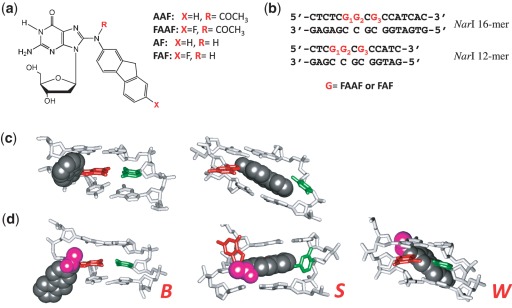
Abstract
Nucleotide excision repair (NER) is a major repair pathway that recognizes and corrects various lesions in cellular DNA. We hypothesize that damage recognition is an initial step in NER that senses conformational anomalies in the DNA caused by lesions. We prepared three DNA duplexes containing the carcinogen adduct N-(2′-deoxyguanosin-8-yl)-7-fluoro-2-acetylaminofluorene (FAAF) at G1, G2 or G3 of NarI sequence (5′-CCG1G2CG3CC-3′). Our 19F-NMR/ICD results showed that FAAF at G1 and G3 prefer syn S- and W-conformers, whereas anti B-conformer was predominant for G2. We found that the repair of FAAF occurs in a conformation-specific manner, i.e. the highly S/W-conformeric G3 and -G1 duplexes incised more efficiently than the B-type G2 duplex (G3∼G1 > G2). The melting and thermodynamic data indicate that the S- and W-conformers produce greater DNA distortion and thermodynamic destabilization. The N-deacetylated N-(2′-deoxyguanosin-8-yl)-7-fluoro-2-aminofluorene (FAF) adducts in the same NarI sequence are repaired 2- to 3-fold less than FAAF: however, the incision efficiency was in order of G2∼G1 > G3, a reverse trend of the FAAF case. We have envisioned the so-called N-acetyl factor as it could raise conformational barriers of FAAF versus FAF. The present results provide valuable conformational insight into the sequence-dependent UvrABC incisions of the bulky aminofluorene DNA adducts.
INTRODUCTION
Adduct formation is an important aspect of DNA damage: if unrepaired, various mutations in DNA could occur (1–3). The presence of mutations on specific oncogenes or tumor suppressor genes may trigger cancer initiation. Human cells are armed with various effective repair pathways to safeguard genomic DNA from continuous assault by exogenous and endogenous sources (4). Nucleotide excision repair (NER) is a major repair pathway that is known for the removal of stretches of bases containing various lesions, including single-base damages, bulky adducts and cross-links, among others (5). Deficiencies in NER are closely associated with the development of several genetic diseases, such as xeroderma pigmentosum that increases the risk of skin cancer due to higher sensitivity to sunlight (6).
The NER pathway in Escherichia coli involves the UvrABC nuclease system and has been studied extensively for understanding DNA damage recognition and incision. E. coli NER is initiated following damage recognition by a dimeric UvrA protein. Next, UvrB protein reaches the damage site, forms a trimer and verifies the damage. Departure of UvrA from the resulting complex recruits UvrC and UvrD proteins, which cleave and remove the lesion-bearing patch of DNA. Finally, DNA polymerase I synthesizes and ligase I seals a new patch to complete the repair process (5,7).
Arylamines are an important class of environmental pollutants that are implicated in the etiology of human cancers, especially of the bladder and liver (1). 2-Acetylaminofluorene was originally developed as an agricultural insecticide, but was later banned due to its strong tumorigenic activity in rat livers (8). It has been used extensively as a model for studying chemical carcinogenesis. In vivo, metabolic activation of AAF produces a highly electrophilic nitrenium ion, which subsequently interacts with DNA to produce two major C8-substituted dG adducts: AAF and AF (Figure 1a) (8,9). In vitro, N-acetylated AAF blocks the activity of high-fidelity polymerases and requires bypass polymerases for a translesion synthesis (TLS), whereas AF only slows down replication (10). In general, the bulky AAF exhibits greater susceptibility towards NER than AF (11), which is known to exist in a sequence-dependent equilibrium between anti B-conformer and syn S-conformer (Figure 1c) (10,12–14). We recently reported that AAF adducts also adopt a sequence-dependent S/B/W-conformational equilibrium (Figure 1d) (15).
Figure 1.
(a) Chemical structures of AAF, FAAF, AF and FAF adducts; (b) sequences of fully paired 16-mer and 12-mer NarI duplexes used in the present study; major groove views of the central trimer segments of (c) the B/S and (d) B/S/W-conformer equilibrium of FAF and FAAF-modified duplexes. The modified dG and the complementary dC are shown in red and green sticks, respectively, and the aminofluorene moiety is highlighted with shiny gray CPK and the N-acetyl with pink CPK. In the B-type conformer, anti-[FAAF/FAF]dG maintains Watson–Crick hydrogen bonds, thereby placing the carcinogen moiety in the major groove. The carcinogens in the S- and W-conformers stack into the helix or wedged into the minor groove, respectively, with the modified dG in the syn conformation.
Local sequence context plays an important role in the repair of arylamine–DNA adducts (16). Fuchs et al. constructed DNA sequences modified with AAF at each of three guanines of the most frequently studied mutational hotspot known as NarI sequence (5′- … CG1G2CG3CC … -3′) and tested their substrate repairability in the E. coli UvrABC and human exonuclease systems. In E. coli, the three AAFs were repaired in a sequence-dependent manner, with relative repair efficiencies of G1:G2:G3 in a ratio of 100:18:66 (17,18). However, different repair efficiencies were observed for the same lesions by the human exonuclease, 38:100:68 for G1, G2 and G3, respectively (18). AAF at G3 of NarI sequence induces ∼100-fold greater frequency of −2 frameshift (−2 deletion) mutations, even though the three guanines exhibit similar chemical reactivities (19). We have shown that the FAF-modified NarI -2 deletion duplex in the 5′–CG1G2CG3*CC-3′ context adopts a single looped-out bulge structure, whereas the 5′–CG1G2CG3*CT–3′ context results in a local conformational heterogeneity (20). These results support the importance of the 3′-next flanking nucleotide to the lesion in modulation of mutation efficiency. The studies verified that the conformational stability of a slipped mutagenic intermediate is a critical determinant for the hotness (up to 30- to 50-fold) of G3 in NarI sequence for −2 frameshift mutation (20–22). Mekhovich et al. (23) found a greater incision rate in E. coli systems when AAF was located at G3 of the NarI sequence (5′—CG1G2CG3*CC—3′) than in a non-NarI sequence (5′—GATG*ATA—3′). Zou et al. (24) have reported that the UvrABC incision efficiency is 70% more in the TG*T than in the CG*C sequence context when adducted with either AF- or AAF lesions.
The NER pathway is characterized by its unique ability to excise a wide array of structurally diverse DNA lesions. The structure of individual adducts per se is not as important as lesion-induced local distortions and destabilizations to trigger a NER response. Examples include disruption of Watson–Crick hydrogen bonding, DNA bending, thermodynamic destabilization, local conformational flexibility and flipped-out bases in the unmodified complementary strand (25,26). However, mechanisms of sequence dependence that control NER efficiencies remained elusive. It could be that the observed local sequence effects described in the previous paragraph for AF and AAF are due to differences in the extent of distortions, which in turn depends on the conformation adopted in a particular sequence context.
