Abstract
The 14-3-3 proteins specifically bind a number of client proteins to influence important pathways, including flowering timing via the photosensory system. For instance, 14-3-3 proteins influence the photosensory system through interactions with Constans (CO) protein. 14-3-3 associations with the photosensory system were further studied in this investigation using 14-3-3 T-DNA insertion mutants to study root and chloroplast development. The 14-3-3 μ T-DNA insertion mutant, 14-3-3μ-1, had shorter roots than the wild type and the difference in root length could be influenced by light intensity. The 14-3-3 ν T-DNA insertion mutants also had shorter roots, but only when grown under narrow-bandwidth red light. Five-day-old 14-3-3 T-DNA insertion and co mutants all had increased root greening compared with the wild type, which was influenced by light wavelength and intensity. However, beyond 10 d of growth, 14-3-3μ-1 roots did not increase in greening as much as wild-type roots. This study reveals new developmental roles of 14-3-3 proteins in roots and chloroplasts, probably via association with the photosensory system.
Keywords: Chloroplast, photosensory system, plastid, 14-3-3 proteins, root development
Introduction
The 14-3-3 proteins are predominantly known for their ability to bind certain phosphorylated proteins to complete phosphoregulation events (Ferl, 1996). In plants, 14-3-3 protein binding activity includes the regulation of key metabolic enzymes, such as nitrate reductase and sucrose synthase (Bachmann et al., 1996; Toroser et al., 1998), and also the activation of the plasma membrane H+-ATPase (Olsson et al., 1998; Fuglsang et al., 1999; Svennelid et al., 1999; Ottmann et al., 2007). Throughout eukaryotes, 14-3-3 proteins are known to bind a multitude of different proteins and, specifically, participate in a wide array of signal transduction regulatory events.
Multicellular eukaryotes contain multiple 14-3-3 protein family members, or isoforms. The number of 14-3-3 isoforms can vary per organism, but Arabidopsis contains 13 expressed 14-3-3 genes (Wu et al., 1997; Rosenquist et al., 2001; Sehnke et al., 2006). The 14-3-3 proteins throughout eukaryotes share sequence homology within the core region of the protein sequence, but sequences of individual isoforms diverge in the amino- and carboxy-terminal tails. The importance of multiple 14-3-3 isoforms per organism remains to be elucidated, however, reverse genetic studies analysing knockout or knock down mutants continue to identify specific roles of individual 14-3-3 isoforms (Su et al., 2001; Sugiyama et al., 2003; Mayfield et al., 2007; Purwestri et al., 2009).
In Arabidopsis, 14-3-3 T-DNA insertion mutants were utilized to find association of the 14-3-3 isoforms μ and υ with both light sensing during early development and time of transition to flowering (Mayfield et al., 2007). Red light-grown 14-3-3 T-DNA insertion mutants exhibit decreased hypocotyl elongation inhibition (Mayfield et al., 2007) reminiscent of, but not identical to Phytochrome B (PHYB) mutants (Koornneef et al., 1980). The 14-3-3 T-DNA insertion mutants also flower later in long-day conditions (16 h of light), but not in short-day conditions (8 h of light) (Mayfield et al., 2007). In rice, the opposite phenotype occurs when manipulating expression of the 14-3-3 isoform, GF14c (Purwestri et al., 2009). Over-expression of GF14c results in delayed flowering, while knockout results in early flowering. These 14-3-3 mutant phenotypes indicate a role in light-related signalling for these particular 14-3-3 isoforms. The relationship of 14-3-3 proteins with light signalling is further supported by the observation that 14-3-3 proteins bind the blue light photoreceptor Phototropin 1 (PHOT1) (Kinoshita et al., 2003) and the flowering timing regulation protein Constans (CO) (Mayfield et al., 2007), as well as downstream targets such as Flowering Timing (FT) and Self-Pruning (SP) (Pnueli et al., 2001).
Light perception is broadly important for phototropic responses, time of flowering, and root development. For example, Red and far-red light influence the directional growth of roots. Roots of some plants, such as maize and rice, grow horizontally in the dark, but grow downward when the plants are irradiated with red or far-red light from above (Takano et al., 2001; Lu et al., 1996). In addition, there is copious expression of photoreceptors in the roots (Pratt and Coleman, 1974; Somers and Quail, 1995; Toth et al., 2001), suggesting a functional relevance for photoreceptors in root development. Root hair development is enhanced by light and mediated by phytochromes (De Simone et al., 2000a, b). Further, both Phytochrome A (PHYA) and PHYB regulate red-light-induced root hair development while far-red-induced root hair development is regulated by PHYA alone (De Simone et al., 2000a).
Photoreceptors also control chloroplast development in roots that are exposed to light. Prolonged exposure of plant roots to light stimulates the development of chloroplasts in the roots (Whatley, 1983). Chloroplast development in the roots is stimulated by red and blue light, with cryptochrome 1 and 2 (CRY1 and CRY2) as the primary regulators of blue light-stimulated chloroplast development and PHYB as the primary regulator in red light (Usami et al., 2004).
This study utilizes the previously described T-DNA insertion mutants of 14-3-3 μ and υ to examine the roles of individual 14-3-3 isoforms in root elongation and chloroplast development, that are controlled by the photosensory system. A 14-3-3μ knockdown mutant containing a T-DNA insertion within the promoter of 14-3-3μ, 14-3-3μ-1 (Mayfield et al., 2007), exhibited shorter roots whose lengths were influenced by various light intensities. The 14-3-3υ knockout T-DNA insertion mutants, 14-3-3υ-1 and 14-3-3υ-2 (Mayfield et al., 2007), each contain a T-DNA insertion within the 14-3-3υ coding region and also had shorter roots than the wild type, but only when grown under narrow bandwidth red light. In addition, the roots of the 14-3-3 T-DNA insertion mutants and CO mutants (co) (co-1; Redei, 1962; Koornneef et al., 1991) exhibited an increase in chloroplast numbers after 5 d of growth.
