Abstract
Bacillus subtilis encodes redox-sensing MarR-type regulators of the OhrR and DUF24-families that sense organic hydroperoxides, diamide, quinones or aldehydes via thiol-based redox-switches. In this article, we characterize the novel redox-sensing MarR/DUF24-family regulator HypR (YybR) that is activated by disulphide stress caused by diamide and NaOCl in B. subtilis. HypR controls positively a flavin oxidoreductase HypO that confers protection against NaOCl stress. The conserved N-terminal Cys14 residue of HypR has a lower pKa of 6.36 and is essential for activation of hypO transcription by disulphide stress. HypR resembles a 2-Cys-type regulator that is activated by Cys14–Cys49′ intersubunit disulphide formation. The crystal structures of reduced and oxidized HypR proteins were resolved revealing structural changes of HypR upon oxidation. In reduced HypR a hydrogen-bonding network stabilizes the reactive Cys14 thiolate that is 8–9 Å apart from Cys49′. HypR oxidation breaks these H-bonds, reorients the monomers and moves the major groove recognition α4 and α4′ helices ∼4 Å towards each other. This is the first crystal structure of a redox-sensing MarR/DUF24 family protein in bacteria that is activated by NaOCl stress. Since hypochloric acid is released by activated macrophages, related HypR-like regulators could function to protect pathogens against the host immune defense.
INTRODUCTION
Reactive oxygen species (ROS) can be generated as a byproduct of respiration. Pathogenic bacteria encounter ROS that are produced as defense by the innate immune system (1,2). During infection activated macrophages release the enzyme myeloperoxidase that utilizes H2O2 to produce the strong oxidant hypochloric acid (HOCl) to kill pathogenic bacteria (3,4). ROS can further generate secondary reactive electrophilic species (RES) (5,6). Bacteria can sense and respond to ROS and RES by expression of dedicated detoxification mechanisms. ROS are sensed by redox-sensitive transcriptional regulators that undergo thiol-disulphide switches leading to activation or inactivation of the transcription factors (7). The OxyR regulator of Escherichia coli is one of the best studied bacterial peroxide-sensors. OxyR is activated by intramolecular disulphide formation resulting in transcription of genes with antioxidant functions (8–11). In addition, the redox-controlled chaperone Hsp33 provides specific protection against HOCl-induced protein aggregation in E. coli (12). Bleach leads to oxidation of the Zn-redox switch centres with subsequent Zn-release, oxidative unfolding, dimerization and activation of Hsp33. Organic peroxides are sensed by the conserved MarR-type repressor OhrR that controls the thiol-dependent peroxidase OhrA (7,13). The OhrR family includes one- and two-Cys OhrR-proteins that differ in their redox-sensing mechanisms. OhrRXc of Xanthomonas campestris is the prototype of the two-Cys family that is oxidized to an intermolecular disulphide between the opposing OhrR subunits (14,15). One-Cys OhrR proteins harbour one conserved N-terminal Cys with the prototype of B. subtilis OhrRBs that is oxidized to S-bacillithiolated OhrR in response to cumene hydroperoxide and HOCl (7,16,17). Redox-sensing MarR-type regulators of pathogenic bacteria include the OhrR-paralogs MgrA and SarZ of Staphylococcus aureus as global regulators for antibiotic resistance, virulence and anaerobiosis, the multidrug-efflux regulator MexR and the oxidative stress response and pigment production regulator OspR of Pseudomonas aeruginosa (7,13,18–23).
In addition, B. subtilis encodes redox-sensing MarR/DUF24-family regulators that sense specifically electrophiles (diamide, quinones or aldehydes) (7,24–28). The paralogous repressors YodB and CatR are inactivated via intermolecular disulphide formation by diamide and quinones resulting in derepression of the azoreductase (AzoR1), nitroreductase (YodC) and thiol-dependent dioxygenase (CatE) catalysing the reduction or ring-cleavage of the electrophiles (24,25,27). Other proteins of the MarR/DUF24 family (HxlR) and MerR/NmlR-families (AdhR) sense aldehydes (formaldehyde and methylglyoxal) via conserved Cys residues (26,28–30). However, the genome of B. subtilis encodes MarR/DUF24 family regulators of unknown functions, including YybR, YdeP, YdzF, YkvN and YtcD (Supplementary Figure S1A).
In this study, we characterize the hypochloric acid-specific regulator YybR (renamed HypR) as a novel MarR/DUF24 transcriptional regulator that positively controls the putative nitroreductase YfkO. HypR resembles a two-Cys-type MarR-type regulator that is activated by Cys14–Cys49′ intersubunit disulphide formation. We present the crystal structure of HypR under reduced and oxidized conditions and provide for the first time insights into the redox-sensing mechanism of a MarR/DUF24 family regulator.
MATERIALS AND METHODS
Bacterial strains and growth conditions
The bacterial strains used were B. subtilis 168 (trpC2), ΔhypR (trpC2,hypR::Cmr), ΔhypO (trpC2,hypO::Cmr), ΔohrA (trpC2,ohrA::Spcr), ΔhypOΔohrA (trpC2,hypO::Cmr,ohrA::Spcr) and the hypRC14S, hypRC49S, hypRC14,49S point mutants (Supplementary Table S1). Bacillus subtilis strains were cultivated under vigorous agitation at 37°C in Belitsky minimal medium described previously (31). Escherichia coli strains were grown in LB for DNA manipulation. The antibiotics were used at the following concentrations: 1 µg/ml erythromycin, 25 µg/ml lincomycin, 5 µg/ml chloramphenicol, 10 µg/ml kanamycin, 100 µg/ml spectinomycin. The compounds used were 2-methylhydroquinone (Acros), diamide (diazinedicarboxylic acid bis(N,N-dimethylamide) (Sigma) and sodium hypochlorite (15% stock solution) (Sigma).
Gene deletions for construction of the hypO mutant were generated using long-flanking-homology polymerase chain reaction (LFH–PCR) as previously described (25). Primers yfkO-F1 and yfkO-F2 were used to amplify the up fragment and primers yfkO-R1 and yfkO-R2 to amplify the down fragment, respectively (Supplementary Table S2). Fragments were amplified and joined together with the chloramphenicol cassette using Pfusion DNA polymerase (Invitrogen) as described (32). Plasmid pCm::spec was used to replace the Cmr cassette with a Specr marker to generate strain ΔohrA::Spec (17). The ΔohrAΔhypO double mutant was constructed by transformation of chromosomal DNA of the hypO mutant into competent cells of the ΔohrA::Spec mutant strain. Integration and deletion of the ohrA and hypO genes were confirmed by PCR.
Construction of the hypRC14S, hypRC49S and hypRC14,49S point mutants
Plasmids pDGhypR, pDGhypRC14S, pDGhypRC49S, pDGhypRC14,49S were produced by using PCR mutagenesis. Using primers yybR-C14S-for1 and yybR-C14S-rev2 and B. subtilis wild-type chromosomal DNA, the hypR gene was amplified by PCR, the PCR product digested with EcoRI and BamHI restriction enzymes and inserted into plasmid pDG795 digested with the same enzymes to generate pDGhypR. For construction of pDGhypRC14S, first PCR was performed in two separate reactions using primers yybR-C14S-for1 with yybR-C14S-rev1 and primers yybR-C14S-for2 with yybR-C14S-rev2 (Supplementary Table S2) and B. subtilis 168 chromosomal DNA as template. The PCR products were hybridized and amplified by a second PCR using primers yybR-C14S-for1 and yybR-C14S-rev2. The PCR products were digested with EcoRI and BamHI and inserted into plasmid pDG795 digested with the same enzymes to generate pDGhypRC14S. For construction of pDGhypRC49S and pDGhypRC14,49S, first PCRs were performed in two separate reactions using primers yybR-C49S-for1 with yybR-C49S-rev1 and primers yybR-C49S-for2 with yybR-C49S-rev2 (Supplementary Table S2) and B. subtilis 168 chromosomal DNA and pDGhypRC14S plasmid DNA as templates, respectively. The PCR products were hybridized and amplified by a second PCR using primers yybR-C49S-for1 and yybR-C49S-rev2. The PCR products from the second PCRs were digested with EcoRI and BamHI and inserted into plasmid pDG795 digested with the same enzymes to generate plasmids pDGhypRC49S and pDGhypRC14,49S. The plasmids pDGhypRC14S, pDGhypRC49S and pDGhypRC14,49S were verified by DNA sequencing and transformed into the thrC locus of the B. subtilis ΔhypR mutant.
Northern blot experiments
Northern blot analyses were performed as described (17) using RNA isolated from B. subtilis wild-type cells before (control) and 10 min after the treatment with 50 µM NaOCl, 1 mM diamide and 0.5 mM MHQ. Hybridizations specific for hypR and hypO were performed with the digoxigenin-labelled RNA probes synthesized in vitro using T7 RNA polymerase from T7 promoter containing internal PCR products of the respective genes using the primer sets yybR-T7for with yybR-T7rev and yfkO-T7for with yfkO-T7rev (Supplementary Table S2).
Primer extension experiments
Primer complementary to the N-terminus encoding region of hypO (yfkO-FT-rev2) and hypR (yybR-C14S-rev1) were 5′-end labelled using T4 polynucleotide kinase (Roche Diagnostics) and 50 µCi [γ32P]-ATP (GE Healthcare). Primer extension analysis was performed using the labelled primers as described previously (25). Sequencing of the corresponding promoter regions was performed using PCR products as templates containing the promoter region of the respective genes amplified with primer set yybR-C14S-for1 and yybR-Cys14S-rev1 and yfkO-FT-for2 and yfkO-FT-rev2 (Supplementary Table S2).
