Abstract
The nuclear hormone receptor peroxisome proliferator activated receptor gamma (PPARγ) is a ligand-activated transcription factor that specifies formation of the adipocyte lineage. PPARγ also serves as a primary target for the treatment of type 2 diabetes, illustrating both its medical relevance as well as the need to understand fundamental aspects of PPARγ expression and function. Here, we characterize molecular changes that occur at the PPARγ2 promoter within the first several hours of adipocyte differentiation in culture. Our results demonstrate that changes in chromatin accessibility at the PPARγ2 promoter and occupancy of the promoter by the c-Fos transcription factor occur within an hour of the onset of differentiation, followed closely by the binding of the CCAAT/Enhancer Binding Protein beta (C/EBPβ) transcription factor. All three events show a remarkable dependency on protein kinase A (PKA) activity. These results reflect novel requirements for the PKA signaling pathway and reinforce the importance of PKA function during the onset of adipocyte differentiation.
Keywords: adipogenesis, PPARγ, Protein Kinase A
INTRODUCTION
White adipose tissue is critical for energy storage and for function as an endocrine organ that secretes hormones that are essential for energy homeostasis. The major components of adipose tissue are adipocytes and their precursors, preadipocytes. While the understanding of adipogenic lineage determination is limited, the regulatory mechanisms involved in terminal differentiation have been examined in detail (see Farmer, 2006; Lefterova and Lazar, 2009; Rosen and MacDougald, 2006 for review). These pathways result in the activation of the master regulators peroxisome proliferator-activated receptor gamma 2 (PPARγ2) and CCAAT/enhancer binding protein alpha (C/EBPα), the induction of adipose-derived hormones, and the massive accumulation of intracellular lipid.
The 3T3-L1 preadipocyte cell line is a widely used in vitro model for the study of adipogenesis (Green and Kehinde, 1975; Smyth et al., 1993). Terminal differentiation of 3T3-L1 preadipocytes is stimulated by treatment with glucocortoids, fetal calf serum (FCS), and insulin. The differentiation process also requires the addition of cyclic adenosine monophophate (cAMP) elevators such as 3-isobutyl-1-methylxanthine (IBMX), a phosphodiesterase inhibitor, or forskolin, a potent activator of adenylyl cyclase (Russell and Ho, 1976). Increased cAMP levels have been shown to be associated with immediate events of differentiation (Cao et al., 1991; Yeh et al., 1995), including the phosphorylation of the cAMP-responsive element binding protein (CREB) and the induction of C/EBPβ expression within 2 hours through CREB-dependent binding (Zhang et al., 2004). The stimulation of CREB is classically mediated through its phosphorylation at serine 133 by the cAMP-dependent protein kinase A (PKA) (Gonzalez and Montminy, 1989) In mammalian cells, PKA is composed of two regulatory (R) subunits and two catalytic (C) subunits, which form a tetrameric R2C2 holoenzyme (Cheng et al., 2008). There are two major C subunit isoforms known as Cα and Cβ (Sugiyama et al., 1997). While the exact functions of PKA during adipogenesis have been recently debated (Cheng et al., 2008; Li et al., 2008; Petersen et al., 2008), it is nonetheless clear that cAMP signaling is involved in adipogenic differentiation.
The changes in gene expression during adipogenesis require epigenetic modifications in the chromatin structure at the regulatory sequences controlling the expression of both transactivators and adipocyte-specific gene products (reviewed in de la Serna et al., 2006; Musri et al., 2007). Two separate classes of proteins, histone modifying enzymes and ATP-dependent chromatin remodeling enzymes modulate chromatin alterations. Histone modifying enzymes posttranslationally and reversibly modify specific amino acids, predominantly in the N and C-terminal ends of the histone residues (reviewed in Berger, 2007). ATP-dependent chromatin remodelers disrupt chromatin structure by perturbing the histone/DNA interactions, which can result in a change in the position of the histone octamer on DNA and changes in the sensitivity of chromatin to nuclease cleavage (reviewed in Clapier and Cairns, 2009; Racki and Narlikar, 2008; Saha et al., 2006). Limited information is known about the changes in chromatin accessibility during the onset of adipogenic gene expression. The first connection was provided by Pedersen et al. (Pedersen et al., 2001), who demonstrated that the ability of C/EBPα to interact with one of the ATPases specific to the SWI/SNF class of ATP-dependent chromatin remodeling enzymes was tightly linked to its ability to turn on adipose-specific genes. This work indicated the importance of the SWI/SNF enzymes in inducing adipogenic gene expression. Others have demonstrated that additional SWI/SNF subunits can interact with adipogenic regulators and contribute to adipocyte-specific gene expression (Caramel et al., 2008; Debril et al., 2004). Previous work from our group demonstrated that the SWI/SNF remodeling enzyme was required for the expression of PPARγ2 during adipogenesis because it promoted the formation and stability of the pol II pre-initiation complex (Salma et al., 2004), again demonstrating that SWI/SNF enzymes were required for adipogenic gene expression. We also showed that the PPARγ2 promoter undergoes changes in chromatin accessibility within 24 hours of the onset of differentiation, which indicates that the gross structural changes precede the expression of PPARγ2 and the rest of the adipogenic gene expression program (Salma et al., 2004). In this study, we show that chromatin accessibility at the PPARγ2 promoter occurs within 1 to 2 hours of the onset of differentiation and coincides with the binding of the transcription factors C/EBPβ and c-Fos. Additionally, using specific PKA inhibitors and knockdown of PKA catalytic subunits, we demonstrate that chromatin accessibility and factor binding at the PPARγ2 promoter are diminished, indicating a cAMP/PKA-dependent role for these early events in adipogenesis.
MATERIALS AND METHODS
Cells and reagents
The following cell lines were purchased from ATCC: 3T3-L1, NIH3T3, Raw264.7, Neuro2a. The retrovirus packaging cell line BOSC23 was described (Pear et al., 1993). LY249002, U0126, SB203580, KT5720, KN93, H89, and forskolin were purchased from Calbiochem. Insulin, dexamethasone, IBMX, dibutyryl-cyclic AMP (db-cAMP), puromycin and polybrene were purchased from Sigma. Restriction enzymes were purchased from New England Biolabs. DNase I was purchased from Promega.
