Summary
Repetitive sequences account for more than 50% of the human genome. Facioscapulohumeral muscular dystrophy (FSHD) is an autosomal-dominant disease associated with reduction in the copy number of the D4Z4 repeat mapping to 4q35. By an unknown mechanism, D4Z4 deletion causes an epigenetic switch leading to de-repression of 4q35 genes. Here we show that the Polycomb group of epigenetic repressors targets D4Z4 in healthy subjects and that D4Z4 deletion is associated with reduced Polycomb silencing in FSHD patients. We identify DBE-T, a chromatin-associated noncoding RNA produced selectively in FSHD patients that coordinates de-repression of 4q35 genes. DBE-T recruits the Trithorax group protein Ash1L to the FSHD locus, driving histone H3 lysine 36 dimethylation, chromatin remodeling, and 4q35 gene transcription. This study provides insights into the biological function of repetitive sequences in regulating gene expression and shows how mutations of such elements can influence the progression of a human genetic disease.
Graphical Abstract
Highlights
► In healthy subjects, the FSHD locus is a Polycomb repressive target ► FSHD patients display loss of Polycomb silencing and gain of Trithorax activation ► DBE-T is a chromatin-bound ncRNA expressed selectively in FSHD patients ► DBE-T recruits Ash1L to the FSHD locus to coordinate long-range gene de-repression
Facioscapulohumeral muscular dystrophy (FSHD) is an autosomal-dominant myopathy associated with a reduced number of D4Z4 repeats, which leads to the expression of a noncoding RNA in a nearby locus. This RNA recruits a Trithorax epigenetic activator, which then switches on the transcription of genes driving FSHD.
Introduction
Facioscapulohumeral muscular dystrophy (FSHD) (MIM 158900) is one of the most common myopathies (Cabianca and Gabellini, 2010). It is an autosomal-dominant disease characterized by progressive wasting of facial, upper arm, and shoulder girdle muscles. In up to 95% of cases, the genetic defect is mapped to the subtelomeric region of chromosome 4q35. Remarkably, FSHD patients do not carry a classical mutation within a protein-coding gene. FSHD is rather caused by deletions reducing the copy number of the 3.3 kb D4Z4 repeat below 11 units. D4Z4 is extremely polymorphic in the general population and belongs to a family of human noncentromerically located tandem repeats termed macrosatellites (Chadwick, 2009). Several FSHD features, such as variability in severity and rate of progression, gender bias in penetrance, asymmetric muscle wasting, and monozygotic twin discordance, strongly suggest the involvement of epigenetic factors (Neguembor and Gabellini, 2010). Accordingly, DNA methylation (van Overveld et al., 2003), histone marks (Bodega et al., 2009; Zeng et al., 2009), and higher order chromatin structure (Bodega et al., 2009; Petrov et al., 2006; Pirozhkova et al., 2008) are altered in FSHD patients. These changes have been associated with the inappropriate de-repression of several 4q35 genes, among which DUX4 is currently the leading FSHD candidate (Gabellini et al., 2002; Lemmers et al., 2010). However, the molecular mechanism underlying the epigenetic switch at the basis of FSHD is currently unknown.
Polycomb (PcG) and Trithorax (TrxG) group proteins act antagonistically in the epigenetic regulation of gene expression. Typically, TrxG counteracts PcG-mediated epigenetic gene silencing. PcG and TrxG factors play crucial roles in many biological aspects such as cell proliferation, stem cell identity, and X inactivation (Schuettengruber et al., 2007). In Drosophila, PcG and TrxG bind to specific DNA regions termed Polycomb/Trithorax response elements (PREs/TREs), constituting a regulated switchable element that influences chromatin architecture and expression of nearby genes (Ringrose and Paro, 2004; Schuettengruber et al., 2007).
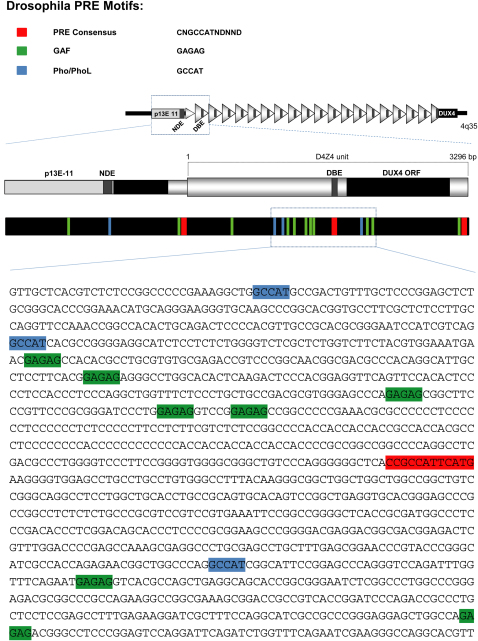
D4Z4 shares several features with PREs/TREs (Figure S1 available online). In healthy subjects, D4Z4 is organized as repressed chromatin and displays the PcG-associated histone mark H3K27me3 (histone H3 lysine 27 tri-methylation) (Bodega et al., 2009; Jiang et al., 2003; van Overveld et al., 2003; Zeng et al., 2009). Interestingly, in FSHD patients loss of repressive marks and 4q35 genes de-repression has been reported (Bodega et al., 2009; Dixit et al., 2007; Gabellini et al., 2002; Rijkers et al., 2004). Each D4Z4 unit contains copies of a motif nearly identical to a conserved sequence (CNGCCATNDNND) found in Drosophila PREs (Gabellini et al., 2002; Mihaly et al., 1998). This sequence overlaps with DBE (D4Z4 binding element), a region necessary and sufficient to confer copy-number-dependent repressive activity (Gabellini et al., 2002) due to its ability to recruit YY1, EZH2, and HMGB2 (human homologs of the Drosophila PcG proteins Pho and E(z) and the PcG recruiter Dsp1, respectively) (Bodega et al., 2009; Déjardin et al., 2005; Gabellini et al., 2002). The region surrounding DBE is enriched in putative binding sites for GAGA factor (Gaf), a DNA-binding protein implicated in PRE function in Drosophila (Busturia et al., 2001; Mishra et al., 2001). Finally, a role for CpG-rich regions in PcG recruitment in mammals has been suggested (Mendenhall et al., 2010), and the region occupied by D4Z4 in healthy subjects is one of the biggest CpG islands of the human genome (Neguembor and Gabellini, 2010).
Figure S1.
The FSHD Locus Displays Similarities with Drosophila Polycomb Response Elements, Related to Figure 1
Occurrence in the FSHD locus of DNA motifs found in Drosophilia PREs/TREs: PRE consensus; Pleiohomeotic/Pleiohomeotic-like (Pho/PhoL) core consensus; GAGAG motif (GAF). A scheme of the FSHD locus is also shown. DNA motifs are highlighted on the DNA sequence in a portion of D4Z4 repeat (GenBank: AF117653.2; 6661-7980 bp).
In Drosophila, the TrxG protein Ash1 plays a crucial role in de-repression of PcG targets (Beisel and Paro, 2011; Papp and Müller, 2006; Schwartz et al., 2010) and can be recruited by ncRNAs (Sanchez-Elsner et al., 2006). Interestingly, ASH1L, the mammalian homolog of fly Ash1, was shown to occupy actively transcribed chromatin (Gregory et al., 2007). However, the exact function of Ash1L and how it is recruited to its targets are poorly understood.
Here, we investigated the possibility that transcription of DBE regulates the epigenetic switch responsible for de-repression of the FSHD locus.
Results
D4Z4 Recruits Polycomb Complexes to Repress 4q35 Genes
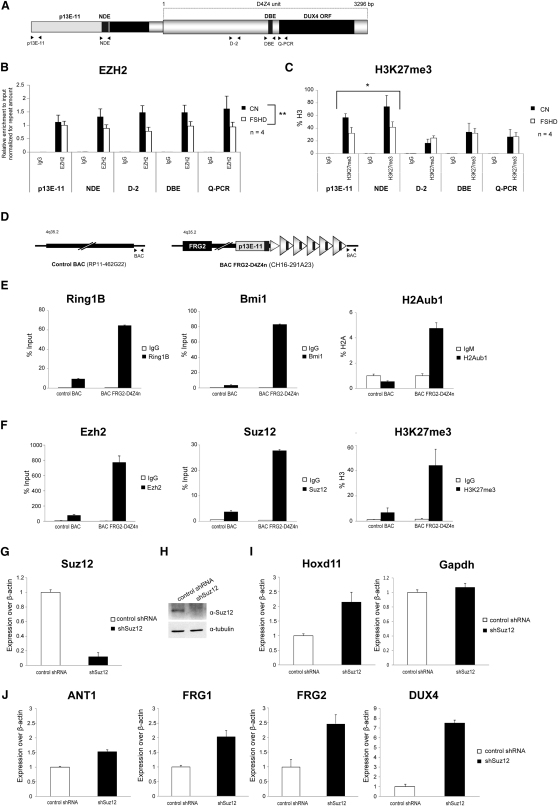
By 3D fluorescence in situ hybridization (3D-FISH), the PcG hallmark H3K27me3 was shown to be reduced selectively on the contracted 4q35 allele in FSHD primary muscle cells (Bodega et al., 2009). This suggested that contraction of D4Z4 repeats could determine a loss of Polycomb silencing in FSHD. We used chromatin immunoprecipitation followed by real-time PCR (ChIP-qPCR) to analyze levels of EZH2 and H3K27me3 in primary muscle cells. In healthy subjects, EZH2 was enriched on the entire D4Z4 unit and on regions immediately proximal to the repeat array (Figure 1B), including an element that we called NDE (for nondeleted element) because it is always maintained in FSHD patients regardless of their type of deletion (Lemmers et al., 2003). H3K27me3 was also enriched on the same regions but showed a peak on NDE (Figure 1C). Intriguingly, in FSHD patients, D4Z4 deletion was associated with a significant reduction of EZH2 at the FSHD locus (Figure 1B) and this translated into a significant decrease of H3K27me3 on the regions immediately proximal to the repeat array, including NDE (Figure 1C).
Figure 1.
D4Z4 Recruits PcG Complexes to Repress 4q35 Genes
(A–C) ChIP for EZH2, H3K27me3, and IgG on primary muscle cells from healthy donors and FSHD patients. A scheme of the FSHD locus primers used for the analysis (A). ChIP analyzed by qPCR for p13E-11, nondeleted element (NDE), and different regions of the D4Z4 repeat. Results are expressed as relative enrichment to input normalized for repeat amount (B), or percentage of total H3 (C). The mean of the signals obtained from four healthy samples or four FSHD patients is shown. The error bars represent SEM. Asterisks indicate statistical significance (p value) as evaluated by two-way repeated-measured ANOVA, respectively p = 0.0051 (B) and p = 0.0497 (C).
(D–F) CHO cells stably transfected with human 4q35 BACs either containing (CH16-291A23) or lacking (RP11-462G22) 4q35 D4Z4 repeats (schematized in D), were analyzed by ChIP for core components of PRC1 and H2Aub1 (E) and PRC2 and H3K27me3 (F). ChIP was analyzed by qPCR with primers for a region common to both BACs (see scheme D). Results are expressed as percentage of input (PcG proteins), percentage of total H3 (H3K27me3), or percentage of H2A, normalized to IgM (H2Aub1). The error bars represent SEM.
(G–J) Chr4/CHO cells stably expressing control shRNA or shSuz12. Knockdown was evaluated by qRT-PCR (G) and immunoblotting (H). Expression of Hoxd11, Gapdh (I) and 4q35 genes ANT1, FRG1, FRG2, and DUX4 was analyzed by qRT-PCR (J) and expressed over β-actin. The error bars represent SEM.
See also Figures S1 and S2.
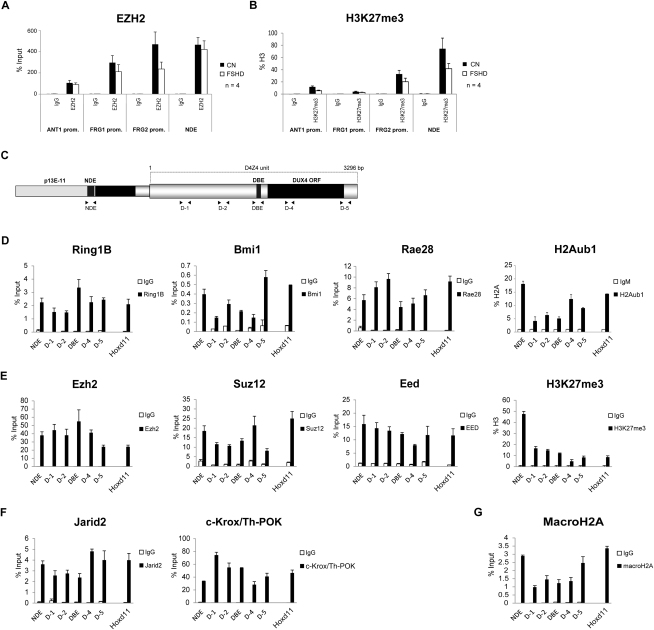
We found that in healthy subjects EZH2 and H3K27me3 were enriched above background at the promoters of ANT1, FRG1, and FRG2, even though they were at lower levels than those in NDE (Figures S2A and S2B). In FSHD patients, a trend toward a reduction in EZH2 and H3K27me3 enrichment at 4q35 genes promoters was observed (Figures S2A and S2B).
Figure S2.
PcG Core Components and the Associated Histone Marks Are Enriched at the FSHD Locus, Related to Figure 1
ChIP-qPCR experiments with antibodies specific for EZH2 (A) and its associated histone mark, H3K27me3 (B), in primary muscle cells from 4 healthy donors and 4 FSHD patients. ChIP was analyzed by qPCR with primers specific for the promoters of 4q35 genes ANT1, FRG1 and FRG2. NDE is shown as comparison of the enrichment in the FSHD locus.
