Abstract
The influenza A(H1N1)pdm09 virus has circulated worldwide and continued to cause complicated infections and deaths. Reports have identified an increased prevalence of the hemagglutinin receptor binding domain D222G mutation in viruses isolated from individuals who have suffered such severe infections, but this association is still unclear. Virus isolated from a nasopharyngeal wash of a severely ill immunocompromised patient at the time of diagnosis contained the D222, but isolates collected later in his course from a bronchoalveolar lavage contained primarily the G222 mutation and was mixed with a minor population of D222. These clinical isolates were compared to a G222 plaque purified virus in the ferret model. The G222 predominant clinical isolate was the most pathogenic in ferrets and developed the most diversity at the 222 amino acid position during infection, suggesting that increased diversity and not a specific polymorphism at HA 222 may be important in predicting pathogenic potential.
Keywords: Influenza A, A(H1N1)pdm09, pathogenesis, D222G, Hemagglutinin, immunocompromised, pandemic, ferrets
Introduction
The 2009 pandemic H1N1 influenza A virus [A(H1N1)pdm09] emerged in March 2009 and caused significant morbidity and mortality worldwide(Bautista et al., 2010). It has continued to circulate post-pandemically in an annual, seasonal pattern along with H3N2 and influenza B viruses. Early during the 2009 pandemic, amino acid polymorphisms in the receptor binding domain of the hemagglutinin gene (HA) were identified(Chutinimitkul et al., 2010; Kilander et al., 2010; WHO, 2010), and an association was made with severe disease or fatal outcomes (Kilander et al., 2010). Specifically, reports have documented an association with severe influenza and isolation of pandemic viruses with position 222 mutations from the consensus Aspartic acid (D) to either Glycine (G) or Asparagine (N) (Baldanti et al., 2010; Baumeister et al., 2010; Drews et al., 2010; Ertek et al., 2010; Ikonen et al., 2010; L’Vov D et al., 2010; Potdar et al., 2010; Puzelli et al., 2010; Wang et al., 2011). Another variant codon at position 222, Glutamic acid (E), has been observed but has not been associated with severe disease (Puzelli et al., 2010). These observations suggested that mutations in this region, particularly the G222 mutation, may be important virological virulence factors associated with severe outcomes in A(H1N1)pdm09 infection. In vivo studies suggest these severe outcomes may be due to the G222 mutation influencing receptor binding specificity leading to a more severe lower respiratory tract infection (Liu et al., 2010; Watanabe et al., 2011). Studies comparing viruses isolated from nasal swabs with matched isolates from bronchoalveolar lavage (BAL) specimens have shown that 222G variants have been isolated in higher percentages from the BAL samples (Baldanti et al., 2010; Chutinimitkul et al., 2010). Matched nasopharyngeal wash and BAL isolates are only available from small numbers of patients, predominantly severely ill ones, so that comparable studies with viruses isolated from patients with mild influenza have not been performed.
Interestingly these HA D222G or D222N mutations have also been identified in primary sequence analysis of cases of pandemic influenza from 1918 (Reid et al., 2003; Sheng et al., 2011) with 222G identified in 5/16 (37.5%) autopsy cases; and a mixture of 222G/222N in a single case. However, case histories, histopathologic changes, and immunohistochemical analyses for viral antigen distribution showed no differences between the 1918 cases with D222 versus G222.
Experimental animal studies with G222 viruses derived by mutagenesis and reverse genetics (Belser et al., 2011) or serial mouse passage (Zheng et al., 2010) have reported enhanced pathogenesis in mice following intranasal inoculation as compared to D222 viruses, but a comparable study in ferrets, showed no significant differences in either pathogenesis or transmissibility (Belser et al., 2011). A study in which matched pairs of isogenic A(H1N1)pdm09 viruses, expressing either D222 or G222, generated conflicting data, with the G222 virus of one matched pair showing enhanced pathogenicity from one set of viruses but not with the other (Watanabe et al., 2011).
Thus, the significance of these HA receptor binding domain polymorphisms is still unclear, and the effects on disease and mechanisms by which they arise have not been fully elucidated. In this report, serially isolated viruses from a previously described immunocompromised patient (Memoli et al., 2010b) with a severe A(H1N1)pdm09 infection were analyzed. The initial isolate, collected from a nasopharyngeal (NP) wash at the time of diagnosis, was noted to encode a D222 in the HA protein. An isolate collected from the lungs by bronchoalveolar lavage (BAL) after the patient developed severe disease on day 24 encoded a G222 in the HA protein, while virus collected from an NP wash at the same time still demonstrated D222. All subsequent viruses isolated from nasal samples throughout this patient’s course of infection were shown to express HA with D222. Given this observation, we performed analyses of quasispecies of the viruses isolated from this patient. Using these data and the viruses isolated, we sought to determine the significance of the changes at position 222 in these isolates and their quasispecies through evaluation of clinical disease, pathogenesis and, transmissibility in the ferret model.
Results
Initial Viral Analysis
The initial sequencing of the initial NP isolate has been previously published and submitted to Genbank (A/Bethesda/NIH106-D0/2009(H1N1), accession numbers:HQ263268-HQ263274, GU571154) (Memoli et al., 2010a; Memoli et al., 2010b). The BAL isolate gene sequences except for the HA and NA were identical to those of the NP wash isolate. The NA gene contained the H275Y mutation, identical to the NA from the resistant virus previously isolated from this patient (accession GU57115) (Memoli et al., 2010b), and the HA gene was found to have a single difference with the NP wash HA sequence at position 222 with a change from D to G.