In the present work, we investigated the role of conformational heterogeneity in the structure–repair relationships of AF and AAF. These two adducts are structurally similar, but differ in the absence and presence, respectively, of an N-acetyl group on the central nitrogen. We prepared oligonucleotides that were site-specifically modified by the fluorine model FAF and FAAF at three different guanines (G1, G2 and G3) of the NarI recognition sequences (Figure 1a and b). We conducted spectroscopic and melting experiments for conformational and thermodynamic analyses. Moreover, we performed NER studies of these adducts using the E. coli UvrABC system. The results present strong structural and thermodynamic evidences for the differential NER efficiencies exhibited by AF and AAF at different guanine residues of the NarI sequence.
MATERIALS AND METHODS
Caution
2-Aminofluorene derivatives are mutagens and suspected human carcinogens and therefore must be handled with caution.
Crude oligodeoxynucleotides (ODN, 10 µmol scale) in desalted form were purchased from Eurofins MWG operon (Huntsville, AL, USA). All HPLC solvents were purchased from Fisher Inc. (Pittsburgh, PA, USA).
Preparation and characterization of FAF- and FAAF-modified ODNs
We previously reported the preparation of 12-mer NarI ODNs (5′-CTCG1G2CG3CCATC-3′), in which each of the three guanine were site-specifically modified by FAF (20). We have also demonstrated that the incorporation of fluorine atom at the longest axis position 7 does not affect the overall conformational and thermal/thermodynamic profiles of AF- or AAF-modified duplexes (14,20). The three FAF-modified NarI sequences were each annealed with a complementary 12-mer sequence (5′-GATGGCGCCGAG-3′) to form fully paired NarI–G1–FAF, NarI–G2–FAF and NarI–G3–FAF duplexes, respectively. These duplexes were thoroughly characterized by 19F-NMR, CD and UV melting experiments (20).
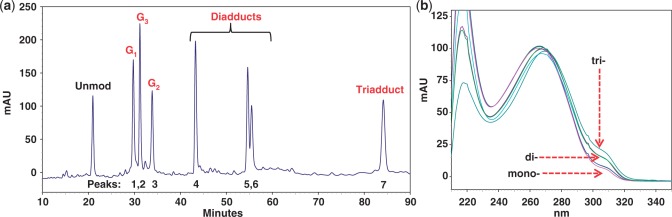
FAAF-modified 16-mer ODN were prepared using the general procedures described previously (15,27). Briefly, approximately 0.5–1 mg of N-acetoxy-N-2-(acetylamino)-7-fluorofluorene dissolved in absolute ethanol was added drop wise to a sodium citrate buffer (pH 6.0) containing 200–250 ODs of unmodified ODN (5′-CTCTCG1G2CG3CCATCAC-3′) and placed in a shaker for 5 min at 37°C. Figure 2a shows a typical reversed-phase HPLC chromatogram derived from the resulting mixture. The FAAF modified oligomers appearing between 28 and 85 min were separated and purified up to >97% purity by repeated injections. The HPLC system consisted of a Hitachi EZChrom Elite HPLC unit with an L2450 diode array detector and a Phenomenex Luna C18 column (150 × 10 mm, 5.0 µm). We employed a gradient system involving 3–15% acetonitrile for 40 min followed by 15–20% and 20–35% acetonitrile for 20 and 40 min, respectively, in pH 7.0 ammonium acetate buffer (100 mM) with a flow rate of 2.0 ml/min.
Figure 2.
(a) Chromatogram of a reaction mixture between 16-mer NarI sequence (5′-CTCTCG1G2CG3CCATCAC-3′) and an activated FAAF (N-acetoxy-N-2-(acetylamino)-7-fluorofluorene). The mono- (G1, G3, G2), di- and tri-FAAF adducts eluted in the 28–35, 42–60 and 84 min were purified by reversed-phase HPLC (see ‘Materials and Methods’ section for gradient condition); (b) online photodiode array UV/Vis spectra of mono-, di- and tri-FAAF adducts.
The three FAAF-modified 16-mer sequences were each annealed with the complementary sequence (5′-GTGATGGCGCCGAGAG-3′) to form fully paired NarI–G1–FAAF, NarI–G2–FAAF and NarI–G3–FAAF duplexes for UV melting, DSC, CD and dynamic 19F-NMR experiments.
LC/MS characterization of FAAF-modified ODNs
Electrospray ionization and quadrupole time-of-flight mass spectrometry was utilized to verify the molecular weights and the position of FAAF attachment of the three oligomers. The 16-mer ODNs were sequenced using 3′–5′ or 5′–3′ exonucleases as described previously for the analysis of modified 12-mers (28). Normally, 1 µg of a particular ODN was combined with 0.01 units of an exonuclease in a 1 mM solution of MgCl2 and incubated for several hours. The digests were separated using a Phenomenex Aqua C18, 1.0 × 50 mm column (5 µm; 120 Å). Solvent A was 5 mM in both ammonium acetate and dimethylbutyl amine. Acetic acid was added to solvent A to adjust the pH to 7.0. Solvent B was 0.1% formic acid in acetonitrile. The flow rate was 100 µl/min and total run time was 20 min. All LC/MS spectra were acquired using a Waters SYNAPT quadrupole time-of-flight mass spectrometer (Milford, MA, USA) operated in the negative ion and V-modes. The measured molecular masses of all three isomeric ODNs were within 0.1 Da of their theoretical monoisotopic mass (5016.9 Da).
UV melting
UV melting data were obtained using a Cary100 Bio UV/VIS spectrophotometer equipped with a 6 × 6 multi-cell block and 1.0 cm path length. Sample cell temperatures were controlled by an in-built Peltier temperature controller. Oligonucleotide duplexes with a concentration range of 0.4–6.4 µM were prepared in solutions containing 0.2 M NaCl, 10 mM sodium phosphate and 0.2 mM EDTA at pH 7.0. Thermomelting curves were constructed by varying temperature of the sample cell (1°C/min) and monitoring absorbance at 260 nm. A typical melting experiment consisted of forward/reverse scans and was repeated five times. Thermodynamic parameters were calculated using the program MELTWIN version 3.5 as described previously (12).
Circular dichroism
Circular dichroism (CD) measurements were conducted on a Jasco J-810 spectropolarimeter equipped with a Peltier temperature controller. Typically, 2 ODs of each strand were annealed with an equimolar amount of a complementary sequence. The samples were dissolved in 400 µl of a neutral buffer (0.2 M NaCl, 10 mM sodium phosphate, 0.2 mM EDTA) and placed in a 1.0 mm path length cell. The samples were heated at 85°C for 5 min and then cooled to 15°C, over a 10 min period to ensure complete duplex formation. Spectra were acquired every 0.2 nm with a 2 s response time from 200 to 400 nm at a rate of 50 nm/min, were the averages of 10 accumulations and were smoothed using 17-point adaptive smoothing algorithms provided by Jasco.
Dynamic 19F-NMR
Approximately 20 ODs of a pure FAAF-modified 16-mer ODN was annealed with an equimolar amount of a complementary sequence to produce a fully paired duplex (Figure 1b). The samples were then dissolved in 300 µl of typical pH 7.0 NMR buffer containing 10% D2O/90% H2O, 100 mM NaCl, 10 mM sodium phosphate and 100 µM EDTA, and filtered into a Shigemi tube through a 0.2 µm membrane filter. All 1H- and 19F-NMR results were recorded using a dedicated 5 mm 19F/1H dual probe on a Bruker DPX400 Avance spectrometer operating at 400.0 and 376.5 MHz, respectively, using acquisition parameters described previously (14, 20, 29). Imino proton spectra at 5°C were obtained using a phase-sensitive jump-return sequence and referenced relative to that of DSS. 19F-NMR spectra were acquired in the 1H-decoupled mode and referenced relative to that of CFCl3 by assigning external C6F6 in C6D6 at −164.9 ppm. One and two-dimensional 19F-NMR spectra were measured between 5 and 60°C with increment of 5–10°C. Temperatures were maintained by a Bruker-VT unit with the aid of controlled boiling liquid N2 in the probe. Line shape simulations were performed as described previously (30) using WINDNMR-Pro (version 7.1.6; J. Chem. Educ. Software Series; Reich, H. J., University of Wisconsin, Madison, WI, USA).