Results are expressed as percentage of input (EZH2), or percentage of total H3 (H3K27me3). The mean of the signals obtained from 4 healthy samples or 4 FSHD patients is shown. The error bars represent SEM.
(C) Scheme of the FSHD locus. The location of the primers used for ChIP-qPCR is shown.
(D-G) ChIP-qPCR experiments with antibodies specific for the core components of PRC1 and H2Aub1 (D), PRC2 and H3K27me3 (E), and Jarid2 and c-Krox/Th-POK (F) and macroH2A (G) in human chr4/CHO. Hoxd11 is shown as positive control. ChIP material was analyzed by qPCR with primers for different regions of the D4Z4 repeat and for NDE (see scheme C). Results are expressed as percentage of input (PcG proteins and macroH2A), percentage of the ChIP signal obtained with anti-H3 antibodies (H3K27me3) or percentage of the ChIP signal obtained with anti-H2A antibodies and normalized for IgM (H2Aub1).
The error bars represent SEM.
The selective study of 4q-located D4Z4 repeats in human samples is challenged by the presence of D4Z4-like sequences on almost all human acrocentric chromosomes (Lyle et al., 1995; Winokur et al., 1994). By sequencing the PCR products of our Polycomb ChIP-qPCRs performed on human samples, we did not find D4Z4-like sequences, suggesting that D4Z4-like repeats elsewhere in the genome are not associated with PcG binding.
Because D4Z4 is a primate-specific repeat (Clark et al., 1996), genetic mouse models of FSHD (displaying for example a different number of D4Z4 repeats) cannot be generated. Nevertheless, we exploited the above limitation by using human/rodent monochromosomal hybrid cells containing, in a CHO background, a single human chromosome 4 derived from a healthy subject (chr4/CHO). In this setting, the only D4Z4 source is the human chromosome 4. Importantly, we confirmed that human chr4/CHO cells display all the epigenetic features typically found at the FSHD locus in healthy subjects including DNA hypermethylation (van Overveld et al., 2003), histone hypoacetilation (Jiang et al., 2003), enrichment of cohesin, HP1γ and H3K9me3 (Zeng et al., 2009), and repression of 4q35 genes (C. Huichalaf and D. Gabellini, unpublished data).
PcG proteins are found in several families of multiprotein complexes. The two main complexes are termed Polycomb Repressive Complex 1 and 2 (PRC1 and PRC2). In mammals, there are multiple versions of PRC1 and PRC2 due to the presence of many alternative subunits (Margueron and Reinberg, 2011). We focused our analysis on core PRC1 and PRC2 components. By ChIP-qPCR in chr4/CHO cells, we found that the PRC1 components Bmi1, Rae28, and Ring1B are enriched on the entire D4Z4 unit and on NDE (Figure S2D). Intriguingly, the typical PRC1 repressive histone mark H2Aub1 (histone H2A mono-ubiquitinated on lysine 119) was enriched on the same regions but showed a peak on NDE (Figure S2D). For PRC2, we analyzed the core subunits Eed, Ezh2, and Suz12. Similarly to PRC1, PRC2 and its repressive mark, H3K27me3, were enriched over the entire FSHD locus (Figure S2E), with H3K27me3 showing a peak on NDE (Figure S2E).
Several proteins collaborate to recruit Polycomb to PREs (Margueron and Reinberg, 2011). Interestingly, Figure S2F shows that two PcG recruiters, Jarid2 (Landeira and Fisher, 2011) and c-Krox/Th-POK (the putative vertebrate homolog of GAGA factor, Busturia et al., 2001; Matharu et al., 2010; Mishra et al., 2001), were enriched at the FSHD locus. Furthermore, the repressive histone variant macroH2A, overlapping locally and functionally with PRC2 (Buschbeck et al., 2009), was enriched at the FSHD locus (Figure S2G). Notably, ChIP-qPCR signals at D4Z4 were comparable to those observed on the bona fide PcG target Hoxd11 (Woo et al., 2010) (Figures S2D–S2G).
One of the features of PREs is their ability to recruit PcG complexes when inserted in ectopic sites. To test this, we introduced in CHO cells human 4q35 bacterial artificial chromosomes (BACs) either containing or lacking D4Z4 repeats (Figure 1D). To directly compare Polycomb recruitment, we performed ChIP-qPCR with primers mapping on the insert-flanking region common to both BACs. As shown in Figures 1E and 1F, we found robust recruitment for PRC1, PRC2, and their associated repressive histone marks selectively in the BAC-containing D4Z4 repeats. This result indicates that D4Z4 repeats are able to initiate de novo recruitment of Polycomb complexes.
Based on our results, we hypothesized that D4Z4 recruits Polycomb complexes to repress 4q35 genes. We tested this directly by performing RNAi-mediated loss of Polycomb. As shown in Figures 1G–1J, Suz12 knockdown caused a de-repression of 4q35 genes comparable to that of the bona fide Polycomb target gene Hoxd11.
Altogether, our results strongly suggest that loss of Polycomb silencing is responsible for de-repression of 4q35 genes in FSHD patients.
Transcription of DBE Occurs Selectively in FSHD Patients and Is Associated with 4q35 Gene De-Repression
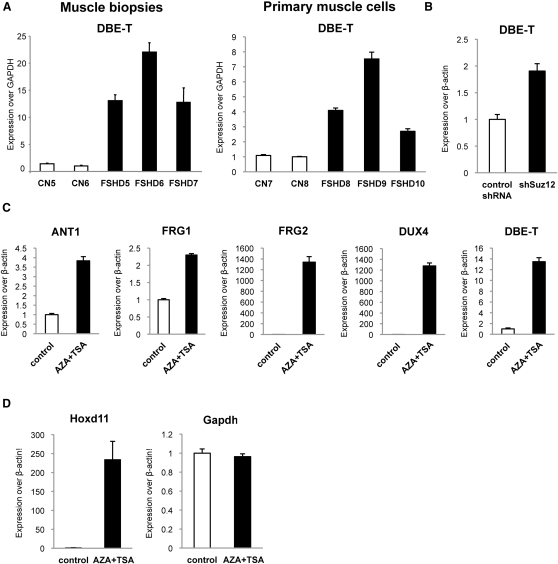
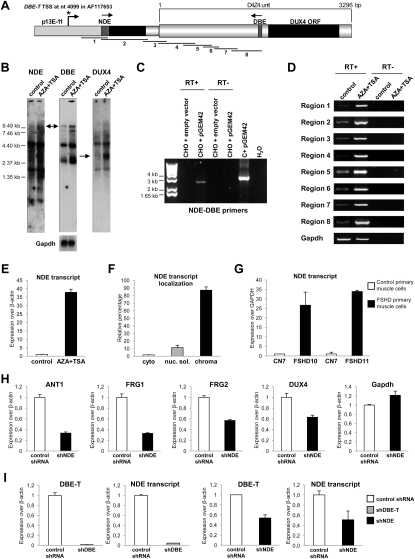
It has been reported that transcription of ncRNAs from PREs could play a role in relief of PcG-mediated silencing in Drosophila (Bae et al., 2002; Lipshitz et al., 1987; Rank et al., 2002; Sanchez-Elsner et al., 2006; Schmitt et al., 2005). Because of the similarities between DBE and PREs, we analyzed transcription from DBE (Figure 1A). We performed real-time RT-PCR (qRT-PCR) on total RNA extracted from muscle biopsies or primary muscle cells from healthy subjects and FSHD patients. Figure 2A shows that DBE was transcribed above background exclusively in FSHD samples to generate an RNA that we named DBE-T (DBE-Transcript). As stated above, D4Z4-like sequences are located on several human chromosomes (Lyle et al., 1995; Winokur et al., 1994). To verify the chromosomal origin of DBE-T, we extensively sequenced the qRT-PCR products from muscle biopsies and primary muscle cells to take advantage of 4q35-specific SNPs. Notably, 97% of the products were 4q35 specific, indicating that DBE-T originated from the FSHD-associated locus. To confirm this, we again took advantage of human chr4/CHO cells. In this system, the de-repression of 4q35 genes due to RNAi-mediated loss of Polycomb (Figure 1J) was associated with DBE-T production (Figure 2B). We have recently found that DNA methylation and histone deacetylation are required for keeping the 4q35 region repressed (C. Huichalaf and D. Gabellini, unpublished data). As a consequence, treatment with AZA (5-Aza-2′-deoxycytidine) plus TSA (Trichostatin A) (inhibitors of DNA methylation and histone de-acetylation, respectively) resulted in de-repression of 4q35 genes (Figure 2C). Intriguingly, also in this case 4q35 gene de-repression was paralleled by production of DBE-T (Figure 2C).
Figure 2.
The DBE Region Is Transcribed Selectively in FSHD Patients or FSHD-Like Conditions
(A) qRT-PCR analysis of DBE-T in muscle biopsies (left) and primary muscle cells (right) from healthy donors and FSHD patients. Results are expressed over GAPDH.
(B) qRT-PCR analysis of DBE-T in control or Suz12 knockdown chr4/CHO cells. Results are expressed over β-actin.
(C) qRT-PCR analysis of 4q35 genes and DBE-T in chr4/CHO cells in the repressed (control) or de-repressed (AZA+TSA) states. Results are expressed over β-actin.
(D) qRT-PCR analysis of Hoxd11 and Gapdh in chr4/CHO cells in the repressed (control) or de-repressed (AZA+TSA) states. Results are expressed over β-actin.
The error bars represent SEM.
See also Figure S3.
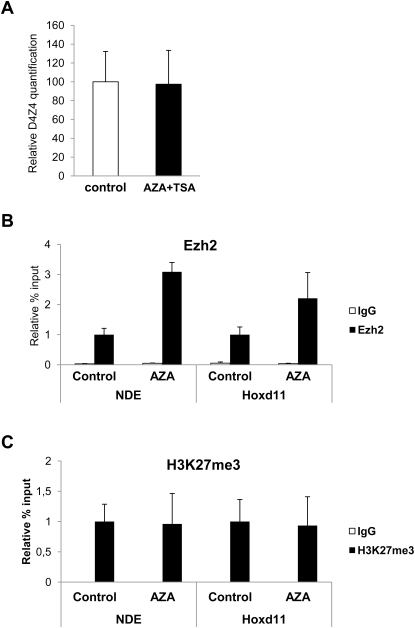
Several elements suggested that AZA plus TSA effects on the FSHD locus were fairly specific. Histone deacetylation has a role in PRC1-mediated chromatin compaction (Eskeland et al., 2010) and TSA treatment causes relief of Polycomb silencing (Garrick et al., 2008). In chr4/CHO cells, AZA plus TSA treatment caused a dramatic de-repression of the bona fide PcG target Hoxd11, whereas the expression of the non-PcG target Gapdh was unaffected, suggesting that loss of PcG silencing was not due to unspecific effects of the treatment (Figure 2D). Finally, AZA treatment did not affect the number of D4Z4 repeats nor reduce PcG binding or H3K27me3 enrichment (Figure S3).
Figure S3.
AZA and TSA Treatments Generate Specific Effects, Related to Figure 2
(A) Relative qPCR analysis showing that the amount of D4Z4 repeats is unaffected by AZA plus TSA treatment. The error bars represent SEM.
(B-C) ChIP-qPCR experiments with antibodies specific for Ezh2 (B) and its associated histone mark, H3K27me3 (C), in chr4/CHO cells treated with AZA. Ezh2 and H3K27me3 signals are not reduced by AZA treatment. ChIP was analyzed by qPCR with primers specific for NDE.
Results are expressed as percentage of input (Ezh2), or percentage of total H3 (H3K27me3). The error bars represent SEM.
DBE-T Regulates 4q35 Chromatin Structure and Gene De-Repression
As shown above, in independent systems DBE-T production is always associated with de-repression of 4q35 genes. To address its biological role, we generated chr4/CHO stable cells expressing a DBE-T shRNA or a control, nonsilencing shRNA. Remarkably, DBE-T knockdown prevented 4q35 gene de-repression caused by AZA plus TSA, strongly suggesting a functional role for this transcript in the positive regulation of 4q35 gene expression (Figure 3A).
Figure 3.
DBE-T Is Required for De-Repression and Topological Reorganization of the 4q35 Region
(A) Control or DBE-T chr4/CHO knockdown cells were treated with AZA+TSA. DBE-T knockdown, 4q35 genes and Gapdh expression were evaluated by qRT-PCR. Results are expressed over β-actin. The error bars represent SEM.
(B) Schematic representation of the region analyzed by 3C. The D1 3C primer, located near DBE, was used as bait and primers spanning the FRG1 genomic locus were tested for interaction. 3C results show the relative frequency of interaction between DBE and FRG1 locus in control and AZA+TSA treated chr4/CHO cells expressing a nonsilencing shRNA (C left) or an shRNA specific for DBE-T (shDBE-T) (C right). The error bars represent SEM.
See also Figures S4 and S5.
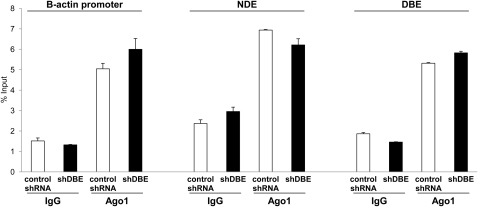
shRNAs can directly cause epigenetic changes by transcriptional gene silencing (TGS) (Turner and Morris, 2010), raising the possibility that DBE-T shRNAs could cause TGS at the FSHD locus. Importantly, treatment with AZA plus TSA prevents TGS because it is strictly dependent on the activity of DNA methyltransferases and histone deacetylases (Morris et al., 2004; Turner and Morris, 2010). Given that we investigated effects of DBE-T shRNAs in cells treated with AZA plus TSA, it was extremely unlikely that TGS could play a role in our findings. To further address this, we analyzed recruitment of Ago1, which is required for TGS (Kim et al., 2006). We used ChIP-qPCR to monitor Ago1 recruitment on NDE, DBE, and β-actin, which is unaffected by treatment with DBE-T shRNAs, as control. As shown in Figure S4, at all regions analyzed Ago1 displayed a very low enrichment that was unaltered by DBE-T shRNAs (Figure S4). These results indicate that DBE-T shRNAs do not cause TGS but only posttranscriptional degradation of the intended target RNA.