Materials and methods
Growth conditions of Arabidopsis seedlings
Arabidopsis seeds were sterilized as previously described by Paul et al. (2001). Thirty to forty seeds of each line were planted onto 0.5× MS vertical plates, which consisted of 0.22% MS salts (Murashige and Skoog, 1962), 0.025% MES buffer, 0.5% sucrose, 0.1% 1000× MS vitamins (Murashige and Skoog, 1962), and 0.8% agar. Nitrate-free media were prepared as above, except that the MS salt mix was substituted for nitrogen-free MS salt mix (Product #MSP007; Caisson Laboratories, Logan, UT), 1 mM ammonium citrate was added and the media was adjusted to a pH of 6.5. After planting, the seed-containing plates were incubated at 4 °C in the dark for 48 h. The plates were then placed under the appropriate light conditions. Constant-light-grown seedlings were grown under a light bank emitting 60–70 μmol m−2 s−1 of light (two Cool White Sylvania bulbs and two Gro-Lux bulbs). Constant red light (630 nm at 25 μmol m−2 s−1), blue light (470 nm at 40 μmol m−2 s−1) or dark-grown plants were exposed to white light for 1 h, after the 48 h cold treatment, and placed under Light Emitting Diodes (LEDs) or in the dark. Treatments included irradiation with specific fluence rates of red light (630 nm; Quantum Devices, Barneveld WI) or blue light (470 nm; Ledtronics Inc, Torrance CA), and a custom design used by Folta et al. (2005).
Characterization of phyB mutants
Seed stocks for the PHYB T-DNA insertion mutant lines Salk_022035 and Salk_069700 were obtained from the Arabidopsis Biological Resource Center. The seeds were planted on wet Fafard Fine-germinating Mix soil and incubated at 4 °C in the dark for 3 d. The seeds were exposed to light for 1 h to promote germination, then placed under ∼20 μmol m−2 s−1 of narrow-bandwidth red light for 4 d. All seedlings were cleared from the soil that did not exhibit reduced hypocotyl elongation inhibition. The remaining seedlings that did exhibit reduced hypocotyl elongation inhibition were later screened for homozygous T-DNA insertion using PCR. A left border T-DNA specific primer (LBb1) and two, T-DNA flanking, PHYB gene-specific primers were used to identify lines that were homozygous for T-DNA insertion into the respective 14-3-3 genes. It is important to add that 100% of the lines, from both Salk_022035 and Salk_069700, selected as exhibiting reduced hypocotyl elongation inhibition were identified as homozygous PHYB T-DNA insertion mutants.
The phenotypes exhibited by Salk_022035 and Salk_069700 in this study were comparable between the two separate lines. In order to make the data easier to convey, only the phenotypic data of Salk_022035 were included in the results and discussion of this study.
Root and chloroplast measurements
Digital images of roots were captured using an Epson Perfection 3170 Photo scanner for further analysis. Root lengths were assessed by measuring root lengths of each Arabidopsis line of wild-type, Res-mu, and 14-3-3μ-1 with the University of Texas Health Science Center San Antonio Dental School (UTHSCSA) Image Tool software (http://ddsdx.uthscsa.edu/dig/itdesc.html). The root measurements collected were standardized and processed in Microsoft Excel.
Visualization of Arabidopsis roots with a fluorescent microscope
Roots were viewed with an Olympus BX51 fluorescent microscope coupled to an Evolution MP cooled charge-coupled device camera with Q-capture 2.60 software (Quantitative Imaging, Burnaby, British Columbia, Canada). Digital photos were taken through a 10× objective and captured with no binning. Regions identical in size were cropped from the digital images taken of roots from each Arabidopsis line. Slides were prepared by mounting the roots in water between a slide and cover slip.
Visualization of Arabidopsis roots with a confocal microscope
Roots from 5-d-old Arabidopsis seedlings were viewed with a confocal LSM5 Pascal Laser Scanning Microscope (Zeiss, Jena, Germany). The light source used was a helium neon laser emitting 543 nm light. The light emitted by the helium neon laser was filtered with a long pass filter resulting in a wavelength of 560 nm, similar to the conditions used to view rhodamine. Images were collected of the red fluorescing chloroplast, as well as Differential Interference Contrast (DIC) images to provide a reference for root size and shape.
Quantification of the red tonal levels of Arabidopsis roots
Four or more fluorescent microscope images of chloroplasts in root tissue were cropped in order to standardize the images to equal sizes using Adobe Photoshop. The standardized images were then processed with the RGB Measure Plus plug-in of Image J. Tonal levels recorded by the RGB Measure Plus plug-in were then processed with Microsoft Excel.
Results
14-3-3μ-1 has shorter roots than wild-type
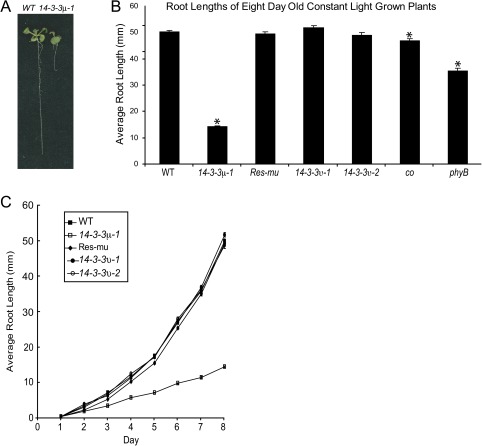
Wild-type and 14-3-3μ-1 were grown on vertical MS media agar plates under constant light conditions and assayed for root lengths after 8 d of growth (Fig. 1). The roots of 14-3-3μ-1, co, and phyB were statistically shorter than wild-type levels (α=9.006×10−61, 0.0011, and 3.744×10−13, respectively). The 14-3-3μ-1 roots were only ∼25% of wild-type root length (Fig. 1A, B), although developmentally similar in the aerial tissue. Wild-type root length was restored by the reintroduction of 14-3-3μ in Res-mu, a 14-3-3μ-1 line that was complemented by the native 14-3-3 μ gene (Mayfield et al., 2007) (Fig. 1B, C). Root lengths of 14-3-3υ-1 and 14-3-3υ-2 were also comparable with that of wild-type roots after 8 d of growth (Fig. 1B, C).
Fig. 1.
Shorter root lengths of 14-3-3μ-1 compared with wild-type root lengths. After 8 d of growth, 14-3-3μ-1 exhibited shorter roots than wild-type (A). The 14-3-3 T-DNA insertion mutants, Res-mu, co, phyB, and the wild type were grown on vertical agar plates in constant light. The average root lengths after 8 d of growth were measured (B). The roots of 14-3-3μ-1 and phyB were ∼30% and ∼70% of the wild-type root length, while other lines were equal in root length to wild-type (B). Error bars represent standard error and asterisks indicate lines with a significant difference in root length compared to wild-type using a t test (B). The root lengths of the 14-3-3 T-DNA insertion mutants and the wild type were measured daily in order to get a time-course of root lengths (C). 14-3-3μ-1 exhibited shorter roots that consistently increase in the deviation from wild-type root lengths through 8 d of growth. Error bars represent standard error (B, C).
