Polo-like kinase 4 (Plk4) plays an essential role in centriole duplication, but recent work led to the conclusion that Plk4 also directly regulates cytokinesis. The consequence of reduced Plk4 levels in human and mouse cells is studied. It is shown that Plk4 controls centriole duplication but does not directly regulate cytokinesis.
Abstract
Centrioles organize the centrosome, and accurate control of their number is critical for the maintenance of genomic integrity. Centrioles duplicate once per cell cycle, and duplication is coordinated by Polo-like kinase 4 (Plk4). We previously demonstrated that Plk4 accumulation is autoregulated by its own kinase activity. However, loss of heterozygosity of Plk4 in mouse embryonic fibroblasts has been proposed to cause cytokinesis failure as a primary event, leading to centrosome amplification and gross chromosomal abnormalities. Using targeted gene disruption, we show that human epithelial cells with one inactivated Plk4 allele undergo neither cytokinesis failure nor increase in centrosome amplification. Plk4 is shown to localize exclusively at the centrosome, with none in the spindle midbody. Substantial depletion of Plk4 by small interfering RNA leads to loss of centrioles and subsequent spindle defects that lead to a modest increase in the rate of cytokinesis failure. Therefore, Plk4 is a centriole-localized kinase that does not directly regulate cytokinesis.
INTRODUCTION
Centrosomes are the major microtubule-organizing center of animal cells and play an important role during mitosis, in which they organize the poles of the bipolar microtubule spindle upon which chromosomes are segregated. Although not strictly essential for the formation of the meiotic/mitotic spindle, whenever they are present centrosomes play a dominant role in guiding spindle formation (Nigg and Raff, 2009; Debec et al., 2010). Extra copies of centrosomes (referred to as centrosome amplification) lead to the formation of multipolar mitotic spindles, which, after centrosome clustering, enrich for inappropriate kinetochore–microtubule interactions, promoting chromosome missegregation (Ganem et al., 2009; Silkworth et al., 2009). Centrosome amplification is frequently observed in aneuploid human tumors, where it is proposed to play a causative role in genome instability and tumor development (Nigg, 2006). One pathway by which cells may acquire extra centrosomes is through a failure of cytokinesis, resulting in the production of tetraploid cells with twice the normal centrosome number (Meraldi et al., 2002; Holland and Cleveland, 2009). Indeed, tetraploidy often precedes the development of aneuploidy and has been shown to promote cellular transformation and tumor formation in murine cells lacking p53 (Fujiwara et al., 2005).
A centrosome is comprised of two centrioles that recruit a proteinaceous matrix called the pericentriolar material (PCM; Tsou and Stearns, 2006; Loncarek and Khodjakov, 2009). Because the centrioles recruit the PCM and determine the number of centrosomes in the cell, centrosome duplication is coupled to centriole duplication. A centriole pair duplicates once per cell cycle before mitosis to create two centrosomes before mitotic entry. In vertebrates and invertebrates the protein kinase Polo-like kinase 4 (Plk4) plays an essential role in controlling centriole duplication; in the absence of Plk4, centriole duplication fails, whereas elevated levels of Plk4 lead to the formation of multiple centrioles in a single cell cycle (Bettencourt-Dias et al., 2005; Habedanck et al., 2005; Kleylein-Sohn et al., 2007; Peel et al., 2007). Determining how the level and activity of Plk4 are controlled during the cell cycle is critical for a better mechanistic understanding of how the correct number of centrioles and centrosomes is maintained in animal cells.
Plk4 is a low-abundance protein and is targeted for destruction by the Skp/Cullin/F-box E3 ligase, which binds to phosphorylated Plk4 through the F-box protein β-TrCP (Cunha-Ferreira et al., 2009; Rogers et al., 2009; Guderian et al., 2010; Holland et al., 2010). Plk4 stability is linked to the activity of the enzyme, with active Plk4 phosphorylating itself to promote its own destruction (Guderian et al., 2010; Holland et al., 2010; Brownlee et al., 2011). This creates a feedback loop that regulates the cellular levels of Plk4 (Holland et al., 2010).
Surprisingly, Plk4 heterozygous mice have been reported to exhibit centrosome amplification and an increased incidence of spontaneous lung and liver tumors (Ko et al., 2005). Murine embryonic fibroblasts (MEFs) derived from Plk4+/− animals were reported to undergo cytokinesis failure after several passages in culture, leading to an enrichment of cells with a (near) tetraploid DNA content (Rosario et al., 2010). Consistent with the generation of tetraploid cells, these Plk4+/− MEFs were also reported to exhibit elevated levels of centrosome amplification, and late-passage MEFs formed tumors when injected into nude mice. The cytokinetic defect observed in Plk4+/− MEFs led to the proposal that Plk4 directly regulates cytokinesis by localizing to the spindle midbody, where it activates RhoA through regulation of the RhoA guanine exchange factor Ect2 (Rosario et al., 2010). However, it is unclear whether Plk4 protein levels are reduced in Plk4+/− cells and, if so, how a partial reduction in the level of Plk4 could directly lead to a failure of cytokinesis.