By performing colony PCR, the initial clinical isolate collected by NP wash was determined to contain a D at position 222 of the HA1 domain with 100% of clones sequenced demonstrating that genotype (Table 1). The isolate collected by BAL during the period of the patient’s most severe illness was determined to be a mixed isolate with quasispecies containing primarily G222 (92%), but a small but significant (8%) percentage of D222 (Table 1). The plaque-purified virus from the BAL specimen was confirmed to contain a pure population of virus with 100% of clones containing G222 (Table 1). No other mutations in the HA gene were noted in the colonies analyzed.
Table 1.
Amino Acid* 222 Variation of Viruses Used for Inoculation.
| NP Clinical Isolate | BAL Clinical Isolate | Plaque Purified Virus | |
|---|---|---|---|
| D | 100% | 8% | 0% |
| G | 0% | 92% | 100% |
| N | 0% | 0% | 0% |
| S | 0% | 0% | 0% |
Aspartic Acid (D), Glycine (G), Asparagine (N), Serine (S)
In Vivo Evaluation in Ferrets
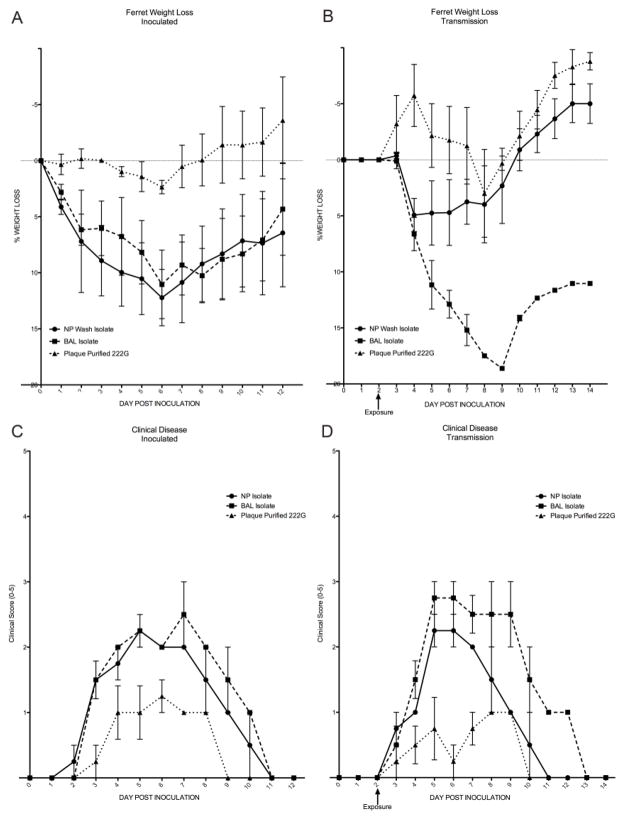
Ferrets inoculated with the NP wash isolate (with D222) developed a moderate course of illness with peak weight loss of 12% on day 6 (Figure 1A) and clinical symptoms on day 5 post inoculation with a mean score 2.25 (Figure 1C), consistent with previous studies (Memoli et al., 2010a). Those animals inoculated with the BAL isolate (D222/G222 mixture) also developed a statistically similar moderate course of illness with a mean peak clinical score of 2.5 (p=0.537) and weight loss of 11% (p=0.792) (Figure 1A, C). This course of illness was significantly different from that of the animals inoculated with the plaque purified 222G containing isolate. In this group, the animals demonstrated minimal illness with a slower progression and significantly lower mean peak clinical score of 1.25 than the other two groups (p = 0.03, 0.017 respectively) (Figure 1C). Weight loss was also significantly less throughout the fourteen days, with mean peak weight loss of 2.36% (p=0.022, 0.031 respectively) and faster recovery (Figure 1A).
Figure 1. Mean weight loss and clinical scores in infected ferrets.
All ferrets demonstrated evidence of clinical disease by clinical score in both the inoculated (C) and transmission groups (D), with the least disease seen in those animals infected with the 222G Plaque Purified Virus. This was consistent with weight loss observed in the inoculated (A) and transmission (B) groups. Error bars represent SEM.
Differences became even more apparent in those animals that were exposed to those that were inoculated. Those animals exposed to either the NP isolate or the BAL isolate both demonstrated moderate clinical illness with a mean peak clinical score of 2–3 by day 3 post exposure, but those animals exposed to the NP isolate recovered more quickly. Ferrets exposed to the BAL isolate were still demonstrating significant illness by day 5 post exposure (day 7 post inoculation) as compared to those exposed to the NP isolate; with one animal progressing to death (Figure 1D). A more significant weight loss was observed in those animals exposed to the BAL isolate, with a mean peak weight loss of 18.62%, as compared to 4.93% in the NP isolate group (p = .004) (Figure 1B). In stark contrast to those exposed by transmission to the clinical nasopharyngeal wash and BAL isolates, animals exposed to the plaque-purified G222 virus demonstrated minimal clinical symptoms, with some animals demonstrating no symptoms (Figure 1D). Weight loss was also minimal with most animals gaining weight throughout their course (Figure 1B).