DSC experiments
Calorimetric measurements of the three FAAF-modified 16-mer duplexes were performed using a Nano-DSC from TA Instruments (Lindon, UT, USA). Prior to temperature scanning, samples were degassed for at least 10 min under house vacuum in a closed vessel. Solutions were loaded, respectively, into the sample and reference cells using a pipette by attaching a small piece of silicone tube at the end of the tip and were purged several times to get rid of air bubbles. After both cells were filled, they were capped and a slight external pressure (∼3 atm) was applied to prevent evaporation of the sample solution. Raw data were collected as microwatts versus temperature. Template–primer solutions were prepared by dissolving desalted samples in a pH 7.0 buffer solution consisting of 20 mM sodium phosphate and 0.1 M NaCl. In a typical scan, a 0.1 mM template–primer solution was scanned against buffer from 15°C to 90°C at a rate of 0.75°C/min. At least five repetitions were obtained. A buffer vs. buffer scan was used as a control and subtracted from the sample scan and normalized for heating rate. This results in base-corrected ΔCpex versus temperature curves. Each transition shows negligible changes in the heat capacities between the initial and final states, thus ΔΔCpex was assumed to be zero. The area of the resulting curve is proportional to the transition heat, which, when normalized for the number of moles of the sample, is equal to the transition enthalpy, ΔH. ΔH is an integration of ΔCpex over temperature T. All sample solutions were 0.1 mM concentration. Tm was the temperature at half the peak area. ΔG and ΔS values have been determined according to the procedures described by Chakrabarti et al. (31).
Substrate construction and UvrABC protein purification
DNA substrates of 55 bp containing a FAAF adduct at each of three guanine residues were constructed as previously described (11,32). Briefly, an FAAF-modified 16-mer ODN (5′-CTCTCG1G2CG3CCATCAC-3′) was ligated with flanking 20-mer ODN (5′-GACTACGTACTGTTACGGCT-3′) and 19-mer ODN (5′-GCAATCAGGCCAGATCTGC-3′) ODN at the 5′- and 3′-end, respectively (Supplementary Figure S1). The 20-mer was 5′-terminally labeled with 32P. The ligation product was purified by urea–PAGE under denaturing conditions. Following the purification, the substrate was annealed to the corresponding complementary strand, and then purified on an 8% native polyacrylamide gel. Similar procedures were employed to construct FAF-modified DNA substrates using 12-mer NarI sequence (5′-CTCG1G2CG3CCATC-3′), which we prepared previously for 19F-NMR/UV/ICD studies (20).
UvrA, UvrB and UvrC proteins were over expressed in E. coli and then purified as previously described (33). The estimated purity of the three proteins was >95%. A Bio-Rad Protein Assay was used to determine the protein concentration with BSA as the standard based on the manufacturer-recommended procedures.
Nucleotide excision assay and quantification of incision products
The 5′-terminally labeled DNA substrates were incised by UvrABC as previously described (11,32). Briefly, the DNA substrates (2 nM) were incubated in the UvrABC reaction buffer (50 mM Tris–HCl, pH 7.5, 50 mM KCl, 10 mM MgCl2, 5 mM DTT) at 37°C in the presence of UvrABC (UvrA, 10 nM; UvrB, 250 nM; and UvrC, 100 nM). The Uvr proteins were diluted and premixed in Uvr storage buffer before addition to the reaction. Aliquots were collected at 0, 5, 10, 15 and 20 min into the reaction. The reaction was terminated by heating at 95°C for 5 min. The products were denatured by addition of formamide loading buffer and heating to 95°C for 5 min, followed by quick chilling on ice. The incision products were then analyzed by electrophoresis on a 12% polyacrylamide sequencing gel under denaturing conditions with TBE buffer.
To quantify the incision products, radioactivity was measured using a Fuji FLA-5000 Image Scanner with MultiGauge V3.0 software. The DNA incised (in fmol) by UvrABC was calculated based on the total molar amount of DNA used in each reaction and the ratio of the radioactivity of incision products to total radioactivity of DNA. At least three independent experiments were performed for determination of the rates of incision.
RESULTS
Model sequences
We previously used FAF-modified 12-mer duplexes (5′-CTCG1G2CG3*CNATC-3′, N = C or T) to probe the impact of flanking and 3′-next flanking sequences on NarI-induced frameshift mutagenesis (20). Initially, we tried to use the same 12-mer NarI sequence for FAAF-modification for sake of comparison and consistency; however, the sequence was unsuitable for FAAF. Although FAAF adduction on the 12-mer NarI sequence was facile, a resulting reaction mixture was difficult to purify on the reverse phase HPLC system (see asterisked peaks in Supplementary Figure S2). Moreover, ligation efficiencies of the FAAF-modified 12-mers, particularly on the G1- and G2-positions, were very low. Accordingly, the length of DNA was increased to 16 (5′-CTCTCGGCGCCATCAC-3′, Figure 1b) by adding two nucleotides (underlined) on either side of the 12-mer. The resulting FAAF-modified 16-mer ODNs were separated well on a reverse-phase HPLC and exhibited excellent ligation efficiencies (see below).
Figure 2a shows an HPLC profile of a work-up mixture after 20 min of reaction. Unreacted control ODN appeared at 21 min, followed by seven FAAF-modified ODNs in three retention time zones: Peaks 1–3 at 28–35 min, Peaks 4–6 at 42–60 min and Peak 7 at 84 min. Online UV (Figure 2b) of the modified ODNs displayed a small shoulder in the 290–320 nm range. The relative absorption intensities (290–320 nm) for the three peak groups were approximately 1:2:3. This finding is reminiscent of AF- or FAF-induced absorption shoulders observed in 290–350 nm, whose intensities correlate consistently with the number of adduct modifications (28). As a result, Peaks 1–3 and 4–6 were assigned as mono- and di-adducts, respectively, and Peak 7 as a tri-adduct. These adducts were characterized by exonuclease digestion/ESI-TOF-MS-MS analyses, as described below. Depending on the location of FAAF, the mono-adducts were designated as NarI–G1, NarI–G2 or NarI–G3, in which G1, G2 and G3 signify the position of the FAAF-modified guanine. Details of the structural characterization and repair of the di- and tri-FAAF adducts will be published separately.
The HPLC elution profile of the FAAF-modified 16-mer ODNs in the present study is similar to that of the AAF-modified 15-mer NarI sequence (5′-TCCTCG1G2CG3CCTCTC-3′) reported by Tan et al. (34). These results indicate that AAF and FAAF are chromatographically comparable, irrespective of sequence length, as long as the common NarI core (underlined) is included in the sequences. This is not surprising since conformational and thermodynamic compatibilities of fluorine containing AF and AAF models have well been documented (20,30,35). A similar elution pattern was observed for the FAF-modified 12-mer and FAAF-modified 16-mer NarI sequences; however, the order of elution of G1 and G3 was reversed (compare Figure 2a with Figure 2 in Ref. 20).