Expression and purification of recombinant His-HypR, His-HypRC14S and His-HypRC49S proteins
Escherichia coli BL21(DE3)pLysS (Invitrogen) was used for overproduction of His-tagged HypR, HypRC14S and HypRC49S proteins. For expression of His-HypR, His-HypRC14S and His-HypRC49S proteins, the hypR coding sequence was amplified by PCR using primers yybR-NdeI-pETfor and yybR-BamHI-pETrev and chromosomal DNA of the B. subtilis wild-type and the hypRC14S and hypRC49S point mutants as templates (Supplementary Table S2). The reverse primer includes the codons for six C-terminal histidine residues. The hypR specific PCR products were digested with NdeI and BamHI and inserted into the pET11b expression plasmid digested with the same enzymes to generate plasmids pEThypR, pEThypRC14S and pEThypRC49S. The hypR, hypRC14S and hypRC49S mutant sequences were verified by DNA sequencing (Supplementary Table S2). Escherichia coli BL21(DE3)pLysS carrying the pEThypR, pEThypRC14S and pEThypRC49S expression plamids was cultured in 1 l LB medium, and 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added at the mid-log phase (OD600 of 0.8) for 2 h. Recombinant His-tagged HypR, HypRC14S and HypRC49S proteins were purified using PrepEaseTM His-Tagged High Yield purification Resin (USB) under native conditions according to the instructions of the manufacturer. HypR was eluted in 50 mM NaH2PO4, 300 mM NaCl, and 250 mM imidazole pH 8.0. The proteins were further purified by anion exchange chromatography (POROS 20 HQ, Applied Biosystems) with 20 mM Tris–HCl pH 7.5 and a 0 to 1 M NaCl gradient followed by dialysis into 10 mM Tris–HCl pH8.0, 100 mM NaCl, 50% glycerol (v/v) and stored at −80°C.
DNA gel mobility shift assays
DNA fragments containing the hypO promoter region were generated by PCR using the primer sets yfkO-FT-for2 and yfkO-FT-rev2 (Supplementary Table S2). Approximately 600 pmol of the purified PCR product was end-labelled using T4 polynucleotide kinase (Roche Diagnostics) and 50 µCi [γ32P]-ATP (GE Healthcare). The labelled hypO-specific promoter probe was purified by ammonium sulphate/ethanol precipitation and 2000 cpm was incubated with different amounts of purified HypR proteins for 10 min at room temperature in EMSA-binding buffer (10 mM Tris–HCl pH 7.5, 100 mM KCl, 5% glycerol (v/v) in the presence of 50 µg/ml of BSA and 5 µg/ml of Salmon sperm DNA. The concentrations of the compounds used for the DNA-binding assays were 1 mM NaOCl, 1 mM diamide, 1 mM DTT. DNA-binding reactions were separated by 4% native polyacrylamide gel electrophoresis in 10 mM Tris–HCl pH 8, 1 mM EDTA buffer, containing 2.5% glycerol at room temperature and constant voltage (250 V) for 15 min. Gels were dried and the radiolabelled bands were visualized using phosphoimaging.
DNase-I footprinting analysis
Primer yfkO-FT-for2 and yfkO-FT-rev2 were each 5′-end labelled using T4 polynucleotide kinase (Roche Diagnostics) and 50 µCi [γ32P]-ATP (GE Healthcare) and purified using ethanol precipitation. DNA probes for hypO (corresponding to positions −160 to +79 relative to TSS), were synthesized by PCR amplification using one 5′-end-labelled primer and the corresponding non-labelled primer, respectively (Supplementary Table S2). Purified PCR fragments that are labelled at one 5′-end were used as sequencing templates. DNA probe purification and DNase I footprinting was performed as described previously (25).
In vitro transcription assay
In vitro transcription assays were performed as described previously (33) using 0.5 µM HypR, HypRC14S and HypRC49S proteins which were added to the in vitro transcription reactions. Templates for a 380-bp PCR product containing the hypO promoter were generated by PCR with primers yfkO-FT-for2 and yfkO-FT-rev3 (Supplementary Table S2), gel purified and used at 0.1 pmol in each reaction. Core E. coli RNA polymerase [Epicentre Biotechnologies (Madison, WI, USA)] was used at 2 pmol per reaction and combined with 8 pmol purified SigmaA protein from B. subtilis (molar ratio 1:4) that was expressed from plasmid pNG590 according to (34). RNAP holoenzyme formation was allowed for 15 min on ice prior to addition to the reactions containing HypR protein that was reduced by DTT or oxidized by diamide and NaOCl. Transcription reactions were performed for 15 min at 37°C and terminated by addition of stop buffer. Reactions were separated on 8.3 M urea–6% polyacrylamide gels and the run-off transcripts visualized by phosphoimaging.
Western blot analysis
Anti-HypR polyclonal rabbit antiserum was generated using purified His-tagged HypR protein. The polyclonal antisera were used for immunoprecipitation experiments and western blot experiments at 1:200 dilution. Protein amounts of 25 µg were loaded onto a 15% SDS–PAGE gel and the western blot analysis was performed as described previously (25).
Immunoprecipitation and non-reducing/reducing diagonal SDS–PAGE analysis
Bacillus subtilis cells were treated with 1 mM diamide and 50 µM NaOCl and harvested with 50 mM iodoacetamide (IAM) to alkylate all reduced thiols. Cells were sonicated and the protein extracts obtained after repeated centrifugation. Immunoprecipitation using HypR-specific antibodies was performed with Dynabeads-ProteinA (Invitrogen) according to the instructions of the manufacturer. The precipitated proteins were eluted by boiling in non-reducing SDS sample buffer (4% SDS; 62.5 mM Tris–HCl pH 8.0, glycerol). Immunoprecipitated HypR protein was separated using the non-reducing/reducing diagonal SDS–PAGE analysis as described previously (35) and subjected to HypR-specific western blot analysis.
Proteome analysis
Preparation of cytoplasmic l-[35S]methionine-labelled proteins from cells treated with 0.5 mM MHQ or 1 mM diamide and separation by 2D gel electrophoresis (2D–PAGE) was perfomed as described (36). The image analysis was performed with the DECODON Delta 2D software (http://www.decodon.com).
MALDI–TOF mass spectrometry of in vitro oxidized HypR protein
The HypR protein was oxidized with NaOCl and alkylated with IAM introducing a mass shift of 57 Da in reduced Cys residues. Then HypR protein was reduced with DTT and alkylated with NEM leading to a mass shift of 125 Da in oxidized Cys. HypR protein was separated by non-reducing SDS–PAGE, digested with trypsin and the peptides were spotted onto MALDI-targets (Voyager DE-STR, PerSeptive Biosystems) and measured using a Proteome-Analyzer 4800 (Applied Biosystems, Foster City, CA, USA) as described (25).
Orbitrap-mass spectrometry of oxidized HypR
Stained gel-bands of immunoprecipitated HypR harvested from wild-type cells or purified His–HypR protein were tryptic digested. Tryptic peptides were separated and measured online by ESI-mass spectrometry using a nanoACQUITY UPLC™ system (Waters, Milford, MA, USA) coupled to an LTQ Orbitrap™ XL mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) as described (17). Post-translational modifications of HypR were identified by searching all MS/MS spectra against a B. subtilis target-decoy protein sequence database extracted from UniprotKB release 12.7 using Sorcerer™-SEQUEST®. The Sequest search was carried out considering the following parameter: parent ion mass tolerance of 10 ppm, fragment ion mass tolerance of 1.00 Da, Methionine oxidation (+15.99492 Da), cysteine carbamidomethylation (+57.021465 Da) and Cys+ICPSITQR peptide (+914.4873 Da) were set as variable modifications. Sequest identifications required at least ΔCn scores >0.10 and XCorr scores more than 2.2, 3.3 and 3.75 for doubly, triply and quadruply charged peptides.
Crystallization, data collection and refinement of reduced HypRC14S and oxidized HypR protein
His-tagged HypRC14S and HypR proteins were purified using Ni-affinity chromatography and anion-exchange chromatography. The HypRC14S protein was purified with β-mercaptoethanol and used for crystal structure determination of reduced HypR. The wild-type HypR protein was oxidized with 1 mM diamide before purification by anion exchange chromatography. Reduced HypRC14S protein was concentrated to 15 mg/ml in 150 mM NaCl, 20 mM Tris–HCl pH 8.0, 5 mM DTT and crystallized from 12% PEG 8000 by the hanging drop vapour diffusion method. Prism-shaped crystals appeared after a few days. Crystals were briefly soaked in 20% PEG 4000, 10% PEG 400 and cryocooled in the N2-stream at 100 K. Oxidized HypR was crystallized from 6% PEG 4000, 0.4 M Li2SO4 in the form of thick hexagonal plates or prisms. Fifteen per cent PEG 8000, 15% PEG 400 were used as cryoprotectant. Diffraction data were collected at beamline BL 14.2 at the synchrotron BESSY in Berlin, Germany (Table 1).
Table 1.