Using targeted gene disruption in RPE1 cells, we demonstrate here that heterozygosity of Plk4 leads to only a slight reduction in the level of centrosome-associated Plk4, consistent with self-regulation of the level of the centrosome-localized kinase (Holland et al., 2010). More important, Plk4+/− human and mouse cells do not undergo cytokinesis failure or exhibit an increase in centrosome amplification. We show Plk4 to be localized exclusively to the centrosome, with none accumulated in the spindle midbody. Depletion of Plk4 by small interfering RNA (siRNA) leads to loss of centrioles and subsequent spindle defects that indirectly lead to a modest increase in the rate of cytokinesis failure. We thus propose that Plk4 is a centriole-localized kinase that does not directly regulate cytokinesis.
RESULTS AND DISCUSSION
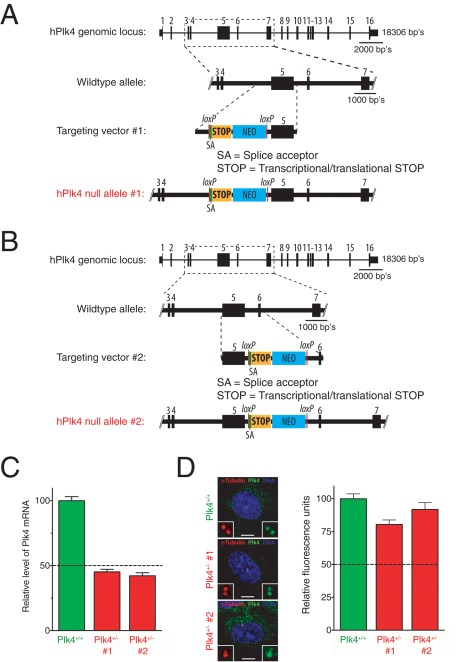
To study the effect of loss of a single allele of Plk4, we used gene targeting to insert a transcriptional/translational stop cassette into intron 4 (null allele 1) or 5 (null allele 2) of the endogenous Plk4 gene in telomerase-immortalized human RPE1 cells (Figure 1, A and B). The transcriptional/translational stop cassette prevents transcription and translation downstream of exon 4 (for null allele 1) or exon 5 (for null allele 2) of the Plk4 gene. Because exon 6-16 of Plk4 contains sequences required for Plk4 recruitment to the centriole and ability to promote centriole duplication, any truncated protein product translated from null allele 1 or 2 would strongly be expected to be nonfunctional (Leung et al., 2002; Habedanck et al., 2005). Quantitative RT-PCR was used to demonstrate that Plk4+/− 1 and 2 cells (each of which carries a single undisrupted copy of the Plk4 gene) have a ∼50% reduction in Plk4 mRNA compared with nontargeted control cells (Figure 1C). Plk4 is a low-abundance protein and, consistent with previous reports, we were unable to detect expression of the endogenous protein by immunoblotting (Supplemental Figure S1A). We therefore used quantitative immunofluorescence to determine the level of Plk4 protein at the centrosome of Plk4+/+ and Plk4+/− cells. An antibody raised against the C-terminus of the human Plk4 protein (amino acids 510–970) detected a centrosome-localized signal by immunofluorescence (Supplemental Figure S1B). This signal was specific for Plk4, as transfection with a pool of siRNAs targeting Plk4 resulted in dramatic reduction in the level of centrosome staining (see Figure 4C later in the paper). Despite ∼50% reduction in mRNA, Plk4+/− cells showed <20% reduction in the level of Plk4 protein localized to the centrosome, suggesting that Plk4 heterozygosity does not lead to haploid levels of the centrosome-associated kinase (Figure 1D).
FIGURE 1:
Generation of heterozygous Plk4 human cells. (A) Schematic showing the targeting strategy used to create a Plk4-null allele in RPE1 cells. A splice acceptor and a transcriptional/translational stop cassette were introduced into intron 4 of the endogenous human Plk4 gene by homologous recombination. (B) As in A, except that the splice acceptor and a transcriptional/translational stop cassette were inserted into intron 5. (C) Quantitative RT-PCR analysis of Plk4 mRNA levels in Plk4+/+ and Plk4+/− RPE1 cell lines. mRNA levels were calculated from two independent sets of Plk4 primers and normalized to two standard genes. Bars represent the mean of six independent replicas, and error bars represent the SEM. (D) Immunofluorescence images show representative Plk4 levels at the centrosome of Plk4+/+ and Plk4+/− RPE1 cell lines. Red, γ-tubulin; green, Plk4; blue, DNA. Scale bar, 5 μm. Graph shows quantitative immunofluorescence analysis of Plk4 protein levels at the centrosome of Plk4+/+ and Plk4+/− RPE1 cell lines. Bars show the mean of >32 cells per condition from three independent experiments. Error bars represent the SEM.
