Abstract
Biological systems use a variety of mechanisms to maintain their functions in the face of environmental and genetic perturbations. Increasing evidence suggests that, among their roles as post-transcriptional repressors of gene expression, microRNAs (miRNAs) help to confer robustness to biological processes by reinforcing transcriptional programs and attenuating aberrant transcripts, and they may in some network contexts help suppress random fluctuations in transcript copy number. These activities have important consequences for normal development and physiology, disease, and evolution. Here we will discuss examples and principles of miRNAs that contribute to robustness in animal systems.
MicroRNAs (miRNAs) are hairpin-derived RNAs ~20–24-nucleotide-long, which post-transcriptionally repress the expression of target genes usually by binding to the 3' UTR of messenger RNA (mRNA). As a class, miRNAs constitute about 1–2% of genes in worms, flies, and mammals (Bartel, 2009). Their regulatory potential is vast: more than 60% of protein-coding genes are computationally predicted as targets based on conserved base-pairing between the 3' UTR and the 5' region of the miRNA, which is called the seed (Friedman et al., 2009). Although many miRNAs and their target binding sites are deeply conserved, which suggests important function, a typical miRNA-target interaction produces only subtle reduction (<2-fold) in protein level, and many miRNAs can be deleted without creating any obvious phenotype. Early observations of miRNA expression profiles revealed that miRNAs tend to be anticorrelated with target gene expression in contiguous developmental stages or tissues (Stark et al., 2005; Farh et al., 2005). Correspondingly, a view emerged that miRNA evolved primarily to play the role of a reinforcer, in that its activities cohere with transcriptional patterns to sharpen developmental transitions and entrench cellular identities. It is also possible that miRNAs buffer fluctuations in gene expression and more faithfully signal outcomes in the context of certain regulatory networks.
Robustness refers to a system’s ability to maintain its function in spite of internal or external perturbations (Kitano, 2004). In biology, such systems can be considered at several levels: a biochemical pathway producing a steady output of a signaling protein; a cluster of cells undergoing patterned differentiation; or an animal surviving periods of food scarcity. Like sophisticated man-made systems, these biological systems use controls such as feedback loops and redundant components to carry on reliably when conditions change or one component fails. Such controls are especially relevant to the development and physiology of multicellular organisms with complex body plans. In these organisms, embryonic cells not only ‘choose’ among many different fates, but they also must ‘remember’ their choice to maintain their cell type identity in the adult. The involvement of miRNAs in regulatory networks that provide developmental robustness is indicated by recent experiments in a variety of model organisms. It is also suggested by three general observations: genes with tissue-specific expression have longer 3' UTRs with more miRNA binding sites (Stark et al., 2005); miRNA expression increases and diversifies over the course of embryonic development (Thomson et al., 2006), as 3' UTRs are lengthened via alternative polyadenylation site choice (Ji et al., 2009); and, the diversity of the miRNA repertoire in animal genomes has increased with increasing organismal complexity (Lee et al., 2007; Heimberg et al., 2008). In this review, we examine the current evidence for how miRNAs contribute to the robustness of biological processes.
Coherent regulation for precise developmental transitions
One of the earliest functions attributed to miRNAs was sharpening developmental transitions by suppressing residual transcripts that were specific to the previous stage. Global gene expression analyses in fly, fish, and mouse have shown that miRNAs and their targets often have mutually exclusive RNA expression across tissues, especially in neighboring tissues derived from common progenitors (Farh et al., 2005; Stark et al., 2005; Sood et al., 2006; Tsang et al., 2007)(Figure 1). This suggests that miRNAs can act to reinforce the transcriptional gene expression program by repressing leaky transcripts.
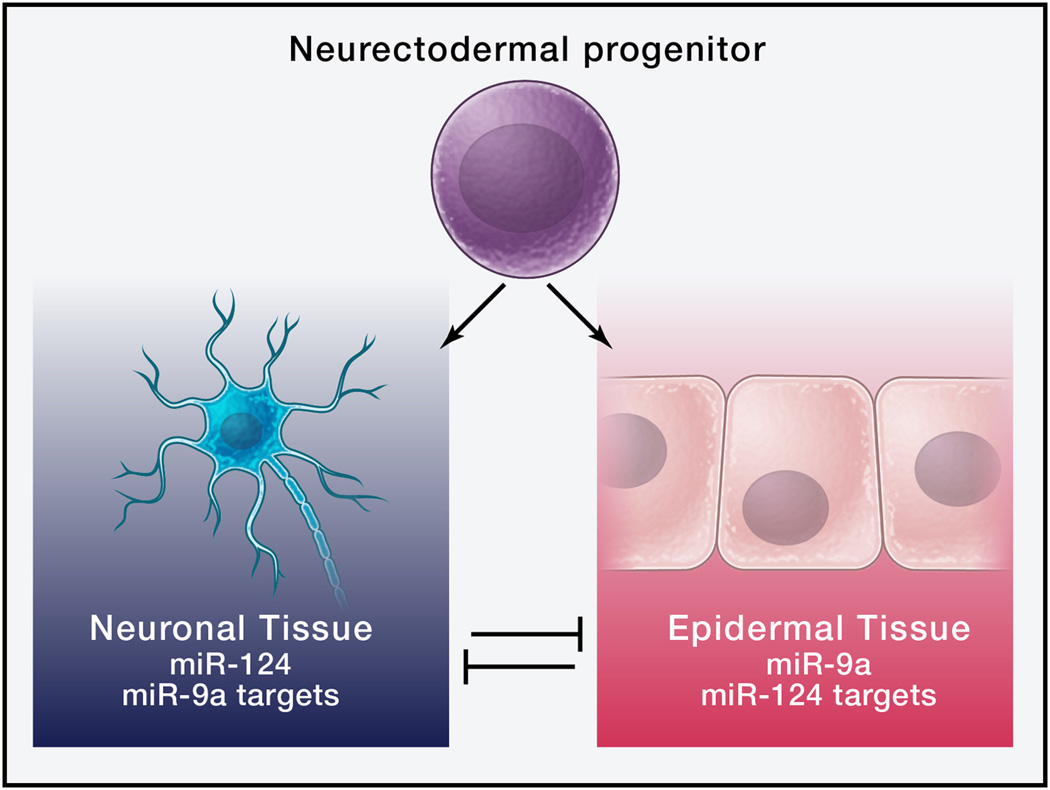
Figure 1.
Anticorrelated expression of miRNAs and targets in developmental transitions. In the Drosophila embryo, neurectodermal progenitors express miR-124 as they differentiate into neurons. Neuronal genes that are induced during this transition tend not to have miR-124 sites, whereas genes expressed in epidermal tissues that are also ectodermal derivatives are enriched for miR-124 sites (Stark et al., 2005). Thus, expression of miR-124 stabilizes the neuronal transition. A reciprocal pattern holds for the ectoderm-specific miR-9a.