Figure S4.
DBE-T shRNA Does Not Cause Direct Transcriptional Gene Silencing, Related to Figure 3
Following AZA+TSA treatment, human chr4/CHO hybrid cells stably expressing a nonsilencing control shRNA or shDBE-T were collected to extract chromatin. ChIP for Ago1 or IgG, as control was performed. Enrichment at the β-actin promoter, NDE and DBE regions was analyzed by qPCR. Results are displayed as percentage of input. The error bars represent SEM.
It has been reported that a chromatin reorganization is associated with 4q35 gene de-repression (Bodega et al., 2009; Petrov et al., 2006; Pirozhkova et al., 2008). In particular, chromatin conformation capture (3C) detected an interaction between D4Z4 and the promoter of the 4q35 gene FRG1 that is reduced in FSHD patients compared to controls (Bodega et al., 2009). We confirmed this interaction in chr4/CHO cells and found that it is reduced when DBE-T is produced and FRG1 is de-repressed (Figure 3C, left, and Figure 2C). Notably, DBE-T knockdown prevented the topological reorganization of the FSHD region characteristic of the de-repressed state (Figure 3C, right). Thus, DBE-T is involved in the regulation of 4q35 higher order chromatin structure and is required for 4q35 gene de-repression.
DBE-T Functions in cis and Is Associated with the Chromatin of the FSHD Locus
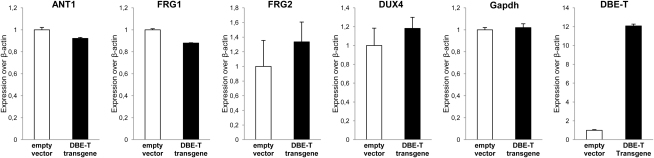
To dissect the role of DBE-T in 4q35 gene de-repression, we asked whether DBE-T ectopic expression could be sufficient to de-repress 4q35 genes. To this aim, chr4/CHO cells were transiently transfected with a construct producing DBE-T. In qRT-PCR assays, DBE-T overexpression was unable to de-repress 4q35 genes in trans (Figure S5).
Figure S5.
DBE-T does not function in trans, related to Figure 3
DBE-T was ectopically overexpressed in human chr4/CHO cells and expression of 4q35 genes and Gapdh, as control, was analyzed by qRT-PCR. Results are shown as expression over β-actin. The error bars represent SEM.
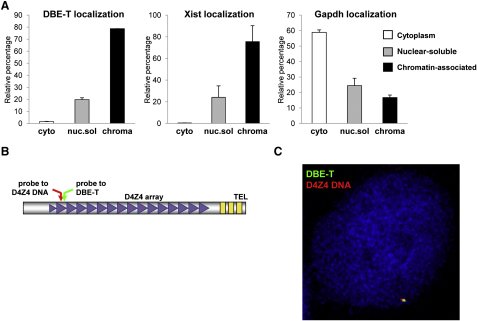
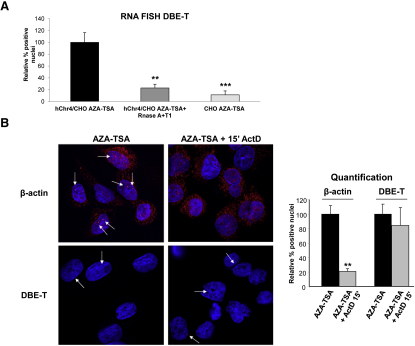
To further elucidate DBE-T mechanism of action, we investigated its subcellular localization by using independent approaches. First, through biochemical fractionation we found that DBE-T was nuclear and mainly chromatin-associated and behaved similarly to the chromatin-bound ncRNA Xist, whereas the protein-coding mRNA Gapdh was preferentially enriched in the cytoplasm, as expected (Figure 4A). Next, we used sequential RNA/DNA FISH to investigate the exact location of DBE-T inside the nucleus. We first hybridized nondenatured cells with LNA oligonucleotides targeting DBE-T. Next, cells were denatured to allow for DNA detection and hybridized with DNA oligonucleotides, mapping to the D4Z4 repeat in order to visualize the 4q35 locus (schematized in Figure 4B). As shown in Figure 4C, DBE-T and D4Z4 signals colocalized in 98.8% of the cells analyzed, indicating that DBE-T is specifically associated with the FSHD locus. Importantly, to derive this conclusion we carried out several controls. First, the DBE-T signal was absent in CHO cells lacking D4Z4 and strongly reduced by RNase A+T1 treatment in chr4/CHO cells (Figure S6A), proving that it corresponded to a specific RNA. Second, using actinomycin D (ActD), we showed that DBE-T is a mature RNA. In fact, actively transcribed genes generate nascent transcripts, appearing as nuclear dots in RNA FISH, which are exquisitely sensitive to ActD. Accordingly, β-actin nascent transcripts quickly disappeared upon ActD treatment. On the contrary, the RNA FISH signals corresponding to DBE-T were unaffected by ActD treatment, strongly indicating that they corresponded to mature RNAs (Figure S6B). Altogether, these results indicate that DBE-T is a mature transcript associated with the chromatin of the FSHD locus.
Figure 4.
DBE-T Is Nuclear, Chromatin-Associated, and Localized to the FSHD Locus
(A) Total RNA from AZA+TSA treated chr4/CHO cells was separated into cytoplasmic, nuclear-soluble, and chromatin-bound fractions. The relative abundance of DBE-T in the different fractions was measured by qRT-PCR. As control, Gapdh and Xist were analyzed. The error bars represent SEM.
(B) Schematic representation of the location of the DBE-T and D4Z4 probes.
(C) Following AZA+TSA treatment, chr4/CHO cells were analyzed by RNA/DNA FISH. A single Z stack acquired with an Olympus IX70 DeltaVision RT Deconvolution System microscope is shown. Signals colocalization was detected in 98.8% of the double positive cells (n = 80) deriving from 3 independent experiments. DBE-T is visualized in green, and the D4Z4 DNA is in red. DAPI is in blue.
See also Figures S6 and S7.
Figure S6.
DBE-T Is a Mature RNA, Related to Figure 4
(A) Human chr4/CHO and CHO cells were collected in the de-repressed state (AZA+TSA). A set of samples were treated with RNase A+T1. RNA FISH quantification of DBE-T positive nuclei was performed in all conditions and it was normalized to signals in human chr4/CHO cells treated with AZA+TSA. The error bars represent SD.
(B) AZA+TSA human chr4/CHO cells were treated (Right panels) or untreated (Left panels) with Actinomycin D at 5 μg/ml for 15′. Hybridization to β-actin (Top panels) or DBE-T (Bottom panels) RNAs was performed. The arrows indicate dots of nuclear RNAs. See Statistical Test section for statistical analysis. The error bars represent SD. DBE-T and β-actin RNAs are in red. DAPI is in blue. The images correspond to a single Z stack acquired with an Olympus IX70 DeltaVision RT Deconvolution System microscope.
(A and B) The asterisks indicate statistically significant differences. A Two-tailed, paired, t test was applied. For DBE-T: human chr4/CHO AZA+TSA n = 4; human chr4/CHO AZA+TSA + Actinomycin D n = 4, p = 0.0914; human Chr4/CHO AZA+TSA + RNase A+T1 n = 4, p = 0.0056; CHO AZA+TSA n = 4, p = 0.0006. For β-actin: human chr4/CHO AZA+TSA n = 4; human chr4/CHO AZA+TSA + Actinomycin D n = 4, p = 0.0012.
DBE-T Is a Long ncRNA Starting from the Region Immediately Proximal to the D4Z4 Repeat Array
NcRNAs greater than 200 nt in length have been shown to be involved in PcG/TrxG function (Margueron and Reinberg, 2011). Interestingly, using either NDE- or DBE-specific probes in northern blot with polyA+ RNA, we detected transcripts as large as 9.8 kb that were upregulated in the de-repressed state (Figure S7B). On the contrary, a DUX4-specific probe detected an ∼3 kb band (in line with the expected size of DUX4 mRNA) not present in the NDE and DBE northern blots (Figure S7B). Taking advantage of the experimental approach previously used for rapid amplification of cDNA ends (RACE) of DUX4 (Kowaljow et al., 2007), we mapped the start of DBE-T upstream of NDE (position 4099 of accession number AF117653, Figure S7A). This result was supported by several independent findings. First, we successfully amplified a single transcript spanning the 2.8 kb from NDE to DBE (Figure S7C). Second, RT-PCR with overlapping primers spanning the ∼3.4 kb from the DBE-T start to the end of the DBE of the first D4Z4 repeat supported a single AZA+TSA inducible long RNA (Figure S7D). Third, like DBE-T, the NDE transcript is transcribed selectively when 4q35 genes are de-repressed (Figure S7E), generates a chromatin-associated RNA (Figure S7F), is overexpressed in FSHD muscle cells compared to controls (Figure S7G), and is 4q specific. Accordingly, NDE stable knockdown in chr4/CHO cells impaired de-repression of 4q35 genes (Figure S7H). Finally, NDE and DBE-T expression was reduced by reciprocal knockdown (Figure S7I).
Figure S7.
DBE-T Is a Long ncRNA Whose Transcription Starts outside the D4Z4 Repeat Array and Encompasses the NDE Region, Related to Figure 4
(A) Schematic representation of the regions analyzed. The asterisk corresponds to the Transcriptional Start Site (TSS) mapped by 5′ RACE. The position indicated is referred to AF117653. The arrows represent the primers used in panel C to amplify a single NDE/DBE-T transcript. Lines and numbers positioned in the lower part of the scheme correspond to the overlapping regions amplified in panel D.
(B) Northern blot assays performed on PolyA+ RNA extracted from human chr4/CHO cells untreated (Control) or treated with AZA+TSA (AZA+TSA). Hybridizations with probes mapping to NDE, DBE, DUX4 and Gapdh, as loading control, are shown.
(C) RT-PCR to amplify a single transcript from NDE to DBE was performed on RNA extracted from CHO cells transfected with a construct carrying the entire AF117653 sequence (pGEM42, derived from an FSHD patient, containing the 4q35 region ranging from upstream p13E-11, two D4Z4 repeats and the distal region, Kowaljow et al., 2007) or with the empty vector as control. As positive control, the pGEM42 plasmid DNA was PCR amplified; RT- and no template reactions were performed as negative controls.
(D) The NDE-DBE region was divided in eight overlapping regions (see panel A) that were amplified by RT-PCR in RNA samples extracted from human chr4/CHO cells untreated (Control) or treated with AZA+TSA (AZA+TSA). As loading control, Gapdh was amplified.
(E) NDE transcription was evaluated in the repressed (control) and de-repressed (AZA+TSA) states by qRT-PCR. Results are shown as expression over β-actin. The error bars represent SEM.
(F) The relative enrichment of the NDE transcript in the indicated subcellular fractions as measured by qRT-PCR is shown. The error bars represent SEM.
(G) Analysis of NDE expression by qRT-PCR in control and FSHD primary muscle cells. Results are shown as expression over GAPDH. The error bars represent SEM.
(H) Expression of 4q35 genes and Gapdh, as control, in human chr4/CHO cells knockdown for NDE transcript (G left) in the de-repressed state (AZA+TSA). Results are shown as expression over β-actin. The error bars represent SEM.
(I) The NDE transcript is downregulated upon DBE-T knockdown and DBE-T is downregulated upon NDE transcript knockdown. Results are shown as expression over β-actin. The error bars represent SEM.
Based on the above results, we propose that DBE-T is a long RNA containing both NDE and DBE. DBE-T is clearly distinct from the protein-coding DUX4 transcript encoded by D4Z4 considering: (1) the northern blot results; (2) that the primers used to analyze DBE-T (Figure S7A) are unable to detect DUX4; and (3) that DBE-T is proximal to D4Z4, whereas only the last, most distal D4Z4 repeat can produce a stable transcript encoding for DUX4 (Dixit et al., 2007; Lemmers et al., 2010).
We believe DBE-T to be nonprotein coding. In fact, even though it contains open reading frames (ORFs) bigger than 100 amino acids, DBE-T is chromatin-associated, and it is generally accepted that protein synthesis occurs only in the cytoplasm (Dahlberg and Lund, 2004). Additionally, DBE-T does not function in trans, arguing against a protein-coding function.
Collectively, our results strongly suggest that a long, chromatin-associated nonprotein-coding RNA (lncRNA) encompassing NDE and DBE regulates gene expression at 4q35.
DBE-T Directly Recruits the TrxG Protein Ash1L to De-Repress 4q35 Genes
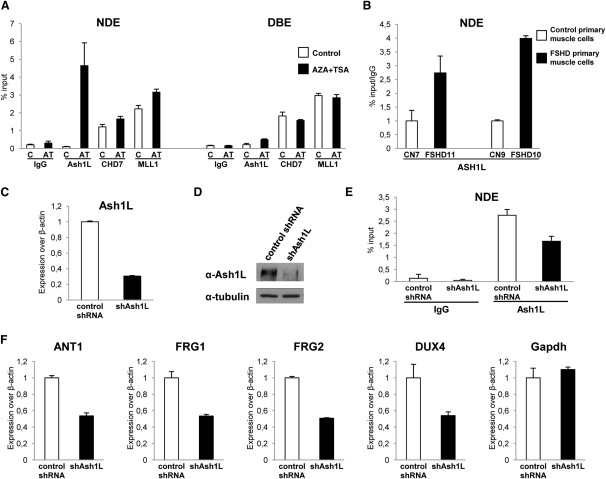
Because ncRNAs were shown to recruit TrxG proteins to target genes (Bertani et al., 2011; Sanchez-Elsner et al., 2006; Wang et al., 2011), we investigated TrxG recruitment to the FSHD locus. The TrxG proteins Chd7 and Mll1 were enriched at similar levels in the repressed and de-repressed states on NDE and DBE regions (Figure 5A). Importantly, our data on Mll1 occupancy parallel what was previously described for its fly homolog Trx, which can co-occupy repressed targets together with PcG proteins (Papp and Müller, 2006; Schuettengruber et al., 2009; Schwartz et al., 2010). On the contrary, the TrxG protein Ash1L was preferentially enriched on NDE specifically in the de-repressed state (Figure 5A). Interestingly, the recruitment of Ash1 is the main determinant that distinguishes the repressed from the de-repressed state in Drosophila as well (Papp and Müller, 2006; Schwartz et al., 2010). Importantly, ChIP-qPCR assays showed that ASH1L is recruited to the FSHD locus preferentially in muscle cells from FSHD patients compared to those from healthy subjects (Figure 5B). Hence, we hypothesized that Ash1L might be involved in de-repression of 4q35 genes. To test this, we generated stable chr4/CHO cells expressing a nonsilencing (control shRNA) or an shRNA specific for Ash1L (shAsh1L). qRT-PCR, immunoblotting and ChIP-qPCR assays indicated that we only partially reduced Ash1L expression and recruitment to the FSHD locus (Figures 5C–5E). Nevertheless, this was sufficient to impair 4q35 gene de-repression (Figure 5F).