Plasmids
A retroviral sh-RNA vector (pSuper-retro-puro) was purchased from OligoEngine (Seattle, WA). Derivatives targeting specific regulatory factors were created by inserting a double stranded oligonucleoties containing a unique 19-nt sequence derived from the mRNA transcript of the gene targeted for suppression as described in the pSuper manual. The pSuper control was made by inserting an oligonucleotide containing a 19-nt nonspecific genomic sequence. The N-19 target sequences used were:
pSuper-c-Fos: 5'-GCG GAG ACA GAT CAA CTT G-3'
pSuper-CEBPβ: 5'-GAC TTC CTC TCC GAC CTC T-3'
pSuper-PKACα: 5'-GTG GTT TGC CAC GAC TGA C-3'
pSuper-PKACβ: 5'-GAG TTT CTA GCC AAA GCC A-3'
3T3-L1 cell differentiation
Cells were cultured in DMEM containing 10%calf serum. For differentiation, two day post-confluent cells were treated with differentiation medium consisting of DMEM containing 10% fetal calf serum, 0.5 mM methylisobutylxanthine (IBMX), 1 μM dexamethasone, and 5 μg /ml insulin for 48 h. The cells were subsequently maintained in DMEM plus 10% fetal calf serum with 5 μg /ml insulin and the medium was replaced every 2 days for the length of the experiment. In some cases, individual components or specified chemicals, instead of the complete component mixture, were added into the differentiation media. In experiments involving chemical inhibitor treatments, the indicated compounds were added one hour prior to the differentiation medium.
Retroviral infection of 3T3-L1 cells
BOSC23 packaging cells were cultured in 100-mm-diameter dishes and transfected at 80% confluence via the FUGENE (Roche) method with 10 μg of the indicated pSUPER plasmid as described previously (Pear et al., 1993). Viral supernatants were harvested 48 h after transfection. 3T3-L1 cells at 50% confluence were infected with virus in Dulbecco's modified Eagle's medium (DMEM) containing 10% calf serum and 4 μg/ml polybrene. 36 hours later, the plates were still subconfluent and were passaged into DMEM plus 10% calf serum containing 2 μg/ml puromycin to select for infected cells. Cells were maintained in a subconfluent state and passaged no more than two times prior to differentiation.
DNase I and restriction enzyme accessibility experiments
The protocols for these assays and for their detection by Southern blot were described (de la Serna et al., 2001; Salma et al., 2004). Detection of accessibility by ligation-mediated PCR was performed as described (de la Serna et al., 2005) except that the PCR primer to detect cleavage at the proximal StuI site in the PPARγ2 promoter was 5'-GTAATGTACCAAGTCTTGCCAAAGCA-3' spanning nucleotides −274 to −251 relative to the PPARγ2 transcription start site. Amplification of input DNA served as a control.
mRNA analysis, Western Blots, Oil Red O staining, and chromatin immunoprecipitation (ChIP)
Conditions for reverse transcription (RT)-PCR for PPARγ2 and Hprt were described (Salma et al., 2004). The following primers were used: PPARγ2 5'-ATG GGT GAA ACT CTG GGA GAT TCT C -3' and 5'-CTT GGA GCT TCA GGT CAT ATT TGT A-3'; Hprt 5'-GTT CTT TGC TGA CCT GCT GGA T-3' and 5'-ATG GCC ACA GGA CTA GAA CAC CTG C-3'.
Preparation of whole cell extract and western blotting was described (de la Serna et al., 2000). 40 μg of whole cell lysate was used for each sample. Antibodies utilized included C/EBPβ (Santa Cruz, sc-7962), c-Fos (sc-7202), PKACα (sc-903) and PKACβ (sc-904), and phosphatidylinositol 3-kinase (06–496; Upstate).
ChIP experiments were performed as described (Salma et al., 2004) except that cross-linking was performed at room temperature for 10 minutes. The primers used amplified the PPARγ2 promoter between −413 and −247 relative to the start site of transcription. Primers for the negative sequence control spanned β-actin sequences from −31 to +135 relative to the start site of transcription. Primer sequences were previously published (Salma et al., 2004). Oil Red O staining was performed as described (Salma et al., 2004).
RESULTS
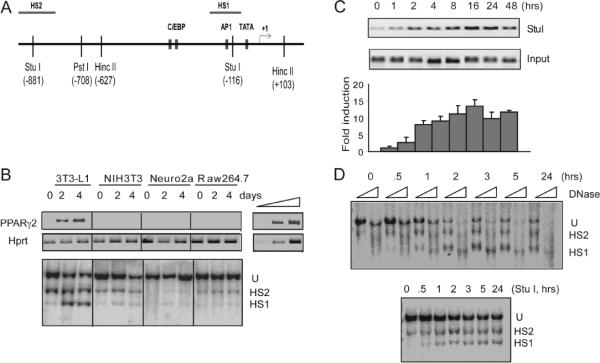
We previously used a DNase I hypersensitivity assay and restriction enzyme accessibility assays to monitor chromatin accessibility changes at the PPARγ2 promoter in differentiating 3T3-L1 cells. This work demonstrated that at the onset of differentiation, there was one constitutively accessible site between the transcription start site (TSS) and 1 kb upstream of the TSS. In addition, there were several sites that were largely inaccessible to nuclease cleavage but became sensitive to cleavage by 24 hours post-differentiation and remained accessible through the end of the time course, which was 6 days post-differentiation (Salma et al., 2004). A schematic diagram of the PPARγ2 promoter is shown in Figure 1A. The specificity of chromatin accessibility at the PPARγ2 locus was confirmed by comparison of accessibility in differentiating 3T3-L1 cells to accessibility in NIH3T3 fibroblasts, Neuro2a neuroblastoma cells, and Raw264.7 macrophage like cells. Differentiating 3T3-L1 cells expressed PPARγ2 whereas each of the other cell lines did not (Fig. 1B top panel, day 0), even when the cells were subjected to the same differentiation protocol as the 3T3-L1 cells (Fig. 1B top panel, days 2 and 4). A Southern blot of Stu I cleaved genomic chromatin showed that cleavage occurred at the HS2 site in both undifferentiated 3T3-L1 cells (day 0) and at 2 and 4 days post-differentiation. In contrast, Stu I cleavage at the HS1 site in 3T3-L1 cells was detectable at day 0 but significantly induced at 2 and 4 days post-differentiation (Fig 1B, lower panel). Cleavage at the HS sites in the other tested cell lines was minimal at each time point tested (Fig 1B, lower panel), indicating that chromatin accessibility at the HS1 site was tissue specific and correlated with PPARγ2 expression.
Figure 1.