Crystallographic data collection and refinement statisticsa
| Reduced HypRC14S (+β-Mercaptoethanol) | Oxidized HypR (+Diamide) | |
|---|---|---|
| PDB entry | 4a5n | 4a5m |
| Data collection | ||
| Space group | P21 | P65 |
| Unit cell parameters | ||
| a, b, c (Å) | 54.93, 68.03, 60.42 | 84.6, 84.6, 358.6 |
| α, β, γ (°) | 90.00, 97.45, 90.00 | 90.00, 90.00, 120.00 |
| Resolution (Å) | 36.46–1.81 (1.90–1.81) | 100.00–3.00 (3.18–3.00) |
| Wavelength (Å) | 0.918 | 0.918 |
| Reflections, unique | 39025 (5528) | 24764 (3457) |
| Multiplicity | 2.3 (2.2) | 5.5 (4.6) |
| Completeness (%) | 96.5 (94.0) | 89.8 (77.4) |
| Mean, I/σ(I) | 8.9 (2.2) | 14.9 (1.4) |
| Mosaicity (°) | 1.15 (Scala) | 0.31 (XDS) |
| Wilson B-factor (Å2) | 23.4 | 82 |
| Rmeas | 0.080 (0.531) | 0.087 (1.089) |
| Refinement | ||
| Resolution | 59.9–1.81 | 45.6–3.0 |
| Protein atoms | 3408 | 6408 |
| Solvent / ligand atoms | 315/20 | 25/37 |
| Average B factor (Å2) | 37.2 | 107.9 |
| Rcryst/Rfree | 0.183/0.223 | 0.207/0.250 |
| r.m.s.d. bond lengths/angles (Å, °) | 0.013/1.53 | 0.008/1.73 |
aValues in parentheses belong to the highest resolution shell.
The structure of reduced HypR was solved by molecular replacement (37) using the YtcD structure from B. subtilis (PDB entry 2 hzt, 49% sequence identity for 133 amino acids). Refmac5 version 5.5.0110 (38) was used to refine using TLS and no NCS restraints (statistics, see Table 1). One ligand per monomer was identified as β-mercaptoethanol. A monomer of reduced HypR was used to solve the structure of the oxidized protein by molecular replacement. Pseudosymmetry of the eight monomers in the asymmetric unit about a 2-fold axis on a causes nearly perfect twinning of the P65 crystals with apparent P6522 symmetry in the diffraction pattern. To reduce the influence of model bias, the density was improved by non-crystallographic symmetry averaging over the eight copies in the asymmetric unit, using the RESOLVE density modification algorithm (39) as implemented in Phenix (40).
Secondary structure elements were assigned by DSSP (41). The dimerization interface was calculated by the PISA server (42). Figures of structural models were made with PyMOL (http://www.pymol.org). The structural data were submitted to the PDB database under the PDB entry 4a5n for reduced HypR and 4a5m for oxidized HypR.
DTNB assay to determine the pKa of Cys14 and Cys49 residues in His-HypR
The purified His-HypRC14S and His-HypRC49S mutant proteins were used to determine the pKa of the Cys14 and Cys49 residues by reaction with 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) generating 2-nitro-5 thiobenzoate. Defined amounts of proteins (4.66 µM HypRC14S or 4.93 µM HypRC49S) reacted with 10–20 µM DTNB in phosphate/citrate buffer (50 mM phosphate, 50 mM citrate, 100 mM NaCl) at various pH values ranging from pH 6 to 8.5. The pH-dependent rate of reactions k(pH) of the HypR Cys mutants with DTNB was measured as time-dependent absorbance change at 410 nm at room temperature using a Cary 50 spectrophotometer (Agilent Technologies, Waldbronn, Germany). The following equation was used for a first fit of the time-dependent absorbance change by non-linear regression:
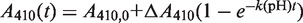 . The resulting pH-dependent rate of reactions k(pH) were used in a second fit to obtain the pKa of the thiol. The first order kinetic constant k(pH) is proportional to the fraction of deprotonated thiol, which is pH dependent:
. The resulting pH-dependent rate of reactions k(pH) were used in a second fit to obtain the pKa of the thiol. The first order kinetic constant k(pH) is proportional to the fraction of deprotonated thiol, which is pH dependent: 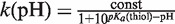 .
.
RESULTS
Identification of HypR (YybR) as hypochlorite-sensing positive regulator of the oxidoreductase HypO (YfkO)
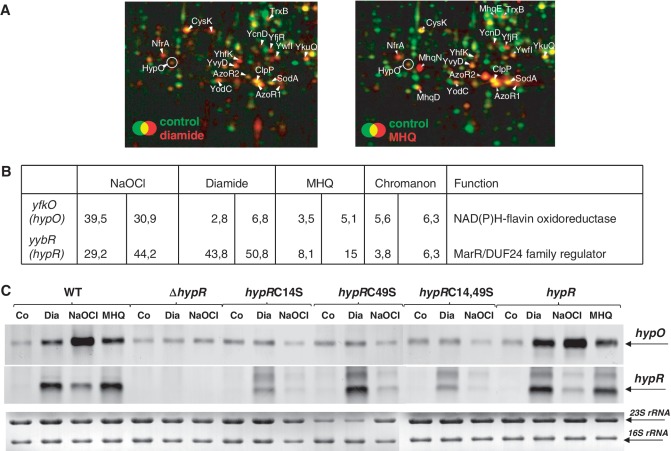
Previous transcriptome analyses revealed that the unknown DUF24-type regulator encoded by yybR responds strongly to diamide, quinones and hypochlorite (Figure 1B) (17,43,44). In addition, several FMN-dependent NAD(P)H oxidoreductase-encoding genes were up-regulated, including yfmJ, ytkL, ywnB, yqiG, yqjM, ywrO, ydeQ, yugJ and yfkO (Figure 1A and B) (43). Some of these redox enzymes are members of the Spx regulon, such as yqiG and yugJ suggesting that these could function in maintenance of the thiol-redox homeostasis (45,46). We were interested if the DUF24-family regulator YybR controls one of these oxidoreductases. Using northern blot analysis we found that the NAD(P)H-flavin oxidoreductase encoded by yfkO was strongly induced in the wild-type but not in the yybR mutant in response to diamide, NaOCl and MHQ stress (Figure 1C). This suggests that yybR positively controls yfkO transcription in response to diamide and NaOCl stress. In addition, yybR transcription is probably also strongly autoregulated by thiol-specific stress conditions. Hence, yybR was renamed as hypochlorite-responsive regulator hypR and yfkO was renamed as hypO. The northern blot analysis further revealed that hypR and hypO are transcribed monocistronically. Using primer extension, the 5′-ends of the hypO and hypR specific transcripts were mapped at T and A, respectively, located 31 and 55 bp upstream of the start codon (Supplementary Figure S2).
Figure 1.
HypO induction in the proteome (A), transcriptional induction of hypO and hypR in the transcriptome (B) and northern blot analysis of hypO and hypR transcription (C) under thiol-specific stress conditions. (A) Cytoplasmic proteins were labelled with 35S-methionine before (control) and after stress exposure and separated by 2D–PAGE as described (36). Close-ups of the overlay proteome images are shown for the wild-type before (green images) and 10 min after exposure to 1 mM diamide (left, red image) or 0.5 mM MHQ (right, red image). Proteins with increased protein synthesis ratios after MHQ and diamide stress including HypO are labelled that were identified from Coomassie-stained 2D gels as described (36). (B) The values represent fold-changes of hypR and hypO induction ratios in two biological replicates of transcriptome experiments of cells treated with 1 mM diamide, 0.5 mM MHQ, 50 µM NaOCl and 100 µM chromanon according to previously published transcriptome data (17,26,44). (C) Northern blot analysis was performed using RNA isolated from B. subtilis wild-type, the ΔhypR mutant and the ΔhypR mutant complemented with hypR, hypRC14S, hypRC49S and hypRC14,49S before (co) and 10 min after treatment with 0.5 mM MHQ, 1 mM diamide and 50 µM NaOCl. The arrows point toward the hypO and hypR specific transcripts. The methylen-blue stained northern blot is shown below as RNA loading control and the 16S and 23S rRNAs are labelled.
The HypR-controlled nitroreductase HypO confers resistance to NaOCl stress
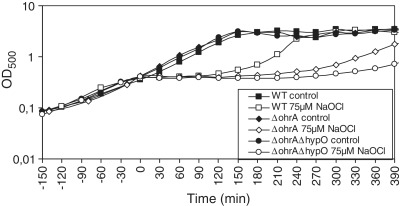
Previous phenotype analyses have shown that the OhrR-controlled OhrA peroxiredoxin is a specific determinant of NaOCl detoxification (17). Recently, it was shown that HypO shows nitroreductase activity in vitro (47). We analysed the growth phenotype of ΔhypR and ΔhypO single and ΔhypOΔohrA double mutants to investigate whether the HypO nitroreductase contributes to NaOCl resistance. No differences in the sensitivities of the ΔhypO and ΔhypR single mutants were detected compared to the wild-type (Supplementary Figure S3A and S3B). However, the growth of the ΔhypOΔohrA double mutant was significantly more impaired than that of the ΔohrA single mutant after treatment with 75 µM NaOCl (Figure 2 and Supplementary Figure S3C). This indicates that HypO also provides protection against NaOCl stress in B. subtilis.
Figure 2.
The OhrA peroxiredoxin and the nitroreductase HypO protect cells against hypochlorite toxicity. Growth phenotype of B. subtilis wild-type (WT), ΔohrA and ΔohrAΔhypO mutant strains that were treated with 75 µM NaOCl at an OD500 of 0.4. The growth curves are representives of at least three independent growth experiments. Two biological replicates are shown in Supplementary Figure S3A–S3C.
Cys14 and Cys49 of HypR are essential for activation of hypO transcription
HypR has two Cys residues (Cys14 and Cys49) and Cys14 is conserved among DUF24-family regulators (Supplementary Figure S1A). We investigated the role of Cys14 and Cys49 of HypR in regulation of hypO transcription in hypRC14S, hypRC49S and hypRC14,49 S mutants in vivo (Figure 1C). Northern blot analysis revealed that hypO transcription was abolished in the hypRC14S, hypRC49S and hypRC14,49 S mutants by diamide and NaOCl in vivo. These results clearly indicate, that both Cys14 and Cys49 of HypR are essential for activation of hypO transcription by disulphide stress in vivo.