Figure 5.
The TrxG Protein Ash1L Is Recruited to the FSHD Locus and De-Represses 4q35 Genes
(A) ChIP for Ash1L, Chd7, Mll1, and IgG on chr4/CHO cells in the repressed (control) or de-repressed (AZA+TSA) states. ChIP was analyzed by qPCR with primers for NDE and DBE and expressed as percentage of input. The error bars represent SEM.
(B) ChIP for ASH1L in control and FSHD primary muscle cells. ChIP was analyzed by qPCR with primers for NDE and expressed as percentage of input normalized to IgG. The error bars represent SEM.
(C–F) Chr4/CHO cells stably expressing control shRNA or shAsh1L. Knockdown after AZA+TSA treatment evaluated by qRT-PCR (C), immunoblotting (D), and ChIP on NDE (E). Expression of 4q35 genes and Gapdh, as control, was analyzed by qRT-PCR (F). Results are expressed over β-actin. The error bars represent SEM.
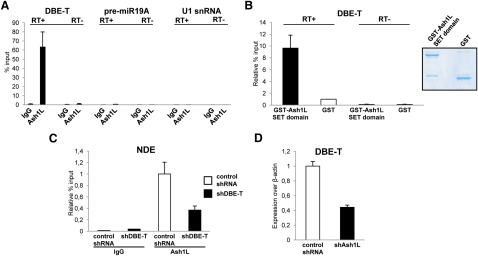
Based on our results, we speculated that DBE-T could play a role in the recruitment of Ash1L to the FSHD locus. To assess this, we performed RNA immunoprecipitation following ultraviolet crosslinking (UV-RIP) with anti-Ash1L antibodies or IgG, as control. DBE-T was significantly enriched in the Ash1L UV-RIP, whereas nonrelated nuclear RNAs such as the precursor of miR19A or the abundant U1 snRNA were not (Figure 6A). Because UV irradiation only identifies direct protein-nucleic-acid interactions (Greenberg, 1979), a direct Ash1L-DBE-T interaction in vivo was suggested. To confirm this, we performed in vitro pull-down experiments by using purified, recombinant GST-Ash1L and in vitro transcribed DBE-T. GST-Ash1L was able to directly interact with DBE-T, whereas no enrichment was obtained by using GST alone (Figure 6B). To investigate whether DBE-T is required for Ash1L recruitment to the FSHD locus, we performed Ash1L ChIP-qPCR in cells knockdown for DBE-T. DBE-T knockdown impaired the recruitment of Ash1L to the FSHD locus (Figure 6C). Collectively, our results strongly indicate that DBE-T functions in cis by directly recruiting Ash1L to the FSHD locus.
Figure 6.
DBE-T Directly Binds the TrxG Protein Ash1L and Recruits It to the FSHD Locus
(A) RNA immunoprecipitation (IP) following UV crosslinking for Ash1L or IgG on AZA+TSA treated chr4/CHO cells. DBE-T or, as control, pre-miR19A and U1 snRNA enrichments were measured by qRT-PCR. The error bars represent SEM.
(B) In vitro RNA-GST pull-down assay showing the interaction between recombinant GST-fused Ash1L SET domain or GST and in vitro transcribed DBE-T. On the right, Coomassie staining of purified recombinant proteins. After RNA recovery, samples were analyzed by qRT-PCR. The error bars represent SEM.
(C) Following AZA+TSA treatment, chr4/CHO cells stably expressing a nonsilencing control shRNA or shDBE-T were analyzed by ChIP for Ash1L or IgG. Enrichment for NDE was analyzed by qPCR and displayed as enrichment relative to input. The error bars represent SEM.
(D) Upon AZA+TSA treatment, control shRNA, and shAsh1L cells were collected to analyze DBE-T expression by qRT-PCR. Results are expressed over β-actin. The error bars represent SEM.
See also Figure S8.
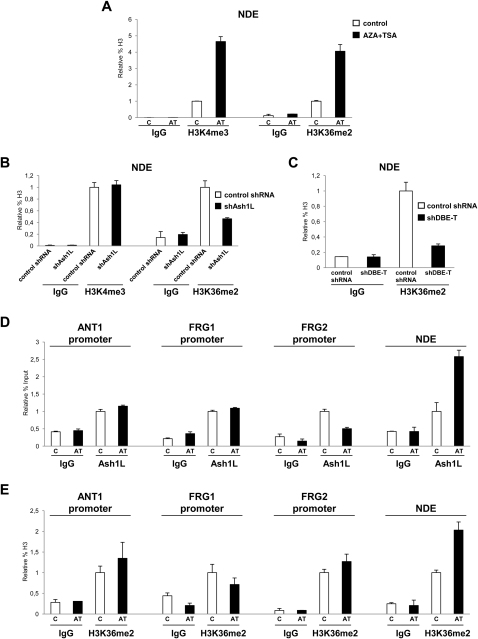
Ash1L is a histone methyltransferase, but there are conflicting reports regarding its enzymatic activity. In particular, the two activating histone marks, H3K4me3 (histone H3 lysine 4 tri-methylation) and H3K36me2 (histone H3 lysine 36 di-methylation), have been ascribed to Ash1L (An et al., 2011; Gregory et al., 2007; Tanaka et al., 2007; Yuan et al., 2011). By ChIP-qPCR, we found that both histone marks are increased in the de-repressed state at the FSHD locus (Figure S8A). Interestingly, Ash1L knockdown caused a decrease in H3K36me2, whereas H3K4me3 was unaffected (Figure S8B). Moreover, H3K36me2 was also decreased by DBE-T knockdown (Figure S8C).
Figure S8.
Ash1L Dimethylates Lysine 36 on Histone H3 at the FSHD Locus, Related to Figure 6
(A) ChIP-qPCR for enrichment on total H3 of H3K4me3, H3K36me2 and IgG, as control, performed on human chr4/CHO cells in the repressed (control) or in the de-repressed (AZA+TSA) state. ChIP material was analyzed by qPCR with primers specific for the NDE region. The error bars represent SEM.
(B) ChIP-qPCR for enrichment on total H3 of H3K4me3, H3K36me2 and IgG, as control, performed in control shRNA or shAsh1L expressing cells in the de-repressed state (AZA+TSA). ChIP material was analyzed by qPCR with primers specific for the NDE region. The error bars represent SEM.
(C) ChIP-qPCR for enrichment on total H3 of H3K36me2 and IgG, as control, in control shRNA or shDBE-T expressing cells in the de-repressed state (AZA+TSA). ChIP material was analyzed by qPCR with primers specific for the NDE region. The error bars represent SEM.
(D) ChIP for Ash1L and IgG, as control, on human chr4/CHO cells in the repressed (control) or de-repressed (AZA+TSA) state. Analysis was performed by qPCR with primers specific for ANT1, FRG1, FRG2 promoters and NDE. Results are expressed as relative percentage of input. The error bars represent SEM.
(E) ChIP-qPCR for enrichment on total H3 of H3K36me2 and IgG, as control, performed on human chr4/CHO cells in the repressed (control) or in the de-repressed (AZA+TSA) state. Analysis was performed by qPCR with primers specific for ANT1, FRG1, FRG2 promoters and NDE. Results are expressed as relative percentage of H3. The error bars represent SEM.
H3K36me2 is also the product of the ordinary process of transcription elongation and does not require Ash1L (Krogan et al., 2003). Although Ash1L and H3K36me2 were detectable above background levels at 4q35 gene promoters, de-repression of 4q35 genes was not associated with an increase in Ash1L or H3K36me2 at their genomic regions (Figures S8D and S8E). Thus, the increase in H3K36me2 observed at the FSHD locus in the de-repressed state was directly due to Ash1L recruitment. Intriguingly, we found that Ash1L positively regulated DBE-T, because DBE-T expression was reduced upon Ash1L knockdown (Figure 6D). Hence, DBE-T and ASH1L could potentially constitute a positive feedback loop that keeps the 4q35 region de-repressed upon D4Z4 deletion.
Altogether, our results indicate that DBE-T directly recruits ASH1L to the FSHD locus to co-ordinate 4q35 gene de-repression.
Discussion
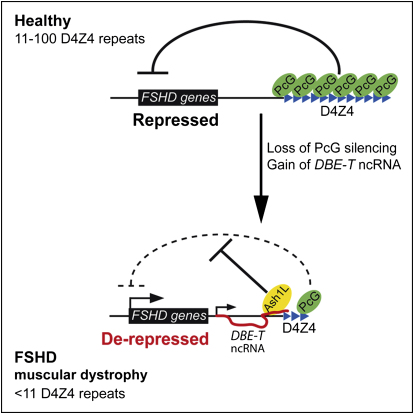
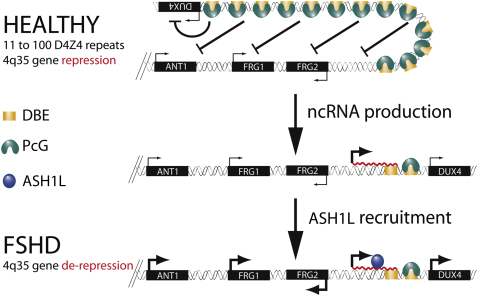
Among the different types of muscle diseases, FSHD is undoubtedly one of the less characterized. Indeed, the molecular events leading to FSHD remain undeciphered (Cabianca and Gabellini, 2010). Based on our results, we propose a model to explain the epigenetic basis of FSHD etiology. In healthy subjects, the presence of many D4Z4 units would result in extensive PcG binding, DNA methylation, histone de-acetylation, and chromatin compaction leading to a repressive chromatin organization (Figure 7). We propose that PcG complexes are recruited first on D4Z4 repeats, and then silencing spreads on the region immediately proximal to the repeat array. In FSHD patients, deletion of D4Z4 repeats results in a critical reduction of PcG silencing, permissive for DBE-T transcription (Figure 7). Once produced at sufficient levels, DBE-T recruits ASH1L, leading to 4q35 gene de-repression (Figure 7). Importantly, we found that Ash1L promotes the expression of DBE-T, implying a positive feedback loop that would sustain 4q35 gene de-repression.
Figure 7.
Model for DBE-T-Mediated 4q35 Gene De-Repression in FSHD
Normal individuals carry multiple D4Z4 copies that are extensively bound by PcG proteins, promoting the maintenance of repressed chromatin at 4q35. In FSHD patients, D4Z4 deletion leads to insufficient binding of PcG, causing the production of DBE-T. DBE-T recruits the TrxG protein ASH1L and promotes a topological reorganization of the FSHD locus leading to de-repression of 4q35 genes.
On chromosomal region 10q26 is located a repeat array almost identical and equally polymorphic to the 4q35 D4Z4 array (Bakker et al., 1995; Cacurri et al., 1998; Deidda et al., 1996). Nevertheless, contraction of D4Z4 repeats on 10q26 is not pathogenic, and FSHD is uniquely linked to 4q35 (Cacurri et al., 1998; Lemmers et al., 1998). It has been shown that H3K9me3 is coregulated on 4q35 and 10q26 D4Z4 repeats (Zeng et al., 2009), implying that nonpathogenic 10q26 contraction should cause loss of H3K9me3 also on 4q35. This suggests that H3K9me3 loss is not directly involved in FSHD. On the contrary, PcG silencing and DNA methylation are reduced selectively at the deleted 4q35 allele in FSHD (Bodega et al., 2009; van Overveld et al., 2003) and better explain the strictly 4q35-linked nature of the disease.
The FSHD locus is epigenetically regulated during normal development. In particular, DUX4 is expressed in pluripotent stem cells and in normal development, and then epigenetically silenced in somatic tissues (Snider et al., 2010). Interestingly, DUX4 has been retained during primate evolution, suggesting that DUX4 might have a normal role in early embryonic muscle development (Snider et al., 2010; Wu et al., 2010). Accordingly, we hypothesize that the PcG/TrxG epigenetic regulation of the FSHD locus could be important in normal muscle biology.
Recent results indicate that RNAi of a single 4q35 gene in an FSHD animal model has a therapeutic value (Bortolanza et al., 2011; Wallace et al., 2011). Nevertheless, the complexity of FSHD could be better explained by envisaging it as a contiguous gene syndrome, where the epigenetic alteration of DUX4, FRG1 and other genes collaborate to determine the final phenotype. Hence, instead of targeting the inappropriate expression of individual 4q35 genes, it could be more effective to target a general 4q35 genes' regulator. Based on our results, it is tempting to speculate that DBE-T is a valid therapeutic target to achieve a general normalization of 4q35 gene expression in FSHD.