The root growth results for 14-3-3μ-1 were in stark contrast to the results for 14-3-3υ insertion mutants. Two separate T-DNA insertion alleles for 14-3-3υ showed wild-type root lengths under constant light conditions. CO also exhibited wild-type root length, while phyB had a slightly decreased root length (Fig. 1B).
The time-course of root length was studied to determine if there was a particular time point that 14-3-3μ-1 root length begins to deviate from that of the wild type and Res-mu, or the two alleles of 14-3-3υ. Root lengths were measured daily over 8 d of growth (Fig. 1C). 14-3-3μ-1 exhibited an overall slower increase in root length throughout the entire 8 d compared with the wild type, with no particular day characterized as a specific point of separation of 14-3-3μ-1 from the wild-type root length. After day 2, when the difference in root length is measurable, the 14-3-3μ-1 root length begins to lag behind, and remain at a slower and constant growth rate, while wild type, Res-mu, and the 14-3-3μ alleles accelerate in growth rate.
The decrease in root length of 14-3-3μ-1 is not due to altered nitrate reductase activity
The 14-3-3 proteins interact with nitrate reductase (NR), as part of the NR inactivation process (Bachmann et al., 1996; Moorhead et al., 1996). In fact, of the 14-3-3 isoforms tested, 14-3-3υ is the third most effective at inactivating nitrate reductase (the 14-3-3μ isoform was not tested in the study) (Bachmann et al., 1996). Because of the 14-3-3 protein association with nitrate reductase inactivation, it was possible that the loss of 14-3-3μ protein resulted in overactive nitrate reductase. Overactive nitrate reductase protein, in combination with nitrate-rich media, could have resulted in a reduced need for root elongation since the roots had adequate nitrate levels available. In order to factor out nitrate reductase activity as the cause of 14-3-3μ-1 decreased root length, plants were grown on nitrate-free media using ammonium-citrate as the only nitrogen source. The 14-3-3μ-1 root lengths were 40% of the wild type, when grown on nitrate-free media for 14 d (see Supplementary Fig. S1 at JXB online), indicating that increased nitrate reductase activity was not the major factor for shorter 14-3-3μ-1 roots.
Differential root length of 14-3-3μ-1 can be partly attributed to the photosensory pathway
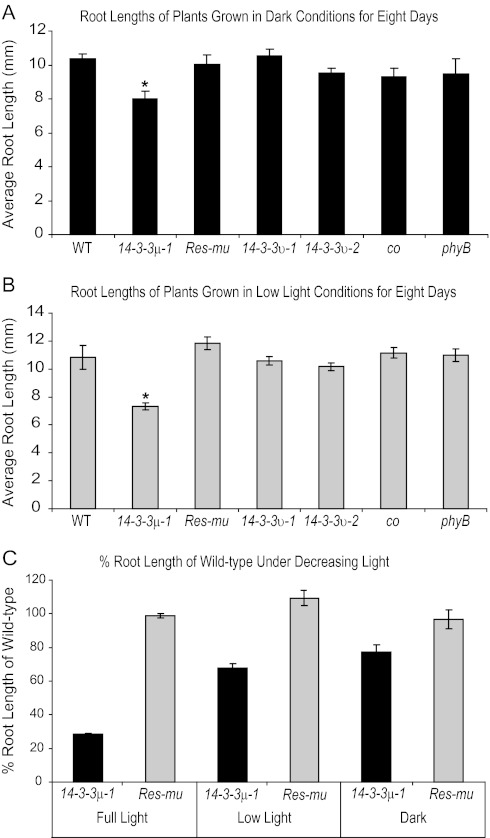
The 14-3-3 T-DNA insertion mutants 14-3-3μ-1, 14-3-3υ-1, and 14-3-3υ-2 all exhibit reduced hypocotyl growth inhibition under narrow-bandwidth red light during early development (Mayfield et al., 2007), similar to PHYB and PHYB-associated protein mutants (Neff and Chory, 1998; Huq et al., 2000). In order to test if 14-3-3μ-1 exhibits light-dependent deviations in root length, as does phyB, the 14-3-3 T-DNA insertion mutants were grown alongside phyB and the wild type under three separate light intensities: full-light (60–70 μmol m−2 s−1), low-light (5 μmol m−2 s−1), and dark growth conditions. A CO mutant, co, was also included in all of the altered light condition experiments because CO is a binding partner of 14-3-3 proteins and is associated with 14-3-3-mediated red-light signalling (Mayfield et al., 2007). Seeds were planted on vertical plates and exposed to light for 1 h to promote germination, then transferred to full-light, low-light, or dark chambers. Root lengths were measured after 8 d (Fig. 2A, B). Under dark conditions, 14-3-3υ-1, 14-3-3υ-2, co, and phyB roots were similar in length to the wild-type roots. 14-3-3μ-1 roots were slightly shorter than wild-type roots when grown in dark and low light conditions. 14-3-3μ-1 roots were ∼80% and ∼68% of wild-type root length in darkness and low light, respectively. By contrast, 14-3-3μ-1 roots were ∼25% of wild-type root length when grown in full-light conditions. As light intensity growth conditions decreased, 14-3-3μ-1 and Res-mu root lengths became more comparable (Fig. 2C). Res-mu root lengths were comparable with the wild type in all three light conditions.
Fig. 2.
The influence of 14-3-3μ-1 root lengths by different light intensities. The 14-3-3 T-DNA insertion mutants, Res-mu, co, phyB, and the wild type were grown on vertical agar plates in dark (A) and low-light (B) conditions for 8 d and root lengths were measured. The roots of 14-3-3μ-1 were ∼77% and ∼68% of the wild-type roots in dark and low-light conditions, respectively. Root lengths of 14-3-3μ-1 and Res-mu were compared as a % of wild-type root lengths when grown in three conditions: full light, low light, and dark (C). Res-mu roots were equal to wild-type root lengths in all treatments. The roots of 14-3-3μ-1 were more comparable to wild-type lengths as the light conditions decreased in intensity. Error bars represent standard error and asterisks indicate lines with a significant difference in root length compared with the wild type using a t test (A, B).