Transcription of hypR is possibly autoinduced in the wild-type and in the hypR complemented ΔhypR mutant strain by disulphide stress. However, the hypR-specific mRNA amounts were also induced in the hypRC14S, hypRC49S and hypRC14,49 S mutants after diamide stress, although hypR transcription was strongly decreased in the hypRC14S and hypRC14,49 S mutants compared to the wild-type (Figure 1C). The response of the HypRC49S protein to disulphide stress could be caused by S-thiolation of the redox-sensitive Cys14 residue, leading to autoinduction of hypR transcription. It is also possible that hypR transcription is controlled by another MarR/DUF24-family paralog (e.g. YdeP or YkvN) in the hypRC14S, hypRC49S and hypRC14,49 S mutants explaining the response of the hypRCys mutants.
Identification of HypR operator sites and effect of disulphide stress on DNA-binding activity of HypR in vitro
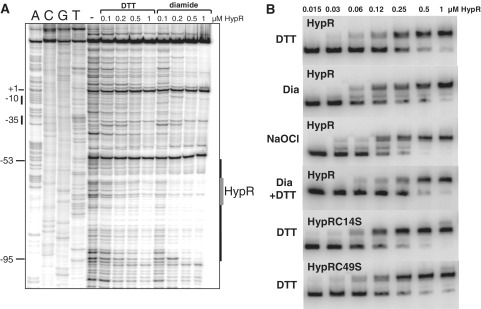
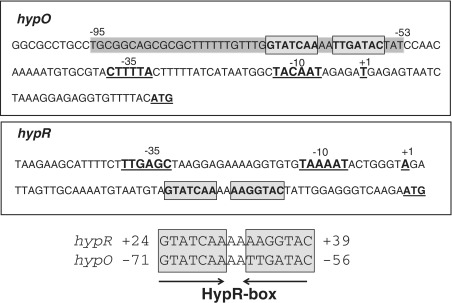
DNase-I footprinting analysis was performed to identify the cis-acting sequences which function as operator sites for HypR binding in the hypO upstream region in vitro. HypR-His protein protected a region upstream of the hypO promoter from positions −53 to −95 relative to the transcription start site (Figure 3A). The protected region contains a 7-2-7 bp inverted repeat GTATCAAAATTGATAC that is also present at positions +24 to +39 downstream of the hypR promoter (Figure 4). The position of this HypO-box confirms the notion that HypR is a positive transcriptional regulator of hypO transcription, but probably represses its own transcription. Furthermore, we were interested if the DNA-binding activity is affected by diamide and NaOCl in vitro and performed gel-shift and DNase-I footprinting analysis of HypR protein under reduced and oxidized conditions. The gel-shift experiments showed binding of HypR to the hypO operator sites at similar affinities under reduced and oxidized conditions (Figure 3B). The calculated dissociation constants (Kd) were 0.18 µM for reduced HypR, 0.14 µM for diamide-oxidized HypR and 0.12 µM for NaOCl-oxidized HypR proteins (Supplementary Figure S4A and S4B). This indicates no significant change in the DNA-binding affinities of reduced and oxidized HypR proteins. Similar Kd values were calculated for reduced HypRC14S and HypRC49S mutant proteins with 0.14 and 0.12 µM, respectively and oxidation caused no significant change in the DNA-binding affinities of the Cys mutant proteins (Supplementary Figure S4A and S4B). However, we observed a change in the mobility of oxidized HypR compared to reduced HypR in the gel-shift assays which was DTT-reversible (Figure 3B). In addition, the DNase-I footprinting analysis showed a higher affinity of oxidized HypR protein to the hypO promoter region indicating an increased DNA-binding activity of oxidized HypR protein in vitro (Figure 3A).
Figure 3.
DNase-I footprinting experiments (A) and gel-shift experiments (B) of purified HypR protein to the hypO promoter in the presence of DTT, diamide and NaOCl. (A) The HypR-protected operator sequence is indicated at the right side of the DNase-I footprint including an 7-2-7 bp inverted repeat with the sequence GTATCAAAATTGATAC that is labelled by arrows in the sequence alignment in Figure 4. The positions relative to the transcriptional start site are shown on the left. Transcription start site is indicated by (+1). For dideoxynucleotide sequencing, the dideoxy nucleotide added in each reaction is indicated above the corresponding lane. HypR protein was treated with 1 mM DTT or 1 mM diamide prior to the DNA-binding reactions. (B) EMSAs were used to analyse the effect of DTT, 1 mM diamide and 100 µM NaOCl on the DNA-binding activity of purified HypR, HypRC14S and HypRC49S proteins to the labelled hypO promoter probe. The HypR protein amounts used for the DNA-binding reactions are indicated. The EMSA experiments are representives of three replicate experiments. The change in the DNA-binding affinity and dissociation constants (Kd) of reduced and oxidized HypR, HypRC14S and HypRC49S proteins are calculated in Supplementary Figure S4A and B.
Figure 4.
Sequence alignment of the HypR-boxes in the hypO and hypR promoter regions. The hypO and hypR promoter sequences (−10 and −35), the transcription start site (+1) and the ATG start codons are underlined in the hypO and hypR upstream regions. The conserved HypR-boxes including the inverted repeats are boxed and indicated by arrows. The HypR protected region identified by the DNase-I footprinting analysis is grey shaded.
Transcription of hypO is activated by HypR after diamide and NaOCl stress in vitro
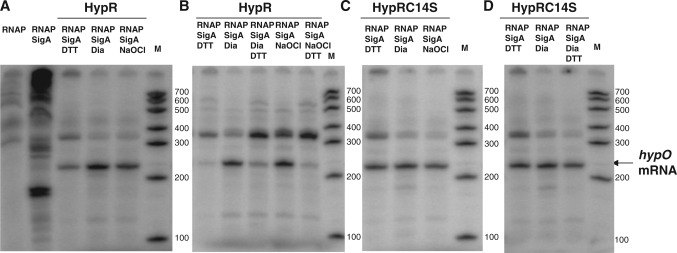
Next, we analysed whether transcription of hypO is activated by oxidized HypR using an in vitro transcription assay. In brief, using E. coli RNA polymerase core enzyme (RNAP) and purified SigmaA and HypR proteins from B. subtilis we performed the in vitro transcription assay for the hypO gene as described previously (33). This assay showed the production of a hypO run-off transcript of the expected size of 220 bp only when HypR was present in the in vitro reactions, indicating that HypR activates transcription of the hypO gene in vitro (Figure 5). Transcription of hypO increased ∼2.5-fold when HypR protein was oxidized by diamide and NaOCl treatment compared to reactions with DTT-reduced HypR (Figure 5A and Supplementary Figure S4C). This indicates that the increased DNA-binding affinity of oxidized HypR protein to the hypO promoter observed in the DNase-I footprinting analyses, results in increased activation of hypO transcription in vitro. In addition, hypO specific transcription ratios decreased in the in vitro transcription reactions containing oxidized HypR protein that was subsequently reduced with DTT (Figure 5B and Supplementary Figure S4C). In contrast, transcription of hypO was not increased by oxidation of HypRC14S mutant protein in vitro and also not affected by oxidized HypRC14S protein that was treated subsequently with DTT (Figure 5C and D; Supplementary Figure S4C). These results show that oxidation of HypR increases activation of hypO transcription by the RNAP in vitro.
Figure 5.
In vitro transcription analysis of hypO in the presence of RNA polymerase holoenzyme (RNAP) and purified HypR (A and B) and HypRC14S proteins (C and D) treated with DTT, diamide or NaOCl. The reactions in (A) and (C) show increased hypO transcription ratios by oxidized HypR but not by oxidized HypRC14S mutant protein. The reactions in (B) and (D) show reduced hypO transcription ratios by oxidized HypR that is subsequently reduced with DTT, but no change in hypO transcription by oxidized HypRC14S that is subsequently reduced with DTT. The in vitro-transcription analyses of hypO are representives of three replicate experiments and the relative transcription ratios were quantified in Supplementary Figure S4C. RNA size standard was generated using the Perfect RNA marker template mix (Novagen). The hypO specific run-off transcript is labelled at a size of 220 bp.
HypR senses diamide and NaOCl by a Cys14–Cys49′ thiol-disulphide switch
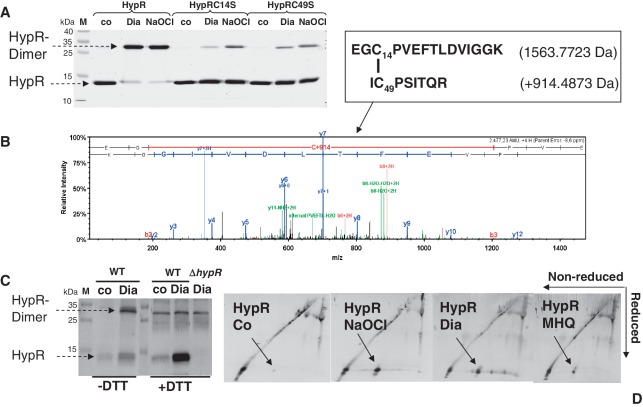
To analyse the oxidative modification of HypR in vitro, purified HypR-His-protein was treated with diamide and NaOCl and separated using non-reducing SDS–PAGE. HypR is reversibly oxidized to intersubunit disulphides by diamide and NaOCl stress since it migrates at the size of the HypR dimer upon oxidation (Figure 6A). In contrast, the majority of HypRC14S and HypRC49S mutant proteins do not form intermolecular disulphides upon oxidation.