Chromatin remodeling at the PPARγ2 promoter occurs shortly after the onset of 3T3-L1 cell differentiation. (A) Schematic drawing of the PPARγ2 promoter. The positions of relevant restriction enzyme sites and DNase I hypersensitive sites that are in the proximal promote regions (HS1) or distal promoter region (HS2) are indicated. The positions of the Southern blot probe and selected transcription factor binding sites are also shown. Sequence numbers are relative to the start site of transcription. (B) Upper panel- mRNA expression of PPARγ2 and Hprt monitored by RT-PCR. A titration of cDNA dilutions is presented to demonstrate the linearity of the reactions. Lower panel- Southern blot showing chromatin accessibility at HS1 and HS2 in different cell types grown to confluence (day 0) or differentiated for 2 or 4 days under standard 3T3-L1 differentiation conditions. (C) A time course of PPARγ2 promoter accessibility at the proximal Stu I (HS1) site in differentiating 3T3-L1 cells as measured by LM-PCR. A representative experiment is shown (upper panel) along with quantification (average +/− standard deviation) from three independent experiments (lower panel). hrs; hours. (D) A time course of PPARγ2 promoter accessibility to DNase I (upper panel; 1 or 5 units per sample) and to Stu I (lower panel) in differentiating 3T3-L1 cells as measured by Southern blot. U; uncleaved DNA hrs; hours.
We next performed a time course of chromatin accessibility in 3T3-L1 cells between 0 and 24 hours post-differentiation. A quantitative analysis of cleavage was performed using ligation mediated polymerase chain reaction (LM-PCR) to amplify cleaved genomic DNA cut at the Stu I site in the HS1 region (Fig. 1C). This experiment demonstrated that robust cleavage was observed 2 hours post-differentiation and that accessibility was slightly increased by 1 hour post-differentiation. Qualitative analysis of accessibility at HS1, as measured by both DNase I and by Stu I cleavage, showed that changes in chromatin structure in this region could be detected by 1 hour post-differentiation and may be occurring within 30 minutes of inducing differentiation (Fig. 1D). Collectively, these data indicate that a specific increase in chromatin accessibility began to occur at the PPARγ2 promoter within minutes of inducing differentiation in the 3T3-L1 cells. This change occurs long before the onset of any adipocyte-specific gene expression, suggesting that the alteration in chromatin structure is likely to be directly related to the induction of signaling pathways induced by the addition of the differentiation cocktail.
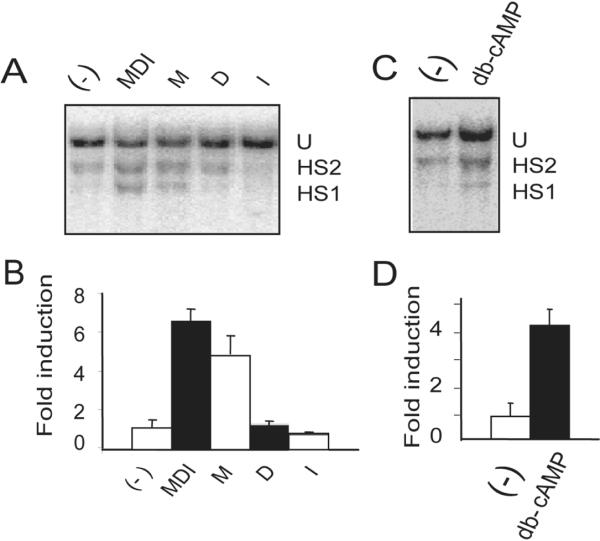
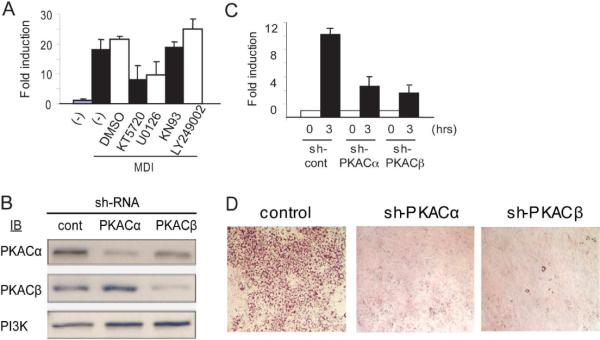
Differentiation of 3T3-L1 preadipocytes is induced after the cells have reached confluence by adding a cocktail of signaling molecules that includes insulin, dexamethasone, and IBMX. To ascertain whether one or more of the signaling pathways activated by this cocktail was required for the change in PPARγ2 promoter accessibility, 3T3-L1 cells were differentiated for 3 hours in media containing FCS alone, FCS containing complete cocktail, or FCS containing only one of the cocktail components. A Southern blot monitoring Stu I cleavage (Fig. 2A) and the quantified LM-PCR assay (Fig. 2B) demonstrate that the inducible accessibility at the HS1 site occurred in the presence of IBMX alone. IBMX increases intracellular cAMP levels and cAMP binding to the PKA regulatory subunits, which releases the activated catalytic subunits to phosphorylate target proteins (reviewed in Skalhegg and Tasken, 2000). To further demonstrate the dependence of chromatin accessibility on cAMP level, we repeated the 3T3-L1 differentiation in the presence of FCS with or without the addition of dibutyryl cAMP (db cAMP), a cell-permeable cAMP analog. Chromatin accessibility was again monitored by both Southern Blot (Fig. 2C) and by quantitative LM-PCR (Fig. 2D). The results show that addition of db-cAMP to FCS was sufficient to induce accessibility at the HS1 site in the PPARγ2 promoter. Further support for the dependence of PPARγ2 promoter chromatin accessibility on cAMP and PKA signaling was obtained through use of chemical inhibitors of various signaling pathways (Fig. 3A). Detection of restriction enzyme cleavage at HS1 by LM-PCR in 3T3 –L1 cells that were pretreated for 1 hour with the inhibitor prior to the addition of complete differentiation cocktail revealed that addition of LY249002, a PI3Kinase inhibitor, and KN93, a calcium calmodulin kinase II inhibitor, had no effect on accessibility. In contrast, KT5720, a PKA inhibitor, decreased accessibility by ~50%. We also tested U0126, a MAP kinase inhibitor. Inhibition of MAP kinase activity caused a decrease in chromatin accessibility similar to what was observed in the presence of the PKA inhibitor.
Figure 2.