14-3-3 T-DNA insertion and co mutant roots develop increased numbers of chloroplasts during early development
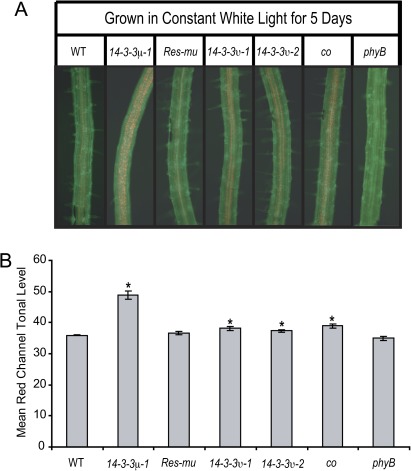
Roots experiencing prolonged light exposure exhibit greening resulting from chloroplast development (Whatley, 1983). PHYB is a positive regulator of chloroplast development in Arabidopsis roots (Usami et al., 2004). Due to similarities of 14-3-3μ-1 and phyB in the influence on root length by light intensity, the 14-3-3 T-DNA insertion mutants were assayed for root chloroplast development along with the wild type, Res-mu, co, and phyB. Plants were grown on vertical agar plates under constant light conditions for 5 d. Roots of the 5-d-old plants were viewed with a fluorescent microscope. The roots were illuminated with blue light (488 nm) and viewed through a long pass filter in order to view the red autofluorescence of chlorophyll.
A large number of chloroplasts were visualized within the roots of the 14-3-3 T-DNA insertion mutants and co (Fig. 3A). Wild-type and Res-mu roots developed chloroplasts, but in smaller numbers compared with the 14-3-3 T-DNA insertion mutants and co. The roots of phyB did not develop any observable chloroplasts under white light. The roots of 14-3-3μ-1 had the most dramatic increase in chloroplast number. The co, 14-3-3υ-1, and 14-3-3υ-2 roots had slightly fewer chloroplasts than 14-3-3μ-1.
Fig. 3.
Increased numbers of chloroplasts in the roots of the 14-3-3 T-DNA insertion mutants and co after 5 d of growth. The 14-3-3 T-DNA insertion mutants, Res-mu, co, phyB, and the wild type were grown on vertical agar plates for 5 d under constant light. Roots were illuminated with a blue light and visualized through a GFP long pass filter with a fluorescent microscope (A). The roots of the 14-3-3 T-DNA insertion mutants and co contained more chloroplasts, which autofluoresce as a red colour. Pictures of roots from each line were cropped to equal sizes and the red tonal levels were measured with the RGB Measure Plus plug-in of the ImageJ program (B). The red tonal levels were higher for 14-3-3μ-1, 14-3-3υ1, 14-3-3υ-2 and co roots compared with the wild type. Error bars represent standard error and asterisks indicate lines with a significant difference in root length compared with the wild type using a t test (B).
The red channel level within the frame of each photograph was also quantified (Fig. 3B). Photo frames of equal size, exposure, and colour adjustment were taken of 4–6 roots from each line. The red channel tonal levels of each photo frame were quantified using the RGB Measure Plus plug-in of the ImageJ software package. Quantification of the red light tonal levels showed that the average red tonal level for the 14-3-3 T-DNA insertion mutants and co were higher than the wild type or Res-mu. T-tests confirmed that red tonal levels of 14-3-3μ-1, 14-3-3υ-1, 14-3-3υ-2, and co root pictures were statistically higher than the wild-type levels (α=0.0000, 0.0067, 0.0127, and 0.0011, respectively). The photo frames of phyB roots contained statistically lower red tonal levels than the wild type, as confirmed by a t test (α=0.0253).
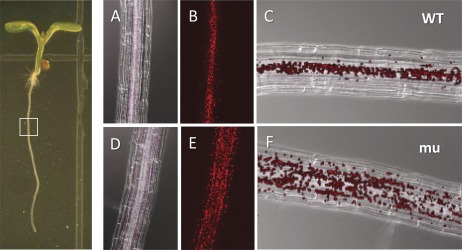
The dramatic increase in chloroplasts in 14-3-3μ-1 roots was also examined with confocal microscopy. Wild-type and 14-3-3μ-1 plants were vertically grown for 5 d under full light conditions (60–70 μmol m−2 s−1), and root sections were observed to determine the distribution of chloroplasts (Fig. 4). The DIC images illustrated that the roots were of the same developmental age, with cells of comparable size, in the wild-type and 14-3-3μ-1 plants (Fig. 4A, D). Fluorescent images of the wild type and 14-3-3μ-1 showed that while the plastid size is comparable for each plant, the quantity of plastids in the 14-3-3μ-1 roots is around 2-fold that of the wild type (Fig. 4B, E).
Fig. 4.
Confocal microscopy of plastid distribution in the roots of wild-type (WT) and 14-3-3μ-1. The left hand panel shows an Arabidopsis seedling; the region of root presented in the right hand panels is indicated with a white box. (A, D) The fluorescing plastids merged with the corresponding DIC image. (B, E) The fluorescing plastids alone. (C, F) The overlay of images (A)+(B) and (D)+(E), respectively.
Narrow-bandwidth red or blue light influence both root length and the numbers of chloroplasts in the roots
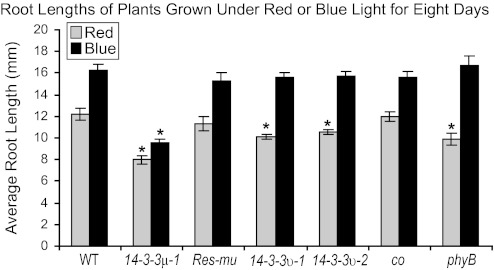
Root lengths were also assayed from plants grown on vertical agar plates for 8 d under narrow-bandwidth red (25 μmol m−2 s−1) or blue (50 μmol m−2 s−1) light (Fig. 5). The 14-3-3μ-1 roots were ∼65% or ∼60% of the length of wild-type roots in red or blue light conditions, respectively. 14-3-3υ-1 and 14-3-3υ-2 roots were similar in length to wild-type roots when grown under blue light, but shorter when grown under red light. The 14-3-3υ-1 and 14-3-3υ-2 roots were ∼83% and ∼86% of the length of wild-type roots, respectively. The differences in root lengths of red-light-grown 14-3-3υ-1 and 14-3-3υ-2 were confirmed to be statistically different by t test (αα=0.0003 and 0.0050, respectively).