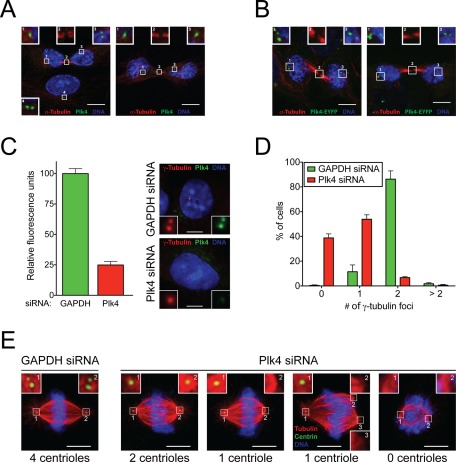
FIGURE 4:
Repression of Plk4 by siRNA results in the loss of centrioles due to impaired duplication. (A) Representative immunofluorescence images showing the localization of endogenous Plk4 in late-telophase cells. Red, α-tubulin; green, Plk4; blue. Scale bar, 5 μm. (B) As in A, except that cells stably overexpressing Plk4-EYFP were used. Red, α-tubulin; green Plk4-EYFP; blue, DNA. Scale bar, 5 μm. (C) Representative immunofluorescence images showing the level of Plk4 at the centrosome 48 h after transfection of Plk4+/− RPE1 cells with Plk4 or GAPDH siRNA. Red, γ-tubulin; Green, Plk4; Blue, DNA. Scale bar = 5 μm. Graph shows the quantitative immunofluorescence analysis of Plk4 protein levels at the centrosome 48 h after cells were transfected with Plk4 or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) siRNA. Note that only cells with at least one centrosome were quantified in this analysis, and thus the level of Plk4 repression is likely to be underestimated. Bars show the mean of >42 cells per condition from two independent experiments. Error bars represent the SEM. (D) The fraction of Plk4+/− RPE1 cells with the indicated number of γ-tubulin foci 48 h after transfection with Plk4 or GAPDH siRNA. Bars show the mean of >180 cells per condition from three independent experiments. Error bars represent the SEM. (E) Representative immunofluorescence images show phenotypes observed in mitotic Plk4+/− RPE1 cells 48 h after transfection with Plk4 or GAPDH siRNA. Red, α-tubulin; green, centrin; blue, DNA. Scale bar, 5 μm.
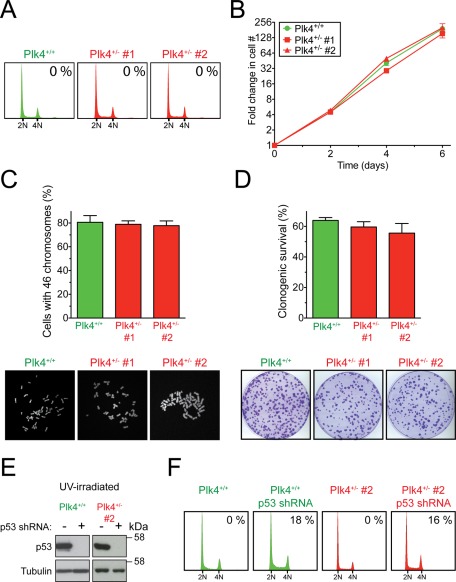
After >30 passages in culture, Plk4+/− RPE1 cells showed no evidence of centrosome amplification or increase in cellular ploidy (Figure 2A). Moreover, when compared with Plk4+/+ cells, Plk4+/− cells grew at the same rate, exhibited similar clonogenic survival, and showed no increase in the fraction of aneuploid cells (Figure 2, B–D). The tumor suppressor protein p53 has been implicated in suppressing the proliferation of polyploid cells, and removal of p53 was reported to increase the fraction of tetraploid Plk4+/− MEFs (Rosario et al., 2010). Nevertheless, depletion of p53 to undetectable levels with a constitutive short hairpin RNA (shRNA) did not yield an increase in the fraction of polyploid Plk4+/+ or Plk4+/− RPE1 cells (Figure 2, E and F). Of interest, p53 depletion resulted in centrosome amplification in ∼15–20% of both Plk4+/+ and Plk4+/− cells, possibly reflecting a role of p53 in regulating centriole duplication (Tarapore et al., 2001; Shinmura et al., 2007).
FIGURE 2:
Plk4+/– human cells do not undergo cytokinesis failure and show no increase in the level of centrosome amplification or aneuploidy. (A) Representative flow cytometry profiles of Plk4+/+ and Plk4+/− RPE1 cells. Numbers at the top right indicate the mean level of centrosome amplification from three independent experiments. (B) To monitor growth rate, Plk4+/+ and Plk4+/− RPE1 cells were counted every 2 d. Values represent the mean of two independent experiments, and error bars represent the SEM. (C) Graph shows the fraction of cells with 46 chromosomes in Plk4+/+ and Plk4+/− RPE1 cell lines. Bars represent the mean of >27 cells per condition from three independent experiments. Error bars represent the SEM. Immunofluorescence images show representative chromosome spreads from the indicated cell lines. (D) The percentage clonogenic survival for the Plk4+/+ and Plk4+/− RPE1 cell lines. Bars represent the mean of at least two independent experiments. Error bars represent the SEM. Images show representative crystal violet–stained colonies for the indicated cell lines. (E) Immunoblot of UV-irradiated Plk4+/+ and Plk4+/− RPE1 cells stably expressing a p53 shRNA. (F) Representative flow cytometry profiles of Plk4+/+ and Plk4+/− RPE1 cells stably expressing a p53 shRNA. Numbers at the top right indicate the mean level of centrosome amplification from two independent experiments.