ESI-QTOF-MS characterization
The molecular weights of all three FAAF-modified ODNs were measured by ESI-QTOF-MS prior to sequence verification by exonuclease digestion. Ionization of ODNs normally occurs by the loss of a proton from a phosphate group in the ODN backbone. As the number of nucleotides in an ODN increases, the average charge state observed in the full scan mass spectra increases as well (28). As shown in Supplementary Figure S3, the ODNs containing 16 nucleotides form (M-4 H)4− ions predominantly unlike the 12-mers studied previously that form (M-3 H)3− primarily upon electrospray (28). Exonucleases cleave terminal deoxynucleotides from the ODN chain until the FAAF-modified nucleotide is exposed at the end of the chain. At that point the digestion reaction slows down significantly. The position of modification is identified (in this case) when the fragment(s) formed by the loss of the unmodified guanine nucleotides is observed in the LC/MS spectra. This is shown in Supplementary Figure S4 for the 3′ digest of the -G1(FAAF)G2CG3- ODN. The ions observed at m/z 659.5 and m/z 989.7 are the (M-3 H)3− and (M-2 H)2− ions formed from the 5′-CTCTCG1(FAAF)-3′ ODN digest fragment. The observation of these ions confirms that this ODN is modified on the G closest to the 5′ end. The LC/MS analysis of the 5′ digest of the second modified ODN to elute is shown in Supplementary Figure S5a and 5b. The exonuclease digestion of this particular reaction product was particularly slow and evidence for endonuclease activity is observed in the mass spectra. All the Y fragments observed in Supplementary Figure S5 are (M-3 H)3− ions. The Y10 and Y9 fragments at m/z 1069.2 (5′-G2CG3(FAAF)CCATCAC-3′) and m/z 959.5 (5′-CG3(FAAF)CCATCAC-3′) are formed by consecutive cleavages of unmodified guanines, confirming that this reaction product is modified on the guanine closest to the 3′ end. LC/MS analysis of the exonuclease digests derived from the third singly-modified ODN to elute indicates that the FAAF group is attached to the central G in the sequence, -G1G2(FAAF)CG3-. LC/MS analysis of the 3′ digest (Supplementary Figure S6) show ions at m/z 1154.7 and m/z 769.5 corresponding to the (M-2 H)2− and (M-3 H)3− ions derived from the 5′-CTCTCG1G2(FAAF)-3′ digest fragment. The mass spectra acquired from the 5′- digest (Supplementary Figure S7) show (M-2 H)2− and (M-3 H)3− ions at m/z 1604.3 and m/z 1069.2 whose masses are consistent with 5′-G2(FAAF)CG3CCATCAC-3′ fragment. No ions formed by the loss of two guanine deoxynucleotides were observed in any of the mass spectra. The observation of ODN fragments with two G's in both 3′ and 5′ digests confirms that the last singly modified ODN to elute from the reaction mixture is modified on the middle G.
Circular dichroism
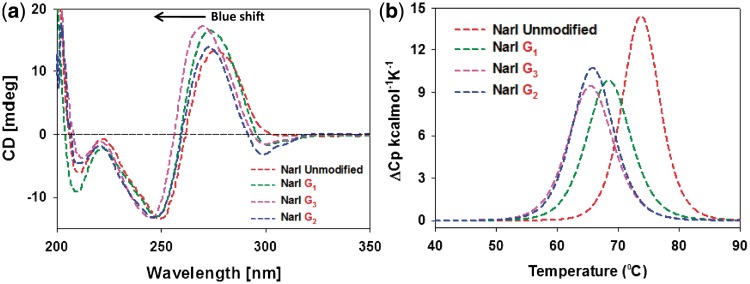
Figure 3a shows an overlay of the CD spectra for the three FAAF-modified NarI–G1, –G2 and –G3 duplexes relative to the unmodified control (red). Unmodified and FAAF-adducted duplexes both displayed a positive and negative ellipticity at around 270 and 250 nm, respectively, which is an S-curve characteristic of a B-form DNA double helix. The modified duplexes displayed significant blue shifts relative to the unmodified duplex, NarI–G3 (6 nm) >> NarI–G1 ∼ G2 (3 nm), indicating adduct-induced DNA bending. A concomitant increase in the positive intensity around 270 nm was noted in the order of G3 ∼ G1 > G2, which could be due to the interaction of the intercalated S-conformeric FAAF with neighboring bases. We noted a similar blue shift and hyperchromic effect for the highly (75%) S-conformeric FAF-modified NarI–G3 duplex (Figure 3b and Table 1 in Ref. 20).
Figure 3.
(a) CD spectral overlays recorded at 15°C and (b) DSC curves recorded in 20 mM phosphate buffer containing 0.1 M NaCl at pH 7.0 of fully paired 16-mer NarI duplexes with FAAF modification at G1 (green), G2 (blue) and G3 (pink).
Table 1.
Thermal and thermodynamic parameters of FAAF modified NarI duplexes obtained from differential scanning calorimetry
| 5′-CTCTCG1G2CG3CCATCAC-3′ | ||||||||
|---|---|---|---|---|---|---|---|---|
| 3′-GAGAGC C GCGGTAGTG-5′ | ||||||||
| −ΔH (kcal/mol) | −ΔS (eu) | −ΔG37°c (kcal/mol) | Tma (°C) | ΔΔHb (kcal/mol) | ΔΔSc (eu) | ΔΔG37°cd (kcal/mol) | ΔTme (°C) | |
| Controlf | 117.5 | 319.1 | 18.6 | 73.9 | – | – | – | – |
| NarI-G1-FAAFf | 95.6 | 260.2 | 14.9 | 68.6 | 21.9 | 58.9 | 3.7 | −5.3 |
| NarI-G2-FAAFf | 98.9 | 272.1 | 14.5 | 66.0 | 18.6 | 47.0 | 4.1 | −7.9 |
| NarI-G3-FAAFf | 92.8 | 254.4 | 13.9 | 65.6 | 24.7 | 64.7 | 4.7 | −8.3 |
aTm values is the temperature at half the peak area.
bΔΔH = ΔH (modified duplex) −ΔH (control duplex).
cΔΔS = ΔS (modified duplex) −ΔS (control duplex).
dΔΔG = ΔG (modified duplex) −ΔG (control duplex).
eΔTm = Tm (modified duplex) −Tm (control duplex).
fThe average standard deviations for −ΔG, −ΔH and Tm are ±0.4, ±3.0 and ±0.4, respectively.
More importantly, FAAF-modified duplexes exhibited sequence-dependent induced CD in the 290–320 nm range (ICD290–320 nm). This finding is reminiscent of ICD290–350 nm, which has been used as a sensitive marker for the FAF-induced S/B/W-conformational heterogeneity (positive for S- and W- and negative for B-conformer) (12, 15, 20, 29, 36). In the present case, however, the FAAF-modified NarI duplexes exhibited negative dips, with the NarI–G2 duplex showing a greater dip than G1 or G3 duplexes (Figure 3a). This result could be due to a higher ratio (57%) of B-conformer for the NarI–G2 duplex.
UV melting experiments
Supplementary Figure S8 shows the UV melting profiles of the three FAAF–NarI duplexes and an unmodified control duplex, all at 6.4 µM. All duplexes showed typical monophasic, sigmoidal, helix–coil transitions with a strong linear correlation (R2 > 0.9) between Tm−1 and lnCt. Thermal and thermodynamic parameters calculated from UV melting are summarized in Supplementary Table S1. As expected, modified duplexes were destabilized thermally and thermodynamically relative to the control duplex. The magnitude of thermal (ΔTm) and thermodynamic (ΔΔG) destabilization was in the order of NarI–G2 ∼ NarI–G3 (−8.7 to −8.8°C, 3.3 to 3.7 kcal/mol, respectively) > NarI–G1 (−4.6°C and 2.0 kcal/mol, respectively).