Figure 6.
HypR is oxidized to Cys14–Cys49′ intersubunit disulphides in response to diamide and NaOCl in vitro (A and B) and in vivo (C and D). (A) His-HypR protein was treated with 1 mM diamide or 100 µM NaOCl, alkylated with 50 mM IAM and subjected to non-reducing SDS–PAGE analysis. The bands corresponding to the oxidized HypR disulphide dimer were tryptically digested and analysed using LTQ-Orbitrap mass spectrometry. (B) The Cys14–Cys49'-intermolecular disulphide-containing peptide was observed as quadruply charged precursor ion at an m/z = 620.3151. The CID MS/MS spectrum of this Cys14–Cys49′-disulphide peptide is shown with b and y ion fragment ions in red and blue. The detailed MS/MS data for this peptide including the Xcorr, ΔCn scores, precursor ion and neutral molecular masses of the peptide and the fragment ion seria are presented in Supplementary Figure S6. (C) 1D-western blot analysis and (D) 2D-diagonal western blot analysis of immunoprecipitated HypR protein purified from wild-type cells before (co) and after exposure to diamide and NaOCl in the presence of 50 mM IAM using HypR-specific polyclonal antibodies. The HypR-specific intersubunit disulphide is indicated by an arrow that was also visible in non-reducing SDS-gels and analysed using Orbitrap LC–MS/MS. The CID MS/MS spectrum, the Xcorr, ΔCn scores, precursor ion and neutral molecular masses of the peptide and fragment ion seria of this Cys14–Cys49′-disulphide peptide of HypR purified from cells in vivo are presented in Supplementary Figure S6.
Using MALDI–TOF–MS, we analysed the Cys-containing tryptic peptides of reduced and oxidized HypR proteins. In the mass spectrum of oxidized HypR the mass peak of 2478.0686 Da corresponds to the Cys14-peptide (EGCPVEFTLDVIGGK) of 1563.7723 Da that is disulphide linked to the Cys49′-peptide (ICPSITQR) of 914.4873 Da (Supplementary Figure S5A and S5B). Detailed CID MS/MS analysis of this 4-fold charged peptide with an m/z = 620.315 confirmed that Cys14 is linked to Cys49' in the HypR intersubunit-disulphide-linked dimer (Figure 6B, Supplementary Figure S6A, S6C, S6D and S6E).
In addition, a differential Cys-alkylation approach by IAM and NEM was used to trap the oxidized Cys-peptides in HypR (Supplementary Figure S5C–S5F). Reduced and oxidized HypR proteins were first alkylated with IAM. The proteins were washed by acetone precipitation, resolved in 8 M urea, followed by DTT reduction of oxidized thiols and alkylation with NEM. Reduced Cys residues should then be IAM alkylated and oxidized Cys residues NEM alkylated. The tryptic Cys14 and Cys49 peptides of reduced and oxidized HypR proteins were analysed by MALDI–TOF–MS. The Cys49 peptide was alkylated predominantly with IAM in reduced HypR (mass peak 974.50 Da) and with NEM in oxidized HypR (mass peak 1042.53 Da). The Cys14 peptide was alkylated mostly with NEM in oxidized HypR (mass peak 1688.87 Da). This differential IAM/NEM thiol-trapping approach confirms that both Cys14 and Cys49 are reversibly oxidized in HypR protein in vitro.
The oxidation of HypR was analysed in vivo by non-reducing western blot analyses (Figure 6C) and the 2D non-reducing/reducing diagonal western blot analysis of immunoprecipitated HypR protein harvested from NaOCl-, diamide- and MHQ-treated cells (Figure 6D). HypR migrates strongly at the right side of the diagonal under disulphide stress in the diagonal western blot (Figure 6D) indicating HypR oxidation to intersubunit disulphides in vivo. Detailed CID MS/MS analysis of the immunoprecipitated HypR dimer identified the Cys14–Cys49′-disulphide-linked peptide in vivo (Supplementary Figure S6B, S6C, S6F and S6G).
Crystal structure of the reduced HypRC14S protein
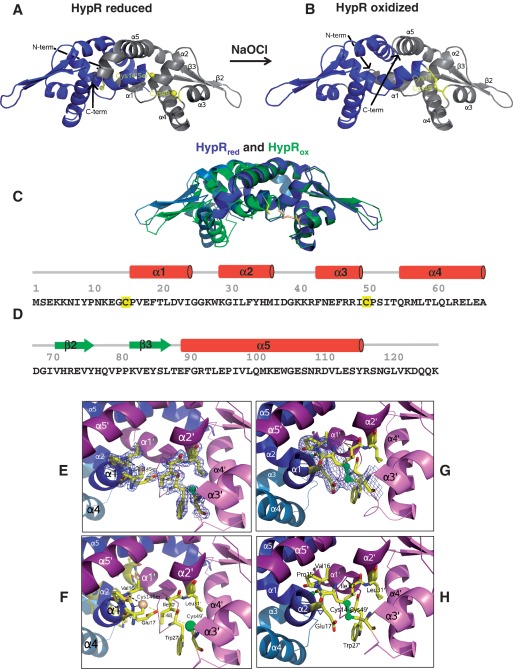
To avoid artificial oxidation of HypR during crystallization, HypRC14S protein was crystallized in the presence of β-mercaptoethanol. The structure was solved using molecular replacement and refined to 1.8 Å resolution. Electron density is visible for residues 13–117 of the native 125 residues (Figure 7). The asymmetric unit contains four similar monomers, organized in 2 biological dimers, 4 mercaptoethanol and 279 water molecules. The structure was refined to an Rcryst of 0.1935 and Rfree of 0.2336. The overall structure of reduced HypR comprises a homodimer adopting a triangular shape with non-crystallographic 2-fold symmetry as found in all MarR-family proteins (48). The structure has the following secondary structure elements: α1(15–23)–α2(28–35)–α3(42–48)–α4(54–66)–β2(70–75)–β3(81–86)–α5(88–114) (Figure 7A and D). The two monomers associate via a large dimer interface of 1600 Å2 provided by helices α1, α2, α5 and their symmetry mates α1′, α2′, α5′. Helix α5 is much longer than in the OhrR-family proteins exemplified by SarZ of S. aureus and OhrR of X. campestris (Supplementary Figure S1B). Pro95 and Gly105 induce kinks at positions 93 and 107 in HypR. The dimer interface significantly differs from that in OhrR structures since α6 is missing in HypR (Supplementary Figures S1B, S7A, S7C and S7D).
Figure 7.
Structures of reduced HypRC14S (A) and oxidized HypR proteins (B), superimposition of reduced and oxidized HypR (C) and secondary structure assignments (D). (A) Reduced HypRC14S dimer with monomers C and D coloured in grey and blue, respectively. Residues Ser14 and Cys49 are shown in yellow as sticks and spheres (Oγ and Sγ, respectively). (B) Oxidized HypR protein with the Cys14–Cys49′ intersubunit disulphide with monomers C and D, labelled in grey and blue, respectively. The intermolecular disulphide bond is shown in yellow in monomer C. (C) Superimposition of the oxidized HypR dimer (light and dark blue) and the reduced HypR dimer (light and dark green). The HypRox/red side view is shown and Cys14 and Cys49′ are shown as yellow sticks. One monomer of each dimer (on the right, dark blue and dark green) is aligned to visualize the differences in the opposing monomers. (D) The secondary structure elements of HypR are α1(15–23), α2(28–35), α3(42–48), α4(54–66), β2(70–75), β3(81–86), α5(88–114) that are shown as red tubes (α-helices) and green arrows (β-sheets). The redox-sensing Cys14 and the Cys49 are labelled in yellow. (E-H) The pocket of the redox-sensitive Cys residues in reduced and oxidized HypR. (E) Electron density map (2Fo–Fc at 1σ in blue) and (F) key interactions between α1 and α3′ helices including the Cys14Ser and Cys49′ residues for reduced HypR. The atoms are coloured in yellow (carbon), dark blue (nitrogen), red (oxygen) and green (sulphur); water molecules are shown as red spheres and hydrogen bonds are labelled as dashed lines. Cys14Ser is shown in orange forming hydrogen bonds with Val16 and Glu17. Cys49′ is in a hydrophobic environment formed by Ile52′, Trp27′, Ile30′, Leu31′, Gln60Cγ′, Ile48′. The distance between Ser14Oγ and Cys49′Sγ that form the intermolecular disulphide bond is 8.48 Å. (G) Electron density map (2Fo–Fc at 1σ in blue) and (H) structure of the Cys14–Cys49′ intersubunit disulphide region in oxidized HypR. Helices α3′ and α4′ on the right are oriented as in Figure 7EF, this emphasizes the movement of α4 on the left.
Each monomer contains a winged helix–turn–helix (wHTH) DNA-binding motif constituted by β2, β3, α3 and α4. Helix α4 is the recognition helix in the DNA-binding domain and the anti-parallel β-sheet of β2 and β3 forms the wing. The HTH motif binds in the major groove, the wing in the minor groove. The β1-strand, that is found in OhrR-like proteins between α2 and α3 (Supplementary Figure S1B) is not explicitely assigned by DSSP, but the β2–β3 sheet forms two main chain hydrogen bonds to Lys40, a shortened version of β1.
The reactivity of the redox-sensing Cys14
The redox-sensing Cys14 of HypR is located at the N-terminus of helix α1 (Figure 7E and F). The Cys14 thiolate is stabilized by hydrogen bonds with Val16N, Glu17N and a water molecule. There are no positive charged amino acid residues in close vicinity of Cys14 that could lower its pKa value; Lys101′ is >10 Å away and hydrogen bonded to Tyr33′. Instead, the reactivity of the Cys14 thiolate is increased by helix α1 at whose N-terminal end Cys14 is located (49,50). The helix dipole lowers the pKa of Cys14 to allow at physiological pH values the formation of the reactive, nucleophilic thiolate susceptible to oxidation.
