60–80%), and an overlapping candidate ER membrane retention signal at the C-terminus of Adgb.
The Human ADGB Gene
The complete human ADGB gene (syn. c6orf103) spans 216 460 bp on chromosome 6q24.3, of which 5 004 bp are coding, and contains 36 exons. A candidate CpG island, possibly involved in ADGB gene regulation, spans the first exon and its 5′ region. Four of the 35 introns are located in the globin domain (fig. 2A and B), of which the intron positions G7.0 (i.e., in “phase 0” between the codons for amino acids 6 and 7 of globin helix G) and B12.2 (in “phase 2”, i.e., between positions 2 and 3 of codon B12) are conserved among other metazoan globin lineages. The calpain domain of the ADGB gene contains eight introns, and none of the exon–intron junctions in human, rat, sea urchin, and Ciona Adgb genes coincide with those in the human calpain-7 gene.
Adgb Expression in Mammalian Tissues
Adgb mRNA expression was quantified by RT-qPCR in eight different mouse tissues (fig. 4A). Expression is most abundant in testes, with a tenfold lower expression in lung, and an ∼100-fold lower expression in the remaining tissues. This result is strongly supported by an in silico expression analysis of different human tissues using the NCBI UniGene, ONCOMINE, and R2 databases (supplementary file S4, Supplementary Material online). During mouse testis development, we observed a strong increase of Adgb expression at postnatal day 25 when postmeiotic spermatids are abundant, and this persisted into adulthood (fig. 4B). Consistent with this evidence for a role of Adgb during the late phases of spermatogenesis, the RT-qPCR experiments detected very low expression levels in different mouse testis-derived cell lines corresponding to Leydig cells, Sertoli cells, spermatogonia, and spermatocytes (supplementary file S4, Supplementary Material online). mRNA in situ hybridization of an Adgb antisense RNA probe to mouse testis cryo-sections showed pronounced signals toward the lumen of the seminiferous tubes, confirming the cellular specificity and expression preference in late spermatogenesis (fig. 4C).
FIG. 4.
Adgb mRNA expression analysis. (A) Quantitative RT-PCR showing Adgb expression in normal mouse tissues. Bars indicate normalized mRNA levels with standard errors of three different mice. (B) Quantitative RT-PCR of Adgb in testes of younger and older mice (d = days, m = months after birth). (C) In situ hybridization of Adgb antisense RNA to mouse testis cryosections (1,2). Dark signal is concentrated towards the lumen of the seminiferous tubules. A hematoxylin/eosin-stained section (3) is shown for comparison. Smooth muscle cells (a), spermatogonia (b), and Sertoli cells (c) are indicated (magnification 40-fold).
Experimental hypoxia of three human cell cultures did not trigger a response in ADGB gene regulation (supplementary file S4, Supplementary Material online), in agreement with already hypoxic conditions within seminiferous tubules (see Discussion). Furthermore, R2 database analysis suggested 4-fold higher expression levels in fertile vs. infertile males (data from Platts et al. 2007; supplementary file S4, Supplementary Material online).
Molecular Evolutionary Analysis of Adgb
Phylogenetic reconstructions based on alignable portions (according to GBLOCKS) of the Adgb protein yielded a tree that is largely consistent with known organismal relationships (fig. 3B). Anomalous phylogenetic placements of Adgb sequences from the trematode worm Schistosoma, three representative arthropods, the echinoderm Strongylocentrotus, and the hemichordate Saccoglossus appear to be attributable to an accelerated rate of amino acid substitution. A Bayesian phylogenetic tree of the Adgb globin domain and different globin lineages (fig. 3C) demonstrates that Adgbs form a well-supported monophyletic group, which shows affinity to the Ngb lineage.
Since genes expressed in testes often show signatures of positive selection (Nielsen et al. 2005), we examined the ratio of non-synonymous to synonymous nucleotide substitutions (dN/dS) for mammalian Adgbs. Pairwise values obtained for the entire Adgb gene region were 0.25 on average, indicating purifying selection and a high level of functional constraint. An analysis of orthologous Adgb sequences from primates revealed higher conservation in the calpain and the globin domain than in unannotated parts of the coding sequence (supplementary file S5, Supplementary Material online). Orthologous comparisons between human and mouse revealed that the amino acid substitution rate of the Adgb globin domain (1.4 × 10−9 substitutions per site per year) was similar to that of Hb subunits (1.0 × 10−9) and considerably higher than that of the highly conserved Ngb and Cygb proteins (0.4 × 10−9 and 0.3 × 10−9, respectively; Burmester et al. 2002).