Intriguingly, this anti-correlative pattern may apply not only to transcription but also to alternative splicing. Drosophila express a cytoplasmic isoform of tropomyosin-1 in the gut, brain and epidermis, but not in muscle, and this isoform is targeted by the muscle-specific miRNA miR-1. In contrast, the three muscle-expressed isoforms lack miR-1 sites, and this trend is conserved across vertebrates (Stark et al., 2005). Thus, a mis-splicing event that generated the cytoplasmic gut/brain/epidermis isoform in muscle cells could be corrected by miRNA-mediated repression.
More recently, sensitive gene expression profiling of cell types in the zebrafish embryo revealed not so much a stark mutual exclusion pattern but rather a tendency for anticorrelated but still overlapping expression of miRNAs and targets (Shkumatava et al., 2009). This suggests that miRNAs play a more prominent role than only reinforcing the patterns dictated by transcriptional regulation. In fact, a strongly transcribed, ubiquitously expressed actin transcript has its levels spatially sculpted by muscle-specific miRNAs in zebrafish (Mishima et al., 2009).
Cell fate switches
A regulatory motif that generates anticorrelated expression, commonly involving miRNAs, is the coherent feed-forward loop (FFL). In a coherent FFL, component A inhibits (or activates) component C and activates (or inhibits, respectively) component B, which is another repressor of component C. This architecture can increase the fidelity of inhibition of the downstream component by acting on it redundantly; that is, a transient loss of component A can be compensated for by the lingering presence of component B.
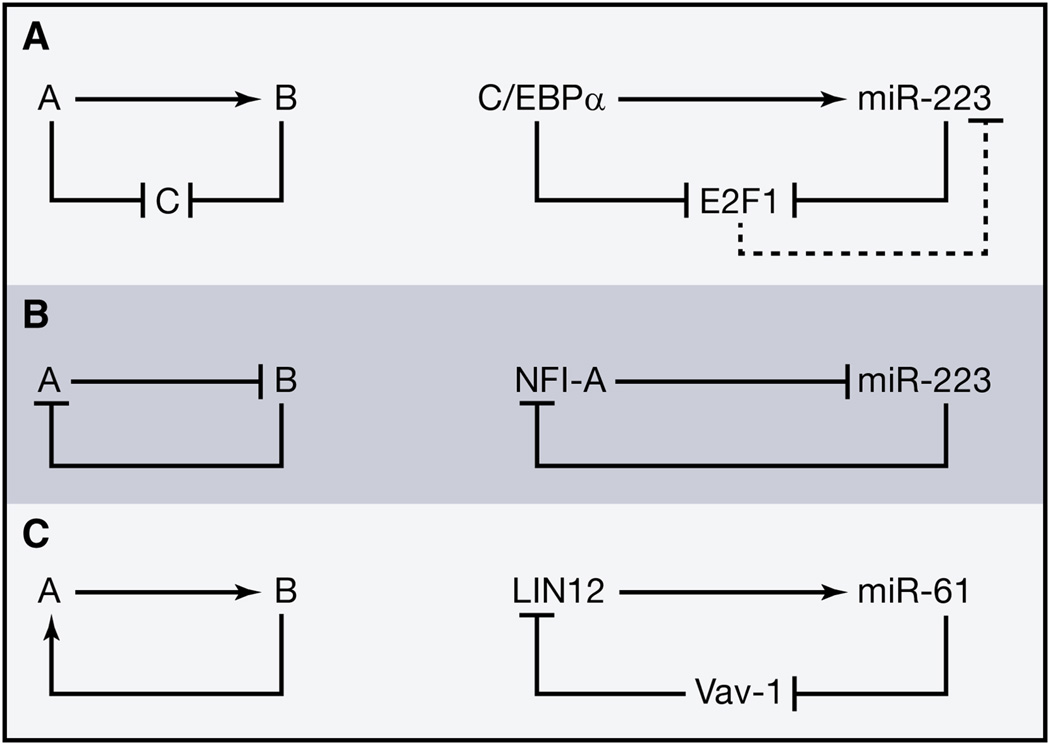
Along with positive and negative feedback loops, this motif is often used in lineage commitment. For example, CCAAT enhancer binding protein alpha (C/EBPα) inhibits transcription of the cell cycle regulator E2F1 during granulopoiesis (Pulikkan et al., 2010). C/EBPα also induces miR-223, which post-transcriptionally represses E2F1. As is often the case, this feedforward loop is interlocked with a feedback loop: E2F1 inhibits production of miR-223 (Figure 2A).
Figure 2.
Network motifs for cell fate switches.
(A) A coherent feedforward loop both directly and indirectly inhibits the cell cycle regulator E2F1 in granulopoiesis.
(B) A mutual negative feedback loop contributes to bistability between myeloid precursors and granulocytes.
(C) A positive feedback loop enforces lineage commitment of nematode “2 degrees” vulval cells.
This example illustrates several principles of miRNA networks in development: 1) In these loops, the miRNA often targets a transcriptional regulator; 2) Combining feedforward with feedback motifs may allow cells to distinguish between transient fluctuations (which should be counteracted) and permanent changes (which should be enhanced or maintained); and 3) There are often other network motifs involving a cell type-specific miRNA that redundantly reinforce the same cell fate decision, as with the mutual negative feedback loop between miR-223 and NFI-A in granulocytes (Fazi et al., 2005). Here the transcription factor NFI-A suppresses expression of the primary miR-223 transcript in undifferentiated myeloid precursors. Upon retinoic acid-induced differentiation into granulocytes, miR-223 accumulates and represses NFI-A, thereby helping to prevent a return to the precursor state (Figure 2B). Mutual negative feedback loops have been shown to underlie bistable genetic switches, as demonstrated by a synthetic genetic toggle switch in E. coli, which can be flipped by a transient cue but is robust to ordinary fluctuations in gene expression (Gardner et al., 2000).
Developmental decisions can also be reinforced by positive feedback loops, in which component A and component B activate each other. For example, the “2 degrees” vulval precursor cell fate is established in the worm when LIN12 directly activates transcription of miR-61, which then represses vav-1, a negative regulator of LIN12 activity (Yoo and Greenwald, 2005) (Figure 2C). In this case the indirect link may build additional control into the lineage decision, as LIN12 expression must be sustained enough for miR-61 to accumulate to sufficiently repress the level of Vav-1 protein in order to allow for adequate LIN12 activity.
Bistability is essential in development, but it can have an adverse function when it is coopted in cancer. For example, the transcription factor ZEB1 induces the epithelial-to-mesenchymal transition, which is important for tissue remodeling during embryonic development. ZEB1 suppresses transcription of miR-200 family members, and the miR-200 family strongly represses ZEB1 (Bracken et al., 2008; Burk et al., 2008). In development, this mutual negative feedback reinforces the mesenchymal cell fate decision. Within carcinomas, some tumor cells lose miR-200 expression and switch to a mesenchymal state, which promotes their ability to metastasize (Gibbons et al., 2009).