Fig. 5.
Root lengths of the 14-3-3 T-DNA insertion mutants are influenced by red and blue light. The 14-3-3 T-DNA insertion mutants, Res-mu, co, phyB, and the wild type were grown on vertical agar plates under constant red or blue light conditions for 8 d and root lengths were measured. The roots of 14-3-3μ-1 were ∼65% and ∼59% of wild-type roots in red and blue light conditions, respectively. The roots of the 14-3-3 υ T-DNA insertion mutants were similar in length to wild-type roots when grown under blue light, but 14-3-3υ-1 and 14-3-3υ-2 were ∼83% and ∼86% of wild-type roots in red light conditions, respectively. Error bars represent standard error and asterisks indicate lines with a significant difference in root length compared with the wild type using a t test.
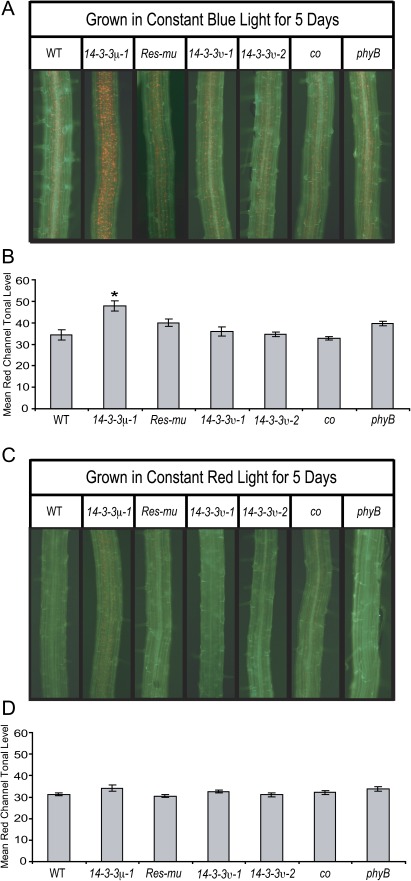
Root chloroplast development can be influenced by growth under different wavelengths of light (Usami et al., 2004). When grown in dark or low light conditions, roots from none of the lines studied developed observable chloroplasts (data not shown). The 14-3-3 T-DNA insertion mutants, co, phyB, Res-mu, and the wild type were also grown under narrow-bandwidth red or blue light to test whether these wavelengths influence root chloroplast development differently among the lines. Plants were grown for 5 d under red or blue light and then the roots were assayed for chloroplasts (Fig. 6). As previously published (Usami et al., 2004), blue light stimulated more root chloroplast development than red light. Furthermore, blue light stimulated more chloroplast development in the roots than white light. The roots of blue-light-grown 14-3-3μ-1 developed a higher number of chloroplasts than any of the other lines (Fig. 6A). Quantification of red tonal levels confirmed that 14-3-3μ-1 roots contain higher red tonal levels than both the wild type and Res-mu (Fig. 6B). The red tonal level of blue light grown 14-3-3μ-1 roots was statistically higher than that of wild-type or Res-mu roots as confirmed by t test (α=0.0012 and 0.0323, respectively).
Fig. 6.
Red and blue light influence the numbers of chloroplasts in the roots. The 14-3-3 T-DNA insertion mutants, Res-mu, co, phyB, and the wild type were grown on vertical agar plates for 5 d under constant red or blue light. Roots were illuminated with a blue light and visualized through a GFP long pass filter with a fluorescent microscope (A, C). All of the lines developed increased numbers of chloroplasts when grown under blue light compared with white light, but 14-3-3μ-1 roots contained higher numbers of chloroplasts than the other lines (A). When grown under red light, the roots of 14-3-3μ-1 and co were the only lines that developed high enough numbers of chloroplasts to be visualized with a fluorescent microscope (C). Pictures of roots from each line were cropped to equal sizes and the red tonal levels were measured with the RGB Measure Plus plug-in of the ImageJ program (B, D). When grown under blue light, the red tonal levels were higher for only 14-3-3μ-1 roots compared with the wild type (B). When grown under red light, the red tonal levels were equal for all lines measured (D). Although chloroplasts could only be visualized for 14-3-3μ-1 and co roots, the differences in tonal levels were not enough to be detected by the RGB Measure Plus program. Error bars represent standard error and asterices indicate lines with a significant difference in root length compared with the wild type using a t test (B, D).
Out of the lines grown under red light, 14-3-3μ-1 and co were the only sets that developed noticeably visible chloroplasts in the roots (Fig. 6C). The small number of chloroplasts in the 14-3-3μ-1 and co roots was not enough to raise the red tonal level of either mutant to a significant difference from that of wild-type roots (Fig. 6D).
Discussion
This study characterizes several novel functions of 14-3-3 proteins in root growth and chloroplast development. The 14-3-3 μ T-DNA insertion mutants exhibited altered root lengths and chloroplast numbers in the roots. The phenotypes exhibited by the 14-3-3 μ T-DNA insertion mutants could all be influenced by growth conditions altered in light intensity or wavelength. The 14-3-3 υ T-DNA insertion mutants exhibited altered root lengths in red light and also had altered numbers of chloroplasts in the roots. Most of the root development phenotypes exhibited by these 14-3-3 T-DNA insertion mutants can be attributed to deficiencies in the photosensory system.
The roots of 14-3-3μ-1 were only ∼25% as long as the roots of the wild type after 8 d of growth under constant light (Fig. 1B). However, the roots of 14-3-3υ-1 and 14-3-3υ-2 are the same length as the wild type after 8 d under constant light. The 14-3-3 μ protein is therefore an important factor in root growth, while 14-3-3 υ does not have an observable effect on root length under constant white light.
The differences in root length in 14-3-3μ-1 are influenced by light intensity (Fig. 2). The root length of 14-3-3μ-1 was more comparable to wild-type roots when grown in the dark, compared with plants grown in full-light conditions (Fig. 2A). Under low-light conditions, the 14-3-3μ-1 root length percentage compared with the wild-type root length was intermediate between the root length percentages under full-light and dark conditions (Fig. 2B, C).