Discussion
A decade after the discovery of vertebrate neuroglobins and cytoglobins (Burmester et al. 2002, 2004), we report the identification, molecular biological, and evolutionary analysis of yet another novel globin type, named androglobin (Adgb). In this protein, the globin fold is embedded in a conserved multi-domain protein structure, emphasizing its evolutionary flexibility. The physiological function of Adgb has yet to be illuminated, but its wide distribution across metazoan phyla, its multidomain chimeric structure, and the circular permutation of the globin domain make it unique among globins in metazoans.
A Rearranged Globin Fused to a Calpain-Related Domain
The Adgb globin domain is the first instance of a naturally occurring circular permutation of α-helices within the globin fold superfamily. This type of rearrangement has been documented in proteins from all kingdoms of life, but it appears to be relatively uncommon (Jung and Lee 2001; Weiner and Bornberg-Bauer 2006). The circular permutation may result from a multistep mutation process, involving an initial tandem duplication of the globin domain, followed by trimming of redundant duplicate regions from the N- and C-termini (Vogel and Morea 2006). Interestingly, protein function appears to tolerate such significant architectural changes: A circularly permutated Mb with an N-terminal C-H helix and a C-terminal A and B helix separated by an eight amino acid linker, comparable to the Adgb permutated globin domain, displays proper folding and heme binding (Ribeiro and Ramos 2005).
Although being rearranged, the globin domain of most (but not all) Adgbs, including that of humans, contains a number of conserved amino acid residues that are known to play critical roles in heme-coordination and gas ligand binding (Perutz 1979). Indeed, our spectroscopic data and molecular modeling indicate that the heme iron atom of the recombinant human ADGB is hexa-coordinated (i.e., bound by two amino acid residues of the globin fold). Such hexa-coordination is observed in many tissue-expressed globins, which, unlike Hb and Mb, presumably perform roles other than O2 delivery. Hexa-coordination in nonsymbiotic plant Hbs, bacterial Hbs, and vertebrate Ngbs and Cygbs follows a bis-histidyl [HisF8]-Fe-[HisE7] binding scheme (Kakar et al. 2010). In Adgbs, by contrast, the hexacoordination probably involves a [HisF8]-Fe-[GlnE7] interaction because of the absence of a distal His. Furthermore, the absence of either B10 or CD1 Tyr in Adgbs precludes a [His]-Fe-[Tyr] hexa-coordination, found recently in a bacterial Hb (Howes et al. 2011). The fact that Adgbs represent an ancient metazoan globin lineage raises the possibility that the precursor of all metazoan globins was hexa-coordinated and that penta-coordination of Hbs and Mbs evolved secondarily, perhaps more than once, to meet the demand for O2 storage and transport in multicellular animals (but see Kakar et al. 2010). While a unified functional interpretation for heme hexacoordination has yet to be obtained, it is often associated with a high autoxidation potential (Dewilde et al. 2001; Geuens et al. 2010). Thus, the Adgb globin domain could play a redox-regulated signaling function, as recently postulated for hexacoordinated vertebrate Ngb (Tiso et al. 2011). Alternatively, the Adgb globin domain might mediate an O2 level-dependent protein activity, analogous to globin-coupled sensors in prokaryotes (Kitanishi et al. 2010).