Subtle repression with adaptive impact
The effect of an individual miRNA on a target’s protein level tends to be subtle, usually less than 2-fold (Baek et al., 2008). Most loss-of-function mutations are recessive; thus, organisms are commonly able to compensate for a 2-fold loss of gene expression. Such differences may even be within the range of random variation in mRNA or protein level between different cells in a genetically identical population or in a given cell at different times. So how do miRNAs and target sites experience selective pressure, and how do miRNAs accomplish any significant regulation? For starters, there are miRNA-target interactions that involve multiple sites for a given target and confer much stronger repression, such as the interaction between the microRNA let-7 and the oncogene HMGA2 (Mayr et al., 2007). More often, different miRNAs work together to co-target a given mRNA, so their combined repressive effect greatly exceeds the individual contributions. On average there are more than four highly conserved seed match sites per UTR considering all miRNAs, and many more sites when more weakly conserved sequences are considered (Friedman et al., 2009). While multiple sites generally show independent additive effects, they can show cooperative effects when in close proximity (~10–40 nt apart) (Bartel, 2009). Multisite target reporters with this type of spacing showed more than 10-fold repression by a moderately expressed endogenous miRNA for a target expressed at low mRNA level (Mukherji et al., 2011).
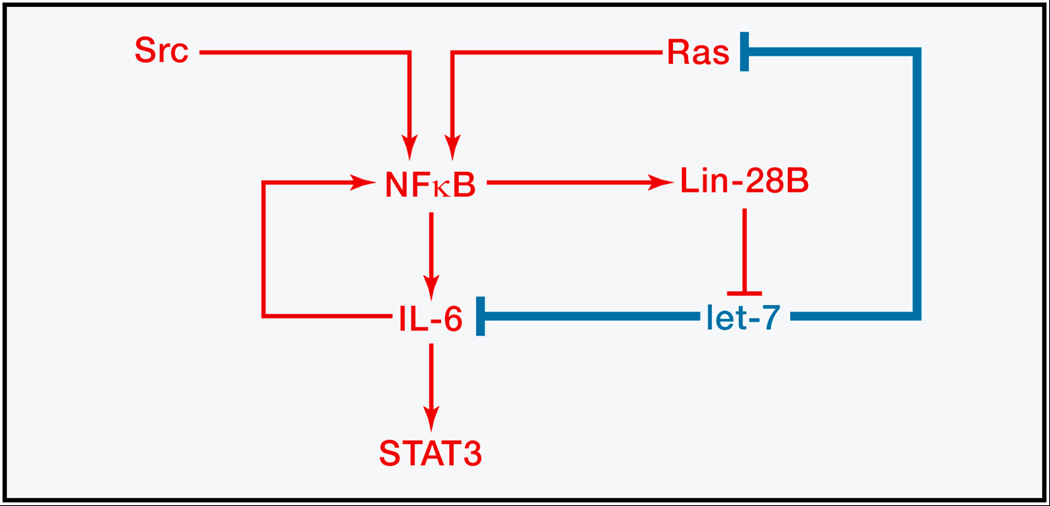
Nevertheless, a small change in the level of protein can sometimes have a large physiological effect, such as when a positive feedback loop amplifies the change. Iliopoulos et al. (2010) recently described a network of feedback loops that flips a switch in cancer. Transient activation of Src or other triggers of NF-κB induces stable transformation of a mammary epithelial cell line. NF-κB transcriptionally activates IL6 and inhibits let-7 family members by activating Lin-28B, which prompts destruction of let-7 precursor RNAs (Figure 3). The ensuing drop in let-7 level derepresses IL6, a direct let-7 target, and IL6 is further activated by derepression of the let-7 target Ras. IL6 feeds back in both an autocrine and paracrine fashion to activate NF-κB, which further inhibits let-7, and it signals through STAT3 to promote cell growth and motility. In normal tissue, a transient inflammatory cue could signal through this pathway to induce cell growth to repair damage, and the miRNA holds the positive feedbacks in check. In cancer, where let-7 is typically down-regulated (Kumar et al., 2008; Dong et al., 2010), the positive feedbacks would go unchecked, and continuous, self-reinforcing proliferation would result. In human tumors the positive feedback loop could be made even stronger by the presence of oncogenic mutations such as v-Src or Ras-V12 (Iliopoulos et al., 2010).
Figure 3.
Positive feedback can amplify small changes. A transient inflammatory cue induces stable malignant transformation through an NF-κB/IL6 positive feedback network that is normally kept in check by let-7. Diagram adapted from (Iliopoulos et al., 2009).
Another mechanism by which a miRNA can increase its impact is by targeting a set of genes that are in a shared pathway or protein complex. Linsley et al. (2007) provided the first experimental demonstration of this principle, showing coordinate regulation of the G0/G1-to-S cell cycle transition by the miR-16 family (Linsley et al., 2007). A statistical analysis of target predictions crossed against functional annotations found such coordinated repression to be prevalent in mammalian genomes (Tsang et al., 2010). By reducing the concentration of several components in a signaling cascade, a miRNA could create significant reductions in signal output over time. On the other hand, by repressing negative regulators in a pathway, a miRNA could increase signal output. In T lymphocytes, miR-181 plays this role by regulating multiple phosphatases downstream of the T cell receptor, and its dynamic expression at different stages of maturation tunes the sensitivity of the pathway to different levels of antigen (Li et al., 2007). Concentrating effects within functional modules is a common feature of robust systems (Kitano, 2004).
Absent and variable phenotypes
In spite of the large numbers of target genes predicted to be affected by miRNA loss of function, gene knockout experiments for individual miRNAs have yielded many disappointing results. In worms, most individual miRNA mutants show no gross phenotype (Miska et al., 2007); the same is true for several of the mouse knockouts generated to date, including miR-21, miR-210, miR-214, miR-206, and miR-143 (Eric Olson, personal communication). A partial explanation for these results resides in the functional redundancy of many miRNAs that share their seed sequence with others. For example, the let-7 family members miR-48, miR-84, and miR-241 operate redundantly to control the L2-to-L3 larval transition in C. elegans (Abbott et al., 2005). Additionally, many miRNAs of different seed families work together to co-target a given gene or set of genes, providing overlapping functions. To generate an observable impairment in the animal, it might be necessary to delete all members of a seed family and also non-seed family members that have a high degree of co-targeting.
It is also possible that a mutant phenotype would only arise upon acute miRNA deletion, if during development miRNA loss can be compensated at the level of gene expression or by one cell type populating a niche to assist an impaired or underpopulated cell type within an organ or system such as the immune system. Use of conditional knockouts or hypomorphs could possibly reveal physiological phenotypes of miRNAs that are not observed with germline nulls because of early lethality or compensation processes.. Along these lines, Smibert et al. (2011) found synaptic transmission defects in photoreceptor neurons of flies with hypomorphic alleles of miRNA core biogenesis genes pasha, drosha, and dcr-1. Even once an organ has developed, miRNAs may be required for maintenance: Dicer loss in the mouse thymic epithelium or the highly structured retina leads to progressive degeneration of tissue architecture (Papadopoulou et al., 2012; Damiani et al., 2008). However, there are several contrary examples in which deletion of Dicer and loss of all miRNAs in mature tissue does not appear to generate a phenotype. Deletion of Dicer in the mouse olfactory system had no apparent phenotype over periods of several months (Choi et al., 2008), whereas the same deletion in developing olfactory tissue led to severe neurogenesis defects.