Along with reduced hypocotyl growth inhibition under narrow-bandwidth red light, phytochromes are implicated in root elongation and gravitropic response (Correll et al., 2003; Correll and Kiss, 2005). The PHYB mutant, phyB, exhibits significantly reduced root elongation rates compared with the wild type when grown in light, but does not exhibit this same reduced root elongation rate when grown in the dark (Correll and Kiss, 2005), which was also observed in this study (Figs 1, 2). The roots of 14-3-3μ-1 were not the same length as wild-type roots in dark grown conditions. Because the roots of 14-3-3μ-1 were still shorter than wild-type roots in dark conditions, the difference in 14-3-3μ-1 root length cannot be solely attributed to deficiencies in light signalling. Future studies will identify additional light-influenced and other developmental differences in 14-3-3μ-1 roots.
Further phenotypic similarity between the 14-3-3 T-DNA insertion mutants and phyB was noticeable under narrow-bandwidth red light conditions (Fig. 5). The roots of phyB grown under red light grow similarly to when they are grown in dark conditions (Correll and Kiss, 2005) as also seen in Figs 2 and 5. This similarity in root lengths under red light and dark conditions was also true for the 14-3-3 T-DNA insertion mutants. The root lengths of 14-3-3μ-1 were ∼8 mm after 8 d of growth in both red light and dark conditions. The root lengths of 14-3-3υ-1 and 14-3-3υ-2 were ∼10 mm after 8 d of growth in both red light and dark conditions. Further similarity to phyB, the root lengths of 14-3-3υ-1 and 14-3-3υ-2 were comparable with the wild type when grown in dark conditions, but shorter when grown under narrow-bandwidth red light.
The 14-3-3 T-DNA insertion mutants also exhibit reduced hypocotyl elongation inhibition under red light in the first 4 d of growth (Mayfield et al., 2007). 14-3-3υ-1 and 14-3-3υ-2 grown under narrow bandwidth red light also exhibit a more vertical directional growth habit than the wild type (Mayfield et al., 2007), similar to phyB (Liscum and Hangarter, 1993; Poppe et al., 1996; Robson and Smith, 1996).
Under blue light, 14-3-3υ-1 and 14-3-3υ-2 root lengths were comparable with wild-type, but 14-3-3μ-1 still exhibited shorter roots (Fig. 5). Wild-type root lengths were ∼36% longer when grown under blue light compared with dark conditions. 14-3-3μ-1 root lengths were ∼17% longer when grown under blue light in comparison to dark conditions. The difference in 14-3-3μ-1 root length when grown in the dark compared with blue light was less distinct than comparing the difference in root length of light- and dark-grown 14-3-3μ-1, indicating that 14-3-3μ-1 is slightly deficient in blue light signalling, a phenotype that is similar, but not as strong as in cry1 mutants (Canamero et al., 2006).
Hypocotyl elongation inhibition is similar for 14-3-3μ-1 and wild-type plants grown under narrow bandwidth blue light (Mayfield et al., 2007). Although 14-3-3 μ was not found to be associated with blue light signalling in the hypocotyl during early development, these root length data indicate that 14-3-3 μ is associated with blue light signalling in the roots during early development. The association of 14-3-3 proteins with blue light has also been seen in broad bean, where the blue light photoreceptor, PHOT1, was identified as a client protein of 14-3-3s (Kinoshita et al., 2003).
The roots of 14-3-3 T-DNA insertion mutants and co exhibit increased chloroplast numbers, or greening, after 5 d of growth on vertical agar plates, while phyB roots exhibit decreased chloroplast numbers (Fig. 3). The development of chloroplasts in the roots can be influenced by both light intensity and different wavelengths of light. Dark-grown plants do not develop enough chloroplasts for detectable levels of root greening, while higher numbers of chloroplasts are developed in the roots of plants grown under blue light than plants grown under red light (Usami et al., 2004). Low-light-grown plants of all lines studied also did not develop enough chloroplasts for visualization in the roots, indicating that a threshold level of light is required for chloroplast development in the roots.
Root greening under red light conditions is primarily induced by PHYB signalling, whereas, under blue light conditions, root greening is mainly induced by CRY1 and CRY2 signalling (Usami et al., 2004). However, phyAphyB mutants are deficient in blue-light-induced chloroplast development, indicating that phytochromes are also important for blue light induced chloroplast development in the roots (Usami et al., 2004). Under blue light, 14-3-3μ-1 exhibited a quantifiable increase in root greening (Fig. 6B), indicating that 14-3-3 μ regulation of chloroplast development in the roots is partly associated with blue light signalling. Differences seen in both root length and greening under blue light will be evaluated in future studies, especially in the potential for 14-3-3 association with CRY1 and its role in root length and greening (Usami et al., 2004; Canamero et al., 2006).
The increased chloroplast numbers in the roots of the 14-3-3 T-DNA insertion mutants and co indicated that 14-3-3 proteins and CO are negative regulators of chloroplast development in the roots. No quantifiable difference in root greening was detected in the roots of any of the lines grown under red light (Fig. 6D), although 14-3-3μ-1 and co were the only lines to develop high enough numbers of chloroplasts to be visualized with a fluorescent microscope (Fig. 6C). Additional work is necessary to further characterize 14-3-3 μ or CO as co-regulators of chloroplast development in the roots via red-light signalling. In fact, this is the first report implicating CO with root development, however, CONSTANS-LIKE3 (COL3) is associated with root development (Datta et al., 2006).
Although PHYB has not been identified as a protein binding partner of 14-3-3 proteins, phytochromes are regulated by phosphorylation, and the association of PHYB with several phosphorylation events presents many situations of possible 14-3-3 protein involvement (Kim et al., 2004). In addition, PHYB and PHYA induce phosphorylation of such proteins as PKS1 and PIF3 (Fankhauser et al., 1999; Al-Sady et al., 2006).
This study presents novel roles of 14-3-3 proteins in root and chloroplast development, as well as new roles of CO in chloroplast development. Transcription levels of CO are increased in early development upon irradiance of 4-d-old dark-grown seedlings with a far-red light treatment (Tepperman et al., 2001), but a function for CO in early development has not been defined. In fact, the increase in chloroplast development in roots is the first reported phenotype for co in early development. The activity of CO in chloroplast development may only be as a component of the activity of CO and the COL proteins.