Chromatin accessibility changes at the PPARγ2 promoter are dependent upon increased cAMP levels. 3T3-L1 cells were differentiated for 3 hours in the presence of DMEM containing 10% fetal calf serum plus insulin alone (I), dexamethasone alone (D), IBMX alone (M), all three (MDI) or no additional additives (−) and subsequently analyzed by (A) Southern blot or (B) LM-PCR for Stu I accessibility at the HS1 site on the PPARγ2 promoter. (C, D) 3T3-L1 cells were differentiated for 3 hours in the presence of DMEM containing 10% fetal calf serum (−) or DMEM containing 10% fetal calf serum plus dibutyryl-cAMP (db-cAMP) and assayed for Stu I accessibility changes at the HS1 site on the PPARγ2 promoter by Southern blot (C) or LM-PCR (D).
Figure 3.
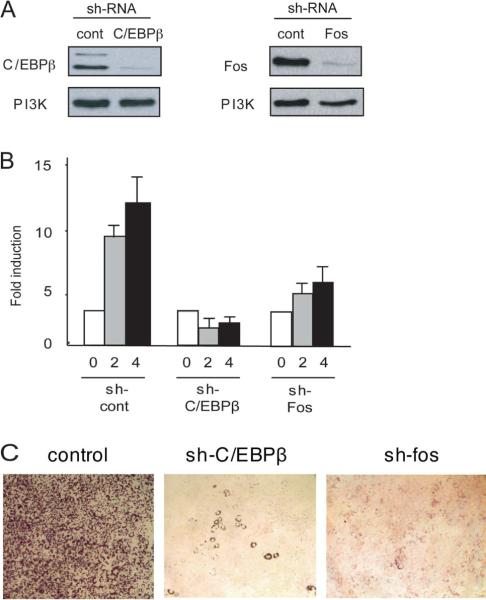
Chromatin accessibility changes at the PPARγ2 promoter are dependent upon the protein kinase A pathway. (A) 2 day post-confluent 3T3-L1 cells were untreated (−) or treated with the indicated compounds for 1 hour prior to the addition of complete differentiation media (MDI) for 3 hours, then assessed for Stu I accessibility at the HS1 region of the PPARγ2 promoter using the LM-PCR assay. (B, C) 3T3-L1 cells were infected with retroviruses expressing shRNA against the indicated protein as described in the Materials and Methods and subsequently differentiated for 3 hours with complete differentiation media. A western blot verifying knockdown of the indicated protein levels is shown in (B). PI3 Kinase levels were monitored as a loading control. cont; control. LM-PCR analysis of Stu I accessibility at the HS1 region of the PPARγ2 promoter is presented in (C). (D) Oil Red O staining of cells infected with the indicated retroviral shRNA vectors and differentiated in complete differentiation media for 6 days.
To directly address whether PKA was involved in generating chromatin accessibility at the PPARγ2 promoter, we infected cells with retroviral vectors encoding shRNAs against PKA catalytic subunits, Cα or Cβ, or encoding a non-specific shRNA control. Cells were collected 3 hours post-differentiation, and Cα and Cβ expression levels were assessed by western blot (Fig. 3B). shRNA against Cα specifically decreased Cα but not Cβ expression. Similarly, shRNA against Cβ reduced the level of Cβ but not Cα. Detection of PPARγ2 promoter cleavage at the Stu I site in HS1 by LM-PCR showed that knockdown of either PKAC subunit decreased accessibility by approximately 65% (Fig. 3C). The requirement for the pKA Cα and Cβ subunits for completion of differentiation was assessed by staining cells with Oil Red O 6 days after the induction of differentiation (Fig. 3D). The results indicate that knockdown of either catalytic subunit caused a significant inhibition of differentiation. Co infection of 3T3-L1 cells with retrovirus encoding shRNA against both Cα and Cβ subunits resulted in cell death. We conclude that PKA signaling contributes to the opening of chromatin structure at the PPARγ2 promoter shortly after the onset of differentiation and to efficient differentiation.
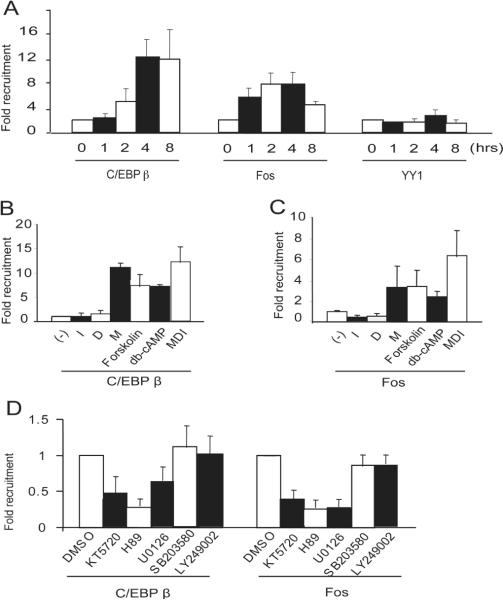
Control of PPARγ2 expression is dependent on a number of transcription factors. C/EBPβ is present in 3T3-L1 preadipocytes and is activated upon differentiation (Wu et al., 1996). We examined C/EBPβ binding to the PPARγ2 promoter by chromatin immunoprecipitation (ChIP) over the first 8 hours of differentiation (Fig. 4A-left). The results show that C/EBPβ binding can be detected 2 hours post-differentiation, with maximal binding achieved by 4 hours post-differentiation. Examination of the PPARγ2 promoter revealed that there is putative activator protein 1 (AP-1) binding site (Fig. 1A). The AP-1 transcription factor consists of a heterodimer of proteins belonging to the c-Fos, c-Jun, Jun dimerizing protein (JDP), and activating transcription factor (ATF) families. The contribution of c-Fos to the expression of specific adipogenic genes has long been recognized (Distel et al., 1987; Distel and Spiegelman, 1988; Spiegelman et al., 1988), however, it has not been linked to regulation of PPARγ2 gene expression during adipogenic differentiation. ChIP experiments determined that c-Fos bound to the promoter as early as 1 hour post-differentiation (Fig 4A, center), whereas an antibody to the transcription factor YY1, which is used here as a negative control, did not immunoprecipitate the PPARγ2 promoter (Fig. 4A, right).
Figure 4.