Viral Shedding and Lung Titers
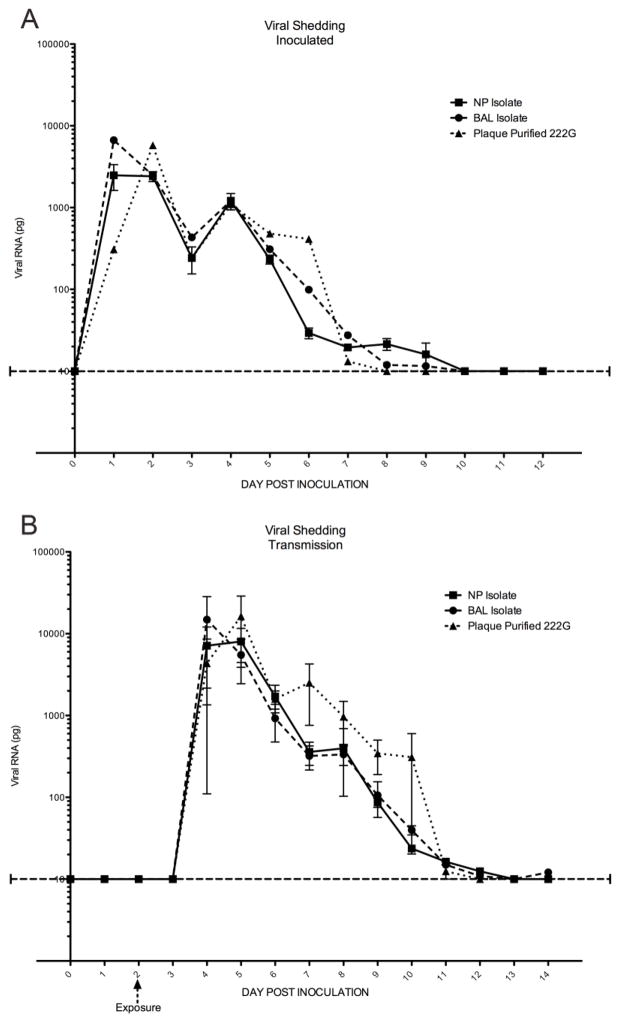
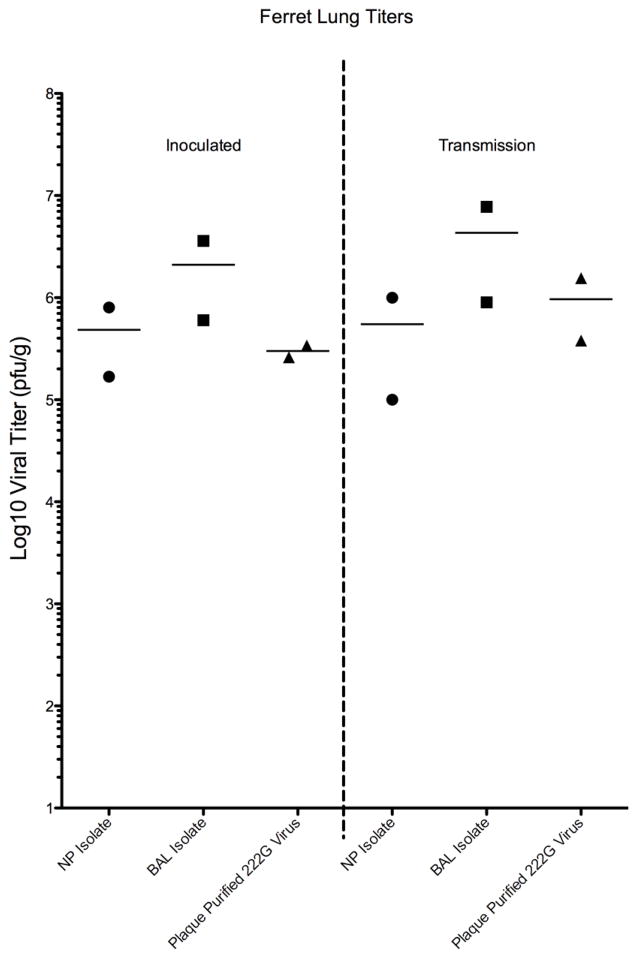
Despite the differences in clinical disease, regardless of virus exposure all animals were found to shed virus nasally and transmission was noted in 100% of the animals exposed to inoculated animals. All animals shed virus for at least 5 days, and no significant difference was noted in the amount of viral shedding (Figure 2). It was noted that those animals inoculated or exposed by transmission to the plaque purified G222 virus reached peak shedding one day later than those exposed to the clinical isolates. In the animals sacrificed on day 5, no significant difference in lung titer was noted between the groups, with virus recovered from the homogenized lungs of all animals at titers of 105 to 107pfu/g.
Figure 2. Mean quantity of viral RNA detected by qPCR in the nasal washes of ferrets after inoculation (A) and by transmission (B).
All animals shed virus during the course of infection to similar titers regardless of virus exposure. 100% transmission was noted as well. Error bars represent SEM.
Pathology
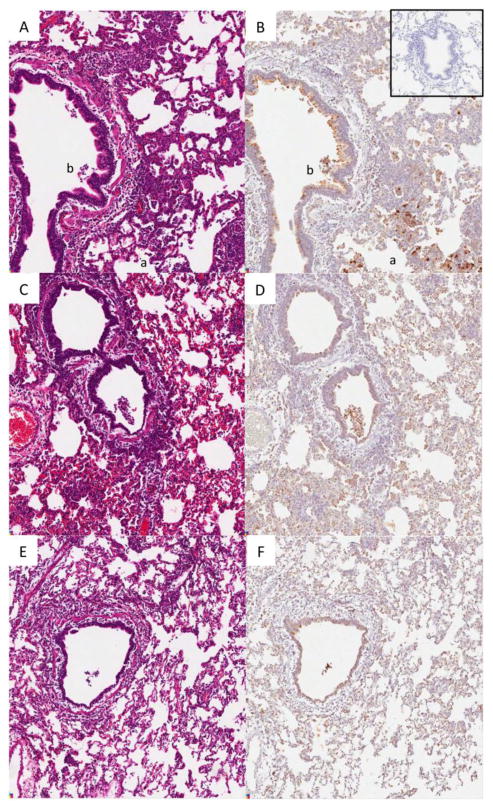
All animals euthanized on day 5 post-exposure demonstrated a range of histopathology equivalent to that described in detail previously (Memoli et al., 2010a). In brief, mild multifocal disease of the nasopharynx, larynx and trachea was observed with laryngitis, tracheitis, and bronchial gland sialadenitis. Within the lungs there was a mild to moderate bronchitis, moderate to severe pan-bronchiolitis, variable amounts of alveolitis and interstitial pneumonia and in the most severe cases patches of acute pneumonia as well as congestion, edema and rarely hemorrhage. Inflammation was mixed lymphohistiocytic and neutrophilic (Figure 3). In the lungs, the bronchial gland sialadenitis was generally more severe than its neighboring bronchitis. Immunohistochemistry revealed multifocal viral antigen labelling in the bronchial epithelium, bronchioles, and alveolar epithelial cells and alveolar macrophages in the areas of focal alveolitis (Figure 3). At the day 12 post-inoculation time point there were rare foci of inflammation within the upper respiratory tract, and occasional foci of resolving bronchointerstitial pneumonias with evidence of repair, as previously described (Memoli et al., 2010a).