In Drosophila, PREs were shown to generate ncRNAs that regulate the epigenetic status of the locus (Bae et al., 2002; Lipshitz et al., 1987; Petruk et al., 2006; Rank et al., 2002; Sanchez-Elsner et al., 2006; Schmitt et al., 2005). Recently, the first example of an activatory lncRNA involved in de-repression of a mammalian PcG target has been provided (Wang et al., 2011). Our results indicate that DBE-T acts directly as a chromatin-associated lncRNA to activate the epigenetic cascade culminating with 4q35 gene de-repression in FSHD. To the best of our knowledge, DBE-T is the first activatory lncRNA involved in a human genetic disease.
ASH1L occupies many active genes (Gregory et al., 2007), and TrxG family proteins are involved in many cell fate decisions in development and disease (Mills, 2010; Smith et al., 2011). Our findings suggest a general function for chromatin-associated lncRNAs in recruiting ASH1L or other chromatin-remodeling complexes to coordinate chromosome structure and gene expression.
A significant portion of the human genome is composed of macrosatellite repeats (Warburton et al., 2008). Although once thought of primarily as “junk,” recent studies indicate that repeated elements play central roles in regulating gene expression at multiple levels (Faulkner et al., 2009; Kaneko et al., 2011; Norris et al., 1995; Shen et al., 2011). Similarly to D4Z4, other repeats display meiotic instability associated with diseases (Bruce et al., 2009; Tremblay et al., 2010), reside within common fragile sites that could contribute to chromosome rearrangements in tumors (Tremblay et al., 2010), and are expressed at high levels in testis and aberrantly expressed in cancer (Gjerstorff and Ditzel, 2008; Ting et al., 2011; Tremblay et al., 2010). Interestingly, in mammals the greatest proportion of PcG-mediated chromatin modifications is located in genomic repeats, and it has been suggested that they could provide a binding platform for PcG proteins (Leeb et al., 2010). Hence, elucidating the role of these elements in setting up functional chromatin states in complex genomes will be of paramount importance in upcoming years.
Experimental Procedures
Mammalian Cell Culture
CHO, HEK293T, human primary muscle cells, and human chromosome 4/CHO hybrid (GM10115) culture, stable knockdown, and treatments with chemicals are described in the Extended Experimental Procedures.
Extended Experimental Procedures.
Human Samples
All procedures were approved by the Fondazione San Raffaele del Monte Tabor Ethical Committee. Human primary myoblasts were obtained from the Fields Center for FSHD of the Rochester Medical Center Dept. of Neurology, NY, USA and the Telethon BioBank of the C. Besta Neurological Institute, Milano, Italy. Muscle biopsies were obtained from the Telethon Neuromuscular Bank of the Department of Neurosciences, University of Padova, Italy. Details of the human samples used are listed in Table S1.
Mammalian Cell Culture
HEK293T and CHO were obtained by ATCC; human chromosome 4/CHO hybrid (GM10115) was obtained from the Coriell Institute for Medical Research. Cells were maintained in DMEM-HIGH (Dulbecco's Modified Eagle's Medium, High Glucose with Sodium Pyruvate and L-Glutamine; EuroClone) supplemented with 10% FBS (Foetal Bovine Serum; EuroClone) and 1% Penicillin/Streptomycin (100 U/ml final concentration; EuroClone). Proline (final concentration 0.2 mM; Sigma) was added to CHO and human chr4/CHO cells. For the generation of stable cell lines infected with pLKO.1 or pTRIPZ lentiviruses, puromycin (6.5 μg/ml; InVivoGen) was added to the normal medium of human chr4/CHO cells in case of pLKO.1 lentiviruses, or to medium containing Tetracycline-negative serum (10%, EuroClone) in case of pTRIPZ. Resistant cells were maintained as a polyclonal population to avoid clone-to-clone variability. For the generation of stable cell lines transfected with either BAC RP11-462G22 or BAC CH16-291A23, CHO cells were cotransfected with pCDNA3.1 vector (Invitrogen) in a ratio 9:1, where BAC constructs where 9 times more concentrated than pCDNA3.1. Positively transfected cells were selected with Neomycin (1,000 μg/ml; InVivoGen) and maintained as a polyclonal population to avoid clone-to-clone variability. Human primary muscle cells from the Telethon BioBank were grown in DMEM-HIGH (Dulbecco's Modified Eagle's Medium, High Glucose with Sodium Pyruvate and L-Glutamine; EuroClone), supplemented with 20% FBS (Foetal Bovine Serum; EuroClone), 1% Penicillin/Streptomycin (100 U/ml final concentration; EuroClone), insulin (10 μg/ml final concentration; Sigma), human basic fibroblast growth factor (25 ng/ml; Peprotech) and epidermal growth factor (10 ng/ml; Peprotech). Human primary muscle cells from the Fields Center for FSHD were grown in F-10 Nutrient Media (Sigma), supplemented with 20% FBS (Foetal Bovine Serum; EuroClone), 1% Penicillin/Streptomycin (100 U/ml final concentration; EuroClone), human basic fibroblast growth factor (10 ng/ml; Peprotech), Dexamethasone (1 μM; Sigma). Muscle cells were induced to differentiate at 70% confluence by replacing the growth medium with a differentiation medium composed of DMEM-HIGH (Dulbecco's Modified Eagle's Medium, High Glucose with Sodium Pyruvate and L-Glutamine; EuroClone) supplemented with 5% Donor Horse Serum (EuroClone), 1% Penicillin/Streptomycin (100 U/ml final concentration; EuroClone), insulin (10 μg/ml final concentration; Sigma). Medium was replaced with fresh differentiation medium every 2 days.
For Doxycycline treatment, human chr4/CHO cells stably infected with pTRIPZ lentiviruses, were seeded at low confluence in tetracycline negative growth medium with Doxycycline (300 ng/ml, Sigma). Fresh medium containing Doxycycline was replaced every day. Cells were collected after 72h of doxycycline treatment. For 5-Aza-2′-deoxycytidine (AZA) and Trichostatin A (TSA) treatment, human chr4/CHO cells were seeded at low confluence in growth medium. The day after, AZA was added to the medium (final concentration 1 μM; Sigma). After 24 hr, the medium was replaced with fresh medium containing 1 μM AZA. After other 24 hr, fresh AZA (final concentration 1 μM) was added to the medium. After 12 hr, TSA was added to the medium (final concentration 1 μM; InVivoGen). Cells were collected after 12 hr to obtain a 72 hr AZA and 12 hr TSA treatment. In case of stable clones, puromycin (final concentration 6.5 μg/ml; InVivoGen) was added to all media.
For Actinomycin D treatment (final concentration 5 μg/ml; Sigma), the drug was added to the medium 15′ before harvesting the cells.
For cell transfection Lipofectamine LTX (Invitrogen) was used. Human chromosome 4/CHO hybrid cells were transfected with pcDNA3 empty vector or with nt 1280-7755 of AF117653 and collected after 48 hr for gene expression analysis. CHO cells were transfected with pGEM42 (Kowaljow et al., 2007) or pGEM-T as control and collected after 24 hr for RNA extraction.
RNAi Vectors
Nonsilencing and Ash1L shRNAs cloned in pLKO.1 or nonsilencing and Suz12 shRNAs cloned in pTRIPZ were directly obtained by Open Biosystems.
DBE and NDE siRNAs were designed by using a program (siRNA Designer, previously available on Promega website) based on parameters specified in the scientific literature (contact Promega for details). SiRNA sequences were cloned into lentiviral pLKO.1 vector to express them as shRNAs driven by the human U6 promoter according to the instructions of the RNAi TRC consortium (http://www.broadinstitute.org/rnai/trc). Briefly, each siRNA sequence was used to generate two complementary oligonucleotides: 5′-CCGG-sense-CTCGAG-antisense-TTTTTG-3′ and the complementary 5′-AATTCAAAAA-sense-CTCGAG-antisense-3′. Oligo pairs were mixed at a final concentration of 1 μg/μl in 1X NEB buffer 2 (New England Biolabs Restriction Endonuclease Reaction Buffer 2; New England Biolabs), denatured 4′ at 95°C and annealed for 10′ at 70°C followed by a slow cool down to RT. The obtained dsDNA fragment has AgeI and EcoRI sticky ends and is ready for cloning into digested pLKO.1 plasmid.
shRNA sequences:
DBE: GCTCACCGCCATTCATGAAGG
NDE: AACGTCACGGACAAGGCCAGA
Nonsilencing: TCTCGCTTGGGCGAGAGTAAG
Ash1L: GCTGGTCATTTATTGCTCAAT
Suz12: TTCTACAAACAGCATACAG
Lentiviruses Production and Transduction
HEK293T cells were seeded in DMEM-HIGH supplemented with 10% FBS without antibiotics in T25 tissue culture flasks (1 flask per infection). The day after, cells at 60%–70% confluence were transfected (Lipofectamine LTX with Plus reagent; Invitrogen) by using these quantities of DNA: 2.925 μg of packaging plasmid (pCMV-dR8.91; http://www.broadinstitute.org/rnai/trc), 325 ng of envelope plasmid (VSV-G/pMD2G; http://www.broadinstitute.org/rnai/trc) and 3.25 μg of hairpin-pLKO.1 or hairpin-pTRIPZ vector. After 5 hr, the medium was replaced with regular growth medium. After 18 hr from transfection, the medium was replaced by a high serum growth medium DMEM-HIGH supplemented with 30% FBS and 1% Penicillin/Streptomycin. Cells were incubated for 24 hr and the medium containing the lentiviral particles was harvested, filtered by using a 0.22 μm filter unit and placed at 4°C for short time storage or −80°C for long-term storage. Fresh high serum growth medium was added to cells. After 24 hr the viral harvesting was repeated and pooled with the previous one.
For viral transduction, human chr4/CHO cells were seeded in T25 tissue culture flasks and infected at 70% confluence. Cells were incubated overnight with the viral supernatant diluted 1:2 with fresh growth medium, supplemented with 0.2 mM proline and polybrene (final concentration 8 μg/ml; Sigma). The day after, cells were passed 1:7. At 24–48 hr postinfection puromycin selection was started and it was continued until all noninfected control cells died (typically, 5 days).
RNA Extraction, RT-PCR, and qRT-PCR Analysis
RNA extraction was performed with Trizol reagent (Invitrogen) followed by purification with RNA spin columns (PureLink RNA MiniKit; Invitrogen) and digestion with DNaseI following the manufacturer's instructions.
For reverse transcription, equal amounts of DNA-free RNA (100 ng-1 μg) were retro-transcribed with SuperScript III First-Strand Synthesis SuperMix (Invitrogen) following the suggested conditions.
RT-PCR assays for the different 8 regions encompassing NDE-DBE and for Gapdh (Gapdh F2-Gapdh R2) were performed by retrotranscribing 2.5 μg of total RNA derived from human chr4/CHO cells treated or untreated with AZA-TSA. For PCR reactions Go Taq Flexi (Promega) or Accuprime (Invitrogen) were used. Conditions varied according to length and base composition. More detailed information on each region is available upon request.
To amplify the single NDE-DBE transcript (primers NDE F+DBE R), 1 μg of total RNA extracted from CHO cells transfected with pGEM42 or pGEM-T as control, was retrotranscribed as previously described. cDNA was PCR amplified by using Expand Long Range dNTPack (Roche). For PCR conditions and amplification steps the manufacturer's instructions were followed, with addition of 6% DMSO in the reaction.
For gene expression analysis, real-time PCR with Sybr GreenER qPCR kit (Invitrogen) was used. GAPDH or β-actin were used as housekeeping genes for sample normalization.
The specificity of the amplified products was monitored by performing melting curves at the end of each amplification reaction.
PCR products obtained from human samples were cloned into pGEM-T vector and sequenced to make sure that they were selectively from the human 4q35 region.
The primers used in qPCR are listed below. In order to allow expression analysis with the ΔΔCT method, all primers were tested and selected for amplification efficiencies ranging between 90%–110%.
Sequences of the primers used are listed in Table S2.
All experiments were repeated at least three times.
RACE
5′ RACE (Invitrogen) was carried out by using 1.5 μg of total RNA extracted from CHO cells transfected with pGEM42. NDE R primer was used in the retrotranscription reaction. A first PCR reaction (Go Taq Flexi, Promega) was performed by using 5RACE1 and Abridged Anchor primer (provided by the kit) with the following conditions: 1 mM MgCl2, 0.4 mM each dNTP and 0.2 μM of each primer (final concentrations). Amplification was performed as follows: initial denaturation 1′ at 95°C, denaturation 30″ at 95°C, annealing 30″ at 60°C, extension 4′ at 72°C. Repeated for 35 cycles. The obtained band was gel purified (QIAGEN) and PCR amplified with 5RACE2 and Abridged Universal Amplification Primer (provided by the kit) with the following conditions: 1.5 mM MgCl2, 0.2 mM each dNTP and 0.2 μM of each primer (final concentrations). Amplification was performed as follows: initial denaturation 1′ at 95°C, denaturation 30″ at 95°C, annealing 30″ at 56°C, extension 1′ at 72°C. Repeated for 35 cycles. The obtained band was sequenced.
RNA Fractionation
AZA+TSA treated human chr4/CHO hybrid cells were detached by treating with 1X Trypsin, counted and centrifuged at RT 168 g for 5′. The pellet was lysed with 175 μl/106 cells of cold RLN1 solution (50 mM Tris HCl pH 8.0; 140 mM NaCl; 1.5 mM MgCl2; 0,5% NP-40; 2mM Vanadyl Ribonucleoside Complex; Sigma; reagent stocks were prepared in RNase-free H20) and incubated 5′ in ice. Next, the suspension was centrifuged at 4°C and 300 g for 2′ and the supernatant, corresponding to the cytoplasmic fraction, was transferred into a new tube and stored in ice. The pellet containing nuclei was extracted with 175 μl/106 cells of cold RLN2 solution (50 mM Tris HCl pH 8.0; 500 mM NaCl; 1.5 mM MgCl2; 0,5% NP-40; 2mM Vanadyl Ribonucleoside Complex; stocks were made in RNase-free H20) and 5′ incubated in ice. The suspension was centrifuged at 4°C and 16360 g for 2′ and the supernatant, corresponding to the nuclear-soluble fraction, was transferred into a new tube and stored in ice. The remaining pellet corresponds to the chromatin-associated fraction.