Finally, a miRNA phenotype may appear only upon the application of certain internal or external stresses. The most well-characterized example of this mechanism is in the Drosophila eye, where miR-7 plays a role in the determination of sensory organs (Li and Carthew, 2005). Loss of miR-7 had little observable impact on the development of the sensory organs under normal, uniform conditions, and expression of the proneural transcription factor Atonal was also detected at wild-type level (Li et al., 2009). But when an environmental perturbation was added during larval development (i.e., fluctuating the temperature between 31°C and 18°C roughly every 90 minutes), the miR-7 mutant eyes showed abnormally low Atonal expression and abnormally high, irregular expression of the antineural transcription factor Yan. Sensory organ precursor (SOP) defects also appeared: some groups of antennal SOPs failed to develop, or developed with abnormal patterning; their cells showed low Atonal levels. The ability of miR-7 to confer developmental robustness against temperature perturbations likely depends on its placement in a network of feedback and feedforward loops with Atonal and Yan (Li et al., 2009; Herranz and Cohen, 2010).
In mice, deletion of the heart muscle-specific miRNA miR-208 has little phenotype under normal conditions but results in a failure to induce cardiac remodeling upon stress (van Rooij et al., 2007). When the mice were treated to induce pressure overload or hypothyroidism, miR-208 activity was required in the cardiomyocytes to upregulate βMHC by targeting the thyroid receptor signaling pathway. The embryonic stem cell-specific miR-290–295 cluster is not required for cell viability until DNA damage stress, upon which it promotes cell survival (Zheng et al., 2011). In worms sensitized by mutations in a variety of regulatory pathways, 25 of 31 deleted miRNAs revealed a mutant phenotype (Brenner et al., 2010); these same deletions in a wild-type background did not produce a phenotype. These examples show the utility of assessing animal systems not only under standard laboratory conditions but also with treatments that mimic the natural hardships and flaws they might experience in the wild.
Some miRNA knockouts show phenotypes with incomplete penetrance. For example, mice deleted for miR-290–295 show partially penetrant embryonic lethality (Medeiros et al., 2011), and flies lacking miR-9a display partially penetrant sensory organ defects (Li et al., 2006). Such variation in phenotypic severity may be attributable to fluctuations in target gene expression occurring during a critical window of development that the miRNA normally suppresses. Raj et al. (2010) demonstrated how incomplete penetrance can be caused by stochastic variation in gene expression combined with threshold effects. Along the same lines, sporadic defects can appear within the organs of an individual mutant animal. For example, flies with miR-263a deleted exhibit random loss of mechanosensory bristles because the miRNA is needed to prevent apoptosis in sensory bristle progenitors during the patterning of the eye (Hilgers et al., 2010).
miRNAs as buffers
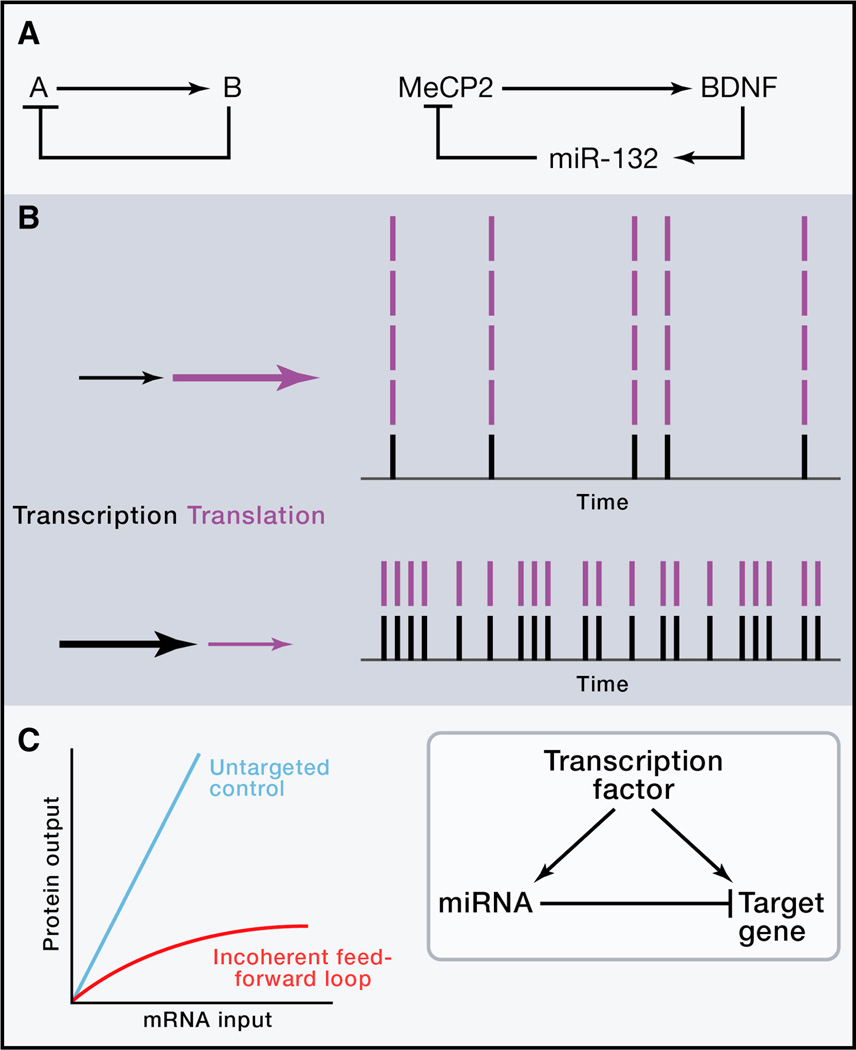
miRNAs have been said to act as “buffers” against variation in gene expression. In what contexts is this true? One way that changes in expression level can be counteracted is the simple negative feedback loop, in which component A activates component B and component B inhibits component A. Genome-wide mapping of interactions between transcription factors and in C. elegans showed enrichment for such feedback loops (Martinez et al., 2008). In mammals, methyl CpG-binding protein 2 (MeCP2) acts through BDNF to induce the neuronal miRNA miR-132, which then feeds back to repress MeCP2 (Klein et al., 2007) (Figure 4A). Homeostasis in the level of MeCP2 expression is important, as over- or under-expression of this regulator causes neurodevelopmental defects.
Figure 4.
miRNAs can reduce noise in gene expression.
(A) A negative feedback loop contributes to homeostasis for MeCP2 protein in neurons.
(B) Post-transcriptional repression to attenuate transcriptional noise. Two transcription-translation strategies to synthesize the same mean level of a protein, here 20 molecules per cell per unit of time. Strong transcription corresponds to more frequent mRNA bursts (black bars). Translation amplifies mRNA bursts into protein bursts (purple bars) so a more consistent protein output occurs when each mRNA produces fewer molecules of protein.