Figure 3. H&Es and influenza antigen IHCs of day 5 post-inoculation ferret lung.
(A) H&E of NP inoculated ferret, (B) IHC of same NP inoculated ferret including inset of influenza antigen labeling of uninfected ferret tissue, (C) H&E of BAL inoculated ferret, (D) IHC of same BAL inoculated ferret, (E) H&E of plaque purified G222 inoculated ferret, (F) IHC of same plaque purified G222 ferret. “a” designates an alveolar lumen and “b” designates a bronchiolar lumen. Original magnification 80x.
When comparing groups of animals, the day 5 post-inoculation pathology coincided with the clinical disease observed. Those animals inoculated with the BAL isolate demonstrated slightly more severe pathology than those inoculated or exposed by transmission to the NP isolate, but were very similar (Figure 3). All of the animals inoculated or exposed by transmission with the BAL or NP isolate demonstrated more diffuse and extensive pathology than those infected with the plaque purified G222 virus. In both the inoculation and transmission groups, these animals demonstrated limited pathology (Figure 3).
One ferret exposed to a ferret inoculated with the BAL isolate died on day 9 post-exposure. Histopathologic analysis revealed a bronchopneumonia with Gram-negative bacilli (data not shown). Gram stains were additionally done on several other acute time-point ferret tissues representing the full range of pathology scores and all were negative.
Sequencing and Quasispecies of Ferret Isolates
More HA genome diversity was noted in those animals exposed by transmission (Table 3), and the BAL isolate developed the most diversity during infections of the animals. This diversity included the identification of an Asparagine (N) at site 222 in virus isolated from the nasal turbinate and a change from G222 to D222 predominance in the lung after transmission (Table 2, 3). The NP wash isolate also developed significant diversity at position 222 during infection of the animals, but still maintained a D predominance throughout both the upper and lower airways (Tables 2, 3). The plaque-purified virus developed the least diversity during infection, with 98–100% of quasispecies maintaining the 222G in both upper and lower airway (Tables 2, 3).
Table 3.
Amino Acid* 222 Variation of Viruses Isolated from Transmission Ferrets.
| NP Clinical Isolate Transmission | BAL Clinical Isolate Transmission | Plaque Purified Virus Transmission | ||||
|---|---|---|---|---|---|---|
| Nasal Turbinate | Lung | Nasal Turbinate | Lung | Nasal Turbinate | Lung | |
| D | 52% | 55% | 41% | 62% | 2% | 0% |
| G | 48% | 45% | 46% | 38% | 98% | 100% |
| N | 0% | 0% | 13% | 0% | 0% | 0% |
| S | 0% | 0% | 0% | 0% | 0% | 0% |
Aspartic Acid (D), Glycine (G), Asparagine (N), Serine (S)
Table 2.
| NP Clinical Isolate Inoculated | BAL Clinical Isolate Inoculated | Plaque Purified Virus Inoculated | ||||
|---|---|---|---|---|---|---|
| Nasal Turbinate | Lung | Nasal Turbinate | Lung | Nasal Turbinate | Lung | |
| D | 100% | 81% | 7% | 13% | 2% | 0% |
| G | 0% | 0% | 93% | 87% | 98% | 100% |
| N | 0% | 17% | 0% | 0% | 0% | 0% |
| S | 0% | 2% | 0% | 0% | 0% | 0% |
Aspartic Acid (D), Glycine (G), Asparagine (N), Serine (S)
Phylogenetic analysis
Intrahost codon usage at site 222 revealed a strong bias towards a particular codon for the different variants isolated from the ferrets. The G222 variants showed a preference for the glycine codon, GGT. The aspartic acid variants, D222, showed a specificity for GAT. A population level analysis of codon usage at amino acid site 222 of HA sequences of 2009 pandemic H1N1 revealed an identical pattern for the G222 and D222 variants. Phylogenetic analysis showed the G222 variants as either sporadic, one-off mutations with no further transmission over time, or rare, small monophyletic clades with limited transmission and very little persistence over time (Supplemental Figure 1). A monophyletic clade containing mostly E222 variants within the HA phylogenetic tree shows a pronounced pattern of persistence and transmission across countries. A similar pattern was found with these E222 variants in the NA phylogenetic tree.
Discussion
The isolation of viruses from this patient containing different HA receptor binding site polymorphisms at position 222 demonstrates that mutations in the HA can occur rapidly in a human host increasing quasispecies diversity with different predominance in the upper and lower respiratory tract. Rapid increase in quasispecies diversity was also seen in the ferret model when clinical isolates were used to inoculate the animals, and a great increase in quasispecies was observed especially after transmission in both the upper and lower airways. In contrast, much less diversity developed after infection with the plaque-purified virus. This was likely due to a lack of significant quasispecies present and/or selective pressures minimizing the ability of the pure G222 population to change. The clinical NP wash isolate that was identified as only containing D222 did develop diversity at this site rapidly, but it is possible that there were pre-existing quasispecies present in this clinical isolate below the limits of detection in this study. This highlights the complex nature of development of these polymorphisms and that multiple host and virological factors are likely involved in their development.