In typical Adgbs, the permutated globin domain is N-terminally flanked by a cysteine protease-like domain, which is phylogenetically related to the catalytic domain of the large subunit of calpain-7, a presumably ancient member of the calpain family (Croall and Ersfeld 2007). Calpains are ubiquitous in eukaryotes, including unicellular eukaryotes, fungi, and plants, and are present in bacteria, but not archaea (Croall and Ersfeld 2007). They function as modulators in various cellular processes and are implicated in pathologic conditions, including muscular dystrophy, diabetes, multiple sclerosis, and neuronal ischemia (Zatz and Starling 2005). Of the 14 human calpains, only calpain-11 is specifically expressed in testis (Ben-Aharon et al. 2006). The Adgb-related calpain-7, however, is ubiquitously expressed in human tissues (Sorimachi et al. 2010), and it may exert proteolytic activity associated with proteins involved in the ESCRT (endosomal sorting complex required for transport) system (Osako et al. 2010). Alignments with human calpain proteins reveal that only the Cys-active site residue is observed at a strictly matching position in the calpain-like domain of Adgb (supplementary file S2, Supplementary Material online). However, the calpain domain of Adgb contains several other His and Asn residues at non-standard positions, suggesting that Adgbs may have a cysteine protease activity. Furthermore, analogous to calpains, which are activated by intracellular calcium (Goll et al. 2003), Adgb activities may also be regulated by Ca2+ levels, via calmodulin binding mediated by the well-conserved 23-residue IQ motif. This IQ motif, which separates the two parts of the globin domain, is flanked on the genomic level by two introns of the same phase (i.e., phase 1), suggesting that the motif may have been acquired by the ancestral Adgb in an exon-shuffling event (Long et al. 1995).
Adgbs: An Evolutionary Ancient Chimeric Globin with Episodic Distribution
Phylogenetic reconstructions and the widespread taxonomic distribution clearly indicate that Adgbs are an ancestral protein lineage. It probably traces back to choanoflagellates, the closest unicellular relatives of metazoans, which contain a marginally recognizable globin domain of unknown functionality in Adgb. Phylogenetic analyses using an artificially concatenated globin domain containing all helices (A-B plus C-H) group Adgb with Ngb, a globin which probably predates the split of protostome and deuterostome animals (Burmester et al. 2000; Roesner et al. 2005). The distinctness of Adgb, however, is emphasized 1) by the finding that in BLAST searches, members of the Adgb family preferentially hit each other and hardly any other globins, and 2) by the divergent Adgb exon-intron gene structure, in which two unusual intron positions are present in addition to the two conserved globin intron positions (Hardison 1996).
In contrast to the rather conserved sequence evolution of the Adgb globin domain in deuterostomes, the putatively orthologous protein domains in protostomes (i.e., in one of two annelids, a trematode, and in three arthropods) are borderline unrecognizable (FUGUE scores Z<6) and possibly non-functional (due to the absence of HisF8). The evolutionary ‘recycling` of a globin fold without a bound heme group is conceivable and has been reported for the stress-responsive gene regulatory protein RsbR in bacteria (Murray et al. 2005). The Adgb gene appears to have been deleted altogether in some protostome taxa, as we detected no trace of the sequence in the genomes of the fly genus Drosophila and in several nematodes including C. elegans. The latter finding is interesting in light of the existence of a large number of globin genes in Nematoda (Hoogewijs et al. 2008). We also note that Adgb is missing in the crustacean D. pulex, although this genome is only in draft status. In contrast, gastropods and another annelid species possess bona fide intact Adgb globin domains. Thus Adgb—and especially its globin domain—appears to be subject to very different levels of functional constraint in different animal lineages.
A Functional Role for Adgb in Mammalian Spermatogenesis?
Our data revealed that Adgb is predominantly expressed in mammalian testis tissue. The developmental expression profile and studies in testis-derived cell lines provide strong indications for Adgb mRNA expression during postmeiotic stages of spermatogenesis, subsequent to the release of spermatozoa into the hypoxic lumen of the seminiferous tubules. The presence of a globin in testis may appear surprising, although vertebrate Ngb mRNA expression was previously reported in spermatogonia and primary spermatocytes (Reuss et al. 2002). However, spermatogenesis requires a delicate balance in its oxidative energy metabolism, between the high O2 consumption by the highly proliferative process of sperm formation and the generation of free radicals toxic to both sperm and hormone-producing testes cells (Aitken and Roman 2008). For protection, spermatogonia are equipped with a dedicated antioxidant defense (Celino et al. 2011). Moreover, the seminiferous tubules are avascular and O2 reaches the luminal region solely by diffusion, making this tissue hypoxic (Wenger and Katschinski 2005). In this environment, globin proteins, which are able either to sense O2 or to detoxify reactive oxygen species, would clearly be advantageous. The lack of hypoxic up-regulation of Adgb mRNA in vitro is in line with a sensing function, but also other roles.