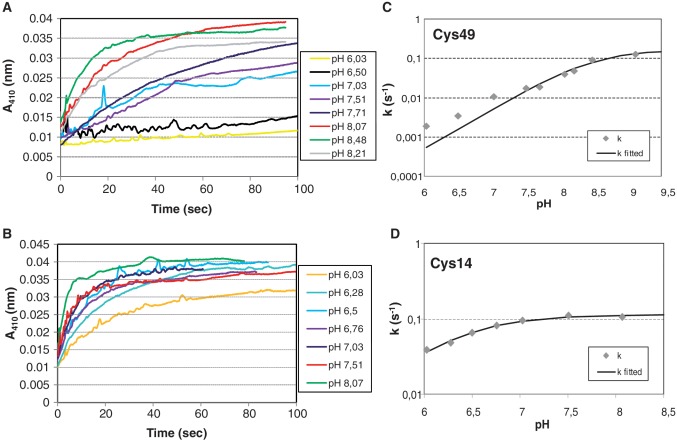
We used the reaction with 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) to analyse the reactivity of both cysteines at different pH values using His-tagged HypRC14S and His-HypRC49S mutant proteins (Figure 8). Spectroscopic measurement of the released 2-nitro-5 thiobenzoate allows determination of the fraction of deprotonated thiol groups and thus the pKa value. The Cys14 thiol has a more acidic pKa of 6.36 ± 0.04 (random error calculated from the second fit) suggesting that Cys14 is present as thiolate anion and a preferred target for oxidation by diamide or NaOCl (Figure 8B). In contrast, the pKa of Cys49 was determined as 8.51 ± 0.07 indicating that Cys49 is present in its protonated thiol form at physiological pH values in the cytoplasm (Figure 8A). The observed pKa value of the thiol group of Cys49 is also in accordance to its localization in a hydrophobic environment provided by Ile52, Trp27, Ile30, Leu31, Gln60Cγ and Ile48. The sulphur atoms of Cys14 (presumed to be at the position of Ser14Oγ) and Cys49′ residues of the opposing subunits are ∼8–9 Å apart in the HypR dimer (9.0, 8.5, 8.0 and 8.3 Å in monomers A, B, C and D, respectively).
Figure 8.
The pH dependence of the reactivity of Cys14 and Cys49 with DTNB. The absorbance at 410 nm at different pH values from 6 to 8 resulting from the release of 2-nitro-5 thiobenzoate after reaction of Cys49 of the HypRC14S (A) and of Cys14 of the HypRC49S (B) mutant proteins with DTNB is plotted over the time. (C and D) The kinetic constants (k) of the Cys49 and Cys14 reactions with DTNB are plotted against the pH and fitted resulting in a pKa of 8.51 for Cys49 (C) and a pKa of 6.36 for Cys14 (D).
Structural changes of HypR upon oxidation to intermolecular disulphides
For crystallization of oxidized HypR protein, His-HypR protein was treated with 1 mM diamide for 15 min and the formation of HypR intersubunit disulphides was verified by non-reducing SDS–PAGE. The structure of oxidized HypR protein could be solved in P65 and P6522 using molecular replacement with reduced HypR as model, and taking twinning into account refined to 3.0 Å resolution in the lower symmetry. Eight monomers are in the asymmetric unit forming four biological dimers. Electron density is visible in all monomers only for residues 14–108 (Figure 7B). Both intermolecular Cys14–Cys49′ and Cys14′–Cys49 disulphide bonds are visible at 1.3 σ in the 2Fo–Fc maps in three of four biological dimers of the asymmetric unit (Figure 7G and H). To confirm that there is useful signal in the data to 3.0 Å resolution, we computed the correlation between observed and calculated structure factor amplitudes in resolution shells, before the highest resolution data had been used in structure refinement, as described by Ling et al. (51) (Supplementary Figure S8D). After refining the molecular replacement solution against data limited to 3.3 Å resolution, there was a good correlation between observed and calculated structure factors to 3.0 Å resolution; in the 3.1–3.0 Å resolution shell, the correlation was 0.21 and the R-factor was 0.45.
Superimposition of the oxidized on the reduced monomer shows a similar overall structure (r.m.s.d. on Cαs: 0.9–1.1 Å) (Figure 7A–C, Supplementary Figure S8A, S8B and S8C). The most significant local difference close to these cysteines is the change of the χ1 angles of Cys14 from 150° in the reduced to −80° in the oxidized form (averages for well determined cysteines). Cys14Sγ does not make hydrogen bonds in the oxidized form. The major changes of HypR upon oxidation are in the quaternary structure. If one of the monomers in the dimer is superimposed, the second monomer rotates 8° around an axis in the helix α5–α5′ interface, the rotation hinge (Supplementary Figure S8). The dimer interface is thus almost unchanged between helices α5 and α5′, but is compressed between helices α1–α2 and α1′–α2′. In the reduced form water molecules separate Gly25N and Gly25′N; in the oxidized form these residues come close. The DNA-binding domains move up to 4 Å towards each other, loops α1–α2, α3, N-terminus of α4 and the wing formed of β2–β3 shift strongly. This is visualized in the change of Cα–Cα′ distances in the reduced compared to the oxidized conformation (Supplementary Figure S9).
Since HypR resembles a transcriptional activator, disulphide bond formation should re-arrange the DNA-binding helices α4 and α4′ in the opposing subunits of the dimer to enable binding to consecutive crevices of the major groove of the DNA duplex under oxidized conditions. This shift of the DNA-binding domains is observed in all crystallographically independent dimers, and visualized in the superimposed structures of reduced and oxidized HypR (Supplementary Figure S8). The spacing between the α4–α4′ helices is 30 Å in reduced HypR and 27 Å in oxidized HypR (measured at the helix axis between residues Leu61 and Arg62). Compared to the related two-Cys-type OhrRXc regulator, HypR oxidation results in only a small domain domain movement of the DNA major groove recognition α4 and α4′ helices.
DISCUSSION
The role of the DUF24 family regulator HypR that controls the HypO nitroreductase
The MarR/DUF24 family is conserved among Gram-positive bacteria, and members of this family have been characterized in Corynebacterium glutamicum and B. subtilis. QorR of C. glutamicum was characterized as a repressor that senses diamide and H2O2 and controls the quinone oxidoreductase QorA (52). Bacillus subtilis encodes eight DUF24-family proteins (HypR, YodB, CatR, HxlR, YdeP, YdzF, YkvN and YtcD) and previous studies revealed that YodB and CatR respond to diamide and quinones by thiol-based mechanisms (7,24–26). Here, we have characterized the novel DUF24-type regulator HypR that is activated by hypochlorite and diamide stress by a thiol-disulphide switch. Recent studies showed that the OhrA peroxiredoxin is a specific determinant of NaOCl resistance (17). HypR positively controls the putative nitroreductase HypO that provides together with OhrA protection against NaOCl stress in B. subtilis. The nitroreductase HypO has been recently structurally characterized (47). Nitroreductases are widely distributed flavoenzymes that catalyse the NAD(P)H-dependent reduction of nitroaromatic and nitroheterocyclic compounds (53). These enzymes have broad substrate specificities and can reduce also azocompounds, quinones, flavins and metal ions (Fe3+ and CrO42–). Previous results suggested that the nitroreductases YodC and MhqN could function as azo- and quinone reductases (27,36,43). However, they could be also involved in the thiol-specific stress response and function as flavoprotein disulphide reductases. Indeed, the flavin-containing NAD(P)H oxidoreductase NfrA of Staphylococcus aureus showed weak disulphide reductase activity and was suggested to function as a thiol-disulphide oxidoreductase under disulphide stress (54). The homologous enzyme NfrA1 of B. subtilis exhibits NADH oxidase activity, catalysing the oxidation of NADH to NAD+, and scavenges high concentrations of H2O2 (55). This suggests that HypO could function to restore the redox balance of the cells by direct detoxification of hypochlorite and diamide or by reduction of disulphide bonds.
Conservation and structural features of the reactive Cys14 pocket of HypR
HypR represents a two-Cys-type MarR-type regulator that is activated by Cys14–Cys49′ intersubunit disulphide formation as confirmed by mass spectrometry. The DNaseI-footprinting analysis showed that oxidized HypR bound with higher affinity to the hypO operator DNA and using an in vitro transcription assay we provide evidence for transcriptional activation of RNAP by oxidized HypR. The crystallographic analysis of reduced and oxidized HypR provides first structural insights into the reactive Cys14 pocket and the conformational changes of HypR upon oxidation that could be required for specific interaction with the operator sites and/or transcription activation by RNAP. Examples for two-Cys type MarR-family repressors that have been structurally characterized include MexR of Pseudomonas aeruginosa and OhrRXc of Xanthomonas campestris (14,15,18,19). Interestingly, while OhrR-proteins and DUF24-family members share only limited sequence similarity, the redox-sensing N-terminal Cys residues align between OhrR and HypR (Supplementary Figure S1B). Mutational analyses indicate that both Cys residues are essential for redox-regulation of HypR consistent with the two-Cys-type redox-switch model. The reactive Cys14 thiolate anion was confirmed using the DTNB assay, revealing a lower pKa value of 6.36 that is similar to that of Cys15 in OhrRBs (with a pKa of 5.2). In both OhrRBs and HypR structures, the N-terminal Cys is located at the N-terminus of helix α1 and the positive macrodipole of helix α1 lowers its pKa value. Sequence alignments indicate that Cys14 is conserved in DUF24 paralogs and is also located at the N-terminus of helix α1 in other DUF24 paralogs (Supplementary Figure S1). It was shown previously that the conserved Cys6 and Cys7 residues in YodB and CatR are required for redox-sensing (24,25). The reactive thiolate anion in OhrR proteins is stabilized by a conserved tyrosine hydrogen-bonding network (15,23,56). In reduced HypR the Cys14 thiolate anion forms hydrogen bonds to Val16N and Glu17N. Interestingly, Glu17 is conserved in DUF24 paralogs suggesting that a conserved hydrogen-bonding network stabilizes the reactive Cys thiolate.