The presence of the H275Y mutation in the neuraminidase (NA) gene of the BAL and plaque purified sample did not seem to have any effect on viral fitness as the BAL isolate, NP isolate, and plaque-purified virus all grew well in culture and grew to similar titers from the lungs of the ferrets (figure 4). In a previously published study we demonstrated that this NP isolate, containing H at position 275, and another NP isolate from later in this same patient’s infection that contained the H275Y mutation maintained fitness and transmissibility in the ferret model (Memoli et al., 2010a). These data in conjunction with the increased presence of H275Y in seasonal H1N1 and increasing evidence that the H275Y mutation in A(H1N1)pdm09 does not effect viral fitness, transmissibility, or pathogenesis (Hamelin et al.; Seibert et al.; van der Vries et al.); it seems likely that the presence of the H275Y mutation did not play a role in the differences observed here.
Figure 4. Viral Titers in Ferret Lung Tissues.
Virus was recovered from all ferret lungs harvested on day 5 post inoculation or exposure (transmission group). Viral titers were similar with no statistical difference in mean titer observed between the groups.
Although in vitro studies have demonstrated that the D222G mutation may alter receptor specificity and cell tropism allowing binding of a broader range of α2-3-linked sialyl receptor sequences on ciliated bronchial epithelial cells and on lung epithelia (Liu et al., 2010; Watanabe et al., 2011), we did not observe any differences in viral antigen distribution, viral titer, or range of histopathology in the upper and lower airways in the ferret model. Additionally, regardless of HA residue 222 sequence, all viruses grew well and were fully transmissible. These data are consistent with a recent study demonstrating similar disease in ferrets infected with a reverse genetics derived mutant virus and a high passage vaccine strain derived from A/CA/04/2009 (H1N1) (Belser et al., 2011).
Although the results of the above studies were similar to what we observed, the current study has some significant differences. By using two very low passage clinical isolates (< 3 passages) and comparing them to a plaque-purified virus, we were able to explore not only the role of the D222G mutation itself, but also that of the described diversity of quasispecies at this position. While we observed differences in quasispecies proportions depending on location in the ferret respiratory tree, it is unclear if this difference in proportion of a certain amino acid at position 222 in the quasispecies present in the upper or lower airways was related to differences in viral binding or cell tropism, or rather reflected stochastic processes related to which viral variant predominated in early infection.
The other significant finding was that the plaque-purified virus containing the G222 caused the least severe illness in ferrets, again suggesting that it is likely that the 222G mutation is not a sole virulence determinant of severe disease as supported by the nonhuman primate infections described by Watanabe et al (Watanabe et al., 2011). Interestingly, in the current study, those viral isolates with more diversity at HA position 222 were the most pathogenic, causing the most clinical disease, weight loss, and histopathology as demonstrated by those animals infected by transmission with the BAL isolate. Those animals developed the most severe clinical course, and the viruses isolated from those animals were the most diverse at position 222, particularly in virus isolated from the lungs. Although those animals infected by exposure to the NP isolate also developed considerable diversity at position 222, they developed slightly less overall disease. This difference may have been due to the fact that the virus causing the infection did not contain any detectable G222 variants; suggesting that the G222 mutation may play a role in enhancing pathogenesis as part of a diverse population.
The least diverse viruses were isolated from the animals infected with the plaque-purified G222 isogenic clone, and these animals demonstrated the least clinical disease and pathology compared to the other 2 diverse infections. This difference in pathogenicity may signify that HA receptor-binding region diversity may allow virus to bind to more diverse cell types in the initial infection, thus spreading more easily throughout the upper and lower airways. This is similar to quasispecies diversity determined neurotrophism and pathogenesis of polioviruses as described by Vignuzzi et al., suggesting that diverse quasispecies populations are the unit of selection, rather than individual variants (Vignuzzi et al., 2006). Thus, diversity at this HA receptor-binding site, rather than specific polymorphisms themselves, may be a better determinant of disease progression than a specific amino acid change.
Although diverse 222 variants are generated at the intrahost level and show a specificity for codon usage bias, phylogenetic analysis of 2009–2011 gene sequence data revealed that G222 variants are a result of sporadic mutations that occur with no forward persistence and very little transmission over time. In contrast, E222 forms a pronounced, monophyletic clade over time. These data suggest that variation at the amino acid site 222 is likely driven by intrahost adaptation, and that the population snap-shot of these variants in the context of phylogenetics is likely not indicative of pathogenic or circulating variants.
Conclusions
Mutations in the HA1 receptor binding domain at position 222 of A(H1N1)pdm09 can develop rapidly as is evident in both human cases and in the ferret model. Mutations such as the D222G mutation likely do not mediate severe disease alone, but may play a role in enhancing pathogenesis as a significant part of a diverse population of quasispecies. Increased diversity of a viral isolate at position 222 seems more important in predicting increased pathogenesis rather than the presence or absence of any specific polymorphism. Although the role of these polymorphisms in disease progression is still unclear, their relationship to pathogenesis is clearly complex, multi-factorial, and likely context dependant. Further clinical and laboratory study will be necessary to clearly elucidate the mechanisms by which quasispecies diversity of the HA1 receptor binding region of A(H1N1)pdm09 can play a role in selection and disease progression.
Materials and Methods
Viruses
After informed consent, the two clinical isolates were collected from the patient in an IRB approved protocol (clinicaltrials.gov identifier: NCT00533182). Viruses were passaged 2 times in MDCK cells after initial isolation. Sequencing of all 8 segments was performed on the clinical NP isolate and the BAL isolate. Colony PCR of the HA gene was performed on both isolates to determine the quasispecies present. A population of virus containing solely the 222G mutation was purified from the BAL isolate using standard plaque purification methods (Rowe and Bau, 1965) in MDCK cells. Colony PCR of the HA gene and sequencing was used to confirm that this plaque-purified isolate contained the 222G and was otherwise identical to the clinical isolate.