Binding of C/EBPβ and c-Fos to the PPARγ2 promoter is dependent upon the PKA signaling pathway. 3T3-L1 cells were differentiated for the indicated number of hours in the presence of complete differentiation media and harvested for ChIP assays. (A) Binding of C/EBPβ and c-Fos to the PPARγ2 promoter. (B, C) 3T3-L1 cells were differentiated for 3 hours in DMEM containing 10% fetal calf serum alone (−), or with insulin (I), dexamethasone (D), IBMX (M), forskalin, db-cAMP, or complete differentiation media (MDI) and harvested for ChIP assays to assess C/EBPβ binding (B) or c-Fos binding (C) to the PPARγ2 promoter. (D) 2 day post-confluent 3T3-L1 cells were treated with the indicated compounds for 1 hour prior to the addition of complete differentiation media for 3 hours, then harvested for ChIP assays to assess the binding of C/EBPβ (left) or c-Fos (right) to the PPARγ2 promoter.
We next assessed the relationship between transcription factor binding and PKA signaling. Both c-Fos and C/EBP binding in the presence of complete differentiation cocktail were compared to a cocktail containing FCS and IBMX, db-cAMP, or forskolin. Forskolin activates adenylyl cyclase and raises intracellular cAMP levels (Seamon et al., 1981). C/EBPβ binding in the presence of either db-cAMP or forskolin was nearly equivalent to the binding observed in the presence of IBMX alone or in the presence of the complete differentiation cocktail (Fig. 4B). c-Fos binding in the presence of IBMX, db-cAMP or forskolin was induced to approximately half the value induced by the complete differentiation cocktail (Fig. 4C). We then assessed transcription factor binding in the presence of chemical inhibitors of various signaling pathways. Neither C/EBPβ nor c-Fos binding was affected by LY249002, a PI3K inhibitor or by SB203580, a p38 inhibitor (Fig. 4D). In contrast, KT5720 and H89, both of which inhibit PKA signaling, decreased C/EBPβ and c-Fos binding to the PPARγ2 promoter, further supporting the idea that PKA signaling contributes to factor binding. We also observed that U0126, a MAP kinase inhibitor, partially inhibited both C/EBPβ and c-Fos binding (Fig. 4D). An important role for MAP kinase modification of C/EBPβ during the early stages of adipogenic differentiation is well-established (Tang et al., 2005). c-Fos is also known to be regulated by MAP kinases through serum response elements and the MAPK transcriptional target Elk1 (Smith et al., 2001). Collectively, the data indicate that the PKA signaling pathway contributes to the ability of C/EBPβ and c-Fos to bind to the PPARγ2 promoter shortly after the onset of differentiation.
The experiments presented above indicate that C/EBPβ binding, c-Fos binding, and chromatin accessibility changes at the PPARγ2 promoter are early events that occur shortly after the initiation of 3T3-L1 differentiation. To better understand the relationship between these events, we infected 3T3-L1 preadipocytes with retroviruses encoding shRNA against either C/EBPβ or c-Fos and examined the consequences on promoter chromatin accessibility. Western blot analysis confirmed the efficiency of the shRNA-mediated knockdown of C/EBPβ and c-Fos levels (Fig. 5A). Upon expression of the control shRNA, a significant increase in chromatin accessibility was observed by 2 hours post-differentiation (Fig. 5B). Knockdown of C/EBPβ completely inhibited the formation of a stable, nuclease-accessible promoter structure. Knockdown of c-Fos partially inhibited chromatin accessibility at the PPARγ2 promoter (Fig. 5B). The data suggest that continued chromatin accessibility is linked to C/EBPβ and c-Fos binding. Finally, we addressed the contribution of C/EBPβ and c-Fos to differentiation. 3T3-L1 cells were infected with the control shRNA retrovirus or the viruses expressing shRNA against C/EBPβ or c-Fos and stained with Oil Red O after 6 days of exposure to the differentiation protocol. As expected, differentiation was greatly reduced when C/EBPβ levels were reduced (Fig. 5C). Reduction of c-Fos levels also significantly inhibited Oil Red O staining (Fig. 5C), underscoring the importance of c-Fos in 3T3-L1 differentiation.
Figure 5.
C/EBPβ and c-Fos binding to the PPARγ2 promoter maintains chromatin accessibility. 3T3-L1 cells were infected with retroviral vectors expressing shRNA against C/EBPβ or c-Fos as described in the Materials and Methods and subsequently differentiated for the indicated number of hours with complete differentiation media. (A) A western blot verifying knockdown of the indicated protein levels at 4 hours post-differentiation. PI3 Kinase levels were monitored as a loading control. cont; control. (B) LM-PCR analysis of Stu I accessibility at the HS1 region of the PPARγ2 promoter at the indicated times in samples expressing the indicated shRNA. hrs; hours. (C) Oil Red O staining of cells infected with the indicated retroviral shRNA vectors and differentiated in complete differentiation media for 6 days.
DISCUSSION
Activation of the PPARγ2 gene is an important transcriptional event that occurs during early 3T3-L1 differentiation. PPARγ2 transcription is robust by 48 hours post-differentiation (Chawla et al., 1994), with transcription initiating between 24 to 48 hours post-differentiation (Chawla et al., 1994) at which time mitotic clonal expansion induced by the differentiation components is completed (Martini et al., 2009). We previously showed that chromatin accessibility changes at the PPARγ2 promoter could be detected 24 hours post-differentiation, suggesting that gross remodeling of the promoter chromatin preceded gene expression. We have since refined this work to demonstrate that accessibility at the PPARγ2 promoter occurs within 1 hour of the onset of differentiation and is maximally altered at 2 hours post-differentiation. Notably, this change in accessibility is also accompanied by recruitment of the C/EBPβ protein to the PPARγ2 promoters. These data indicate that the chromatin remodeling events at the PPARg2 promoter are closely linked to facilitated binding of C/EBPβ. Moreover, the data presented here and in our previous study (Salma et al., 2006) demonstrate that even though adipogenic transcription does not occur until after mitotic clonal expansion in 3T3-L1 cells, signaling to chromatin and to transcription factors such as C/EBPβ occurs with minimal delay following the onset of differentiation, as C/EBPβ binds to multiple adipogenic genes within 2 to 4 hours post-differentiation. Thus while mechanisms exist to restrict adipocyte specific transcription until the completion of mitotic clonal expansion, the rate limiting step is neither the opening of PPARγ2 chromatin structure nor the binding of C/EBPβ to binding sites in the PPARγ2 promoter.