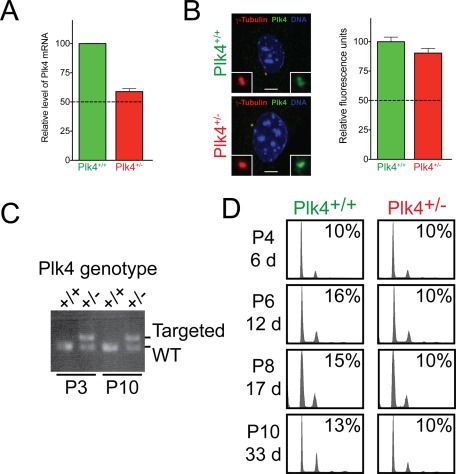
Because we were unable to observe a phenotypic consequence of Plk4 heterozygosity in human RPE1 cells, we reexamined the reported cytokinetic defects in Plk4+/− MEFs that were obtained from the authors of a previous study (Rosario et al., 2010). Use of quantitative RT-PCR revealed that Plk4+/− MEFs had close to a 50% reduction in Plk4 mRNA; however, quantitative immunofluorescence showed this resulted in <10% reduction in the level of Plk4 protein associated with the centrosome (Figure 3, A and B). Moreover, in contrast with previous reports, we were unable to find any evidence of elevated centrosome amplification or polyploidization in Plk4+/− MEFs, even after a prolonged period in culture (Figure 3, C and D). Taken together, our data demonstrate that heterozygosity of Plk4 does not lead to polyploidization and centrosome amplification in mouse or human cells.
FIGURE 3:
Plk4+/– MEFs show no evidence of cytokinesis failure or increased levels of centrosome amplification after a prolonged duration in culture. (A) Quantitative PCR analysis of Plk4 mRNA levels in Plk4+/+ and Plk4+/− MEFs. mRNA levels were calculated from five independent sets of Plk4 primers and normalized to two standard genes. Bars represent the mean of three independent replicas, and error bars represent the SEM. (B) Immunofluorescence images show representative Plk4 levels at the centrosome of Plk4+/+ and Plk4+/− MEFs. Red, γ-tubulin; green, Plk4; blue, DNA. Scale bar, 5 μm. Graph shows the quantitative immunofluorescence analysis of Plk4 protein levels at the centrosome of the indicated cells. Bars show the mean of >146 cells per condition from three independent experiments. Error bars represent the SEM. (C) PCR genotyping of passage 3 (P3) and passage 10 (P10) Plk4+/+ and Plk4+/− MEFs. (D) Representative flow cytometry profiles of Plk4+/+ and Plk4+/− MEFs at various passages after derivation. Experiments were performed twice on two independent Plk4+/+ and Plk4+/− MEF cell lines. Numbers on the left indicate the passage number (P) and the days (d) cells were in culture when analysis was performed. Numbers at the top right indicate the mean level of centrosome amplification from two independent experiments.
We next reexamined the reported localization of Plk4 to the spindle midbody, where it has been proposed to control the activity of RhoA to directly regulate cytokinesis (Rosario et al., 2010). As expected, staining with our Plk4 antibody revealed that endogenous Plk4 was localized at centrosomes of human cells; however, midbody staining was never observed (Figure 4A). This is consistent with a previous study that also reported an inability to detect midbody associated Plk4 (Habedanck et al., 2005). This lack of Plk4 midbody staining was not due to epitope masking, because an overexpressed enhanced yellow fluorescent protein (EYFP)–tagged Plk4 transgene also localized to the centrosome, with none at the spindle midbody (Figure 4B).
Because heterozygosity of Plk4 does not substantially reduce the level of centrosome-bound Plk4 (Figure 1, C and D), we carefully reexamined the phenotype(s) caused by substantial depletion of Plk4 with small interfering RNA (siRNA). Quantitative immuno-fluorescence showed Plk4 levels at the centrosome were reduced by >75% at 2 d after depletion of Plk4 by siRNA. This is an underestimate of the level of Plk4 depletion, as we were only able to quantify centrosome-localized Plk4 levels in cells with at least one centrosome, and 39% of Plk4 depleted cells contained no centrosomes at 2 d after depletion (Figure 4D). Staining with γ-tubulin antibodies to monitor centrosomes revealed that 86% of control siRNA cells showed two γ-tubulin foci, whereas 92% of cells depleted of Plk4 contained one or no γ-tubulin foci (Figure 4D). Analysis of mitotic figures revealed that control siRNA-treated cells exhibited normal bipolar spindles with two centrioles at each pole (Figure 4E). By contrast, depletion of Plk4 resulted in a number of mitotic spindle abnormalities: 48 h after Plk4 depletion a minority of cells had only a single centriole at each pole of a bipolar spindle, and many cells possessed only a single centriole at one pole (Figure 4E). In these cells the spindle was asymmetrically shaped, with the poorly focused acentriolar pole lacking astral microtubules and having a shorter half-spindle length (Figure 4E). In some cases, the acentriolar pole was multipolar, and occasionally aberrant spindles lacking centrioles altogether were observed (Figure 4E). Thus, consistent with previous reports, cells depleted of Plk4 undergo a stepwise loss of centrioles but continue to cycle (Bettencourt-Dias et al., 2005; Habedanck et al., 2005).