In conclusion, the discovery of a novel, phylogenetically ancient chimeric globin lineage, whose expression is associated with late stages of spermatogenesis in mammals, adds even more diversity to the vertebrate globin family. Although the molecular functions of the Adgb remain to be determined, the finding of higher Adgb expression levels in spermatozoa from fertile vs. infertile males underscores the putative functional importance of Adgb during spermatogenesis and its potential biomedical relevance. On the level of gene and protein evolution, the adaptive consequences of a rearranged globin fold are most interesting to study.
Supplementary Material
Supplementary files S1–S5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjourals.org/).
Acknowledgments
We thank Roland H. Wenger for helpful discussions. D.H. is supported by a postdoctoral Marie Curie IEF Fellowship from the European Commission. T.H. acknowledges funding by a University of Mainz internal research grant (Center for Computational Sciences, SRFN). L.M. and S.D. would like to acknowledge the Fund for Scientific Research (FWO) for financial support. F.G. is a PhD fellow of the FWO. J.F.S. acknowledges funding from the National Institutes of Health (R01 HL087216) and the National Science Foundation (IOS-0949931).
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Aitken RJ, Roman SD. Antioxidant systems and oxidative stress in the testes. Adv Exp Med Biol. 2008;636:154–171. doi: 10.1007/978-0-387-09597-4_9. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashford D, Chothia C, Lesk AM. Determinants of a protein fold. Unique features of the globin amino acid sequences. J Mol Biol. 1987;196:199–216. doi: 10.1016/0022-2836(87)90521-3. [DOI] [PubMed] [Google Scholar]
- Ben-Aharon I, Brown PR, Shalgi R, Eddy EM. Calpain 11 is unique to mouse spermatogenic cells. Mol Reprod Dev. 2006;73:767–773. doi: 10.1002/mrd.20466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmester T, Ebner B, Weich B, Hankeln T. Cytoglobin: a novel globin type ubiquitously expressed in vertebrate tissues. Mol Biol Evol. 2002;19:416–421. doi: 10.1093/oxfordjournals.molbev.a004096. [DOI] [PubMed] [Google Scholar]
- Burmester T, Haberkamp M, Mitz S, Roesner A, Schmidt M, Ebner B, Gerlach F, Fuchs C, Hankeln T. Neuroglobin and cytoglobin: genes, proteins and evolution. IUBMB Life. 2004;56:703–707. doi: 10.1080/15216540500037257. [DOI] [PubMed] [Google Scholar]
- Burmester T, Weich B, Reinhardt S, Hankeln T. A vertebrate globin expressed in the brain. Nature. 2000;407:520–523. doi: 10.1038/35035093. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Celino FT, Yamaguchi S, Miura C, Ohta T, Tozawa Y, Iwai T, Miura T. Tolerance of spermatogonia to oxidative stress is due to high levels of Zn and Cu/Zn superoxide dismutase. PLoS One. 2011;6:e16938. doi: 10.1371/journal.pone.0016938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croall DE, Ersfeld K. The calpains: modular designs and functional diversity. Genome Biol. 2007;8:218. doi: 10.1186/gb-2007-8-6-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewilde S, Kiger L, Burmester T, Hankeln T, Baudin-Creuza V, Aerts T, Marden MC, Caubergs R, Moens L. Biochemical characterization and ligand binding properties of neuroglobin, a novel member of the globin family. J Biol Chem. 2001;276:38949–38955. doi: 10.1074/jbc.M106438200. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs C, Burmester T, Hankeln T. The amphibian globin gene repertoire as revealed by the Xenopus genome. Cytogenet Genome Res. 2006;112:296–306. doi: 10.1159/000089884. [DOI] [PubMed] [Google Scholar]