The present reverse-genetic study establishes novel, isoform-specific roles of 14-3-3 proteins in Arabidopsis root and chloroplast development. 14-3-3 proteins are increasingly recognized as a major node in the plant interactome, an interaction node that includes CO and other signalling molecules related to light responses (AIMC, 2011). Further, the roles of 14-3-3 proteins in root and chloroplast development are controlled by light intensity or wavelength. 14-3-3 TDNA insertion mutants also exhibit reduced hypocotyl elongation and time to flowering, compared with the wild type (Mayfield et al., 2007). Both the hypocotyl elongation and flowering timing phenotypes are, in part, due to 14-3-3 binding/influence of CO (Mayfield et al., 2007). The study presented here also characterizes CO as a potential factor in root chloroplast development of Arabidopsis during early development. Further work will be necessary to understand the mechanism of 14-3-3 control of both root and chloroplast development and how the association of 14-3-3 proteins and CO factors into these processes.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Overactivity of nitrate reductase can not be attributed to the shorter root lengths of 14-3-3μμ-1.
Acknowledgments
We would like to thank Drs Kevin Folta and Amit Dhingra for their time and intellectual input. We would also like to thank Dr Kevin Folta for the use of his light sources and other various pieces of equipment. We would like to thank members of the laboratory of Dr Robert Ferl that assisted with this project. The authors recognize support from NASA NNX07AH270 and NSF MCB-0114501.
References
- AIMC. Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333:601–607. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sady B, Ni W, Kircher S, Schafer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Molecular Cell. 2006;23:439–446. doi: 10.1016/j.molcel.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Bachmann M, Huber JL, Athwal GS, Wu K, Ferl RJ, Huber SC. 14-3-3 proteins associate with the regulatory phosphorylation site of spinach leaf nitrate reductase in an isoform-specific manner and reduce dephosphorylation of Ser-543 by endogenous protein phosphatases. FEBS Letters. 1996;398:26–30. doi: 10.1016/s0014-5793(96)01188-x. [DOI] [PubMed] [Google Scholar]
- Canamero RC, Bakrim N, Bouly JP, Garay A, Dudkin EE, Habricot Y, Ahmad M. Cryptochrome photoreceptors cry1 and cry2 antagonistically regulate primary root elongation in Arabidopsis thaliana. Planta. 2006;224:995–1003. doi: 10.1007/s00425-006-0280-6. [DOI] [PubMed] [Google Scholar]
- Correll MJ, Coveney KM, Raines SV, Mullen JL, Hangarter RP, Kiss JZ. Phytochromes play a role in phototropism and gravitropism in Arabidopsis roots. Advances in Space Research. 2003;31:2203–2210. doi: 10.1016/s0273-1177(03)00245-x. [DOI] [PubMed] [Google Scholar]
- Correll MJ, Kiss JZ. The roles of phytochromes in elongation and gravitropism of roots. Plant and Cell Physiology. 2005;46:317–323. doi: 10.1093/pcp/pci038. [DOI] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi GH, Deng XW, Holm M. Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. The Plant Cell. 2006;18:70–84. doi: 10.1105/tpc.105.038182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Simone S, Oka Y, Inoue Y. Effect of light on root hair formation in Arabidopsis thaliana phytochrome-deficient mutants. Journal of Plant Research. 2000a;113:63–69. [Google Scholar]
- De Simone S, Oka Y, Nishioka N, Tadano S, Inoue Y. Evidence of phytochrome mediation in the low-pH-induced root hair formation process in lettuce (Lactuca sativa L. cv. Grand Rapids) seedlings. Journal of Plant Research. 2000b;113:45–53. [Google Scholar]
- Fankhauser C, Yeh KC, Lagarias JC, Zhang H, Elich TD, Chory J. PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science. 1999;284:1539–1541. doi: 10.1126/science.284.5419.1539. [DOI] [PubMed] [Google Scholar]
- Ferl RJ. 14-3-3 Proteins and signal transduction. Annual Review of Plant Physiology and Plant Molecular Biology. 1996;47:49–73. doi: 10.1146/annurev.arplant.47.1.49. [DOI] [PubMed] [Google Scholar]
- Folta KM, Koss LL, McMorrow R, Kim H-H, Kenitz JD, Wheeler R, Sager JC. Design and fabrication of adjustable red-green-blue LED light arrays for plant research. BMC Plant Biology. 2005;5:17. doi: 10.1186/1471-2229-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang AT, Visconti S, Drumm K, Jahn T, Stensballe A, Mattei B, Jensen ON, Aducci P, Palmgren MG. Binding of 14-3-3 protein to the plasma membrane H+-ATPase AHA2 involves the three C-terminal residues Tyr(946)-Thr-Val and requires phosphorylation of Thr(947) Journal of Biological Chemistry. 1999;274:36774–36780. doi: 10.1074/jbc.274.51.36774. [DOI] [PubMed] [Google Scholar]
- Huq E, Tepperman JM, Quail PH. GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2000;97:9789–9794. doi: 10.1073/pnas.170283997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Shen Y, Han YJ, Park JE, Kirchenbauer D, Soh MS, Nagy F, Schafer E, Song PS. Phytochrome phosphorylation modulates light signaling by influencing the protein–protein interaction. The Plant Cell. 2004;16:2629–2640. doi: 10.1105/tpc.104.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Emi T, Tominaga M, Sakamoto K, Shigenaga A, Doi M, Shimazaki K. Blue-light- and phosphorylation-dependent binding of a 14-3-3 protein to phototropins in stomatal guard cells of broad bean. Plant Physiology. 2003;133:1453–1463. doi: 10.1104/pp.103.029629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP. Genetic control of light inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Zeitschrift für Pflanzenphysiologie. 1980;100:147–160. [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Molecular and General Genetics. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- Liscum E, Hangarter RP. Genetic evidence that the red-absorbing form of phytochrome B modulates gravitropism in. Arabidopsis thaliana. Plant Physiology. 1993;103:15–19. doi: 10.1104/pp.103.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YT, Hidaka H, Feldman LJ. Characterization of a calcium/calmodulin-dependent protein kinase homolog from maize roots showing light-regulated gravitropism. Planta. 1996;199:18–24. doi: 10.1007/BF00196876. [DOI] [PubMed] [Google Scholar]