We also observed rapid binding of c-Fos to the PPARγ2 promoter following the stimulation of 3T3-L1 differentiation. Analysis of putative transcription factor binding sites in the sequences upstream of the PPARγ2 mRNA start site revealed a possible interaction site for c-Fos, and a ChIP assay confirmed its presence on the promoter. c-Fos can bind to DNA as a heterodimer with other immediate early proteins such as c-Jun (Halazonetis et al., 1988; Kaminska et al., 2000; Porte et al., 1997). Preliminary attempts to ChIP c-Jun to the PPARγ2 promoter were not successful (data not shown). This could easily be due to the limitations of the antibody used to test for c-Jun binding or to the possibility that a different dimerization partner might be functioning with c-Fos at this site (Kaminska et al., 2000). The mechanisms by which c-Fos contributes to PPARγ2 activation and adipogenesis in general remain to be further detailed, but the overall importance of c-Fos for 3T3-L1 differentiation was clearly demonstrated by the observation that knockdown of c-Fos significantly inhibited differentiation. It has been long known that immediate early genes such as c-Fos, c-Myc, c-Jun, and Krox-20 are rapidly stimulated during adipogenesis (reviewed in Cornelius et al., 1994; Ntambi and Young-Cheul, 2000), but to date only roles for Krox-20 (Birsoy et al., 2008; Chen et al., 2005) and c-Myc (Heath et al., 2000a; Heath et al., 2000b) have been described. This study adds c-Fos to the list of immediate early proteins that play a functional role in adipocyte differentiation.
The activating functions of C/EBPβ during adipogenesis are well documented (reviewed in Darlington et al., 1998). Presumably, c-Fos also plays a role in stimulating PPARγ2 expression. However, the data presented here raise the possibility that both C/EBPβ and c-Fos also contribute to PPARγ2 activation via an additional mechanism. Targeted knockdown of c-Fos and C/EBPβ greatly reduced the PPARγ2 accessibility, indicating that both may be serving a novel function during the early stages of adipogenesis, which is to help maintain an open chromatin conformation at the PPARγ2 promoter. Both c-Fos and C/EBPβ are involved in the recruitment of p300/CBP to their target promoters (Preston et al., 2000; Schwartz et al., 2003). We propose that the presence of C/EBPβ and c-Fos at the PPARγ2 promoter facilitates chromatin modifications that ultimately lead to gene expression and that continued occupancy of the C/EBP and potentially the AP-1 site are necessary to maintain chromatin accessibility as differentiation progresses.
While it is apparent that the chromatin accessibility change that occurs at the PPARγ2 promoter is rapid, it remains unclear which chromatin remodeling enzyme or complex is mediating this function. Our previous data indicated that the Brg1 and Brm ATPases that are the enzymatic subunits of the SWI/SNF family of chromatin remodelers were not targeted to the PPARγ2 promoter until 24 to 48 hours into the adipogenic program (Salma et al., 2004). The presence of the remodeling enzymes correlated with the completion of a stable pol II transcription initiation complex in both 3T3-L1 cells and in NIH3T3 fibroblasts driven to adipogenic differentiation by ectopic expression of C/EBPα and the addition of the appropriate adipogenic signaling cocktail. We proposed that SWI/SNF enzymes were modulating more subtle changes in chromatin in the area of the transcription start site. It remains possible that one or both of the SWI/SNF ATPases could be functioning at the PPARγ2 promoter immediately after differentiation. However, in the absence of direct evidence that SWI/SNF enzymes are present when the observed changes in PPARγ2 promoter chromatin accessibility occur, we undertook a series of ChIP experiments using available antibodies to other known ATP-dependent chromatin remodelers (data not shown). None of these enzymes could be localized to the PPARγ2 promoter. Thus the enzyme responsible for the remodeling events observed immediately after differentiation was not identified.
The rapidity of the changes in PPARγ2 promoter chromatin structure and transcription factor binding indicated that these events are not a consequence of induced transcription but instead result from protein modification. Thus, the components of the cocktail used to signal differentiation of the 3T3-L1 cells were the most likely candidates for mediating the observed molecular changes at the PPARγ2 promoter. Drop-out experiments quickly implicated the IBMX component as the factor responsible for chromatin accessibility changes, and since IBMX increases intracellular levels of cyclic AMP and activates PKA, we employed a combination of enzymatic inhibitors and directed knockdown of the catalytic subunits of PKA to conclusively demonstrate the requirement for PKA signaling in PPARγ2 promoter chromatin structural changes. Specifically, the loss of either catalytic subunit of PKA significantly inhibited the chromatin accessibility at the PPARγ2 promoter that was observed upon induction of differentiation. We also showed that the loss of PKA catalytic subunits via shRNA knockdown blocked adipogenic differentiation, reflecting the critical nature of PKA activity during differentiation. In addition to inhibition of PKA catalytic activity, our results could also reflect a loss of interaction with PKA scaffolding proteins (Levin et al., 2006). Regardless, to the best of our knowledge, this report is the first to indicate that PKA signaling is responsible for altering promoter chromatin accessibility during adipogenesis.
Although a cAMP analog added to the media in place of the standard differentiation cocktail was sufficient to generate changes in PPARγ2 promoter accessibility, we noted that a MAP kinase inhibitor also limited remodeling of the PPARγ2 promoter and partially inhibited recruitment of c-Fos and C/EBPβ. These data are consistent with a reduction in PPARγ2 expression in the presence of a MEK inhibitor (Prusty et al., 2002), and with the extensive evidence that MEK signaling activates C/EBPβ (Farmer, 2005).
Finally, it is important to note that while our studies suggest a direct role for PKA signaling in chromatin remodeling, cAMP signaling in adipocytes also involves activation of Epacs (exchange proteins directly activated by cAMP)(Ji et al., 2010; Martini et al., 2009; Petersen et al., 2008). With the advent of Epac selective analogs (Martini et al., 2009; Petersen et al., 2008) and Epac-1 specific knockdowns (Ji et al., 2010), it will be possible to assess whether rapid chromatin remodeling occurs in an Epac dependent or independent manner to further understand the role of cAMP signaling during adipocyte differentiation.
ACKNOWLEDGMENTS
We thank C. Baron for assistance with the figures and Y.-J. Hu and K. Imbalzano for comments on the manuscript. This work was supported by grants from the National Institutes of Health to ANI (DK079239 and DK084278) and to SEL (F32DK082263). ANI is a member of the UMass Diabetes Endocrinology Research Center, which is supported by DERC grant 5P30DK32520.