(C) Uncoupling of target protein output from mRNA input (left) by means of an incoherent feedforward loop (right) in which miRNA and target mRNA are transcriptionally co-induced. Cartooned from (Bleris et al., 2011) data.
A less intuitive way of buffering was reported for the zebrafish miR-430, which targets both the Nodal agonist squint and antagonist lefty. Although this motif does not provide a way to counteract an increase in agonist concentration without also knocking down the antagonist, it was observed that overexpression of either agonist or antagonist mRNA was tolerated in the presence of miR-430 but not in its absence (Choi et al., 2007). Similarly, miR-430 represses both the secreted ligand Sdf1a and its sequestration receptor Cxcr7b, primordial germ cell migration factors in the gonad. With this circuit, small reductions in gene expression of either the ligand or its inhibitory receptor were permitted without reducing the accuracy of cell migration (Staton et al., 2011).
Random fluctuations in protein levels over time and between clonally identical cells arise from several sources. Intrinsic noise refers to variation arising from stochastic events including promoter binding, mRNA decay, translation, and protein degradation (Raser and O’Shea, 2005). The degree to which a protein level fluctuates around its mean may be influenced by the rates at which transcription and translation occur. Transcription occurs in stochastic bursts (Blake et al., 2006), and higher transcription rates correlate with lower noise (Paulsson, 2004). Translation events amplify the mRNA bursts (Paulsson, 2004; Pedraza and van Oudenaarden, 2005), such that noise increases linearly with the rate of translation (Ozbudak et al., 2002). Thus, by transcribing a gene at a high rate (with more frequent mRNA bursts) and simultaneously reducing its translation rate using miRNAs, cells should reduce fluctuations in target protein number (Figure 4B). More precisely, because translational repression typically accounts for only ~20% of miRNA-mediated repression (Guo et al., 2010), the dominant effect is the reduction of a gene’s overall translation events by reducing the lifetime of the target mRNA in the cytoplasm. In agreement with this, Levine et al. (2007) reported lower variance in the expression of targets of bacterial small non-coding regulatory RNAs (sRNAs), which induce target mRNA degradation.
Extrinsic noise refers to variation arising from differences such as transcription factor or ribosome concentration or cell cycle stage. It has been proposed that extrinsic noise for a given gene could be reduced by co-regulated production of a miRNA that targets the gene, in a motif called an incoherent feedforward loop (FFL) (Hornstein and Shomron, 2006) (Figure 4C). A transient increase in transcription factor activity would propagate to an increase in target mRNA transcription but be counteracted by increased miRNA; a decrease in transcription factor activity and target mRNA production would be accompanied by posttranscriptional derepression of the target. Thus, protein output could be largely uncoupled from fluctuations in transcription factor concentration or activity. This noise reduction effect was achieved in a computational model of an incoherent FFL in two different scenarios: 1) the transcription factor transcribes a given amount of target mRNA, and a miRNA knocks down target expression by 40% (a typical amount), compared to no miRNA involvement; and 2) the transcription factor transcribes a higher level of target mRNA, and the miRNA represses protein expression, compared to generating the same mean protein output by combining weaker transcription with lack of miRNA involvement (Osella et al., 2011). Recently, Bleris et al. (2011) tested the incoherent FFL using synthetic reporters and an intron-embedded synthetic miRNA in mammalian cells. In contrast to a control reporter lacking the miRNA binding site, the incoherent FFL target’s protein output was largely insensitive to the variation in mRNA input levels generated by transfection with a range of plasmid DNA copy number (Figure 4C). We observed the same result from a similar reporter system (Ebert and Sharp, personal communication). There are a few natural examples of such “pure” incoherent FFLs, where an intron-embedded miRNA targets its host gene, e.g. miR-26b produced from the ctdsp2 pre-mRNA represses this REST cofactor gene during neurogenesis (Dill et al., 2012).
There are some caveats to the expected buffering effect of the incoherent FFL and the negative feedback loop. The change in miRNA concentration and the repression mediated by the miRNA must be rapid compared to the activity of the target gene product; if the miRNA response is slow, then it will create pulses and dips in target gene expression that return to the mean level faster than in the absence of repression but only slightly dampen the amplitude of the fluctuation. At the level of phenotypic output, whether the noise dampening is relevant depends on how the protein’s fluctuations propagate through its network of interactions, and whether the fluctuations are already smoothed out by virtue of the protein being long-lived, or are otherwise corrected by regulated protein activity or localization. It is also necessary to note that the incoherent FFL is probably not acting in isolation from other regulatory events: a target gene is likely regulated by more than just the transcription factor that also induces the pri-miRNA; the miRNA is likely regulated by more than just the transcription factor that also induces the target gene; and the target gene may be repressed by other miRNAs or other post-transcriptional regulators too. More sophisticated models will be needed to determine whether noise reduction can occur in more complex conditions.
Why use miRNAs?
miRNAs are surely not the only regulatory factors that contribute to system robustness. Whole-genome bioinformatic analysis of worm and fly reveal transcription factors enriched in feedforward loops as well (Gerstein et al., 2010; the modENCODE consortium et al., 2010). Compared to transcriptional regulators though, miRNAs do have some distinguishing features that may make them well-suited in this role. As post-transcriptional regulators acting in the cytoplasmic compartment, miRNAs can intervene late in the pipeline of gene expression to counteract variation from the upstream processes of transcription, splicing, and nuclear export. They are able to regulate transcripts in special compartments, such as maternally deposited transcripts in the early embryo (Giraldez et al., 2006; Bushati et al., 2008) and locally translated transcripts of dendrites far from the cell body of neurons. They can also be present at high concentrations (10,000s of molecules per cell) by virtue of being very stable (e.g., the heart muscle-specific miR-208 has an in vivo half-life of more than one week) (van Rooij et al., 2007). This is consistent with theoretical constraints indicating the need for many more molecules of a regulator to achieve a small reduction in the noise of a target gene (Lestas et al., 2010). miRNA expression profiling from progressive stages of T-lymphocyte development found that the total number of miRNAs expressed per cell changed in parallel with changes in total cellular RNA content, suggesting that global miRNA levels are tuned to the translational capacity of the cell (Neilson et al., 2007).