Total RNA was extracted by using PureLink RNA MiniKit (Invitrogen) following the manufacturer's instructions for the extraction from aqueous solutions for the cytoplasmic and nuclear-soluble fractions, whereas the chromatin-associated fraction was considered as a pellet. The samples were treated with on column DNaseI, washed and then eluted in 30 μl RNase-free water.
RNA/DNA FISH
Preparation of FISH Probes. For D4Z4 DNA and β-actin transcript detection, 4 different DNA aminoallyl-modified oligonucleotides were used (the Midland Certified Reagent Company). Each oligo was resuspendend in 100 μl of H20 and purified with chloroform extraction (1vol). DNA was then precipitated with 1/10 vol of 3 M NaAc and 2.5 vol of 100% EtOH for 30′ at −80°C. After centrifuging 15′ at 16,300 g, the pellet was washed with 70% EtOH, centrifuged again 5′ at 16300 g, air-dried and resuspended in 150 μl of H20. After DNA quantification, 5 μg of each of the 4 oligos were pooled together and H20 was added to a final volume of 20 μl. To this, 12 μl of NaHCO3 25 mg/ml and 8 μl of Alexa Fluor 555 (resuspended at 30 μg/μl in DMSO; Invitrogen) were added. After a brief vortex, the samples were let stand in the dark at RT overnight to allow the coupling of the fluorochrome. The day after, the oligos were purified by using QIAquick Nucleotide Removal kit (QIAGEN) and eluted in 40 μl of Rnase free H20. This corresponds to the stock solution. We calculated the relative efficiency of labeling measuring the base/dye ratio as described by Invitrogen's protocol of labeling with amine-reactive reagent. For the D4Z4 probe we obtained a base/dye ratio of 18.22, whereas for the β-actin RNA probe we obtained a base/dye ratio of 15.20.
For DBE transcript detection, 3 FAM-labeled LNA oligonucleotides were used (Exiqon). Each LNA was resuspended in RNase-free H20 at 2 μg/μl. To prepare a stock solution, an aliquot of each was pooled together and RNase-free H20 was added to reach a dilution of 1:4 for each LNA.
To perform FISH, the oligo and LNA probes were diluted 1:100 and 1:500, respectively, in hybridization mix (50% deionized formamide [Sigma], 2X SSC [recipe of 20X SSC from Sambrook and Russel (2001)], 30 mM phosphate buffer, 10% dextran sulfate; [pH 7.0]) containing salmon sperm ssDNA (final concentration 700 ng/μl; Sigma), human Cot1 DNA (final concentration 300 ng/μl; Invitrogen) and Vanadyl Ribonucleoside Complex (final concentration 2 mM; Sigma), denatured 5′ at 80°C and incubated at 37°C for at least 20′ before use.
Hybridization. Hybridization was performed essentially as described in Custódio et al., 2006.
Human chr4/CHO or CHO cells were seeded on glass coverslips and, following treatment with AZA+TSA, were briefly washed twice with PBS and fixed with 4% PFA (Electron Microscopy Sciences) for 10′ at RT. Next, three washes of 5′ with PBS and a rinse with 70% ice cold EtOH were performed. Slides were stored in 70% ice-cold EtOH at −20°C. After at least an overnight incubation, slides were rehydrated in PBS for 5′ twice. Next, cells were permeabilized with a 10′ incubation with gentle swirl in 0.5% Triton X-100, 2 mM Vanadyl Ribonucleoside Complex (not used if RNase treatment was following) in PBS. Then, three washes of 5′ in PBS and two washes of 5′ in 2X SSC, 0.05% Tween 20 (National Diagnostics) were performed. In control experiments requiring an RNase treatment at this step, cells were incubated with RNase A+T1 (final concentrations 20 μg/ml RNase A and 50 U/ml RNase T1; Fermentas) in 10 mM Tris-HCl (pH 7.5), 300 mM NaCl, 5 mM EDTA (pH 7.5) for 1 hr at 37°C in humid chamber. At the end of the treatment, cells were washed twice with 2X SSC, 0.05% Tween 20. Before hybridization, cells were incubated for 30′ at RT in 2X SSC, 0.05% Tween 20, supplemented with 1% BSA (Sigma), 1 μg/μl tRNA (Sigma) and 2 mM Vanadyl Ribonucleoside Complex in a humid chamber. The blocking solution was carefully removed and the denatured LNA probe (see previous section) 1:500 was added. Samples were incubated in the dark in a humid chamber at 37°C overnight. The day after, two washes of 30′ at 37°C with 1X SSC, 0.025% Tween 20, 50% deionized formamide, pH 7.0 were performed. Next, after 3 × 5′ washes with 2X SSC, 0.05% Tween 20 (2 performed at 37°C and 1 performed at RT), slides were mounted with ProLong Gold antifade reagent with DAPI (Invitrogen) if simply RNA FISH was performed. Alternatively, for sequential RNA/DNA FISH experiments, cells were denatured at 80°C for 30′ in prewarmed 1X SSC, 0.025% Tween 20, 50% deionized formamide pH 7.0. Next, cells were hybridized with denatured oligonucleotides to D4Z4 1:100 for 3 hr. Next, the same washes described previously after hybridization to RNA were performed. Slides were mounted with ProLong Gold antifade reagent with DAPI. Analyses were performed on single Z stacks acquired with an Olympus IX70 DeltaVision RT Deconvolution System microscope. All experiments were repeated at least three times. Probe sequences are listed in Table S3.
Chromatin Immunoprecipitation
Human chr4/CHO cells were seeded in 5–15 15 cm dishes, whereas human primary myoblasts were seeded in 2–3 10 cm dishes. In order to collect chromatin, cells were briefly washed once in PBS and immediately fixed for 10′ at RT in 1% formaldehyde in PBS (from a 37.5% formaldehyde/10% methanol stock; Sigma). After formaldehyde quenching with Glycine (final concentration 125 mM) for 2′, cells were washed three times for 5′ in PBS with gentle swirl. Cells were harvested by using a silicon scraper and cold PBS. Cells were collected in 50 ml tubes and centrifuged at 1350 g for 5′ at 4°C. The resulting pellet was resuspended in 3 ml of PBS every 3 × 15 cm dishes of starting material, and aliquoted in 15 ml tubes. Human primary myoblasts were kept in a single 15 ml tube. Cells were centrifuged at 1,350 g for 5′ at 4°C. Each pellet was then lysed in 5 ml of LB1 solution (50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP40, 0.25% Triton X-100) and incubated 10′ in ice. Next, the samples were centrifuged at 1350 g for 5′ at 4°C. The resulting pellet was washed in 5 ml of LB2 solution (10 mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA) with gentle swirl 10′ at RT. Next, samples were centrifuged at 1350 g for 5′ at 4°C and the resulting pellet was lysed in 3 ml of LB3 (10 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-Deoxycholate, 0.5% N-laurylsarcosine). LB1, LB2 and LB3 were supplemented with protease inhibitor (Complete EDTA-free Protease Inhibitor Cocktail Tablets; Roche). Lysates were sonicated with Bioruptor (Diagenode). Briefly, 1 ml aliquots of LB3 lysates were put in 15 polystirene tubes and were sonicated for 10′ (medium intensity 30″ on 30″ off). An aliquot (55 μl) of the sonicated material was collected to determine the quality of the chromatin by adding 0.1M NaHCO3, 1% SDS (100 μl), 5 μl of Proteinase K (20 mg/ml in 50 mM Tris-HCl pH 8.0, 10 mM CaCl2, 50% glycerol; Promega) and incubate 1 hr at 55°C for cross-link reversal. Next, samples were precipitated with 5M LiCl (3.2 μl) and 1 ml 100% EtOH by centrifuging for 30′ at 4°C at 16,360 g. The pellet was washed in 70% EtOH and centrifuged for 10′ at 4°C at 16,360 g. After air dry, the pellet was resuspended in H2O and loaded on 1% agarose gel for electrophoresis. We considered good a chromatin enriched in fragments of 500-300 bp. The samples were quantified with Nanodrop spectrophotometer to determine the concentration of chromatin. Before ChIP, Triton X-100 was added to chromatin samples at a final concentration of 1% and a clarification step of 10′ at 16,360 g at 4°C followed. Supernatants were transferred into a new tube and were precleared with 1/50 vol of Dynabeads protein G (Invitrogen) rotating for 2 hr at 4°C. Fifty to one hundred micrograms of chromatin were used for each ChIP to a protein target, whereas 5–10 μg of chromatin were used for each ChIP to histones. For each ChIP, 100 μl of Dynabeads protein G (50 μl in ChIPs to histones) were washed three times with 0.5% BSA in PBS and incubated with 10 μg of antibody (5 μg in ChIPs to histones) in 250 μl of 0.5% BSA in PBS for 2–3 hr on rotation at 4°C. Next, the beads-antibody complex was washed three times with 0.5% BSA in PBS and resuspended in 50 μl of 0.5% BSA in PBS. Precleared chromatin and beads-antibody complexes were incubated on rotation overnight at 4°C. The day after, before starting the washes, 5% of the total ChIP volume was taken from the control IgG supernatant as input fraction. Next, six washes of 5′ on rotation at 4°C in RIPA buffer were performed (50 mM HEPES-KOH pH 7.6, 500 mM LiCl, 1 mM EDTA, 1% NP-40, 0.7% Na-Deoxycholate). An additional wash of 5′ on rotation at 4°C in TE buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA) with 50 mM NaCl was performed. Next, samples were centrifuged for 3′ at 1,000 g at 4°C. The supernatant was discarded and to the beads-antibody-chromatin complex were added 240 μl of elution buffer (TE buffer with 2% SDS). Samples were incubated in a thermo mixer for 15′ at 65°C, shaking, and then centrifuged for 1′ at RT at 16,360 g. The elute supernatant was transferred to a new tube and samples, together with the input fraction to which 3 vol of elution buffer were added, were cross-linked reverted overnight at 65°C. For samples purification, the QIAquick PCR purification kit was used (QIAGEN), following the manufacturer recommendations. DNA was eluted in 70 μl of TE buffer and 1–2 μl were used in qPCR with Sybr GreenER qPCR kit (Invitrogen). Chromatin Immunoprecipitation for H2Aub1 and IgM as control was performed according Stock et al. (2007) with 20 μg of chromatin produced and quantified as previously described. All experiments were repeated at least three times.
Antibodies used are listed in Table S5.
ChIP q-PCR Analysis
To determine the enrichment obtained by using nonhistone antibodies, we normalized ChIP-qPCR data for input chromatin (reported as % input in the figures). For histone modification antibodies, we expressed the enrichment as percentage of total histone (reported as % H3 or % H2A in the figures). To normalize for the highly variable amount of D4Z4 repeats present in human samples, the EZH2 ChIP-qPCR signal shown in Figure 1 was normalized for the input signal of five different single copy regions. Primers used are listed in Table S2.
RNA Immunoprecipitation
This assay was carried out mainly as described in Jeon and Lee, 2011. Briefly, cells were seeded in 15 cm dishes (1 for each RNA IP). After treatment with AZA+TSA, cells were UV cross-linked with 100,000 μJ/cm2 twice (interval of 1′ between the two irradiations) on ice. Lysed in 0.5% NP40, 0.5% Na Deoxycholate, 300 U/ml Superase Inhibitor (Ambion), Protease inhibitor in PBS pH 7.9 and put on rotation for 25′ at 4°C. Samples were treated with 30 U of Turbo DNase (Ambion) and incubated 15′ at 37°C. After centrifuging 5′ at 1,350 g at 4°C, the supernatant was used in RNA IP. For each RNA IP 100 μl of Dynabeads Protein G, with 10 μg of anti Ash1L antibody (Sigma) or IgG (Jackson lab) as control, were used. Before performing the RNA IP, 10% of the supernatant was saved as input. The antibody-conjugated beads were added to the samples and put on rotation at 4°C over night. Beads were washed three times (5′ at 4°C) with PBS supplemented with 1% NP40, 0.5% Na Deoxycholate, additional 150 mM NaCl (final 300 mM), and 1:200 Superase inhibitor (Ambion). Beads were resuspended in 100 μl of PBS + 10X DNase buffer and 3 U (1.5 μl) of Turbo DNase (Ambion) was added. Samples were incubated 30′ on rotation at 37°C. Beads were washed three times (5′ at 4°C) with PBS supplemented with 1% NP40, 0.5% Na Deoxycholate, 10 mM EDTA, additional 150 mM NaCl (total 300 mM), and 1:200 Superase inhibitor (Ambion). RNA was eluted with 100 μl of 100 mM Tris HCl (pH 7.5), 50 mM NaCl, 10 Mm EDTA, 100 μg Proteinase K, 0.5% SDS for 30′ at 55°C, with shaking. Eluate was centrifuged at 16100 g at RT and the supernatant was collected. Samples were purified with Trizol LS following the manufacturer's instructions. After RNA resuspension in 21.5 μl of RNase-free water, samples were treated with DNase (Turbo DNase Ambion) and 5 μl of DNA-free RNA were used in RT reactions with Superscript III supermix (Life technologies). All experiments were repeated at least three times.
In Vitro RNA Pulldown Assay
Recombinant GST-fusion proteins were prepared as described previously (Tanaka et al., 2008). Briefly, Rosetta host cells (Novagen) were transformed with pGEX-6P vectors (Amersham) with or without Ash1L SET domain (including pre-SET and post-SET regions, as described in Tanaka et al., 2007) and induced with 1 mM isopropyl b-D-1-thiogalactopyranoside. Next, bacteria were resuspended in a buffer containing 30 mM Tris-HCl pH 8.0, 1 mM EDTA, 20% sucrose for 10 min and in ice-cold water for 30 min. Subsequently, cells were lysed in 20 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 0.1 mg/ml lysozyme, 1 × Protease Inhibitor Cocktail (Roche), and 0.001% b-mercaptoethanol for 30 min on ice. After one freeze-thawing cycle at −80°C, cell lysates were cleared by ultracentrifugation at 20,000 rpm for 90′ by using Avanti HP-25 (Beckman). Supernatants were incubated with Glutathione Agarose beads (Sigma) for 2 hr at 4°C on rotation. Protein/beads complexes were quantified by SDS-PAGE by using a BSA standard curve.