- Geuens E, Hoogewijs D, Nardini M, et al. (16 co-authors) Globin-like proteins in Caenorhabditis elegans: in vivo localization, ligand binding and structural properties. BMC Biochem. 2010;11:17. doi: 10.1186/1471-2091-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll DE, Thompson VF, Li H, Wei W, Cong J. The calpain system. Physiol Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- Hankeln T, Ebner B, Fuchs C, et al. (23 co-authors) Neuroglobin and cytoglobin in search of their role in the vertebrate globin family. J Inorg Biochem. 2005;99:110–119. doi: 10.1016/j.jinorgbio.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Hardison RC. A brief history of hemoglobins: plant, animal, protist, and bacteria. Proc Natl Acad Sci U S A. 1996;93:5675–5679. doi: 10.1073/pnas.93.12.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann FG, Opazo JC, Storz JF. Gene cooption and convergent evolution of oxygen transport hemoglobins in jawed and jawless vertebrates. Proc Natl Acad Sci U S A. 2010;107:14274–14279. doi: 10.1073/pnas.1006756107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogewijs D, De Henau S, Dewilde S, Moens L, Couvreur M, Borgonie G, Vinogradov SN, Roy SW, Vanfleteren JR. The Caenorhabditis globin gene family reveals extensive nematode-specific radiation and diversification. BMC Evol Biol. 2008;8:279. doi: 10.1186/1471-2148-8-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes BD, Giordano D, Boechi L, et al. (14 co-authors) The peculiar heme pocket of the 2/2 hemoglobin of cold-adapted Pseudoalteromonas haloplanktis TAC125. J Biol Inorg Chem. 2011;16:299–311. doi: 10.1007/s00775-010-0726-y. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Jung J, Lee B. Circularly permuted proteins in the protein structure database. Protein Sci. 2001;10:1881–1886. doi: 10.1110/ps.05801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakar S, Hoffman FG, Storz JF, Fabian M, Hargrove MS. Structure and reactivity of hexacoordinate hemoglobins. Biophys Chem. 2010;152:1–14. doi: 10.1016/j.bpc.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitanishi K, Kobayashi K, Kawamura Y, Ishigami I, Ogura T, Nakajima K, Igarashi J, Tanaka A, Shimizu T. Important roles of Tyr43 at the putative heme distal side in the oxygen recognition and stability of the Fe(II)-O2 complex of YddV, a globin-coupled heme-based oxygen sensor diguanylate cyclase. Biochemistry. 2010;49:10381–10393. doi: 10.1021/bi100733q. [DOI] [PubMed] [Google Scholar]
- Kugelstadt D, Haberkamp M, Hankeln T, Burmester T. Neuroglobin, cytoglobin, and a novel, eye-specific globin from chicken. Biochem Biophys Res Commun. 2004;325:719–725. doi: 10.1016/j.bbrc.2004.10.080. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. (13 co-authors) Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lesk AM, Chothia C. How different amino acid sequences determine similar protein structures: the structure and evolutionary dynamics of the globins. J Mol Biol. 1980;136:225–270. doi: 10.1016/0022-2836(80)90373-3. [DOI] [PubMed] [Google Scholar]
- Long M, Rosenberg C, Gilbert W. Intron phase correlations and the evolution of the intron/exon structure of genes. Proc Natl Acad Sci U S A. 1995;92:12495–12499. doi: 10.1073/pnas.92.26.12495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, et al. (27 co-authors) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JW, Delumeau O, Lewis RJ. Structure of a nonheme globin in environmental stress signaling. Proc Natl Acad Sci U S A. 2005;102:17320–17325. doi: 10.1073/pnas.0506599102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Bustamante C, Clark AG, et al. (13 co-authors) A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osako Y, Maemoto Y, Tanaka R, Suzuki H, Shibata H, Maki M. Autolytic activity of human calpain 7 is enhanced by ESCRT-III-related protein IST1 through MIT-MIM interaction. FEBS J. 2010;277:4412–4426. doi: 10.1111/j.1742-4658.2010.07822.x. [DOI] [PubMed] [Google Scholar]
- Papadopoulos JS, Agarwala R. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- Perutz MF. Regulation of oxygen affinity of hemoglobin: influence of structure of the globin on the heme iron. Annu Rev Biochem. 1979;48:327–386. doi: 10.1146/annurev.bi.48.070179.001551. [DOI] [PubMed] [Google Scholar]