Structural changes of HypR upon oxidation to intersubunit disulphides
MexR and OhrRXc sense oxidative stress by intermolecular disulphide bond formation of two Cys residues of opposing subunits that are >15 Å apart. With this large distance, remarkable re-arrangements are required to bring the Cys residues together for disulphide bond formation. Upon oxidation, OhrRXc undergoes conformational changes in the α6–α6′ helices of the dimer interface leading to rigid body rotation of the HTH motifs and dissociation of OhrRXc from the operator DNA (15). Compared to the distance of 15.5 Å between Cys22 and Cys127′ in OhrRXc, the distance of 8–9 Å between Cys14 and Cys49′ in HypR is rather small. Thus, small structural changes are sufficient for disulphide bond formation and activation of HypR. Oxidation of the Cys14 thiolate to sulphenylchloride by hypochlorite could abolish the negative charge on the sulphur and break the hydrogen bonding network. Rotation of the Cys14 side chain about χ1 can then bring the sulphur closer to Cys49′. The subsequent formation of the disulphide pulls helices α1 and α2 closer to their counterparts, helices α1′ and α2′. The move might be supported by the helix dipole of α1 attracting the Cys49′ thiolate. At the dimer's 2-fold axis Gly25N and Gly25′N are bridged by a water molecule and 6.5 Å apart in the reduced form, but only 4 Å apart with no water molecule in the oxidized form. The ionic interaction between Asp21Oδ and Lys26′Nε is only observed in the oxidized form (distance ∼ 3.5 Å), but not in the reduced form (∼ 9 Å). This interaction could temporarily stabilize the oxidized conformation until the disulphide bond is formed.
Disulphide bond formation moves the α4 and α4′ helices of HypR ∼4 Å towards each other causing minor changes in the spacing of the DNA-binding domains. Similarly, small conformational changes between the reduced and oxidized conformations were observed for MexR, that is inactivated by oxidation to Cys30–Cys62′ intersubunit disulphides (19). In oxidized MexR, neither the spacing of the major groove binding α4–α4′ helices, nor the orientation of the dimerization domain was changed. Instead, oxidation causes rigid body rotation of the α2′ and α3′ helices resulting in steric clashes of α2′ and the disulphide bond with the DNA backbone that lead to dissociation of the MexR repressor from the operator DNA (19). In contrast to the MexR repressor, the DUF24-family regulator HypR is activated by disulphide bond formation and local domain movements in the wHTH region might be sufficient for recognition of the operator sites and recruitment of the RNAP to initiate transcription. We did not measure a significant change of the Kd values of reduced and oxidized HypR proteins in the gel-shift assays. Instead, we observed a different mobility of oxidized HypR in the gel-shift assays and higher affinity of oxidized HypR to the operator DNA in the DNase-I footprinting analysis. Oxidized HypR also resulted in ∼2-fold transcriptional induction of hypO in an in vitro-transcription assay. Thus, the 4 Å movements of the α4 and α4′ major groove recognition helices of HypR upon oxidation suggest a different DNA-binding mode of oxidized HypR leading to recruitment of the RNAP to initiate transcription at the hypO promoter. In addition, the strong autoinduction of HypR itself by disulphide stress suggests that also increased amounts of oxidized HypR are required for transcriptional induction of the hypO gene. We are aware that the structure of oxidized HypR in the DNA-binding state needs to be resolved to shed light into the specific interaction of oxidized HypR with the operator DNA and into the mechanism of transcriptional activation. Thus, our ongoing studies are directed to further explore the molecular details of transcriptional activation by crystallization of HypR-operator-DNA complexes under oxidized conditions.
ACCESSION NUMBERS
PDB codes: 4a5n, 4a5m.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–2, Supplementary Figures 1–9 and Supplementary References [32,39,40,57].
FUNDING
Deutsche Forschungsgemeinschaft (AN 746/2-1 to H.A.); Wellcome Trust (UK) (grant 082961 to R.J.R.). Funding for open access charge: Deutsche Forschungsgemeinschaft (AN 746/2-1).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Dana Clausen for excellent technical assistance and Birthe Steigemann for the performance of the DTNB assay for pKa determination of Cys14 and Cys49. The authors wish to thank Kazuo Kobayashi for providing the transcription factor mutant strains ΔhypR and ΔohrR and Peter Lewis for the SigmaA expression plasmid pNG590. The authors further wish to thank David Noone and Kevine Devine for the advice on in vitro transcription assays. The Helmholtz-Zentrum Berlin-Electron storage ring BESSY II is acknowledged for provision of synchrotron radiation at beamline BL14.2.
REFERENCES
- 1.Imlay JA. Pathways of oxidative damage. Annu. Rev. Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 2.Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies MJ. Myeloperoxidase-derived oxidation: mechanisms of biological damage and its prevention. J. Clin. Biochem. Nutr. 2011;48:8–19. doi: 10.3164/jcbn.11-006FR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hawkins CL, Pattison DI, Davies MJ. Hypochlorite-induced oxidation of amino acids, peptides and proteins. Amino Acids. 2003;25:259–274. doi: 10.1007/s00726-003-0016-x. [DOI] [PubMed] [Google Scholar]
- 5.Marnett LJ, Riggins JN, West JD. Endogenous generation of reactive oxidants and electrophiles and their reactions with DNA and protein. J. Clin. Invest. 2003;111:583–593. doi: 10.1172/JCI18022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rudolph TK, Freeman BA. Transduction of redox signaling by electrophile-protein reactions. Sci. Signal. 2009;2:re7. doi: 10.1126/scisignal.290re7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antelmann H, Helmann JD. Thiol-based redox switches and gene regulation. Antioxid Redox. Signal. 2011;14:1049–1063. doi: 10.1089/ars.2010.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 9.Lee C, Lee SM, Mukhopadhyay P, Kim SJ, Lee SC, Ahn WS, Yu MH, Storz G, Ryu SE. Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat Struct Mol Biol. 2004;11:1179–1185. doi: 10.1038/nsmb856. [DOI] [PubMed] [Google Scholar]
- 10.Choi H, Kim S, Mukhopadhyay P, Cho S, Woo J, Storz G, Ryu SE. Structural basis of the redox switch in the OxyR transcription factor. Cell. 2001;105:103–113. doi: 10.1016/s0092-8674(01)00300-2. [DOI] [PubMed] [Google Scholar]
- 11.Kim SO, Merchant K, Nudelman R, Beyer WF, Jr., Keng T, DeAngelo J, Hausladen A, Stamler JS. OxyR: a molecular code for redox-related signaling. Cell. 2002;109:383–396. doi: 10.1016/s0092-8674(02)00723-7. [DOI] [PubMed] [Google Scholar]
- 12.Winter J, Ilbert M, Graf PC, Ozcelik D, Jakob U. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell. 2008;135:691–701. doi: 10.1016/j.cell.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen PR, Brugarolas P, He C. Redox signaling in human pathogens. Antioxid. Redox. Signal. 2011;14:1107–1118. doi: 10.1089/ars.2010.3374. [DOI] [PubMed] [Google Scholar]
- 14.Panmanee W, Vattanaviboon P, Poole LB, Mongkolsuk S. Novel organic hydroperoxide-sensing and responding mechanisms for OhrR, a major bacterial sensor and regulator of organic hydroperoxide stress. J. Bacteriol. 2006;188:1389–1395. doi: 10.1128/JB.188.4.1389-1395.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newberry KJ, Fuangthong M, Panmanee W, Mongkolsuk S, Brennan RG. Structural mechanism of organic hydroperoxide induction of the transcription regulator OhrR. Mol. Cell. 2007;28:652–664. doi: 10.1016/j.molcel.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 16.Lee JW, Soonsanga S, Helmann JD. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc. Natl Acad. Sci. USA. 2007;104:8743–8748. doi: 10.1073/pnas.0702081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chi BK, Gronau K, Mäder U, Hessling B, Becher D, Antelmann H. S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol. Cell. Proteomics. 2011;10:M111 009506. doi: 10.1074/mcp.M111.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen H, Hu J, Chen PR, Lan L, Li Z, Hicks LM, Dinner AR, He C. The Pseudomonas aeruginosa multidrug efflux regulator MexR uses an oxidation-sensing mechanism. Proc. Natl Acad. Sci. USA. 2008;105:13586–13591. doi: 10.1073/pnas.0803391105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H, Yi C, Zhang J, Zhang W, Ge Z, Yang CG, He C. Structural insight into the oxidation-sensing mechanism of the antibiotic resistance of regulator MexR. EMBO Rep. 2010;11:685–690. doi: 10.1038/embor.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen PR, Bae T, Williams WA, Duguid EM, Rice PA, Schneewind O, He C. An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat. Chem. Biol. 2006;2:591–595. doi: 10.1038/nchembio820. [DOI] [PubMed] [Google Scholar]