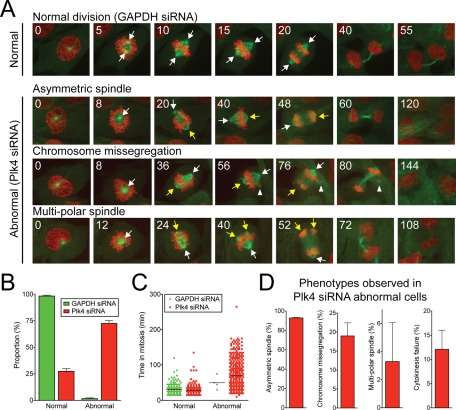
To establish whether substantial Plk4 depletion resulted in a failure of cytokinesis, we used time-lapse microscopy to follow mitotic divisions in Plk4-depleted RPE1 cells coexpressing EYFP–α-tubulin and histone H2B–monomeric red fluorescent protein (RFP). Ninety-eight percent of cells treated with control siRNA progressed through mitosis normally, with an average time of 31 min (Figure 5, A–C, and Supplemental Movie S1). However, 73% of cells depleted of Plk4 divided abnormally and delayed in mitosis, taking on average 69 min to divide. The vast majority (93%) of the Plk4-depleted cells that behaved abnormally divided in a bipolar manner with an asymmetric spindle containing one acentriolar pole (Figures 5A, yellow arrow, and 5D and Supplemental Movie S2).
FIGURE 5:
Depletion of Plk4 by siRNA does not result in a primary cytokinesis defect. (A) Time-lapse images of Plk4+/− RPE1 cells stably expressing histone H2B-mRFP (red) and EYFP-tubulin (green). Filming began 48 h after transfection with Plk4 or GAPDH siRNA. Numbers at the top left refer to the time in minutes after nuclear envelope breakdown (NEBD). White arrows show the position of normal spindle poles, and yellow arrows mark the position of acentriolar spindle poles (Figure 4C). White arrowhead marks the position of an ectopic, acentriolar bundle of microtubules, which attach to and lead to the missegregation of a small number of chromosomes. (B) Quantification of the proportion of cells dividing normally or abnormally in time-lapse movies of Plk4 or GAPDH siRNA–transfected Plk4+/− RPE1 cells. Cells were filmed for 48 h beginning 24 h after transfection with siRNA. Bars show the mean of >241 cells per condition from two independent experiments. Error bars represent the SEM. (C) The time from NEBD to anaphase for the cells filmed in A. Line represents the mean. (D) Graphs quantifying the proportion of abnormal Plk4 siRNA–transfected cells exhibiting the indicated phenotypes. Bars show the mean of two independent experiments. Error bars represent the SEM.
However, in a small fraction of cells (3%) the poorly focused acentriolar pole split into two, causing a multipolar division (Figures 5A, yellow arrow, and 5D and Supplemental Movie S4). Plk4-depleted cells also occasionally formed ectopic tubulin foci that were not incorporated into the spindle. In 19% of abnormally dividing Plk4-depleted cells, these ectopic foci acted as microtubule organizing centers and resulted in chromosome missegregation (Figure 5A, white arrowhead, and Supplemental Movie S3). However, despite the mitotic spindle abnormalities caused by the loss of centrioles, 88% of abnormally dividing cells depleted of Plk4 underwent normal cytokinesis and completed abscission (Figure 5D). Of importance, the 12% of Plk4-depleted cells that failed cytokinesis did so likely as a result of mitotic spindle abnormalities or chromosome missegregation events that resulted in chromatin left in the cleavage furrow.
Thus, we have demonstrated that a substantial reduction in Plk4 protein levels results in loss of centrioles, confirming the essential role of Plk4 in centriole duplication (Bettencourt-Dias et al., 2005; Habedanck et al., 2005). Cytokinesis failure, however, is seen only as an indirect consequence of the spindle abnormalities caused by loss of centrioles following loss of Plk4.
MATERIALS AND METHODS
Cell culture
hTERT RPE-1 and DLD-1 cells were maintained at 37°C in a 5% CO2 atmosphere with 21% oxygen. hTERT RPE-1 cells were grown in DMEM:F12 medium containing 10% tetracycline-free fetal bovine serum (FBS; Clontech, Mountain View, CA), 0.348% sodium bicarbonate, 100 U/ml penicillin, 100 U/ml streptomycin, and 2 mM l-glutamine, whereas Flp-In TRex-DLD-1 cells were grown in DMEM containing 10% tetracycline-free FBS (Clontech), 100 U/ml penicillin, 100 U/ml streptomycin, and 2 mM l-glutamine. Primary MEFs were grown in DMEM supplemented with 15% FBS, 0.1 mM nonessential amino acids (Life Technologies, Carlsbad, CA), 1 μM 2-mercaptoethanol (Specialty Media, Phillipsburg, NJ), 1 mM sodium pyruvate (Life Technologies), 2 mM glutamine, and 50 μg/ml penicillin/streptomycin in a 37°C humidified incubator with 10% CO2 and 3% oxygen. Low-oxygen conditions were used to extend the cycling time of the primary cells (Parrinello et al., 2003). RPE-1 cells were used in all experiments except in Figure 3, where MEFs were used, and Figure 4B, where DLD-1 cells were used.