- Mayfield JD, Folta KM, Paul AL, Ferl RJ. The 14-3-3 proteins mu and upsilon influence transition to flowering and early phytochrome response. Plant Physiology. 2007;145:1692–1702. doi: 10.1104/pp.107.108654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead G, Douglas P, Morrice N, Scarabel M, Aitken A, MacKintosh C. Phosphorylated nitrate reductase from spinach leaves is inhibited by 14-3-3 proteins and activated by fusicoccin. Current Biology. 1996;6:1104–1113. doi: 10.1016/s0960-9822(02)70677-5. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Neff MM, Chory J. Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiology. 1998;118:27–35. doi: 10.1104/pp.118.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A, Svennelid F, Ek B, Sommarin M, Larsson C. A phosphothreonine residue at the C-terminal end of the plasma membrane H+-ATPase is protected by fusicoccin-induced 14-3-3 binding. Plant Physiology. 1998;118:551–555. doi: 10.1104/pp.118.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottmann C, Marco S, Jaspert N, et al. Structure of a 14-3-3 coordinated hexamer of the plant plasma membrane H+-ATPase by combining X-ray crystallography and electron cryomicroscopy. Molecular Cell. 2007;25:427–440. doi: 10.1016/j.molcel.2006.12.017. [DOI] [PubMed] [Google Scholar]
- Paul AL, Daugherty CJ, Bihn EA, Chapman DK, Norwood KL, Ferl RJ. Transgene expression patterns indicate that spaceflight affects stress signal perception and transduction in arabidopsis. Plant Physiology. 2001;126:613–621. doi: 10.1104/pp.126.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L, Gutfinger T, Hareven D, Ben-Naim O, Ron N, Adir N, Lifschitz E. Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. The Plant Cell. 2001;13:2687–2702. doi: 10.1105/tpc.010293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppe C, Hangarter RP, Sharrock RA, Nagy F, Schafer E. The light-induced reduction of the gravitropic growth-orientation of seedlings of Arabidopsis thaliana (L.) Heynh. is a photomorphogenic response mediated synergistically by the far-red-absorbing forms of phytochromes A and B. Planta. 1996;199:511–514. doi: 10.1007/BF00195180. [DOI] [PubMed] [Google Scholar]
- Pratt LH, Coleman RA. Phytochrome distribution in etiolated grass seedlings as assayed by an indirect antibody-labelling method. American Journal of Botany. 1974;61:195–202. [Google Scholar]
- Purwestri YA, Ogaki Y, Tamaki S, Tsuji H, Shimamoto K. The 14-3-3 protein GF14c acts as a negative regulator of flowering in rice by interacting with the florigen Hd3a. Plant and Cell Physiology. 2009;50:429–438. doi: 10.1093/pcp/pcp012. [DOI] [PubMed] [Google Scholar]
- Redei GP. Supervital mutants of Arabidopsis. Genetics. 1962;47:443–460. doi: 10.1093/genetics/47.4.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson PR, Smith H. Genetic and transgenic evidence that phytochromes A and B act to modulate the gravitropic orientation of Arabidopsis thaliana hypocotyls. Plant Physiology. 1996;110:211–216. doi: 10.1104/pp.110.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenquist M, Alsterfjord M, Larsson C, Sommarin M. Data mining the Arabidopsis genome reveals fifteen 14-3-3 genes. Expression is demonstrated for two out of five novel genes. Plant Physiology. 2001;127:142–149. doi: 10.1104/pp.127.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnke PC, Laughner B, Cardasis H, Powell D, Ferl RJ. Exposed loop domains of complexed 14-3-3 proteins contribute to structural diversity and functional specificity. Plant Physiology. 2006;140:647–660. doi: 10.1104/pp.105.073916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Quail PH. Temporal and spatial expression patterns of PHYA and PHYB genes in Arabidopsis. The Plant Journal. 1995;7:413–427. doi: 10.1046/j.1365-313x.1995.7030413.x. [DOI] [PubMed] [Google Scholar]
- Su TT, Parry DH, Donahoe B, Chien CT, O'Farrell PH, Purdy A. Cell cycle roles for two 14-3-3 proteins during Drosophila development. Journal of Cell Science. 2001;114:3445–3454. doi: 10.1242/jcs.114.19.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama A, Miyagi Y, Komiya Y, Kurabe N, Kitanaka C, Kato N, Nagashima Y, Kuchino Y, Tashiro F. Forced expression of antisense 14-3-3beta RNA suppresses tumor cell growth in vitro and. in vivo. Carcinogenesis. 2003;24:1549–1559. doi: 10.1093/carcin/bgg113. [DOI] [PubMed] [Google Scholar]
- Svennelid F, Olsson A, Piotrowski M, Rosenquist M, Ottman C, Larsson C, Oecking C, Sommarin M. Phosphorylation of Thr-948 at the C terminus of the plasma membrane H+-ATPase creates a binding site for the regulatory 14-3-3 protein. The Plant Cell. 1999;11:2379–2391. doi: 10.1105/tpc.11.12.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano M, Kanegae H, Shinomura T, Miyao A, Hirochika H, Furuya M. Isolation and characterization of rice phytochrome A mutants. The Plant Cell. 2001;13:521–534. doi: 10.1105/tpc.13.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH. Multiple transcription-factor genes are early targets of phytochrome A signaling. Proceedings of the National Academy of Sciences, USA. 2001;98:9437–9442. doi: 10.1073/pnas.161300998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toroser D, Athwal GS, Huber SC. Site-specific regulatory interaction between spinach leaf sucrose-phosphate synthase and 14-3-3 proteins. FEBS Letters. 1998;435:110–114. doi: 10.1016/s0014-5793(98)01048-5. [DOI] [PubMed] [Google Scholar]
- Toth R, Kevei E, Hall A, Millar AJ, Nagy F, Kozma-Bognar L. Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiology. 2001;127:1607–1616. doi: 10.1104/pp.010467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami T, Mochizuki N, Kondo M, Nishimura M, Nagatani A. Cryptochromes and phytochromes synergistically regulate Arabidopsis root greening under blue light. Plant and Cell Physiology. 2004;45:1798–1808. doi: 10.1093/pcp/pch205. [DOI] [PubMed] [Google Scholar]
- Whatley JM. The ultrastructure of plastids in roots. International Review of Cytology. 1983;85:175–220. [Google Scholar]
- Wu K, Lu G, Sehnke P, Ferl RJ. The heterologous interactions among plant 14-3-3 proteins and identification of regions that are important for dimerization. Archives of Biochemistry and Biophysics. 1997;339:2–8. doi: 10.1006/abbi.1996.9841. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.