Differential scanning calorimetry
Figure 3b shows differential scanning calorimetry (DSC) plots of excess heat capacity Cpex versus temperature for the FAAF–NarI duplexes relative to the unmodified control. Table 1 summarizes the thermal and thermodynamic parameters derived from these DSC curves. Consistent with the UV melting data, the NarI–G3 duplex was most destabilized (ΔΔH = 24.7 kcal/mol, ΔΔG37°C = 4.7 kcal/mol, ΔTm = −8.3°C), followed by NarI–G2 (ΔΔG37°C = 4.1 kcal/mol, ΔTm = −7.9°C). NarI–G1 was the least affected (ΔΔG37°C = 3.7 kcal/mol, ΔTm = −5.3°C).
Differences in the thermal and thermodynamic destabilizations must have arisen from the differences in the S/B/W-conformational characteristics. The most S-conformeric (61%) NarI–G3 duplex causes disturbance of Watson–Crick base pairing, resulting in enthalpy reduction (ΔΔH = 24.7 kcal/mol) (Table 2). However, the large entropy (ΔΔS = 64.7 eu) compensates for the enthalpy, thus resulting in the overall free energy loss of ΔΔG37°C = 4.7 kcal/mol (37). In contrast, NarI–G1 and –G2 duplexes possess higher populations of B-conformer (46 and 57%, respectively), thus exhibiting lower differences in the enthalpy values (ΔΔH = 21.9 kcal/mol and ΔΔH = 18.6 kcal/mol, respectively) (Table 2). The relatively small enthalpy differences observed for the G1 and G2 duplexes could be attributed to the presence of S- and W-conformers in addition to B-conformer. As expected, entropy compensation was less in these two duplexes (NarI–G1, ΔΔS = 58.9 eu and NarI–G2, ΔΔS = 47.0 eu), yielding similar overall free energies (NarI–G1, ΔΔG = 3.7 kcal/mol and NarI–G2, ΔΔG = 4.1 kcal/mol).
Table 2.
Conformational heterogeneity (B/S/W), thermal destabilization and relative percent incision rates of FAAF- and FAF-modified NarI duplexes
| NarI Duplexes | Population Ratiosa (%) |
ΔTmb (°C) | Relative incision ratec (%) | ||
|---|---|---|---|---|---|
| B | S | W | |||
| NarI-G1-FAAF | 46 | 34 | 20 | −5.3 | 93 |
| NarI-G2-FAAF | 57 | 15 | 9 | −7.9 | 32 |
| NarI-G3-FAAF | 13 | 61 | 26 | −8.3 | 100 |
| NarI-G1-FAF | 42 | 58 | – | −9.4 | 44 |
| NarI-G2-FAF | 69 | 31 | – | −6.8 | 43 |
| NarI-G3-FAF | 35 | 65 | – | −8.3 | 25 |
aThe percent population ratios were calculated at 5°C on the basis of line simulations.
bΔTm = Tm (modified duplex) − Tm (control duplex).
cPercent incision rate of modified duplexes with respect to NarI-G3-FAAF (100%).
S/B/W conformational heterogeneity
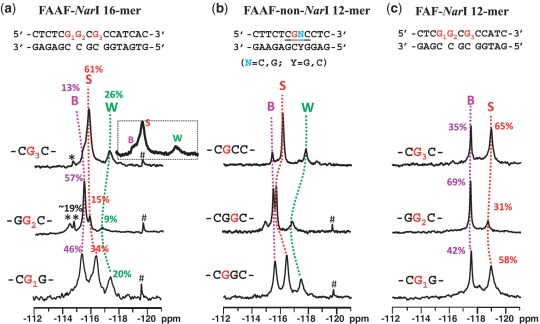
Figure 4a shows the 19F-NMR spectra of FAAF–NarI 16-mer G1-, G2- and G3-duplexes measured at 5°C, in which 19F signals are in slow chemical exchange. These NarI-FAAF duplexes exhibited three to five 19F signals, each representing a particular conformation. The percent population ratios shown were calculated on the basis of line simulations as shown in Supplementary Figure S9. Assignments of the different 19F signals of each duplex were necessary to carry out meaningful structure-activity-relationship studies. The signal assignments in Figure 4a were made initially on the basis of chemical exchange, ring current effect and chemical shift pattern recognition as have been done for a number of FAAF- and FAF-adducts in various sequence contexts (30,35). It has been demonstrated that AF and AAF adducts adopt the S/B- and S/B/W-conformational equilibrium, respectively (Figure 1c and d) and their 19F chemical shifts are independent of overall sequence and its length, but strongly rely on the nature of the bases flanking the lesion (15,30). The major 19F signals in Figure 4a correlate well with the S/B/W-profiles reported previously for FAAF adducts (15), i.e. B-, S- and W-conformers going from downfield to upfield, i.e. −115.0 to −115.5, −115.5 to −117.0 and −117.0 to −118.0 ppm, respectively. Additional signals were observed in the −114.0∼−115.0 ppm range for the NarI–G2 and –G3 duplexes (Figure 4a, see Supplementary Table S2 for exact chemical shifts). Consistent with this observation, their proton spectra displayed a mixture of broad imino signals arising not only from those involved in Watson–Crick hydrogen bonds (12–14 ppm), but also from the lesion site and its vicinity (11–12 ppm) (Supplementary Figure S10).
Figure 4.
19F-NMR spectra of (a) FAAF-modified NarI 16-mer, (b) FAAF-modified non-NarI 12-mer and (c) FAF-modified NarI 12-mer duplexes at 5°C. *unknown conformers; #impurity.
To complement the 19F signal assignments, we additionally conducted a set of comparative spectral analyses using three FAAF-12-mer duplexes in the non-NarI sequences (Figure 4b) in otherwise identical flanking sequence contexts (CG*G, GG*C and CG*C context for G1, G2 and G3, respectively). The top trace in Figure 4b is the 19F-NMR spectrum of a FAAF-modified 12-mer duplex (5′-CTTCTCG*CCCTC-3′), whose S/B/W conformational profiles have been well characterized (15). It should be noted that this non-NarI 12-mer duplex contains the identical CG*C flanking sequence context as the 16-mer NarI–G3–FAAF duplex. Comparison of the two spectra (i.e. top traces of Figure 4a and b) revealed a parallel trend both in terms of chemical shifts and population ratios (Table 2 and Supplementary Table S2), supporting the conformational assignments. This is consistent with our previous findings that the electronic environment for the 19F signals of AF and AAF adducts are strongly modulated by the nature of flanking bases (15, 30). Similarly, we prepared two additional FAAF-modified non-NarI 12-mer duplexes (5′-CTTCTCG*GCCTC-3′ and 5′-CTTCTCGG*CCTC-3′) with the same flanking base contexts (underlined) as the NarI–G1 and –G2 duplexes, respectively. Figure 4a and b compares the 19F-NMR spectra of all three NarI 16-mer and non-NarI 12-mer duplexes side by side. The 19F signal profiles, as indicated by dotted lines (pink, B; red, S; green, W) for G1 → G2 → G3 of each sequence context, match quite well overall despite of slight variations observed in chemical shifts and population ratios, particularly for the GG*C sequence context. Whereas the S- and W-conformer signals were prone to shift, the B-conformer signal appeared to be steady at −115.5 ppm. This trend is more apparent in Supplemental Figure S11, in which the two FAAF-modified sequence series (NarI-16-mer versus non-NarI-12-mer) are compared in a pair for each –CG*G-, -GG*C- and –CG*C- sequence contexts. It is plausible that the carcinogen moiety in the major groove of the B-conformer is not subjected to the ring current effect, as the S- and W-conformers would be (14). We were unable to identify the minor signals (asterisked) in the 16-mer NarI–G2– (<19%) and –G3–FAAF duplexes, although their downfield shifts relative to the B-conformer imply B-like conformers, in which the fluorine containing carcinogen moiety is exposed.