LITERATURE CITED
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Birsoy K, Chen Z, Friedman J. Transcriptional regulation of adipogenesis by KLF4. Cell Metab. 2008;7(4):339–347. doi: 10.1016/j.cmet.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Umek RM, McKnight SL. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991;5(9):1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- Caramel J, Medjkane S, Quignon F, Delattre O. The requirement for SNF5/INI1 in adipocyte differentiation highlights new features of malignant rhabdoid tumors. Oncogene. 2008;27(14):2035–2044. doi: 10.1038/sj.onc.1210847. [DOI] [PubMed] [Google Scholar]
- Chawla A, Schwarz EJ, Dimaculangan DD, Lazar MA. Peroxisome proliferator-activated receptor (PPAR) gamma: adipose-predominant expression and induction early in adipocyte differentiation. Endocrinology. 1994;135(2):798–800. doi: 10.1210/endo.135.2.8033830. [DOI] [PubMed] [Google Scholar]
- Chen Z, Torrens JI, Anand A, Spiegelman BM, Friedman JM. Krox20 stimulates adipogenesis via C/EBPbeta-dependent and -independent mechanisms. Cell Metab. 2005;1(2):93–106. doi: 10.1016/j.cmet.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Cheng X, Ji Z, Tsalkova T, Mei F. Epac and PKA: a tale of two intracellular cAMP receptors. Acta Biochim Biophys Sin (Shanghai) 2008;40(7):651–662. doi: 10.1111/j.1745-7270.2008.00438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Cornelius P, MacDougald OA, Lane MD. Regulation of adipocyte development. Annu Rev Nutr. 1994;14:99–129. doi: 10.1146/annurev.nu.14.070194.000531. [DOI] [PubMed] [Google Scholar]
- Darlington GJ, Ross SE, MacDougald OA. The role of C/EBP genes in adipocyte differentiation. J Biol Chem. 1998;273(46):30057–30060. doi: 10.1074/jbc.273.46.30057. [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Carlson KA, Hill DA, Guidi CJ, Stephenson RO, Sif S, Kingston RE, Imbalzano AN. Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol Cell Biol. 2000;20(8):2839–2851. doi: 10.1128/mcb.20.8.2839-2851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Carlson KA, Imbalzano AN. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat Genet. 2001;27(2):187–190. doi: 10.1038/84826. [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, Imbalzano AN. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol Cell Biol. 2005;25(10):3997–4009. doi: 10.1128/MCB.25.10.3997-4009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat Rev Genet. 2006;7(6):461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- Debril MB, Gelman L, Fayard E, Annicotte JS, Rocchi S, Auwerx J. Transcription factors and nuclear receptors interact with the SWI/SNF complex through the BAF60c subunit. J Biol Chem. 2004;279(16):16677–16686. doi: 10.1074/jbc.M312288200. [DOI] [PubMed] [Google Scholar]
- Distel RJ, Ro HS, Rosen BS, Groves DL, Spiegelman BM. Nucleoprotein complexes that regulate gene expression in adipocyte differentiation: direct participation of c-fos. Cell. 1987;49(6):835–844. doi: 10.1016/0092-8674(87)90621-0. [DOI] [PubMed] [Google Scholar]
- Distel RJ, Spiegelman BM. Involvement of fos as a trans-acting factor in adipogenic gene expression. Prog Clin Biol Res. 1988;284:187–209. [PubMed] [Google Scholar]
- Farmer SR. Regulation of PPARgamma activity during adipogenesis. Int J Obes (Lond) 2005;29(Suppl 1):S13–16. doi: 10.1038/sj.ijo.0802907. [DOI] [PubMed] [Google Scholar]
- Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4(4):263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59(4):675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- Green H, Kehinde O. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell. 1975;5(1):19–27. doi: 10.1016/0092-8674(75)90087-2. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Georgopoulos K, Greenberg ME, Leder P. c-Jun dimerizes with itself and with c-fos, forming complexes of different DNA binding affinities. Cell. 1988;55(5):917–924. doi: 10.1016/0092-8674(88)90147-x. [DOI] [PubMed] [Google Scholar]
- Heath VJ, Gillespie DA, Crouch DH. Inhibition of adipocyte differentiation by cMyc is not accompanied by alterations in cell cycle control. Biochem Biophys Res Commun. 2000a;269(2):438–443. doi: 10.1006/bbrc.2000.2316. [DOI] [PubMed] [Google Scholar]
- Heath VJ, Gillespie DA, Crouch DH. Inhibition of the terminal stages of adipocyte differentiation by cMyc. Exp Cell Res. 2000b;254(1):91–98. doi: 10.1006/excr.1999.4736. [DOI] [PubMed] [Google Scholar]
- Ji Z, Mei FC, Cheng X. Epac, not PKA catalytic subunit, is required for 3T3-L1 preadipocyte differentiation. Front Biosci (Elite Ed) 2010;2:392–398. doi: 10.2741/e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminska B, Pyrzynska B, Ciechomska I, Wisniewska M. Modulation of the composition of AP-1 complex and its impact on transcriptional activity. Acta Neurobiol Exp (Wars) 2000;60(3):395–402. doi: 10.55782/ane-2000-1358. [DOI] [PubMed] [Google Scholar]
- Lefterova MI, Lazar MA. New developments in adipogenesis. Trends Endocrinol Metab. 2009;20(3):107–114. doi: 10.1016/j.tem.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Levin AM, Coroneus JG, Cocco MJ, Weiss GA. Exploring the interaction between the protein kinase A catalytic subunit and caveolin-1 scaffolding domain with shotgun scanning, oligomer complementation, NMR, and docking. Protein Sci. 2006;15(3):478–486. doi: 10.1110/ps.051911706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wang D, Zhou Y, Zhou B, Yang Y, Chen H, Song J. Protein kinase A suppresses the differentiation of 3T3-L1 preadipocytes. Cell Res. 2008;18(2):311–323. doi: 10.1038/cr.2008.12. [DOI] [PubMed] [Google Scholar]
- Martini CN, Plaza MV, Vila Mdel C. PKA-dependent and independent cAMP signaling in 3T3-L1 fibroblasts differentiation. Mol Cell Endocrinol. 2009;298(1–2):42–47. doi: 10.1016/j.mce.2008.10.023. [DOI] [PubMed] [Google Scholar]
- Musri MM, Gomis R, Parrizas M. Chromatin and chromatin-modifying proteins in adipogenesis. Biochem Cell Biol. 2007;85(4):397–410. doi: 10.1139/O07-068. [DOI] [PubMed] [Google Scholar]