Ferrets
Twenty four influenza A virus seronegative, 5-month-old, male ferrets were housed in pairs in separate cages with individual airflow. The ferrets were divided into six groups of four ferrets each. Three groups were inoculated intranasally with 105 plaque forming units (pfu) of virus, one with the original nasopharyngeal wash clinical isolate, one with the BAL isolate, and one with the plaque-purified 222G containing virus. After 48 hours, the other three groups were exposed to the inoculated ferrets by placing one naive ferret in each cage with one inoculated ferret. Daily weights, temperatures, nasopharyngeal washes and clinical scores were performed as previously described (Memoli et al., 2009).
Two ferrets from each of the inoculated groups and transmission groups were euthanized on day 5 post inoculation or exposure respectively. On day 12 post-inoculation and day 12 post-exposure all remaining animals in each respective group were euthanized. Whole lungs, trachea, and rostral nasal turbinates were harvested. All animal experiments were performed following NIH Institutional Animal Care and Use Committee approved protocols and guidelines.
Quantification of virus
Nasopharyngeal washes were analyzed using one step real-time RT-PCR for the influenza A virus matrix 1 gene to quantify viral shedding as previously described (Krafft et al., 2005). Standard plaque assays were performed in MDCK cells (Szretter, Balish, and Katz, 2006) on lung tissues that were weighed and homogenized in sterile cold Leibovitz’s medium (Gibco L-15, Invitrogen Corporation Carlsbad, CA) containing 1X penicillin, streptomycin, and amphotericin B (Gibco Anti-Anti, Invitrogen Corporation Carlsbad, CA). All lung titers were measured as plaque forming units per gram of tissue (pfu/gm).
Sequence analysis
Viral RNA was extracted directly from the nasal turbinates and lungs of the animals on day 5 and evaluated using colony PCR techniques to determine the prevalence of mutations at position 222 of the HA1 domain in quasispecies. 30 colonies from each were chosen for analysis. Comparisons were made to the original isolates used for inoculation. Custom overlapping primers were used to sequence the full HA gene for each clone (primers available upon request).
Histopathologic Analysis
Tissue samples from every ferret including nasal cavity, larynx, trachea and lung were fixed in 10% neutral buffered formalin, and processed using standard protocols. Hematoxylin-and-eosin (H&E) stains were performed on all tissues using standard methods. Tissue Gram stains were performed on selected tissues. Immunohistochemistry for influenza A flu antigen distribution, using goat polyclonal anti-H1N1 (ab20841, Abcam, Cambridge, MA), was completed on a subset of the tissues that represented all acute time point ferrets’ lungs and a representative piece of nasal cavity, larynx and trachea for each virus from both inoculated and transmission animals using standard methods.
Two pathologists reviewed all slides. Lung pathology was assessed taking into account the pathology severity as well as its distribution and degree of lung involvement in the examined tissues.
Phylogenetic Analysis
Codon analysis of viruses isolated from nasal turbinates and lungs of ferrets at day 5 was compared to the codon usage of site 222 of available HA sequences of A(H1N1)pdm09. Phylogenetic analysis was performed on 1500 HA (1701nt) and 680 NA (1410nt) sequences downloaded from the Influenza Virus Resource (http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html) and/or GenBank. The best-fit GTR+I+Γ4 model of nucleotide substitution was determined using ModelTest 3.7 (Posada and Crandall, 1998), and resulting parameter estimates were imported into PAUP* (Swofford, 2003) to create maximum likelihood (ML) trees through TBR branch-swapping (parameter values available upon request). Amino acid and codon differences at the variable amino acid site 222 in the HA protein were recorded using MacClade (Maddison and Maddison, 1989).
Supplementary Material
Horizontal branch lengths are drawn to a scale of nucleotide substitutions per site. The tree is rooted in the sequence, A/Brevig/1918/H1N1. Those sequences with the G222 change are in magenta, N222 are in red and E222 are in orange.
Acknowledgments
This work was supported by the intramural funds of the NIH and the NIAID. We thank the Comparative Medicine Branch (NIAID, NIH) for assistance with animal studies. We also thank the Histopathology Laboratory at the College of Veterinary Medicine, North Carolina State University for assistance with the immunohistochemical analysis.