Gene targeting
To generate the Plk4-null allele 1 and 2 targeting construct, a transcriptional/translational stop unit was created by gene synthesis (GenScript, Piscataway, NJ) and inserted into the pBluescript derivative pNY to create a loxP-STOP-neoR-loxP cassette. The transcriptional/translational stop is composed of the human adenovirus type 3/5 splice acceptor, 849 base pairs of the SV40 polyadenylation signal region, and a translation initiation signal followed by an in-frame stop codon and a 5′ donor splice site. Taq polymerase (Invitrogen, Carlsbad, CA) was used to amplify fragments of the human Plk4 locus from genomic RPE1 DNA. Error-free clones were identified or created by QuikChange site-directed mutagenesis (Agilent Technologies, Santa Clara, CA), and 5′ and 3′ arms were PCR amplified and cloned on either side of a central loxP-STOP-neoR-loxP cassette in pNY. The entire insert was then excised via NotI digestion and ligated to the pAAV vector backbone. The final construct was fully sequenced to verify its integrity. Procedures for preparation of infectious AAV particles, transduction of hTERT RPE1 cells, and isolation of properly targeted clones was performed as described previously (Berdougo et al., 2009).
Generation of stable cell lines and shRNA/siRNA treatment
RFP-tagged histone H2B (H2B-mRFP) and EYFP-tubulin was introduced into Plk4+/− RPE1 cells using retroviral delivery as described previously (Shah et al., 2004). Stable integrates were selected in 5 μg/ml puromycin and 10 μg/ml blasticidin S and single clones isolated using fluorescence-activated cell sorting (FACSVantage; Becton Dickinson, Franklin Lakes, NJ). A p53 shRNA was introduced into cells using lentiviral delivery and single clones isolated using fluorescence activated cell sorting (Tiscornia et al., 2003). For siRNA treatment, 1.5 × 105 cells were plated in a six-well plate or 35-mm poly-l-lysine–coated glass bottom culture dishes (MatTek, Ashland, MA) and duplexed siRNAs introduced using Lipofectamine RNAiMax (Invitrogen). A pool of four siRNAs directed against Plk4 5′-GAAAUGAACAGGUAUCUAA-3′, 5′-GAAACAUCCUUCUAUCUUG-3′, 5′-GUGGAAGACUCAAUUGAUA-3′, and 5′-GGACCUUAUUCACCAGUUA-3′) and a single siRNA directed against glyceraldehyde-3-phosphate dehydrogenase (5′-UGGUUUACAUGAUCCAAUA-3′) were purchased from Dharmacon (Lafayette, CO). Cells were processed for immunofluorescence microscopy or live-cell imaging 48 h after transfection.
Antibody production
A C-terminal Plk4 fragment (amino acids 510–970) was cloned into a pET-23b bacterial expression vector (Novagen, EMD4Biosciences, Gibbstown, NJ) containing a C-terminal hexahistidine tag. Recombinant protein was purified from Escherichia coli using Ni–nitrilotriacetic acid beads (Qiagen, Valencia, CA) and used for immunization (Moravian Biotechnology, Brno, Czech Republic)
Antibody techniques
For immunoblot analysis protein samples were separated by SDS–PAGE, transferred onto nitrocellulose membranes (Bio-Rad, Hercules, CA), and then probed with DM1A (mouse anti–α-tubulin, 1:5000) Plk4 (rabbit, 1:500), or p53 (mouse, 1:100; Calbiochem, La Jolla, CA) antibody. Immunofluorescence was performed as previously described (Holland et al., 2010) using the following antibodies: GTU-88 (mouse anti–γ-tubulin, 1:250; Abcam, Cambridge, MA), Plk4 (rabbit, 1:250), DM1A (mouse anti–α-tubulin, 1:1000), and centrin (mouse, 1:1000; a kind gift of Jeffrey Salisbury, Mayo Clinic Foundation, Rochester, MN). YFP was visualized directly using a GFP filter set. Immunofluorescence images were collected using a Deltavision Core system (Applied Precision, Issaquah, WA) controlling an interline charge-coupled device (CCD) camera (Cool Snap, Roper Scientific Photometrics, Photometrics, Tucson, AZ). Images were collected using a 100×, 1.4 numerical aperture (NA), or a 60×, 1.4 NA, oil objective at 0.2 μm z-sections and subsequently deconvolved. Maximum-intensity two-dimensional (2D) projections were assembled for each image using softWoRx (Applied Precision). For quantification of Plk4 signal intensity, undeconvolved 2D maximum-intensity projections were saved as unscaled 16-bit TIFF images and signal intensities determined using MetaMorph (Molecular Devices, Sunnyvale, CA). A 20 × 20 pixel box and a larger 25 × 25 pixel box was drawn around the centrosome (marked with γ-tubulin staining). Integrated Plk4 intensity in the smaller box was calculated by subtracting the mean fluorescence intensity in the area between the two boxes (mean background) from the mean Plk4 intensity in the smaller box and multiplying by the area of the smaller box.