- Ntambi JM, Young-Cheul K. Adipocyte differentiation and gene expression. J Nutr. 2000;130(12):3122S–3126S. doi: 10.1093/jn/130.12.3122S. [DOI] [PubMed] [Google Scholar]
- Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci U S A. 1993;90(18):8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen TA, Kowenz-Leutz E, Leutz A, Nerlov C. Cooperation between C/EBPalpha TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev. 2001;15(23):3208–3216. doi: 10.1101/gad.209901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RK, Madsen L, Pedersen LM, Hallenborg P, Hagland H, Viste K, Doskeland SO, Kristiansen K. Cyclic AMP (cAMP) mediated stimulation of adipocyte differentiation requires the synergistic action of Epac- and cAMP-dependent protein kinase-dependent processes. Mol Cell Biol. 2008;28(11):3804–3816. doi: 10.1128/MCB.00709-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porte D, Oertel-Buchheit P, John M, Granger-Schnarr M, Schnarr M. DNA binding and transactivation properties of Fos variants with homodimerization capacity. Nucleic Acids Res. 1997;25(15):3026–3033. doi: 10.1093/nar/25.15.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston GA, Srinivasan D, Barrett JC. Apoptotic response to growth factor deprivation involves cooperative interactions between c-fos and p300. Cell Death Differ. 2000;7(2):215–226. doi: 10.1038/sj.cdd.4400637. [DOI] [PubMed] [Google Scholar]
- Prusty D, Park BH, Davis KE, Farmer SR. Activation of MEK/ERK signaling promotes adipogenesis by enhancing peroxisome proliferator-activated receptor gamma (PPARgamma) and C/EBPalpha gene expression during the differentiation of 3T3-L1 preadipocytes. J Biol Chem. 2002;277(48):46226–46232. doi: 10.1074/jbc.M207776200. [DOI] [PubMed] [Google Scholar]
- Racki LR, Narlikar GJ. ATP-dependent chromatin remodeling enzymes: two heads are not better, just different. Curr Opin Genet Dev. 2008;18(2):137–144. doi: 10.1016/j.gde.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7(12):885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- Russell TR, Ho R. Conversion of 3T3 fibroblasts into adipose cells: triggering of differentiation by prostaglandin F2alpha and 1-methyl-3-isobutyl xanthine. Proc Natl Acad Sci U S A. 1976;73(12):4516–4520. doi: 10.1073/pnas.73.12.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR. Mechanisms for nucleosome movement by ATP-dependent chromatin remodeling complexes. Results Probl Cell Differ. 2006;41:127–148. doi: 10.1007/400_005. [DOI] [PubMed] [Google Scholar]
- Salma N, Xiao H, Imbalzano AN. Temporal recruitment of CCAAT/enhancer-binding proteins to early and late adipogenic promoters in vivo. J Mol Endocrinol. 2006;36(1):139–151. doi: 10.1677/jme.1.01918. [DOI] [PubMed] [Google Scholar]
- Salma N, Xiao H, Mueller E, Imbalzano AN. Temporal recruitment of transcription factors and SWI/SNF chromatin-remodeling enzymes during adipogenic induction of the peroxisome proliferator-activated receptor gamma nuclear hormone receptor. Mol Cell Biol. 2004;24(11):4651–4663. doi: 10.1128/MCB.24.11.4651-4663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C, Beck K, Mink S, Schmolke M, Budde B, Wenning D, Klempnauer KH. Recruitment of p300 by C/EBPbeta triggers phosphorylation of p300 and modulates coactivator activity. Embo J. 2003;22(4):882–892. doi: 10.1093/emboj/cdg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seamon KB, Padgett W, Daly JW. Forskolin: unique diterpene activator of adenylate cyclase in membranes and in intact cells. Proc Natl Acad Sci U S A. 1981;78(6):3363–3367. doi: 10.1073/pnas.78.6.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalhegg BS, Tasken K. Specificity in the cAMP/PKA signaling pathway. Differential expression,regulation, and subcellular localization of subunits of PKA. Front Biosci. 2000;5:D678–693. doi: 10.2741/skalhegg. [DOI] [PubMed] [Google Scholar]
- Smith ER, Smedberg JL, Rula ME, Hamilton TC, Xu XX. Disassociation of MAPK activation and c-fos expression in F9 embryonic carcinoma cells following retinoic acid-induced endoderm differentiation. J Biol Chem. 2001;276(34):32094–32100. doi: 10.1074/jbc.M105009200. [DOI] [PubMed] [Google Scholar]
- Smyth MJ, Sparks RL, Wharton W. Proadipocyte cell lines: models of cellular proliferation and differentiation. J Cell Sci. 1993;106(Pt 1):1–9. doi: 10.1242/jcs.106.1.1. [DOI] [PubMed] [Google Scholar]
- Spiegelman BM, Distel RJ, Ro HS, Rosen BS, Satterberg B. fos protooncogene and the regulation of gene expression in adipocyte differentiation. J Cell Biol. 1988;107(3):829–832. doi: 10.1083/jcb.107.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama H, Chen P, Hunter MG, Sitkovsky MV. Perturbation of the expression of the catalytic subunit C alpha of cyclic AMP-dependent protein kinase inhibits TCR-triggered secretion of IL-2 by T helper hybridoma cells. J Immunol. 1997;158(1):171–179. [PubMed] [Google Scholar]
- Tang QQ, Gronborg M, Huang H, Kim JW, Otto TC, Pandey A, Lane MD. Sequential phosphorylation of CCAAT enhancer-binding protein beta by MAPK and glycogen synthase kinase 3beta is required for adipogenesis. Proc Natl Acad Sci U S A. 2005;102(28):9766–9771. doi: 10.1073/pnas.0503891102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Bucher NL, Farmer SR. Induction of peroxisome proliferator-activated receptor gamma during the conversion of 3T3 fibroblasts into adipocytes is mediated by C/EBPbeta, C/EBPdelta, and glucocorticoids. Mol Cell Biol. 1996;16(8):4128–4136. doi: 10.1128/mcb.16.8.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh WC, Cao Z, Classon M, McKnight SL. Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev. 1995;9(2):168–181. doi: 10.1101/gad.9.2.168. [DOI] [PubMed] [Google Scholar]
- Zhang JW, Klemm DJ, Vinson C, Lane MD. Role of CREB in transcriptional regulation of CCAAT/enhancer-binding protein beta gene during adipogenesis. J Biol Chem. 2004;279(6):4471–4478. doi: 10.1074/jbc.M311327200. [DOI] [PubMed] [Google Scholar]