- 21.Chen PR, Nishida S, Poor CB, Cheng A, Bae T, Kuechenmeister L, Dunman PM, Missiakas D, He C. A new oxidative sensing and regulation pathway mediated by the MgrA homologue SarZ in Staphylococcus aureus. Mol. Microbiol. 2009;71:198–211. doi: 10.1111/j.1365-2958.2008.06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lan L, Murray TS, Kazmierczak BI, He C. Pseudomonas aeruginosa OspR is an oxidative stress sensing regulator that affects pigment production, antibiotic resistance and dissemination during infection. Mol. Microbiol. 2010;75:76–91. doi: 10.1111/j.1365-2958.2009.06955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poor CB, Chen PR, Duguid E, Rice PA, He C. Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus. J. Biol. Chem. 2009;284:23517–23524. doi: 10.1074/jbc.M109.015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chi BK, Albrecht D, Gronau K, Becher D, Hecker M, Antelmann H. The redox-sensing regulator YodB senses quinones and diamide via a thiol-disulfide switch in Bacillus subtilis. Proteomics. 2010;10:3155–3164. doi: 10.1002/pmic.201000230. [DOI] [PubMed] [Google Scholar]
- 25.Chi BK, Kobayashi K, Albrecht D, Hecker M, Antelmann H. The paralogous MarR/DUF24-family repressors YodB and CatR control expression of the catechol dioxygenase CatE in Bacillus subtilis. J. Bacteriol. 2010;192:4571–4581. doi: 10.1128/JB.00409-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen TT, Eiamphungporn W, Mäder U, Liebeke M, Lalk M, Hecker M, Helmann JD, Antelmann H. Genome-wide responses to carbonyl electrophiles in Bacillus subtilis: control of the thiol-dependent formaldehyde dehydrogenase AdhA and cysteine proteinase YraA by the MerR-family regulator YraB (AdhR) Mol. Microbiol. 2009;71:876–894. doi: 10.1111/j.1365-2958.2008.06568.x. [DOI] [PubMed] [Google Scholar]
- 27.Leelakriangsak M, Huyen NT, Töwe S, van Duy N, Becher D, Hecker M, Antelmann H, Zuber P. Regulation of quinone detoxification by the thiol stress sensing DUF24/MarR-like repressor, YodB in Bacillus subtilis. Mol. Microbiol. 2008;67:1108–1124. doi: 10.1111/j.1365-2958.2008.06110.x. [DOI] [PubMed] [Google Scholar]
- 28.Yurimoto H, Hirai R, Matsuno N, Yasueda H, Kato N, Sakai Y. HxlR, a member of the DUF24 protein family, is a DNA-binding protein that acts as a positive regulator of the formaldehyde-inducible hxlAB operon in Bacillus subtilis. Mol. Microbiol. 2005;57:511–519. doi: 10.1111/j.1365-2958.2005.04702.x. [DOI] [PubMed] [Google Scholar]
- 29.Potter AJ, Kidd SP, McEwan AG, Paton JC. The MerR/NmlR family transcription factor of Streptococcus pneumoniae responds to carbonyl stress and modulates hydrogen peroxide production. J. Bacteriol. 2010;192:4063–4066. doi: 10.1128/JB.00383-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McEwan AG, Djoko KY, Chen NH, Counago RL, Kidd SP, Potter AJ, Jennings MP. Novel bacterial MerR-like regulators their role in the response to carbonyl and nitrosative stress. Adv. Microb. Physiol. 2011;58:1–22. doi: 10.1016/B978-0-12-381043-4.00001-5. [DOI] [PubMed] [Google Scholar]
- 31.Stülke J, Hanschke R, Hecker M. Temporal activation of beta-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J. Gen. Microbiol. 1993;139:2041–2045. doi: 10.1099/00221287-139-9-2041. [DOI] [PubMed] [Google Scholar]
- 32.Hayashi K, Kensuke T, Kobayashi K, Ogasawara N, Ogura M. Bacillus subtilis RghR (YvaN) represses rapG and rapH, which encode inhibitors of expression of the srfA operon. Mol. Microbiol. 2006;59:1714–1729. doi: 10.1111/j.1365-2958.2006.05059.x. [DOI] [PubMed] [Google Scholar]
- 33.Bisicchia P, Lioliou E, Noone D, Salzberg LI, Botella E, Hübner S, Devine KM. Peptidoglycan metabolism is controlled by the WalRK (YycFG) and PhoPR two-component systems in phosphate limited Bacillus subtilis cells. Mol. Microbiol. 2010;75:972–989. doi: 10.1111/j.1365-2958.2009.07036.x. [DOI] [PubMed] [Google Scholar]
- 34.Johnston EB, Lewis PJ, Griffith R. The interaction of Bacillus subtilis sigmaA with RNA polymerase. Protein Sci. 2009;18:2287–2297. doi: 10.1002/pro.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pöther DC, Liebeke M, Hochgräfe F, Antelmann H, Becher D, Lalk M, Lindequist U, Borovok I, Cohen G, Aharonowitz Y, et al. Diamide triggers mainly S Thiolations in the cytoplasmic proteomes of Bacillus subtilis and Staphylococcus aureus. J. Bacteriol. 2009;191:7520–7530. doi: 10.1128/JB.00937-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Töwe S, Leelakriangsak M, Kobayashi K, Van Duy N, Hecker M, Zuber P, Antelmann H. The MarR-type repressor MhqR (YkvE) regulates multiple dioxygenases/glyoxalases and an azoreductase which confer resistance to 2-methylhydroquinone and catechol in Bacillus subtilis. Mol. Microbiol. 2007;66:40–54. doi: 10.1111/j.1365-2958.2007.05891.x. [DOI] [PubMed] [Google Scholar]
- 37.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terwilliger TC. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- 40.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 42.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 43.Antelmann H, Hecker M, Zuber P. Proteomic signatures uncover thiol-specific electrophile resistance mechanisms in Bacillus subtilis. Exp. Rev. Proteomics. 2008;5:77–90. doi: 10.1586/14789450.5.1.77. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen VD, Wolf C, Mäder U, Lalk M, Langer P, Lindequist U, Hecker M, Antelmann H. Transcriptome and proteome analyses in response to 2-methylhydroquinone and 6-brom-2-vinyl-chroman-4-on reveal different degradation systems involved in the catabolism of aromatic compounds in Bacillus subtilis. Proteomics. 2007;7:1391–1408. doi: 10.1002/pmic.200700008. [DOI] [PubMed] [Google Scholar]
- 45.Zuber P. Management of oxidative stress in Bacillus. Annu. Rev. Microbiol. 2009;63:575–597. doi: 10.1146/annurev.micro.091208.073241. [DOI] [PubMed] [Google Scholar]
- 46.Nakano S, Kuster-Schock E, Grossman AD, Zuber P. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc. Natl Acad. Sci. USA. 2003;100:13603–13608. doi: 10.1073/pnas.2235180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prosser GA, Patterson AV, Ackerley DF. uvrB gene deletion enhances SOS chromotest sensitivity for nitroreductases that preferentially generate the 4-hydroxylamine metabolite of the anti-cancer prodrug CB1954. J. Biotechnol. 2010;150:190–194. doi: 10.1016/j.jbiotec.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 48.Perera IC, Grove A. Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J. Mol. Cell. Biol. 2010;2:243–254. doi: 10.1093/jmcb/mjq021. [DOI] [PubMed] [Google Scholar]
- 49.Hol WG, van Duijnen PT, Berendsen HJ. The alpha-helix dipole and the properties of proteins. Nature. 1978;273:443–446. doi: 10.1038/273443a0. [DOI] [PubMed] [Google Scholar]
- 50.Wada A. The alpha-helix as an electric macro-dipole. Adv. Biophys. 1976:1–63. [PubMed] [Google Scholar]
- 51.Ling H, Boodhoo A, Hazes B, Cummings MD, Armstrong GD, Brunton JL, Read RJ. Structure of the shiga-like toxin I B-pentamer complexed with an analogue of its receptor Gb3. Biochemistry. 1998;37:1777–1788. doi: 10.1021/bi971806n. [DOI] [PubMed] [Google Scholar]
- 52.Ehira S, Ogino H, Teramoto H, Inui M, Yukawa H. Regulation of quinone oxidoreductase by the redox-sensing transcriptional regulator QorR in Corynebacterium glutamicum. J. Biol. Chem. 2009;284:16736–16742. doi: 10.1074/jbc.M109.009027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roldan MD, Perez-Reinado E, Castillo F, Moreno-Vivian C. Reduction of polynitroaromatic compounds: the bacterial nitroreductases. FEMS Microbiol. Rev. 2008;32:474–500. doi: 10.1111/j.1574-6976.2008.00107.x. [DOI] [PubMed] [Google Scholar]
- 54.Streker K, Freiberg C, Labischinski H, Hacker J, Ohlsen K. Staphylococcus aureus NfrA (SA0367) is a flavin mononucleotide-dependent NADPH oxidase involved in oxidative stress response. J. Bacteriol. 2005;187:2249–2256. doi: 10.1128/JB.187.7.2249-2256.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cortial S, Chaignon P, Iorga BI, Aymerich S, Truan G, Gueguen-Chaignon V, Meyer P, Morera S, Ouazzani J. NADH oxidase activity of Bacillus subtilis nitroreductase NfrA1: insight into its biological role. FEBS Lett. 2010;584:3916–3922. doi: 10.1016/j.febslet.2010.08.019. [DOI] [PubMed] [Google Scholar]
- 56.Hong M, Fuangthong M, Helmann JD, Brennan RG. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol. Cell. 2005;20:131–141. doi: 10.1016/j.molcel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 57.Guerout-Fleury AM, Shazand K, Frandsen N, Stragier P. Antibiotic-resistance cassettes for Bacillus subtilis. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.