To produce in vitro transcribed DBE-T, a fragment containing nt 4317–7463 of AF117653 was cloned into pGEM-T (Promega). The construct was linearized and DBE-T was transcribed in vitro by using T7 RNA polymerase (Promega). Transcripts were treated with RNase-free DNase (Promega) for 1 hr at 37°C, TRIzol (Invitrogen) purified, treated with TURBO DNase (Ambion) and renatured by heating 2′ at 90°C and slow-cooling at room temperature. Next, 300 ng of RNA per pull-down were precleared by a 30′ incubation at room temperature with Glutathione Agarose beads. 1/10 of this material was saved and TRIzol purified as input. RNA was incubated with two micrograms of GST-Ash1L SET domain or GST protein-beads complexes at room temperature for 1 hr in PBS containing 2 mM MgCl2, 0.2 mM ZnCl2, 1mM DTT, 100 U/ml RNase Inhibitor, 0.1 mg/ml yeast tRNA (Sigma), 0.05% BSA, and 0.2% NP40. Beads were washed with the same incubation buffer supplemented with additional 150 mM NaCl (total 300 mM NaCl) five times with 4′ washes at room temperature. The recovered RNA was TRIzol purified and 1/3 was analyzed by qRT-PCR. All experiments were repeated at least three times.
SDS-PAGE and Immunoblotting
After SDS-PAGE electrophoresis, transfers were performed by using the iBlot dry blotting system following the manufacturer recommended conditions (Invitrogen). After transfer, the filters were prehybridized in 5% nonfat milk (Regilait) in TBS-Tween 0.1% solution (Tris Buffered Saline: 10 mM TrisHCl pH 7.4, 140 mM NaCl, Tween-20; National Diagnostics) for Tubulin and Suz12 or in 5% BSA (Sigma) in PBS 0.2% Tween-20 for Ash1L western, for 1h at RT or overnight at 4°C. Incubation with the primary antibody was performed overnight at 4°C in 5% nonfat milk TBS-Tween 0.1% solution for Tubulin (1:20,000; Sigma T9026) and Suz12 (1:300; Abcam) or in 2% BSA in PBS 0.2% Tween-20 for Ash1L (1:2,000; Bethyl Laboratories A301-749A). Filters were then washed three times at RT (10′ each) with TBS-Tween 0.1% for Tubulin and Suz12, or PBS 0.2% Tween-20 for Ash1L. HRP-conjugated secondary antibody (1:20,000; Jackson ImmunoResearch Laboratories) was incubated for 1-2 hr at RT in 5% nonfat milk TBS-Tween 0.1% solution for Tubulin and Suz12 or in 2% BSA in PBS 0.2% Tween-20 for Ash1L. Filters were washed again as described, and for detection they were incubated for 5 min with horseradish peroxidase (HRP) chemiluminescent substrate (SuperSignal West Pico Chemiluminescent Substrate; Thermo scientific).
Chromosome Conformation Capture
3C was performed essentially as described in Bodega et al., 2009. Briefly, a total of 50 × 106 human chr4/CHO cells for each treatment were lysed and nuclei were cross-linked with 1% formaldehyde for 10′ at room temperature. Digestion was performed with 600–800 U of PvuII (50U/ml, NEB, R0151M) at 37°C overnight with constant agitation.
An appropriate amount of DNA that would amplify within the linear range was subsequently used for the experiments. A total of 33 rounds of PCR amplification were used. PCR products were run on 2% agarose gels and quantified by Typhoon (GE Healthcare).
Primers used are listed in Table S4.
Northern Blotting
One microgram of polyA+ RNA from control or AZA+TSA treated human chr4/CHO cells was analyzed by northern blotting by using the Northern Max-Gly kit (Ambion) following manufacturer's instructions. Filters were hybridized with probes to NDE, DBE, DUX4 homeobox region and Gapdh.
q-PCR Quantification of Relative Amount of D4Z4 Repeats
Chromatin DNA deriving from 5 independent control and AZA+TSA treatment experiments on human chr4/CHO cells were analyzed by qPCR to DBE in order to detect signal from D4Z4 repeats and normalized to a single copy region located on the human chromosome 4, namely SSLP. The graph is expressed as relative amount of D4Z4 repeats. The error bars represent standard error of the mean (SEM).
Primers used are listed in Table S2.
Characterization of BAC CH16-291A23
The BAC clone 291A23 containing D4Z4 repeats was isolated by using D4Z4 as probe to screen the CHORI-16 BAC library generated from sheared DNA (Osoegawa et al., 2007). The BAC extremities were sequenced with SP6 and T7 oligonucleotides. The number of D4Z4 repeats was inferred based on restriction digestion with EcoRI and XhoI enzymes and by q-PCR normalized to a single copy region, as described above.
RNA Extraction, RT-PCR, qRT-PCR, Northern Blotting, and RACE
RNA fractionation, RNA extraction, reverse transcription, real-time PCR, northern blotting, and RACE are described in the Extended Experimental Procedures (DBE-T accession number is JQ639078).
RNA/DNA FISH
RNA/DNA FISH was performed essentially as described in Custodio et al. (2006). Probe sequences and experimental details are described in the Extended Experimental Procedures.
Chromatin Immunoprecipitation
Assays were performed with normal IgG or the indicated antibodies. Precipitated DNA was measured by qPCR. Primer sets and methods are described in the Extended Experimental Procedures.
RNA ImmunoPrecipitation
This assay was carried out mainly as described previously (Jeon and Lee, 2011). The detailed method is described in the Extended Experimental Procedures.
In Vitro RNA Pull-down Assay
Recombinant GST-fusion proteins were prepared as described previously (Tanaka et al., 2008). In vitro RNA pull-down assay was essentially carried out as previously described (Jeon and Lee, 2011). For a detailed protocol refer to the Extended Experimental Procedures.
Chromosome Conformation Capture
Chromosome Conformation Capture (3C) was performed essentially as described in Bodega et al. (2009). Primer sequences and methods are described in the Extended Experimental Procedures.
Statistical Analyses
For EZH2 and H3K27me3 ChIP-qPCR in human samples, two-way repeated-measured ANOVA was used.
For RNA FISH, a two-tailed, paired, t test was used.
Acknowledgments
We thank W. Bickmore, M. Carmo-Fonseca, C. Carvalho, G. Cavalli, M. Crippa, J. Mattick, T. Misteli, K. Morris, and V. Orlando for helpful discussions. Anti-Rae28 antibody was a kind gift of Y. Takihara. Part of this work was carried out in Alembic (Advanced Light and Electron Microscopy Bio-Imaging Centre), San Raffaele Scientific Instutute. This study is a partial fulfillment of Valentina Casa's PhD in Molecular Medicine, Program in Neuroscience, San Raffaele University, Milan, Italy. The Ginelli lab is supported by Telethon (GGP07078) and Association Française contre les Myopathies (AFM) (13160 and 14464). Support for the Gabellini lab came from the European Research Council (ERC), the Italian Ministry of Health, the Muscular Dystrophy Association USA (MDA), AFM, the FSHD Global Research Foundation, and the Facio Scapulo Humeral Muscular Dystrophy Society. D.G. is a Dulbecco Telethon Institute Assistant Scientist.
Published online: April 26, 2012
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, eight figures, and five tables and can be found with this article online at doi:10.1016/j.cell.2012.03.035.
Accession Numbers
The GenBank accession number for DBE-T reported in this paper is JQ639078.
Supplemental Information
References
- An S., Yeo K.J., Jeon Y.H., Song J.J. Crystal structure of the human histone methyltransferase ASH1L catalytic domain and its implications for the regulatory mechanism. J. Biol. Chem. 2011;286:8369–8374. doi: 10.1074/jbc.M110.203380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae E., Calhoun V.C., Levine M., Lewis E.B., Drewell R.A. Characterization of the intergenic RNA profile at abdominal-A and Abdominal-B in the Drosophila bithorax complex. Proc. Natl. Acad. Sci. USA. 2002;99:16847–16852. doi: 10.1073/pnas.222671299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker E., Wijmenga C., Vossen R.H., Padberg G.W., Hewitt J., van der Wielen M., Rasmussen K., Frants R.R. The FSHD-linked locus D4F104S1 (p13E-11) on 4q35 has a homologue on 10qter. Muscle Nerve. 1995;2:S39–S44. [PubMed] [Google Scholar]
- Beisel C., Paro R. Silencing chromatin: comparing modes and mechanisms. Nat. Rev. Genet. 2011;12:123–135. doi: 10.1038/nrg2932. [DOI] [PubMed] [Google Scholar]
- Bertani S., Sauer S., Bolotin E., Sauer F. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol. Cell. 2011;43:1040–1046. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bodega B., Ramirez G.D., Grasser F., Cheli S., Brunelli S., Mora M., Meneveri R., Marozzi A., Mueller S., Battaglioli E., Ginelli E. Remodeling of the chromatin structure of the facioscapulohumeral muscular dystrophy (FSHD) locus and upregulation of FSHD-related gene 1 (FRG1) expression during human myogenic differentiation. BMC Biol. 2009;7:41. doi: 10.1186/1741-7007-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolanza S., Nonis A., Sanvito F., Maciotta S., Sitia G., Wei J., Torrente Y., Di Serio C., Chamberlain J.R., Gabellini D. AAV6-mediated systemic shRNA delivery reverses disease in a mouse model of facioscapulohumeral muscular dystrophy. Mol. Ther. 2011;19:2055–2064. doi: 10.1038/mt.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce H.A., Sachs N., Rudnicki D.D., Lin S.G., Willour V.L., Cowell J.K., Conroy J., McQuaid D.E., Rossi M., Gaile D.P. Long tandem repeats as a form of genomic copy number variation: structure and length polymorphism of a chromosome 5p repeat in control and schizophrenia populations. Psychiatr. Genet. 2009;19:64–71. doi: 10.1097/YPG.0b013e3283207ff6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschbeck M., Uribesalgo I., Wibowo I., Rué P., Martin D., Gutierrez A., Morey L., Guigó R., López-Schier H., Di Croce L. The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat. Struct. Mol. Biol. 2009;16:1074–1079. doi: 10.1038/nsmb.1665. [DOI] [PubMed] [Google Scholar]
- Busturia A., Lloyd A., Bejarano F., Zavortink M., Xin H., Sakonju S. The MCP silencer of the Drosophila Abd-B gene requires both Pleiohomeotic and GAGA factor for the maintenance of repression. Development. 2001;128:2163–2173. doi: 10.1242/dev.128.11.2163. [DOI] [PubMed] [Google Scholar]
- Cabianca D.S., Gabellini D. The cell biology of disease: FSHD: copy number variations on the theme of muscular dystrophy. J. Cell Biol. 2010;191:1049–1060. doi: 10.1083/jcb.201007028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacurri S., Piazzo N., Deidda G., Vigneti E., Galluzzi G., Colantoni L., Merico B., Ricci E., Felicetti L. Sequence homology between 4qter and 10qter loci facilitates the instability of subtelomeric KpnI repeat units implicated in facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 1998;63:181–190. doi: 10.1086/301906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick B.P. Macrosatellite epigenetics: the two faces of DXZ4 and D4Z4. Chromosoma. 2009;118:675–681. doi: 10.1007/s00412-009-0233-5. [DOI] [PubMed] [Google Scholar]
- Clark L.N., Koehler U., Ward D.C., Wienberg J., Hewitt J.E. Analysis of the organisation and localisation of the FSHD-associated tandem array in primates: implications for the origin and evolution of the 3.3 kb repeat family. Chromosoma. 1996;105:180–189. doi: 10.1007/BF02509499. [DOI] [PubMed] [Google Scholar]
- Custodio N., Carvalho C., Carneiro T., Carmo-Fonseca M. Cell Biology – a Laboratory Manual. Elsevier Science USA; USA: 2006. In situ hybridization for simultaneous detection of DNA, RNA and protein. [Google Scholar]
- Dahlberg J.E., Lund E. Does protein synthesis occur in the nucleus? Curr. Opin. Cell Biol. 2004;16:335–338. doi: 10.1016/j.ceb.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Deidda G., Cacurri S., Piazzo N., Felicetti L. Direct detection of 4q35 rearrangements implicated in facioscapulohumeral muscular dystrophy (FSHD) J. Med. Genet. 1996;33:361–365. doi: 10.1136/jmg.33.5.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déjardin J., Rappailles A., Cuvier O., Grimaud C., Decoville M., Locker D., Cavalli G. Recruitment of Drosophila Polycomb group proteins to chromatin by DSP1. Nature. 2005;434:533–538. doi: 10.1038/nature03386. [DOI] [PubMed] [Google Scholar]
- Dixit M., Ansseau E., Tassin A., Winokur S., Shi R., Qian H., Sauvage S., Mattéotti C., van Acker A.M., Leo O. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc. Natl. Acad. Sci. USA. 2007;104:18157–18162. doi: 10.1073/pnas.0708659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskeland R., Freyer E., Leeb M., Wutz A., Bickmore W.A. Histone acetylation and the maintenance of chromatin compaction by Polycomb repressive complexes. Cold Spring Harb. Symp. Quant. Biol. 2010;75:71–78. doi: 10.1101/sqb.2010.75.053. [DOI] [PubMed] [Google Scholar]
- Faulkner G.J., Kimura Y., Daub C.O., Wani S., Plessy C., Irvine K.M., Schroder K., Cloonan N., Steptoe A.L., Lassmann T. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- Gabellini D., Green M.R., Tupler R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110:339–348. doi: 10.1016/s0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- Garrick D., De Gobbi M., Samara V., Rugless M., Holland M., Ayyub H., Lower K., Sloane-Stanley J., Gray N., Koch C. The role of the polycomb complex in silencing alpha-globin gene expression in nonerythroid cells. Blood. 2008;112:3889–3899. doi: 10.1182/blood-2008-06-161901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjerstorff M.F., Ditzel H.J. An overview of the GAGE cancer/testis antigen family with the inclusion of newly identified members. Tissue Antigens. 2008;71:187–192. doi: 10.1111/j.1399-0039.2007.00997.x. [DOI] [PubMed] [Google Scholar]