Footnotes
Conflict of Interest:
The authors report no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baldanti F, Campanini G, Piralla A, Rovida F, Braschi A, Mojoli F, Iotti G, Belliato M, Conaldi PG, Arcadipane A, Pariani E, Zanetti A, Minoli L, Emmi V. Severe outcome of influenza A/H1N1/09v infection associated with 222G/N polymorphisms in the haemagglutinin: a multicentre study. Clin Microbiol Infect. 2010;17(8):1166–9. doi: 10.1111/j.1469-0691.2010.03403.x. [DOI] [PubMed] [Google Scholar]
- Baumeister E, Palacios G, Cisterna D, Solovyov A, Hui J, Savji N, Bussetti AV, Campos A, Pontoriero A, Jabado OJ, Street C, Hirschberg DL, Rabadan R, Alonio V, Molina V, Hutchison S, Egholm M, Lipkin WI. Molecular characterization of severe and mild cases of influenza A (H1N1) 2009 strain from Argentina. Medicina (B Aires) 2010;70(6):518–23. [PMC free article] [PubMed] [Google Scholar]
- Bautista E, Chotpitayasunondh T, Gao Z, Harper SA, Shaw M, Uyeki TM, Zaki SR, Hayden FG, Hui DS, Kettner JD, Kumar A, Lim M, Shindo N, Penn C, Nicholson KG. Clinical aspects of pandemic 2009 influenza A (H1N1) virus infection. N Engl J Med. 2010;362(18):1708–19. doi: 10.1056/NEJMra1000449. [DOI] [PubMed] [Google Scholar]
- Belser JA, Jayaraman A, Raman R, Pappas C, Zeng H, Cox NJ, Katz JM, Sasisekharan R, Tumpey TM. Effect of D222G Mutation in the Hemagglutinin Protein on Receptor Binding, Pathogenesis and Transmissibility of the 2009 Pandemic H1N1 Influenza Virus. PLoS One. 2011;6(9):e25091. doi: 10.1371/journal.pone.0025091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chutinimitkul S, Herfst S, Steel J, Lowen AC, Ye J, van Riel D, Schrauwen EJ, Bestebroer TM, Koel B, Burke DF, Sutherland-Cash KH, Whittleston CS, Russell CA, Wales DJ, Smith DJ, Jonges M, Meijer A, Koopmans M, Rimmelzwaan GF, Kuiken T, Osterhaus AD, Garcia-Sastre A, Perez DR, Fouchier RA. Virulence-associated substitution D222G in the hemagglutinin of 2009 pandemic influenza A(H1N1) virus affects receptor binding. J Virol. 2010;84(22):11802–13. doi: 10.1128/JVI.01136-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews SJ, Pabbaraju K, Wong S, Tokaryk KL, May-Hadford J, Lee B, Tellier R, Louie M. Surveillance of autopsy cases for D222G substitutions in haemagglutinin of the pandemic (H1N1) 2009 virus in Alberta, Canada. Clin Microbiol Infect. 2010;17(4):582–4. doi: 10.1111/j.1469-0691.2010.03341.x. [DOI] [PubMed] [Google Scholar]
- Ertek M, Durmaz R, Guldemir D, Altas AB, Albayrak N, Korukluoglu G. Epidemiological, demographic, and molecular characteristics of laboratory-confirmed pandemic influenza A (H1N1) virus infection in Turkey, May 15–November 30, 2009. Jpn J Infect Dis. 2010;63(4):239–45. [PubMed] [Google Scholar]
- Hamelin ME, Baz M, Abed Y, Couture C, Joubert P, Beaulieu E, Bellerose N, Plante M, Mallett C, Schumer G, Kobinger GP, Boivin G. Oseltamivir-resistant pandemic A/H1N1 virus is as virulent as its wild-type counterpart in mice and ferrets. PLoS Pathog. 6(7):e1001015. doi: 10.1371/journal.ppat.1001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonen N, Haanpaa M, Ronkko E, Lyytikainen O, Kuusi M, Ruutu P, Kallio-Kokko H, Mannonen L, Lappalainen M, Ziegler T, Julkunen I. Genetic diversity of the 2009 pandemic influenza A(H1N1) viruses in Finland. PLoS One. 2010;5(10):e13329. doi: 10.1371/journal.pone.0013329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilander A, Rykkvin R, Dudman SG, Hungnes O. Observed association between the HA1 mutation D222G in the 2009 pandemic influenza A(H1N1) virus and severe clinical outcome, Norway 2009–2010. Euro Surveill. 2010;15(9) doi: 10.2807/ese.15.09.19498-en. [DOI] [PubMed] [Google Scholar]
- Krafft AE, Russell KL, Hawksworth AW, McCall S, Irvine M, Daum LT, Connoly JL, Reid AH, Gaydos JC, Taubenberger JK. Evaluation of PCR testing of ethanol-fixed nasal swab specimens as an augmented surveillance strategy for influenza virus and adenovirus identification. J Clin Microbiol. 2005;43(4):1768–75. doi: 10.1128/JCM.43.4.1768-1775.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Vov DK, Iashkulov KB, Prilipov AG, Burtseva EI, Shchelkanov M, Shliapnikova OV, Poglazov AB, Sadykova GK, Dzhambinov SD, Fediakina IT, Bushkieva B, L’Vov DN, Zhuravleva MM, Al’khovskii SV, Samokhvalov EI, Trushakova SV, Lavrishcheva VV, Vereshchagin NN, Mikhaliaeva LB, Darbakova TA, Limanskaia OS, Dzhaparidze NI, Imkenova LN, Ledenev Iu A, Boldyreva VV, Ivanov LI, Zdanovskaia NN. Detection of amino acid substitutions of asparaginic acid for glycine and asparagine at the receptor-binding site of hemagglutinin in the variants of pandemic influenza A/H1N1 virus from patients with fatal outcome and moderate form of the disease. Vopr Virusol. 2010;55(3):15–8. [PubMed] [Google Scholar]
- Liu Y, Childs RA, Matrosovich T, Wharton S, Palma AS, Chai W, Daniels R, Gregory V, Uhlendorff J, Kiso M, Klenk HD, Hay A, Feizi T, Matrosovich M. Altered receptor specificity and cell tropism of D222G hemagglutinin mutants isolated from fatal cases of pandemic A(H1N1) 2009 influenza virus. J Virol. 2010;84(22):12069–74. doi: 10.1128/JVI.01639-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. Interactive analysis of phylogeny and character evolution using the computer program MacClade. Folia Primatol (Basel) 1989;53(1–4):190–202. doi: 10.1159/000156416. [DOI] [PubMed] [Google Scholar]