Figure 4c shows the 19F-NMR spectra of FAF-modified 12-mer duplexes with the same NarI sequence contexts, which have been thoroughly characterized (20). The B/S conformer population ratios were determined to be 42%:58%, 69%:31%, 35%:65% for FAF-modified NarI–G1, –G2 and –G3, respectively, at 5°C (Table 2) (20). Although the chemical shift difference (0.4–1.0 ppm) for the B and S conformer of FAAF (Figure 4a) is significantly smaller than that (∼1.5 ppm) of the FAF counterparts, their overall S/B ratios appear to match (Figure 4a and c). The B/S/W population ratios for the FAAF–NarI–G1, –G2 and –G3 16-mer duplexes were 46:34:20, 57:15:9 and 13:61:26, respectively (Table 2). In both the FAAF- (Figure 4a) and FAF- (Figure 4c) NarI duplexes, the population of S-conformer decreased in the order of G3 > G1 > G2 and that of the B-conformer decreased in the reverse order, G2 > G1 >G3. This comparative analysis was based on the assumption that structurally similar FAAF would experience similar sequence effects on their conformational profiles as observed by FAF in different sequence contexts of the NarI sequence (20). As expected, the aminofluorene-induced B/S-heterogeneity is strongly dependent on the nature of the flanking sequences, regardless of whether the lesion has the bulky acetyl group on the central nitrogen linking the carcinogen and the modified guanine, thus validating our assumption.
Dynamic 19F-NMR
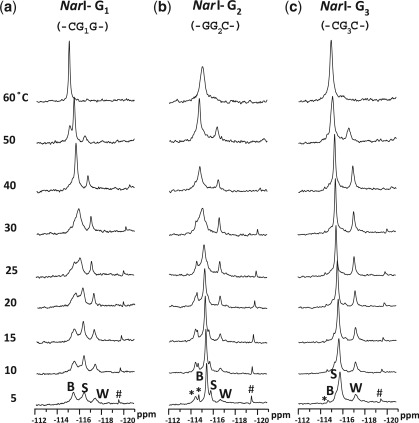
Figure 5 shows the 19F-NMR spectra of the three FAAF–NarI duplexes as a function of temperature (5–60°C). Whereas the three 19F signals in each duplex were in slow exchange at 5°C, the two downfield B- and S-signals became exchange broadened, giving rise to coalescent signals at around 30, 40 and 25°C for G1, G2 and G3, respectively. In all cases, the merged signals coalesced with the upfield W-signal at around 60°C. All three NarI duplexes showed relatively strong off-diagonal contour peaks of the major signals in the exchange spectra (data not shown), confirming their chemical exchanges.
Figure 5.
Dynamic 19F-NMR spectra of fully paired 16-mer NarI duplexes. FAAF modification at (a) G1, (b) G2 and (c) G3. *unknown conformers; #impurity.
UvrABC incisions of FAAF-adducts on NarI sequence
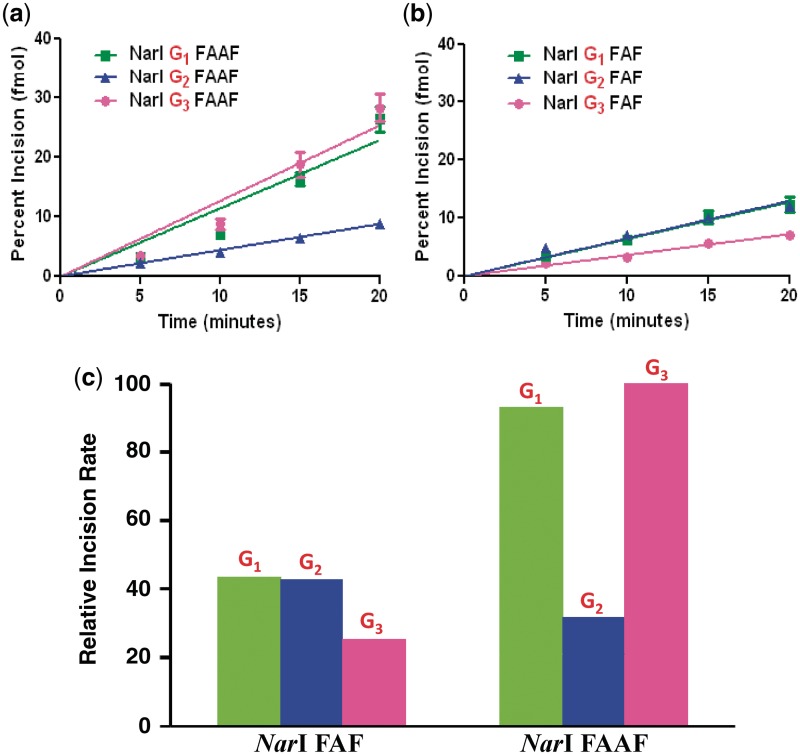
Figure 6 shows the kinetic assay results, in which 55-mer FAAF-modified DNA duplex substrates were incised by UvrABC nuclease. These substrates were radioactively labeled at the 5′-end of the adducted strand. The major incision products can be seen as 18-mer (NarI–G1), 19-mer (NarI–G2) or 21-mer (NarI–G3) separated on a urea–PAGE gel under denaturing conditions (Supplementary Figure S12). The incision occurred at the eighth phosphate bond 5′ to the modified nucleotide, which is consistent with the previously reported results of UvrABC incision (11,32).
Figure 6.
Absolute percent incision rates of (a) FAAF and (b) FAF–NarI duplexes modified at G1, G2 and G3; (c) percent incision rates histogram of FAF and FAAF at different positions relative to NarI–G3 FAAF as 100%.
Quantitative analysis of the incision indicated that the substrates were incised at different efficiencies, depending on where the damage site was located in the sequence (Figure 6). Specifically, the N-acetylated FAAF adducts at NarI–G1 and NarI–G3 displayed similar rates of incision, whereas NarI–G2 had a much lower rate of incision, G3 (100%) ≥ G1 (93%) > G2 (32%) (Figure 6c, Table 2). For comparison, we also determined the UvrABC incision of FAF adducts in the same NarI sequence context. As shown in Figure 6b and c, the N-deacetylated FAF adducts in the same NarI sequence context were repaired 2 - to 3-fold less than FAAF. Despite having similar B/S-conformer profiles (Figure 4 and Table 2) the incision efficiency of FAF adducts at the three different sites in the NarI sequence followed the order of G1 (44%) ≈ G2 (43%) > G3 (25%), where the percentages were calculated relative to FAAF NarI–G3 (which was the most efficiently incised).