Live-cell microscopy
Cells were seeded into 35-mm, poly-l-lysine–coated glass-bottom culture dishes (MatTek) and 24 h later transferred to CO2-independent media supplemented with 10% FBS, 100 U/ml penicillin, 100 U U/ml streptomycin, and 2 mM l-glutamine. For bulk population analysis, cells were maintained at 37°C in an environmental control station and images collected using a Deltavision RT system with a 40×, 1.35 NA, oil lens at 5-min time intervals. For each time point, 4 × 3 μm z-sections were acquired for GFP and RFP and maximum-intensity projection created using softWoRx (Applied Precision).
For high-resolution spinning-disk fluorescence microscopy, cells were imaged using a 60×, 1.4 NA, Plan Apochromat oil lens on a spinning disk confocal mounted on a Nikon (Melville, NY) TE2000-E inverted microscope equipped with a solid-state laser combiner (ALC)—491- and 561-nm lines—a Yokogawa CSU10 head, and a CCD Clara camera (Andor Technology, South Windsor, CT). Acquisition parameters, shutters, and focus were controlled by iQ 1.10.0 software (Andor Technology). The 5 × 2 μm z-sections were acquired at 5-min time intervals for GFP and RFP and maximum-intensity projection created using MetaMorph. Movies were assembled and analyzed using QuickTime (Apple, Cupertino, CA).
Cell biology
To prepare cells for flow cytometry, cell pellets were fixed in cold 70% EtOH for 24 h, washed once in PBS, and suspended in PBS supplemented with 10 μg/ml RNase A and 50 μg/ml propidium iodide. Samples were incubated at room temperature for 30 min and analyzed on a flow cytometer (LSRII; BD Biosciences, San Diego, CA). Chromosome spreads were carried out as previously described (Holland and Taylor, 2006). For clonogenic assays 500 cells were seeded in a 10-cm2 culture dish and left to grow for ∼2 wk until colonies were visible by eye. Cells were fixed in methanol for 10 min at room temperature and colonies stained with crystal violet (Sigma-Aldrich, St. Louis, MO). The percentage clonogenic survival was determined by dividing the number of colonies formed by the number of cells initially seeded in the dish.
Quantitative real-time PCR
Total RNA was extracted from cell pellets using TRIzol (Invitrogen) and cDNA generated using random hexamers and Superscript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Quantitative real-time PCR for mouse and human Plk4 was performed using the iQ SYBR green supermix (Bio-Rad) on the IQ5 multicolor real-time PCR detection system (Bio-Rad). Analysis was performed using the IQ5 optical system software (version 2.1, Bio-Rad). All reactions were carried out in duplicate using two or five independent sets of primers for human or mouse Plk4, respectively, and expression values were normalized to two control genes: either TATA box–binding protein (TBP) and eukaryotic elongation factor 1 (EEF1) for human cells or actin gamma 1 (ACTG1) and cyclophilin for mouse cells. The fold changes in mRNAs expression were calculated as previously described (Pfaffl, 2001), and expression values were expressed as a percentage of the average expression of the Plk4+/+ cells. The following primers were used: human Plk4-1, forward, 5′- GGT GGT AGA GGT TTT CCT CTT GC-3′; human Plk4-1, reverse, 5′-CAG CAC CAG GAG AAT TCT CC ATC-3′; human Plk4-2, forward, 5′-CCT AAG GCC TTA TCA CCT CCT CC-3′; human Plk4-2 reverse, 5′-CTG AAC CCA CAC AGC TCC ACT AG-3′; mouse Plk4-1, forward, 5′-GGA GAG GAT CGA GGA CTT TAA GG-3′; mouse Plk4-1, reverse, 5′-CCA GTG TGT ATG GAC TCA GCT C-3′; mouse Plk4-2, forward, 5′-GTG CAT CGG GGA GAG GAT CGA-3′; mouse Plk4-2, reverse, 5′-CAA GAC AGA GGG GTG TTT CAA CTG-3′; mouse Plk4-3, forward, 5′- GTG CTT CAG ATA TCG AGT GAT GGG-3′; mouse Plk4-3, reverse, 5′- GGA AGC ATA CTG ATA TTT CCG CCA G-3′; mouse Plk4-4, forward, 5′-GGA CCA TGC TAA TGA GGG TCA C-3′; mouse Plk4-4, reverse, 5′-GAA AGC GGC ACT ATT CAC GCT C-3′; mouse Plk4-5, forward, 5′-GAG CGT GAA TAG TGC CGC TTT C-3′; mouse Plk4-5, reverse, 5′-TGA ACC CAC ACA GCT CCG CTA G-3′.
Supplementary Material
Acknowledgments
We thank Caroline Swallow (Samuel Lunenfeld Research Institute, Toronto, Canada) for providing the Plk4+/+ and Plk4+/− MEFs, Prasad Jallepalli (Memorial Sloan-Kettering Cancer Center, New York, NY) for providing reagents for AAV-mediated gene targeting, and Bryan Tsou (Memorial Sloan-Kettering Cancer Center) for providing the Plk4AA-doxycline–inducible RPE1 cells. We thank the University of California, San Diego, Neuroscience Microscopy Shared Facility (P30 NS047101). This work was supported by Grant GM29513 from the National Institutes of Health to D.W.C, who receives salary support from the Ludwig Institute for Cancer Research. A.J.H. is supported by a Leukemia and Lymphoma Society Special Fellowship. D.F. is supported by a European Molecular Biology Organization (EMBO) long-term fellowship, and B.V. is supported by a Human Frontier Science Program (HFSP) postdoctoral fellowship.