- Memoli MJ, Davis AS, Proudfoot K, Chertow DS, Hrabal RJ, Bristol T, Taubenberger JK. Multidrug-resistant 2009 pandemic influenza A(H1N1) viruses maintain fitness and transmissibility in ferrets. J Infect Dis. 2010a;203(3):348–57. doi: 10.1093/infdis/jiq067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memoli MJ, Hrabal RJ, Hassantoufighi A, Eichelberger MC, Taubenberger JK. Rapid selection of oseltamivir- and peramivir-resistant pandemic H1N1 virus during therapy in 2 immunocompromised hosts. Clin Infect Dis. 2010b;50(9):1252–5. doi: 10.1086/651605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memoli MJ, Tumpey TM, Jagger BW, Dugan VG, Sheng ZM, Qi L, Kash JC, Taubenberger JK. An early ‘classical’ swine H1N1 influenza virus shows similar pathogenicity to the 1918 pandemic virus in ferrets and mice. Virology. 2009;393(2):338–45. doi: 10.1016/j.virol.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14(9):817–8. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Potdar VA, Chadha MS, Jadhav SM, Mullick J, Cherian SS, Mishra AC. Genetic characterization of the influenza A pandemic (H1N1) 2009 virus isolates from India. PLoS One. 2010;5(3):e9693. doi: 10.1371/journal.pone.0009693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puzelli S, Facchini M, De Marco MA, Palmieri A, Spagnolo D, Boros S, Corcioli F, Trotta D, Bagnarelli P, Azzi A, Cassone A, Rezza G, Pompa MG, Oleari F, Donatelli I. Molecular surveillance of pandemic influenza A(H1N1) viruses circulating in Italy from May 2009 to February 2010: association between haemagglutinin mutations and clinical outcome. Euro Surveill. 2010;15(43) doi: 10.2807/ese.15.43.19696-en. [DOI] [PubMed] [Google Scholar]
- Reid AH, Janczewski TA, Lourens RM, Elliot AJ, Daniels RS, Berry CL, Oxford JS, Taubenberger JK. 1918 influenza pandemic caused by highly conserved viruses with two receptor-binding variants. Emerg Infect Dis. 2003;9(10):1249–53. doi: 10.3201/eid0910.020789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe WP, Bau SG. Studies of adenovirus SV40 hybrid viruses. II. Defectiveness of the hybrid particles. J Exp Med. 1965;122(5):955–66. doi: 10.1084/jem.122.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibert CW, Kaminski M, Philipp J, Rubbenstroth D, Albrecht RA, Schwalm F, Stertz S, Medina RA, Kochs G, Garcia-Sastre A, Staeheli P, Palese P. Oseltamivir-resistant variants of the 2009 pandemic H1N1 influenza A virus are not attenuated in the guinea pig and ferret transmission models. J Virol. 84(21):11219–26. doi: 10.1128/JVI.01424-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng ZM, Chertow DS, Ambroggio X, McCall S, Przygodzki RM, Cunningham RE, Maximova OA, Kash JC, Morens DM, Taubenberger JK. Autopsy series of 68 cases dying before and during the 1918 influenza pandemic peak. Proc Natl Acad Sci U S A. 2011;108(39):16416–21. doi: 10.1073/pnas.1111179108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods) Sinauer Associates; Sunderland (MA): 2003. Version 4 ed. [Google Scholar]
- Szretter KJ, Balish AL, Katz JM. Influenza: propagation, quantification, and storage. Curr Protoc Microbiol. 2006;Chapter 15(Unit 15G):1. doi: 10.1002/0471729256.mc15g01s3. [DOI] [PubMed] [Google Scholar]
- van der Vries E, Veldhuis Kroeze EJ, Stittelaar KJ, Linster M, Van der Linden A, Schrauwen EJ, Leijten LM, van Amerongen G, Schutten M, Kuiken T, Osterhaus AD, Fouchier RA, Boucher CA, Herfst S. Multidrug resistant 2009 A/H1N1 influenza clinical isolate with a neuraminidase I223R mutation retains its virulence and transmissibility in ferrets. PLoS Pathog. 7(9):e1002276. doi: 10.1371/journal.ppat.1002276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439(7074):344–8. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Dwyer DE, Soedjono M, Shi H, Matlho K, Ratnamohan M, Blyth C, McPhie K, Cunningham AL, Saksena NK. Evidence of the circulation of pandemic influenza (H1N1) 2009 with D222D/G/N/S hemagglutinin polymorphisms during the first wave of the 2009 influenza pandemic. J Clin Virol. 2011 doi: 10.1016/j.jcv.2011.08.023. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Shinya K, Watanabe S, Imai M, Hatta M, Li C, Wolter BF, Neumann G, Hanson A, Ozawa M, Yamada S, Imai H, Sakabe S, Takano R, Iwatsuki-Horimoto K, Kiso M, Ito M, Fukuyama S, Kawakami E, Gorai T, Simmons HA, Schenkman D, Brunner K, Capuano SV, 3rd, Weinfurter JT, Nishio W, Maniwa Y, Igarashi T, Makino A, Travanty EA, Wang J, Kilander A, Dudman SG, Suresh M, Mason RJ, Hungnes O, Friedrich TC, Kawaoka Y. Avian-type receptor-binding ability can increase influenza virus pathogenicity in macaques. J Virol. 2011;85(24):13195–203. doi: 10.1128/JVI.00859-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Preliminary review of D222G amino acid substitution in the haemagglutinin of pandemic influenza A (H1N1) 2009 viruses. Wkly Epidemiol Rec. 2010;85(4):21–2. [PubMed] [Google Scholar]
- Zheng B, Chan KH, Zhang AJ, Zhou J, Chan CC, Poon VK, Zhang K, Leung VH, Jin DY, Woo PC, Chan JF, To KK, Chen H, Yuen KY. D225G mutation in hemagglutinin of pandemic influenza H1N1 (2009) virus enhances virulence in mice. Exp Biol Med (Maywood) 2010;235(8):981–8. doi: 10.1258/ebm.2010.010071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Horizontal branch lengths are drawn to a scale of nucleotide substitutions per site. The tree is rooted in the sequence, A/Brevig/1918/H1N1. Those sequences with the G222 change are in magenta, N222 are in red and E222 are in orange.