- Greenberg J.R. Ultraviolet light-induced crosslinking of mRNA to proteins. Nucleic Acids Res. 1979;6:715–732. doi: 10.1093/nar/6.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory G.D., Vakoc C.R., Rozovskaia T., Zheng X., Patel S., Nakamura T., Canaani E., Blobel G.A. Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes. Mol. Cell. Biol. 2007;27:8466–8479. doi: 10.1128/MCB.00993-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y., Lee J.T. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146:119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G., Yang F., van Overveld P.G., Vedanarayanan V., van der Maarel S., Ehrlich M. Testing the position-effect variegation hypothesis for facioscapulohumeral muscular dystrophy by analysis of histone modification and gene expression in subtelomeric 4q. Hum. Mol. Genet. 2003;12:2909–2921. doi: 10.1093/hmg/ddg323. [DOI] [PubMed] [Google Scholar]
- Kaneko H., Dridi S., Tarallo V., Gelfand B.D., Fowler B.J., Cho W.G., Kleinman M.E., Ponicsan S.L., Hauswirth W.W., Chiodo V.A. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Villeneuve L.M., Morris K.V., Rossi J.J. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struct. Mol. Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- Kowaljow V., Marcowycz A., Ansseau E., Conde C.B., Sauvage S., Matteotti C., Arias C., Corona E.D., Nunez N.G., Leo O. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul. Disord. 2007;17:611–623. doi: 10.1016/j.nmd.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Krogan N.J., Kim M., Tong A., Golshani A., Cagney G., Canadien V., Richards D.P., Beattie B.K., Emili A., Boone C. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeira D., Fisher A.G. Inactive yet indispensable: the tale of Jarid2. Trends Cell Biol. 2011;21:74–80. doi: 10.1016/j.tcb.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M., Pasini D., Novatchkova M., Jaritz M., Helin K., Wutz A. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev. 2010;24:265–276. doi: 10.1101/gad.544410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers R.J., Osborn M., Haaf T., Rogers M., Frants R.R., Padberg G.W., Cooper D.N., van der Maarel S.M., Upadhyaya M. D4F104S1 deletion in facioscapulohumeral muscular dystrophy: phenotype, size, and detection. Neurology. 2003;61:178–183. doi: 10.1212/01.wnl.0000078889.51444.81. [DOI] [PubMed] [Google Scholar]
- Lemmers R.J., van der Maarel S.M., van Deutekom J.C., van der Wielen M.J., Deidda G., Dauwerse H.G., Hewitt J., Hofker M., Bakker E., Padberg G.W., Frants R.R. Inter- and intrachromosomal sub-telomeric rearrangements on 4q35: implications for facioscapulohumeral muscular dystrophy (FSHD) aetiology and diagnosis. Hum. Mol. Genet. 1998;7:1207–1214. doi: 10.1093/hmg/7.8.1207. [DOI] [PubMed] [Google Scholar]
- Lemmers R.J., van der Vliet P.J., Klooster R., Sacconi S., Camaño P., Dauwerse J.G., Snider L., Straasheijm K.R., van Ommen G.J., Padberg G.W. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329:1650–1653. doi: 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipshitz H.D., Peattie D.A., Hogness D.S. Novel transcripts from the Ultrabithorax domain of the bithorax complex. Genes Dev. 1987;1:307–322. doi: 10.1101/gad.1.3.307. [DOI] [PubMed] [Google Scholar]
- Lyle R., Wright T.J., Clark L.N., Hewitt J.E. The FSHD-associated repeat, D4Z4, is a member of a dispersed family of homeobox-containing repeats, subsets of which are clustered on the short arms of the acrocentric chromosomes. Genomics. 1995;28:389–397. doi: 10.1006/geno.1995.1166. [DOI] [PubMed] [Google Scholar]
- Margueron R., Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matharu N.K., Hussain T., Sankaranarayanan R., Mishra R.K. Vertebrate homologue of Drosophila GAGA factor. J. Mol. Biol. 2010;400:434–447. doi: 10.1016/j.jmb.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Mendenhall E.M., Koche R.P., Truong T., Zhou V.W., Issac B., Chi A.S., Ku M., Bernstein B.E. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 2010;6:e1001244. doi: 10.1371/journal.pgen.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaly J., Mishra R.K., Karch F. A conserved sequence motif in Polycomb-response elements. Mol. Cell. 1998;1:1065–1066. doi: 10.1016/s1097-2765(00)80107-0. [DOI] [PubMed] [Google Scholar]
- Mills A.A. Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat. Rev. Cancer. 2010;10:669–682. doi: 10.1038/nrc2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra R.K., Mihaly J., Barges S., Spierer A., Karch F., Hagstrom K., Schweinsberg S.E., Schedl P. The iab-7 polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and pleiohomeotic for silencing activity. Mol. Cell. Biol. 2001;21:1311–1318. doi: 10.1128/MCB.21.4.1311-1318.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris K.V., Chan S.W., Jacobsen S.E., Looney D.J. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- Neguembor M.V., Gabellini D. In junk we trust: repetitive DNA, epigenetics and facioscapulohumeral muscular dystrophy. Epigenomics. 2010;2:271–287. doi: 10.2217/epi.10.8. [DOI] [PubMed] [Google Scholar]
- Norris J., Fan D., Aleman C., Marks J.R., Futreal P.A., Wiseman R.W., Iglehart J.D., Deininger P.L., McDonnell D.P. Identification of a new subclass of Alu DNA repeats which can function as estrogen receptor-dependent transcriptional enhancers. J. Biol. Chem. 1995;270:22777–22782. doi: 10.1074/jbc.270.39.22777. [DOI] [PubMed] [Google Scholar]
- Papp B., Müller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov A., Pirozhkova I., Carnac G., Laoudj D., Lipinski M., Vassetzky Y.S. Chromatin loop domain organization within the 4q35 locus in facioscapulohumeral dystrophy patients versus normal human myoblasts. Proc. Natl. Acad. Sci. USA. 2006;103:6982–6987. doi: 10.1073/pnas.0511235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S., Sedkov Y., Riley K.M., Hodgson J., Schweisguth F., Hirose S., Jaynes J.B., Brock H.W., Mazo A. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127:1209–1221. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirozhkova I., Petrov A., Dmitriev P., Laoudj D., Lipinski M., Vassetzky Y. A functional role for 4qA/B in the structural rearrangement of the 4q35 region and in the regulation of FRG1 and ANT1 in facioscapulohumeral dystrophy. PLoS ONE. 2008;3:e3389. doi: 10.1371/journal.pone.0003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rank G., Prestel M., Paro R. Transcription through intergenic chromosomal memory elements of the Drosophila bithorax complex correlates with an epigenetic switch. Mol. Cell. Biol. 2002;22:8026–8034. doi: 10.1128/MCB.22.22.8026-8034.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijkers T., Deidda G., van Koningsbruggen S., van Geel M., Lemmers R.J., van Deutekom J.C., Figlewicz D., Hewitt J.E., Padberg G.W., Frants R.R., van der Maarel S.M. FRG2, an FSHD candidate gene, is transcriptionally upregulated in differentiating primary myoblast cultures of FSHD patients. J. Med. Genet. 2004;41:826–836. doi: 10.1136/jmg.2004.019364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L., Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Sanchez-Elsner T., Gou D., Kremmer E., Sauer F. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science. 2006;311:1118–1123. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- Schmitt S., Prestel M., Paro R. Intergenic transcription through a polycomb group response element counteracts silencing. Genes Dev. 2005;19:697–708. doi: 10.1101/gad.326205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B., Chourrout D., Vervoort M., Leblanc B., Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B., Ganapathi M., Leblanc B., Portoso M., Jaschek R., Tolhuis B., van Lohuizen M., Tanay A., Cavalli G. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol. 2009;7:e13. doi: 10.1371/journal.pbio.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz Y.B., Kahn T.G., Stenberg P., Ohno K., Bourgon R., Pirrotta V. Alternative epigenetic chromatin states of polycomb target genes. PLoS Genet. 2010;6:e1000805. doi: 10.1371/journal.pgen.1000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Lin L., Cai J.J., Jiang P., Kenkel E.J., Stroik M.R., Sato S., Davidson B.L., Xing Y. Widespread establishment and regulatory impact of Alu exons in human genes. Proc. Natl. Acad. Sci. USA. 2011;108:2837–2842. doi: 10.1073/pnas.1012834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E., Lin C., Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 2011;25:661–672. doi: 10.1101/gad.2015411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider L., Geng L.N., Lemmers R.J., Kyba M., Ware C.B., Nelson A.M., Tawil R., Filippova G.N., van der Maarel S.M., Tapscott S.J., Miller D.G. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 2010;6:e1001181. doi: 10.1371/journal.pgen.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Katagiri Z., Kawahashi K., Kioussis D., Kitajima S. Trithorax-group protein ASH1 methylates histone H3 lysine 36. Gene. 2007;397:161–168. doi: 10.1016/j.gene.2007.04.027. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Nakayama Y., Taniguchi M., Kioussis D. Regulation of early T cell development by the PHD finger of histone lysine methyltransferase ASH1. Biochem. Biophys. Res. Commun. 2008;365:589–594. doi: 10.1016/j.bbrc.2007.10.159. [DOI] [PubMed] [Google Scholar]
- Ting D.T., Lipson D., Paul S., Brannigan B.W., Akhavanfard S., Coffman E.J., Contino G., Deshpande V., Iafrate A.J., Letovsky S. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331:593–596. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay D.C., Alexander G., Jr., Moseley S., Chadwick B.P. Expression, tandem repeat copy number variation and stability of four macrosatellite arrays in the human genome. BMC Genomics. 2010;11:632. doi: 10.1186/1471-2164-11-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A.M., Morris K.V. Controlling transcription with noncoding RNAs in mammalian cells. Biotechniques. 2010;48:ix–xvi. doi: 10.2144/000113442. [DOI] [PubMed] [Google Scholar]
- van Overveld P.G., Lemmers R.J., Sandkuijl L.A., Enthoven L., Winokur S.T., Bakels F., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat. Genet. 2003;35:315–317. doi: 10.1038/ng1262. [DOI] [PubMed] [Google Scholar]
- Wallace L.M., Garwick-Coppens S.E., Tupler R., Harper S.Q. RNA interference improves myopathic phenotypes in mice over-expressing FSHD region gene 1 (FRG1) Mol. Ther. 2011;19:2048–2054. doi: 10.1038/mt.2011.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B.R., Protacio A., Flynn R.A., Gupta R.A. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton P.E., Hasson D., Guillem F., Lescale C., Jin X., Abrusan G. Analysis of the largest tandemly repeated DNA families in the human genome. BMC Genomics. 2008;9:533. doi: 10.1186/1471-2164-9-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winokur S.T., Bengtsson U., Feddersen J., Mathews K.D., Weiffenbach B., Bailey H., Markovich R.P., Murray J.C., Wasmuth J.J., Altherr M.R. The DNA rearrangement associated with facioscapulohumeral muscular dystrophy involves a heterochromatin-associated repetitive element: implications for a role of chromatin structure in the pathogenesis of the disease. Chromosome Res. 1994;2:225–234. doi: 10.1007/BF01553323. [DOI] [PubMed] [Google Scholar]
- Woo C.J., Kharchenko P.V., Daheron L., Park P.J., Kingston R.E. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.L., Tsai M.S., Wong S.H., Hsieh-Li H.M., Tsai T.S., Chang W.T., Huang S.L., Chiu C.C., Wang S.H. Characterization of genomic structures and expression profiles of three tandem repeats of a mouse double homeobox gene: Duxbl. Dev. Dyn. 2010;239:927–940. doi: 10.1002/dvdy.22210. [DOI] [PubMed] [Google Scholar]
- Yuan W., Xu M., Huang C., Liu N., Chen S., Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 2011;286:7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W., de Greef J.C., Chen Y.Y., Chien R., Kong X., Gregson H.C., Winokur S.T., Pyle A., Robertson K.D., Schmiesing J.A. Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD) PLoS Genet. 2009;5:e1000559. doi: 10.1371/journal.pgen.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplemental References
- Ohtsubo, M., Yasunaga, S., Ohno, Y., Tsumura, M., Okada, S., Ishikawa, N., Shirao, K., Kikuchi, A., Nishitani, H., Kobayashi, M., and Takihara, Y. (2008). Polycomb-group complex 1 acts as an E3 ubiquitin ligase for Geminin to sustain hematopoietic stem cell activity. Proc. Natl. Acad. Sci. USA 105, 10396–10401. [DOI] [PMC free article] [PubMed]
- Osoegawa, K., Vessere, G.M., Li Shu, C., Hoskins, R.A., Abad, J.P., de Pablos, B., Villasante, A., and de Jong, P.J. (2007). BAC clones generated from sheared DNA. Genomics 89, 291–299. [DOI] [PMC free article] [PubMed]
- Sambrook, J., and Russel, D.W. (2001). Molecular cloning: a laboratory manual (Cold Spring Harbor, NY: Cold Harbor Spring Laboratory Press).
- Stock, J.K., Giadrossi, S., Casanova, M., Brookes, E., Vidal, M., Koseki, H., Brockdorff, N., Fisher, A.G., and Pombo, A. (2007). Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat. Cell Biol. 9, 1428–1435. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.