DISCUSSION
It is well known that DNA sequence is a major determining factor for repair outcomes of site-specifically modified bulky DNA lesions. In this study, we examined the conformational heterogeneity and thermodynamics of FAAF and FAF at three different guanine positions (G1, G2 and G3) of the well-known NarI recognition sequence. Moreover, we obtained NER data of these adducts using the E. coli UvrABC system. Table 2, which summarizes the S/B/W solution conformational heterogeneity, Tm and NER efficiency results, presents strong evidence that the NER repair efficiencies of AAF and AF adducts in the NarI sequences are modulated by their conformational and thermodynamic properties.
FAAF-induced B/S/W-conformational heterogeneity
Our combined 19F-NMR/ICD results show that FAAF adduct in a well-known mutational hotspot NarI sequence exist in a mixture of B/S/W conformers with varying populations (Figure 4a, Table 2). A greater population of syn-glycosidic S-(61%) and W-(26%) conformers was observed in NarI–G3, in which the lesion is flanked with C on both 5′- and 3′-ends (-CG3*C-). This result is consistent with the preferred syn-conformation adopted by duplexes modified by AF (20), 2-amino-3-methylimidazo(4,5-f)quinolone (IQ) (38–40) and 2-amino-1-methyl-6-phenylimid-azo(4,5-b)pyridine (PHIP) (41), with the same -CG*C- contexts, in either NarI or non-NarI sequences (20,38,39,41). The mostly syn NarI–G3 duplex appeared to be distorted, bent or possibly formed a B–Z junction, as evidenced by a significant blue shift and hyperchromic effect in CD (Figure 3a) (42). The latter was probably due to the п–п stacking interaction between the intercalated aminofluorene and flanking base pairs. On the other hand, the NarI–G2 duplex (-G1G2*C-) exhibited largely the anti-B-conformer (57%) along with S- (15%), W- (9%) and two unidentified minor conformers (∼19%). In comparison to NarI–G3, the NarI–G2 duplex exhibited smaller blue shift and hyperchromic effect (Figure 3a), suggesting lesser disturbance of the double helical DNA structure.
These results indicate the heterogeneous nature of AAF in the NarI sequence and are consistent with a previous CD study that showed a major DNA distortion for AAF at G3 adduct compared to G1 and G2 (42). Similarly, Veaute et al. (43) conducted a DNase I footprint study on the NarI sequence and showed that AAF at the G2 position inhibits DNase I digestion of DNA at up to five bases in the modified strand and four bases in the complementary strand. In contrast, inhibition at the G1 and G3 positions was extended to eight and six bases, respectively in the modified strand.
The G3 and G2 duplexes are chemically isomeric, differing only in the direction of the G:C base pair at the 5′-position (e.g. C:G → G:C). Such a polarity swap is clearly responsible for the rather dramatic conformational shift from S- (61 to 15%) to B- (13 to 57%) and W-conformation (26 to 9%) (Table 2). A similar polarity switch at the 3′-end of the NarI–G1 duplex resulted in varying degrees of conformational shift in S- (61 to 34%), B- (13 to 46%) and W-conformation (26 to 20%).
As expected, FAAF-modification at the three guanines of the 16-mer NarI sequence resulted in thermal (ΔTm = −5.3 to −8.3°C) and thermodynamic (ΔΔG37°C = 3.7–4.7 kcal/mol) destabilization relative to the unmodified control duplex (Table 1). The destabilizing effect of the FAAF modification was sequence-dependent and was related to the S/B/W-conformational profile. As summarized in Tables 1 and 2, the highly S-conformeric NarI–G3 duplex (61%) promoted lesion stacking and disrupted the lesion site Watson–Crick base pairs, resulting in thermal (ΔTm = −8.3°C) and enthalpic destabilization (ΔΔH = 24.7 kcal/mol). In contrast, the highly B-conformeric (57%) NarI–G2 duplex exerted less enthalpy change (ΔΔH = 18.6 kcal/mol). As expected, NarI–G1 (46% B, 34% S) produced an intermediate change in enthalpy (ΔΔH = 21.9 kcal/mol). In all cases, however, enthalpy–entropy compensation resulted in a small overall difference (∼1 kcal/mol) in thermodynamic destabilization (Table 1). A similar case could occur if the W-conformer was considered as a thermodynamic destabilizer, G3 (26%) > G1 (20%) > G2 (9%). We previously studied three fully paired FAF-modified 12-mer duplexes in the same NarI sequence contexts (20). The UV melting results showed that FAF modification destabilizes the duplexes (ΔTm = −6.8 to −9.4°C, ΔΔG = 4.2–4.6 kcal/mol) similarly. The highly S-conformeric (65%) NarI–G3 duplex resulted in thermal destabilization (ΔTm = −8.3°C), whereas the highly B-conformeric (69%) NarI–G2 duplex exerted less destabilization (ΔTm = −6.8°C) (Table 2) (20).
Conformation-specific nucleotide excision repair
The E. coli UvrABC system displayed significant differences in repair of the FAAF adduct at each guanine position (G1, G2 and G3) of the NarI sequence. The NarI–G2 duplex showed considerably lower efficiency than NarI–G1 or NarI–G3, [G3 (100%) ≥ G1 (93%) > G2 (32%)] (Figure 6). It is clear from Table 2 that these NER results are in good agreement with the order of the S-conformer population [G3 (61%) > G1 (34%) > G2 (15%)], but are in exactly the reverse order of the population of B-conformer, [G2 (57%) > G1 (46%) > G3 (13%)]. This data suggest that the S-conformation is recognized and incised by E. coli NER dominantly over the B-conformation. We reported previously a similar conformation-specific NER results on a series of FAF-modified duplexes (16, 30).
The carcinogen in the highly S-conformer NarI–G3 is base-displaced at the lesion site, thus resulting in a major disturbance in the DNA helical structure (ΔTm = −8.3°C, ΔΔG37°C = 4.7 kcal/mol) (Table 2). This finding is contrasted with the 57% B-conformer NarI–G2 duplex, which maintains Watson–Crick base pairs at the lesion site (ΔTm = −7.9°C, ΔΔG37°C = 4.1 kcal/mol). Similar correlations could be made with either the W-conformer alone, G3 (26%) > G1 (20%) > G2 (9%) or the syn-conformation (combined S and W), G3 (87%) > G1 (54%) > G2 (24%).
For comparison, we also determined the UvrABC incisions of FAF-adducts in the same NarI sequence context. The two lesions revealed a similar B/S conformer heterogeneity in the NarI sequence context (Figure 4). Therefore, the expectation was that FAF would show a similar NER profile as FAAF, i.e. S-/W-conformer promotes NER over B-conformer. However, the NER results revealed that incision efficiency was in the order of G1 ≈ G2 > G3 (Figure 6, Table 2). At first, this result appears to be in line with the B-conformer population. It should be noted that FAF is repaired consistently 2- to 3-fold less than FAAF (Figure 6c, Table 2). This result is a general trend reported in the literature, although much greater differences in incision efficiency between AF and AAF have been noted (11, 44). As a result, the difference between G1 and G2 of FAF is not statistically significant (P = 0.83), but their difference with G3 is significant (P < 0.0001).