In terms of buffering gene expression noise, the simulations for incoherent FFL regulation showed that using a transcriptional repressor instead of a miRNA was not as effective in dampening fluctuations in the output of the target protein (Osella et al., 2011). For suppressing intrinsic noise, there are other posttranscriptional repressive mechanisms besides miRNA targeting that might reduce the impact of mRNA bursts. For example, AU-rich elements reduce the lifetime of an mRNA, upstream open reading frames and weak noncanonical Kozak sequences reduce the efficiency of translation initiation (Calvo et al., 2009), and rare codons and secondary structures slow translation elongation. Importantly though, these ubiquitously acting cis-regulatory elements cannot provide the tunability that arises from mixing different combinations of miRNA binding sites with different cell type-specific miRNA milieus.
miRNAs, robustness, and evolution
Although miRNAs may in some contexts act as buffers of gene expression, there is, to date, only one well-characterized example of a general mutation buffering agent. The chaperone Hsp90 assists the folding of client proteins such that it can compensate for point mutations in the protein coding regions of client genes (Rutherford and Lindquist, 1998). In doing so Hsp90 acts as a capacitor of phenotypic variation, storing cryptic genetic variation until environmental stress temporarily overwhelms Hsp90 and reveals the mutant proteins, allowing them to affect phenotypes and become substrates for selection. Do miRNAs potentiate cryptic genetic variation in the regulatory elements of their target genes? The ability of the miRNA to compensate for otherwise elevated target protein levels could allow such mutations to accrue without selective penalty. Analogous to Hsp90, transient and reversible loss of miRNA activity due to stress (Leung et al., 2011) could unleash the mutated gene products for exposure to natural selection. The emergence of non-lethal mutations that give diverse phenotypes is one requirement for evolvability, the generation of genetic diversity that can be selected (Kitano, 2004). In this way, miRNAs might contribute to evolvability.
On timescales of days to years, compromised miRNA activity and enhanced evolvability could have important implications for cancer. miRNAs are globally depleted in tumors relative to their normal tissue counterparts (Lu et al., 2005), and tumor growth is accelerated in models of global miRNA depletion, such as knockdown of components of the miRNA biogenesis pathway (Kumar et al., 2007) or heterozygous deletion of Dicer (Kumar et al., 2009). In addition, 3' UTRs are frequently shortened in tumors via alternative polyadenylation site choice (Sandberg et al., 2008; Mayr and Bartel, 2009). The combined effect of these trends should be widespread derepression of miRNA target genes and also potentially un-buffering of gene expression, which could increase the heterogeneity and plasticity of the tumor cell population. A tumor may be analogous to a clonal population of bacteria or yeast where noise in the population adapts them to unpredictably changing environmental conditions (Acar et al., 2008; Cağatay et al., 2009). For cancer cells, these spatially and temporally changing conditions could include increasingly hypoxic tumor cores, new microenvironments for metastases, or on-and-off chemotherapy regimens. The consequence of their noise-driven adaptability would be that a fraction of the cells persist through almost any condition.
At longer evolutionary timescales, miRNAs could have a role in enhancing evolvability by the process of canalization. Canalization refers to evolved robustness: a trait that is canalized presents itself consistently among different individuals of a species in spite of environmental or genetic perturbations (Hornstein and Shomron, 2006). miRNAs may contribute to canalization by reducing random fluctuations in target gene expression, stabilizing signaling decisions, and helping to produce distinct developmental outcomes. In doing so, they should tighten the linkage between genotype and phenotype, thereby increasing the heritability of traits (Peterson et al., 2009). The more heritable a trait is, the more efficiently selection can act on it.
Threshold effects and endogenous miRNA competitors
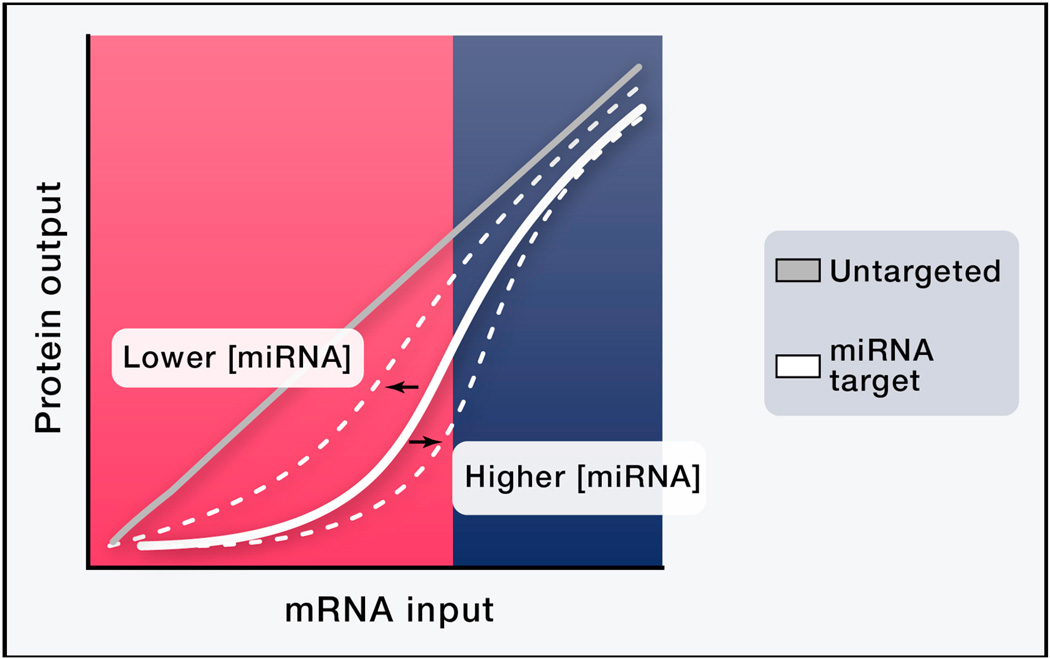
Recently it was shown that miRNA target genes have an mRNA expression threshold below which the gene is efficiently repressed and above which it can overwhelm the available miRNA (Mukherji et al., 2011). The threshold level is determined by the available miRNA concentration, whereas the steepness of the transition is determined by the strength and number of miRNA binding sites in the target (Figure 5). For a miRNA (miRNA-20 in HeLa cells) present at about 2000 copies per cell, a target gene with seven strong binding sites appears to begin this transition at around 60 copies per cell of target mRNA (Mukherji et al., 2011). Because mRNA degradation (~ 5 fold) is the primary result of miRNA regulation in this system, the target mRNA level per cell at the threshold is ~10 copies per cell. The threshold is due to titration of the miRNA available to interact with mRNAs containing the seed region. Because the cell contains hundreds, if not thousands, of different mRNAs interacting with the same seed, summation of background and the target mRNA accounts for the threshold. Different tissues or a particular tissue under different conditions exhibit different miRNA expression profiles and thus have different thresholds for a given target gene.
Figure 5.
miRNA-target interaction produces non-linear target protein output. Below a certain threshold of target mRNA production, the target is strongly repressed (pink). Above the threshold, repression is weaker and the target can exert competitive “sponge”-like effects (blue). The position of the threshold depends on miRNA concentration. Cartooned from (Mukherji et al., 2011) data.
This threshold effect could make a strong impact during a developmental transition in which a miRNA is upregulated and its pool of target genes are down-regulated in the anticorrelated pattern, as described by Stark et al. (2005) and Farh et al. (2005). At this point the miRNA’s effective concentration and, therefore its potency, could greatly increase for a small number of functionally important targets. On the other hand, the noise-buffering function of incoherent feed-forward loops could be significantly compromised by cross-talk from other targets outside the loop if those targets are themselves highly expressed or drastically fluctuating (Osella et al., 2011).