- Platts AE, Dix DJ, Chemes HE, et al. (11 co-authors) Success and failure in human spermatogenesis as revealed by teratozoospermic RNAs. Hum Mol Genet. 2007;16:763–773. doi: 10.1093/hmg/ddm012. [DOI] [PubMed] [Google Scholar]
- Reuss S, Saaler-Reinhardt S, Weich B, Wystub S, Reuss MH, Burmester T, Hankeln T. Expression analysis of neuroglobin mRNA in rodent tissues. Neuroscience. 2002;115:645–656. doi: 10.1016/s0306-4522(02)00536-5. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro EA, Jr., Ramos CH. Circular permutation and deletion studies of myoglobin indicate that the correct position of its N-terminus is required for native stability and solubility but not for native-like heme binding and folding. Biochemistry. 2005;44:4699–4709. doi: 10.1021/bi047908c. [DOI] [PubMed] [Google Scholar]
- Roesner A, Fuchs C, Hankeln T, Burmester T. A globin gene of ancient evolutionary origin in lower vertebrates: evidence for two distinct globin families in animals. Mol Biol Evol. 2005;22:12–20. doi: 10.1093/molbev/msh258. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Shi J, Blundell TL, Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J Mol Biol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- Sorimachi H, Hata S, Ono Y. Expanding members and roles of the calpain superfamily and their genetically modified animals. Exp Anim. 2010;59:549–566. doi: 10.1538/expanim.59.549. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Steinway SN, Dannenfelser R, Laucius CD, Hayes JE, Nayak S. JCoDA: a tool for detecting evolutionary selection. BMC Bioinformatics. 2010;11:284. doi: 10.1186/1471-2105-11-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobl S, Fernandez-Catalan C, Braun M, et al. (12 co-authors) The crystal structure of calcium-free human m-calpain suggests an electrostatic switch mechanism for activation by calcium. Proc Natl Acad Sci U S A. 2000;97:588–592. doi: 10.1073/pnas.97.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiso M, Tejero J, Basu S, et al. (13 co-authors) Human neuroglobin functions as a redox-regulated nitrite reductase. J Biol Chem. 2011;286:18277–18289. doi: 10.1074/jbc.M110.159541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov SN, Hoogewijs D, Bailly X, Arredondo-Peter R, Gough J, Dewilde S, Moens L, Vanfleteren JR. A phylogenomic profile of globins. BMC Evol Biol. 2006;6:31. doi: 10.1186/1471-2148-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov SN, Hoogewijs D, Bailly X, Arredondo-Peter R, Guertin M, Gough J, Dewilde S, Moens L, Vanfleteren JR. Three globin lineages belonging to two structural classes in genomes from the three kingdoms of life. Proc Natl Acad Sci U S A. 2005;102:11385–11389. doi: 10.1073/pnas.0502103102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov SN, Hoogewijs D, Vanfleteren JR, Dewilde S, Moens L, Hankeln T. Evolution of the globin superfamily and its function. In: Nagai M, editor. Hemoglobin: recent developments and topics. Kerala (India): Research Signpost; 2011. pp. 232–254. [Google Scholar]
- Vinogradov SN, Moens L. Diversity of globin function: enzymatic, transport, storage, and sensing. J Biol Chem. 2008;283:8773–8777. doi: 10.1074/jbc.R700029200. [DOI] [PubMed] [Google Scholar]
- Vogel C, Morea V. Duplication, divergence and formation of novel protein topologies. Bioessays. 2006;28:973–978. doi: 10.1002/bies.20474. [DOI] [PubMed] [Google Scholar]
- Weiner J, 3rd, Bornberg-Bauer E. Evolution of circular permutations in multidomain proteins. Mol Biol Evol. 2006;23:734–743. doi: 10.1093/molbev/msj091. [DOI] [PubMed] [Google Scholar]
- Wenger RH, Katschinski DM. The hypoxic testis and post-meiotic expression of PAS domain proteins. Semin Cell Dev Biol. 2005;16:547–553. doi: 10.1016/j.semcdb.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Yu NY, Wagner JR, Laird MR, et al. (11 co-authors) PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatz M, Starling A. Calpains and disease. N Engl J Med. 2005;352:2413–2423. doi: 10.1056/NEJMra043361. [DOI] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.