Abbreviations used:
- PBS
phosphate-buffered saline
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-12-1043) on March 28, 2012.
REFERENCES
- Berdougo E, Terret ME, Jallepalli PV. Functional dissection of mitotic regulators through gene targeting in human somatic cells. Methods Mol Biol. 2009;545:21–37. doi: 10.1007/978-1-60327-993-2_2. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK, Carmo N, Balloux F, Callaini G, Glover DM. SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Brownlee CW, Klebba JE, Buster DW, Rogers GC. The protein phosphatase 2A regulatory subunit Twins stabilizes Plk4 to induce centriole amplification. J Cell Biol. 2011;195:231–243. doi: 10.1083/jcb.201107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha-Ferreira I, Rodrigues-Martins A, Bento I, Riparbelli M, Zhang W, Laue E, Callaini G, Glover DM, Bettencourt-Dias M. The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr Biol. 2009;19:43–49. doi: 10.1016/j.cub.2008.11.037. [DOI] [PubMed] [Google Scholar]
- Debec A, Sullivan W, Bettencourt-Dias M. Centrioles: active players or passengers during mitosis? Cell Mol Life Sci. 2010;67:2173–2194. doi: 10.1007/s00018-010-0323-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guderian G, Westendorf J, Uldschmid A, Nigg EA. Plk4 trans-autophosphorylation regulates centriole number by controlling betaTrCP-mediated degradation. J Cell Sci. 2010;123:2163–2169. doi: 10.1242/jcs.068502. [DOI] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Lan W, Niessen S, Hoover H, Cleveland DW. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J Cell Biol. 2010;188:191–198. doi: 10.1083/jcb.200911102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Taylor SS. Cyclin-B1-mediated inhibition of excess separase is required for timely chromosome disjunction. J Cell Sci. 2006;119:3325–3336. doi: 10.1242/jcs.03083. [DOI] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Ko MA, Rosario CO, Hudson JW, Kulkarni S, Pollett A, Dennis JW, Swallow CJ. Plk4 haploinsufficiency causes mitotic infidelity and carcinogenesis. Nat Genet. 2005;37:883–888. doi: 10.1038/ng1605. [DOI] [PubMed] [Google Scholar]
- Leung GC, Hudson JW, Kozarova A, Davidson A, Dennis JW, Sicheri F. The Sak polo-box comprises a structural domain sufficient for mitotic subcellular localization. Nat Struct Biol. 2002;9:719–724. doi: 10.1038/nsb848. [DOI] [PubMed] [Google Scholar]
- Loncarek J, Khodjakov A. Ab ovo or de novo? Mechanisms of centriole duplication. Mol Cells. 2009;27:135–142. doi: 10.1007/s10059-009-0017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P, Honda R, Nigg EA. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53-/- cells. EMBO J. 2002;21:483–492. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA. Origins and consequences of centrosome aberrations in human cancers. Int J Cancer. 2006;119:2717–2723. doi: 10.1002/ijc.22245. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol. 2003;5:741–747. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel N, Stevens NR, Basto R, Raff JW. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr Biol. 2007;17:834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GC, Rusan NM, Roberts DM, Peifer M, Rogers SL. The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J Cell Biol. 2009;184:225–239. doi: 10.1083/jcb.200808049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario CO, Ko MA, Haffani YZ, Gladdy RA, Paderova J, Pollett A, Squire JA, Dennis JW, Swallow CJ. Plk4 is required for cytokinesis and maintenance of chromosomal stability. Proc Natl Acad Sci USA. 2010;107:6888–6893. doi: 10.1073/pnas.0910941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah JV, Botvinick E, Bonday Z, Furnari F, Berns M, Cleveland DW. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr Biol. 2004;14:942–952. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
- Shinmura K, Bennett RA, Tarapore P, Fukasawa K. Direct evidence for the role of centrosomally localized p53 in the regulation of centrosome duplication. Oncogene. 2007;26:2939–2944. doi: 10.1038/sj.onc.1210085. [DOI] [PubMed] [Google Scholar]
- Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS ONE. 2009;4:e6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarapore P, Horn HF, Tokuyama Y, Fukasawa K. Direct regulation of the centrosome duplication cycle by the p53-p21Waf1/Cip1 pathway. Oncogene. 2001;20:3173–3184. doi: 10.1038/sj.onc.1204424. [DOI] [PubMed] [Google Scholar]
- Tiscornia G, Singer O, Ikawa M, Verma IM. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc Natl Acad Sci USA. 2003;100:1844–1848. doi: 10.1073/pnas.0437912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou MF, Stearns T. Controlling centrosome number: licenses and blocks. Curr Opin Cell Biol. 2006;18:74–78. doi: 10.1016/j.ceb.2005.12.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.