For protein-coding targets it has been hypothesized that some genes with conserved miRNA binding sites act as bona fide targets in one cell type or condition but as miRNAsequestering agents (“pseudotargets”) in other cell types or conditions (Seitz, 2009). Protein-coding genes that have miRNA binding sites but are knocked down without any functional consequence for their own protein have also been classified as neutral targets (Bartel, 2009). It is not controversial that miRNAs partition among all possible cytoplasmic target RNAs, including noncoding RNAs that have binding sites. The more overall target sites available in the transcriptome, the lower the effective concentration of a miRNA; this dilution effect was shown quantitatively by Arvey et al. (2010) and Garcia et al. (2011).
What is less evident is whether an RNA that contains miRNA binding sites has been selected to act as a miRNA target decoy for the regulatory benefit of other genes. A potential decoy RNA’s effectiveness must depend not only on how its expression compares to the miRNA’s abundance but also on the abundance of other target mRNAs in the cell. For example, the median target abundance for conserved vertebrate miRNA families is over 3000 binding sites in the genome (Garcia et al., 2011), which may correspond to thousands of competing binding sites in the transcriptome of a typical cell. The kind of gene that could exert meaningful effects by competing for miRNA is precisely the kind that, over the course of evolution, has tended to avoid taking on miRNA binding sites: housekeeping genes and highly expressed tissue-specific genes known as “antitargets” (Farh et al., 2005).
Recently there have been reports of endogenous transcripts that appear to act as natural miRNA decoys, called “competitive endogenous RNAs” (ceRNAs) (Poliseno et al., 2010; Cesana et al., 2011; Karreth et al., 2011; Sumazin et al., 2011; Tay et al., 2011). The most direct primary evidence for ceRNAs was the finding that the expression of a target of a particular combination of miRNAs was further suppressed when another such target gene in the cell was knocked down by siRNA. These results suggest that there is a highly delicate balance between the levels of miRNA seed families and their total pool of target mRNAs. However, what is lacking from these reports is a quantitative analysis of the molecules involved: how many molecules per cell of ceRNAs are competing for how many molecules of the miRNAs, against how many molecules of other target mRNAs? For reference, artificial miRNA target decoys or sponges that compete by binding to a miRNA family through seed sequence execute their function when expressed in the hundreds to thousands of copies per cell (Ebert et al., 2007). Karreth et al. (2011) and Tay et al. (2011) are able to observe miRNA inhibition when they use the format of these synthetic miRNA sponges in experiments that overexpress the 3' UTR of the purported ceRNA using transient plasmid transfection and a strong viral promoter rather than the endogenous ceRNA’s regulatory elements. But would a deletion of the miRNA binding sites from a ceRNA locus generate the same effect? While it is easy to see how large target ensembles can influence miRNA potency, it remains to be seen how modulation of individual ceRNA candidates could be causing significant effects simply by a miRNA titration mechanism.
Concluding remarks
Multicellular organisms must manage the tasks of development and physiology in unpredictable, changing environments and with imperfect genetic and biochemical components. Random noise in gene expression must be dampened or, as in the case of some cell fate decisions, harnessed in a system control network to designate one fate or another among neighboring cells. Robustness goes beyond the job of keeping one state the same in the face of perturbations. In development, it can mean not sending a signal until the right time, and then sending it strongly and irreversibly. Although miRNAs act to confer accuracy and uniformity to developmental transitions, the loss of a miRNA may result not in catastrophic defects but rather in imprecise, variable phenotypes. If other feedback or back-up mechanisms are in place, then the loss of robustness may only be detected by applying additional perturbations. The addition of miRNAs to metazoan genomes over time and the diversity of miRNA repertoires among different tissues of developing animals suggest that miRNAs are involved in reinforcing developmental decisions to make organismal complexity reliable and heritable from one generation to the next.
Acknowledgments
We thank David Bartel, Shankar Mukherji, Anthony Leung, and anonymous reviewers for critical reading and helpful suggestions. We thank Mary Lindstrom for assistance making figures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acar M, Mettetal JT, van Oudenaarden A. Stochastic switching as a survival strategy in fluctuating environments. Nat. Genet. 2008;40:471–475. doi: 10.1038/ng.110. [DOI] [PubMed] [Google Scholar]
- Arvey A, Larsson E, Sander C, Leslie CS, Marks DS. Target mRNA abundance dilutes microRNA and siRNA activity. Mol. Syst. Biol. 2010;6:363. doi: 10.1038/msb.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek J, Villen C, Shin FD, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bail S, Swerdel M, Liu H, Jiao X, Goff LA, Hart RP, Kiledjian M. Differential regulation of microRNA stability. RNA. 2010;16:1032–1039. doi: 10.1261/rna.1851510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake WJ, Balázsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ. Phenotypic consequences of promoter-mediated transcriptional noise. Mol. Cell. 2006;24:853–865. doi: 10.1016/j.molcel.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Bleris L, Xie Z, Glass D, Adadey A, Sontag E, Benenson Y. Synthetic incoherent feedforward circuits show adaptation to the amount of their genetic template. Mol. Syst. Biol. 2011;7:519. doi: 10.1038/msb.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- Brenner JL, Jasiewicz KL, Fahley AF, Kemp BJ, Abbott AL. Loss of individual microRNAs causes mutant phenotypes in sensitized genetic backgrounds in C. elegans. Curr. Biol. 2010;20:1321–1325. doi: 10.1016/j.cub.2010.05.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Stark A, Brennecke J, Cohen SM. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr. Biol. 2008;18:501–506. doi: 10.1016/j.cub.2008.02.081. [DOI] [PubMed] [Google Scholar]
- Cağatay T, Turcotte M, Elowitz MB, Garcia-Ojalvo J, Süel GM. Architecture-dependent noise discriminates functionally analogous differentiation circuits. Cell. 2009;139:512–522. doi: 10.1016/j.cell.2009.07.046. [DOI] [PubMed] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl. Acad. Sci. USA. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi PS, Zakhary L, Choi WY, Caron S, Alvarez-Saavedra E, Miska EA, McManus M, Harfe B, Giraldez AJ, Horvitz HR, Schier AF, Dulac C. Members of the miRNA-200 family regulate olfactory neurogenesis. Neuron. 2008;57:41–55. doi: 10.1016/j.neuron.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- Damiani D, Alexander JJ, O'Rourke JR, McManus M, Jadhav AP, Cepko CL, Hauswirth WW, Harfe BD, Strettoi E. Dicer inactivation leads to progressive functional and structural degeneration of the mouse retina. J. Neurosci. 2008;28:4878–4887. doi: 10.1523/JNEUROSCI.0828-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q, Meng P, Wang T, Qin W, Qin W, Wang F, Yuan J, Chen Z, Yang A, Wang H. MicroRNA let-7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2. PLoS One. 2010;5:e10147. doi: 10.1371/journal.pone.0010147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Meth. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- Gerstein MB, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons DL, Lin W, Creighton CJ, Rizvi ZH, Gregory PA, Goodall GJ, Thilaganathan N, Du L, Zhang Y, Pertsemlidis A, Kurie JM. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23:2140–2151. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466 doi: 10.1038/nature09267. 835-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimberg AM, Sempere LF, Moy VN, Donoghue PC, Peterson KJ. MicroRNAs and the advent of vertebrate morphological complexity. Proc. Natl. Acad. Sci. USA. 2008;105:2946–2950. doi: 10.1073/pnas.0712259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz H, Cohen SM. MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes Dev. 2010;24:1339–1344. doi: 10.1101/gad.1937010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V, Bushati N, Cohen SM. Drosophila microRNAs 263a/b confer robustness during development by protecting nascent sense organs from apoptosis. PLoS Biol. 2010;8:e1000396. doi: 10.1371/journal.pbio.1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstein E, Shomron N. Canalization of development by microRNAs. Nat. Genet. 2006;38(Suppl):S20–S24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc. Natl. Acad. Sci. USA. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H. Biological robustness. Nat. Rev. Genet. 2004;5:826–837. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat. Neurosci. 2007;10:1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natl. Acad. Sci. USA. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, Kirsch DG, Golub TR, Jacks T. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CT, Risom T, Strauss WM. Evolutionary conservation of microRNA regulatory circuits: an examination of microRNA gene complexity and conserved microRNA-target interactions through metazoan phylogeny. DNA Cell Biol. 2007;26:209–218. doi: 10.1089/dna.2006.0545. [DOI] [PubMed] [Google Scholar]
- Lestas I, Vinnicombe G, Paulsson J. Fundamental limits on the suppression of molecular fluctuations. 2010;467:174–178. doi: 10.1038/nature09333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AKL, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell. 2011;42:489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine E, Zhang Z, Kuhlman T, Hwa T. PLoS Biol. 2007;5:e229. doi: 10.1371/journal.pbio.0050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, Klein LO, Davis MM, Chen CZ. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Li X, Carthew RW. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell. 2005;123:1267–1277. doi: 10.1016/j.cell.2005.10.040. [DOI] [PubMed] [Google Scholar]
- Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137:273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, Chau N, Cleary M, Jackson AL, Carleton M, Lim L. Transcripts Targeted by the MicroRNA-16 family cooperatively regulate cell cycle progression. Mol. Cell Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Barrasa MI, Hammell M, Sequerra R, Doucette-Stamm L, Roth FP, Ambros VR, Walhout AJ. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev. 2008;22:2535–2549. doi: 10.1101/gad.1678608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros LA, Dennis LM, Gill ME, Houbaviy H, Markoulaki S, Fu D, White AC, Kirak O, Sharp PA, Page DC, Jaenisch R. Mir-290-295 deficiency in mice results in partially penetrant embryonic lethality and germ cell defects. Proc. Natl. Acad. Sci. USA. 2011;108:14163–14168. doi: 10.1073/pnas.1111241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y, Abreu-Goodger C, Staton AA, Stahlhut C, Shou C, Cheng C, Gerstein M, Enright AJ, Giraldez AJ. Zebrafish miR-1 and miR-133 shape muscle gene expression and regulate sarcomeric actin organization. Genes Dev. 2009;23:619–632. doi: 10.1101/gad.1760209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, Bartel DP, Ambros VR, Horvitz HR. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- modENCODE Consortium. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji S, Ebert MS, Zheng GZ, Tsang JS, Sharp PA, van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nat. Genet. 2011;43:854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson JR, Zheng GX, Burge CB, Sharp PA. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 2007;21:578–589. doi: 10.1101/gad.1522907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osella M, Bosia C, Corá D, Caselle M. The role of incoherent microRNA-mediated feedforward loops in noise buffering. PLoS Comput. Biol. 2011;7:e1001101. doi: 10.1371/journal.pcbi.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat. Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- Paulsson J. Summing up the noise in gene networks. Nature. 2004;427:415–418. doi: 10.1038/nature02257. [DOI] [PubMed] [Google Scholar]
- Pedraza JM, van Oudenaarden A. Noise propagation in gene networks. Science. 2005;307:1965–1969. doi: 10.1126/science.1109090. [DOI] [PubMed] [Google Scholar]
- Peterson KJ, Dietrich MR, McPeek MA. MicroRNAs and metazoan macroevolution: insights into canalization, complexity, and the Cambrian explosion. Bioessays. 2009;31:736–747. doi: 10.1002/bies.200900033. [DOI] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulou AS, Dooley J, Linterman MA, Pierson W, Ucar O, Kyewski B, Zuklys S, Hollander GA, Matthys P, Gray DH, De Strooper B, Liston A. The thymic epithelial microRNA network elevates the threshold for infection-associated thymic involution via miR-29a mediated suppression of the IFN-α receptor. Nat. Immunol. 2011;13:181–187. doi: 10.1038/ni.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulikkan JA, Dengler V, Peramangalam PS, Peer Zada AA, Müller-Tidow C, Bohlander SK, Tenen DG, Behre G. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood. 2010;115:1768–1778. doi: 10.1182/blood-2009-08-240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Rifkin SA, Andersen E, van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463:913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK. Noise in gene expression: origins, consequences, and control. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H. Redefining microRNA targets. Curr. Biol. 2009;19:870–873. doi: 10.1016/j.cub.2009.03.059. [DOI] [PubMed] [Google Scholar]
- Shkumatava A, Stark A, Sive H, Bartel DP. Coherent but overlapping expression of microRNAs and their targets during vertebrate development. Genes Dev. 2009;23:466–481. doi: 10.1101/gad.1745709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P, Bejarano F, Wang D, Garaulet DL, Yang JS, Martin R, Bortolamiol-Becet D, Robine N, Hiesinger PR, Lai EC. A Drosophila genetic screen yields allelic series of core microRNA biogenesis factors and reveals post-developmental roles for microRNAs. RNA. 2011;17:1997–2010. doi: 10.1261/rna.2983511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc. Natl. Acad. Sci. USA. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3'UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Staton AA, Knaut H, Giraldez AJ. miRNA regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration. Nat. Genet. 2011;43:204–211. doi: 10.1038/ng.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, Rajbhandari P, Bansal M, Guarnieri P, Silva J, Califano A. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–381. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang JS, Ebert MS, van Oudenaarden A. Genome-wide dissection of microRNA functions and cotargeting networks using gene set signatures. Mol. Cell. 2010;38:140–153. doi: 10.1016/j.molcel.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- Yoo AS, Greenwald I. LIN-12/Notch activation leads to microRNA-mediated down-regulation of Vav in C. elegans. Science. 2005;310:1330–1333. doi: 10.1126/science.1119481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GX, Ravi A, Calabrese JM, Medeiros LA, Kirak O, Dennis LM, Jaenisch R, Burge CB, Sharp PA. A latent pro-survival function for the mir-290-295 cluster in mouse embryonic stem cells. PLoS Genet. 2011;7:e1002054. doi: 10.1371/journal.pgen.1002054. [DOI] [PMC free article] [PubMed] [Google Scholar]