The incision differences between FAAF and FAF seem to suggest that, in addition to the sequence-dependent adduct conformation, the acetyl group in FAAF may play a role in DNA damage recognition by UvrABC. The only structural difference between FAF and FAAF is the absence of a bulky acetyl group on the linking nitrogen of the former (Figure 1a). It has been documented that N-acetylated FAAF adducts in fully paired duplexes produce a mixture of complex S/B/W-conformers, whereas N-deacetylated FAF adopts a simple exchangeable S/B-equilibrium (15, 30). Thus, it is clear that the N-acetyl group is responsible for generating up to 26% W-conformer in the NarI sequence (Figure 4). The bulkiness of the acetyl group with its possibility for cis and trans rotamer transitions about the amide bond (14, 15, 45) may facilitate the repositioning of the fluorenyl rings into the minor groove from the S conformation. This conformational rearrangement is relatively straightforward since it does not require a change in the glycosidic bond, which is syn in both cases. We observed a good correlation between the proportion of W-conformation and NER efficiency of FAAF.
Moreover, although FAF and FAAF have similar S/B-conformational profile (Figure 4), the N-acetyl group in the latter could act as a ‘conformational locker’ to raise the energy barriers among conformers. Such a scenario, i.e. higher energy barriers of FAAF vs. FAF, is plausible and might contribute to a greater disturbance in DNA, and thus greater repair. By contrast, the N-deacetylated FAF adopts a facile interchangeable B/S-equilibrium (<2 kcal/mol) that triggers weaker binding affinities with the damage-recognition protein UvrA. A recent crystal study indicated that the UvrA dimer does not contact the lesion site directly, but rather binds DNA regions on both sides of the modification and primarily recognizes adduct-induced unwinding, bending and deformity in the overall DNA structure (25). Furthermore, DNA damage recognition in E. coli NER is achieved through a sequential 2-step mechanism (46). The initial step is to recognize the adduct-induced distorted DNA structure. After strand opening at the damage site, the DNA adduct structure is further recognized or verified in a second step, which may facilitate the flipping of the adducted nucleotide (47,48). Therefore, it is possible that, for FAF, the second step of recognition plays a more important role than the first step, whereas the first step is a dominate recognition for FAAF.
The order of NER efficiencies described here is roughly consistent with bacterial NER data on AAF adducts embedded in a similar NarI sequence (17): G1 (100%), G3 (66%) and G2 (18%). Sequence dependence was also found in human NER of AAF adducted in the NarI sequence (18). In contrast to the E. coli NER data, however, the AAF adduct at G2 (100%) was found to be more repairable, followed by G3 (68%) and G1 (38%). Despite differences in the nature of proteins involved in prokaryotic and eukaryotic NER, the two systems show similar involvement of β-hairpin intrusion as damage recognition factors (49). Liu et al. (50) found a general qualitative trend toward similar relative NER incision efficiencies for 65% of bulky benzo[a]pyrene and equine estrogen substrates. Similar to bacterial UvrA, Rad4 (XPC) in yeast also recognizes helical distortion to sense DNA damage; unlike bacteria, yeast use a base-flipping mechanism for repair (26). Therefore, the efficiency of repair depends not only on the damage recognition step, but also on other factors, such as ease of base flipping.
In summary, our structural and thermodynamic data provide valuable conformational insights into the sequence-dependent UvrABC incisions of the bulky FAF and FAAF adducts in the NarI sequence context. Repair of the bulky N-acetylated FAAF adduct seems to occur in a conformation-specific manner, i.e. the highly S/W-conformeric G3 and G1 duplexes incised considerably more efficiently than the G2 duplex (G3 ∼ G1 > G2) (Table 2). These results were supported by melting and thermodynamic data. Not surprisingly, FAF was repaired 2- to 3-fold less than FAAF; however, the order of incision efficiencies was the reverse of that in the FAAF case. We considered the so-called N-acetyl factor and lesion-specific recognition mechanism for the different orders of incision for FAF and FAAF. Finally, the temperature dependence of the S/B/W-conformational equilibria of the FAAF-adducts in the NarI sequence could provide valuable opportunities for conformation-specific NER utilizing thermophilic UvrABC proteins (51). Taken together, the results of this study demonstrate the complexity of NER mechanisms of bulky DNA lesions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables S1–S2, Supplementary Figures S1–S12.
FUNDING
National Institutes of Health (Grant number R01CA098296); RI-INBRE Research Core Facility supported by the National Center for Research Resources (in part); National Institutes of Health (Grant number P20 RR016457). Funding for open access charge: National Institutes of Health (Grant number R01CA098296).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Luch A. Nature and nurture - lessons from chemical carcinogenesis. Nat. Rev. Cancer. 2005;5:113–125. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- 2.Melchior WB, Jr, Marques MM, Beland FA. Mutations induced by aromatic amine DNA adducts in pBR322. Carcinogenesis. 1994;15:889–899. doi: 10.1093/carcin/15.5.889. [DOI] [PubMed] [Google Scholar]
- 3.Neumann HG. Aromatic amines in experimental cancer research: tissue-specific effects, an old problem and new solutions. Crit. Rev. Toxicol. 2007;37:211–236. doi: 10.1080/10408440601028603. [DOI] [PubMed] [Google Scholar]
- 4.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T, editors. DNA Repair and Mutagenesis. 2nd edn. Washington: ASM Press; 2006. [Google Scholar]
- 5.Truglio JJ, Croteau DL, Van Houten B, Kisker C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem. Rev. 2006;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 6.Lehmann AR. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie. 2003;85:1101–1111. doi: 10.1016/j.biochi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Van Houten B. Nucleotide excision repair in Escherichia coli. Microbiol. Rev. 1990;54:18–51. doi: 10.1128/mr.54.1.18-51.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heflich RH, Neft RE. Genetic toxicity of 2-acetylaminofluorene, 2-aminofluorene and some of their metabolites and model metabolites. Mutat. Res. 1994;318:73–114. doi: 10.1016/0165-1110(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 9.Beland FA, Kadlubar FF. Handbook of Experimental Pharmacology. Spring, Heidelberg: 1990. [Google Scholar]
- 10.Cho BP. Dynamic conformational heterogeneities of carcinogen-DNA adducts and their mutagenic relevance. J. Environ. Sci. Health C. Environ. Carcinog. Ecotoxicol. Rev. 2004;22:57–90. doi: 10.1081/LESC-200038217. [DOI] [PubMed] [Google Scholar]
- 11.Luo C, Krishnasamy R, Basu AK, Zou Y. Recognition and incision of site-specifically modified C8 guanine adducts formed by 2-aminofluorene, N-acetyl-2-aminofluorene and 1-nitropyrene by UvrABC nuclease. Nucleic Acids Res. 2000;28:3719–3724. doi: 10.1093/nar/28.19.3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meneni SR, D'Mello R, Norigian G, Baker G, Gao L, Chiarelli MP, Cho BP. Sequence effects of aminofluorene-modified DNA duplexes: thermodynamic and circular dichroism properties. Nucleic Acids Res. 2006;34:755–763. doi: 10.1093/nar/gkj480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel DJ, Mao B, Gu Z, Hingerty BE, Gorin A, Basu AK, Broyde S. Nuclear magnetic resonance solution structures of covalent aromatic amine-DNA adducts and their mutagenic relevance. Chem. Res. Toxicol. 1998;11:391–407. doi: 10.1021/tx9702143. [DOI] [PubMed] [Google Scholar]
- 14.Zhou L, Rajabzadeh M, Traficante DD, Cho BP. Conformational heterogeneity of arylamine-modified DNA: 19F NMR evidence. J. Am. Chem. Soc. 1997;119:5384–5389. [Google Scholar]
- 15.Patnaik S, Cho BP. Structures of 2-acetylaminofluorene modified DNA revisited: insight into conformational heterogeneity. Chem. Res. Toxicol. 2010;23:1650–1652. doi: 10.1021/tx100341u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meneni S, Shell SM, Zou Y, Cho BP. Conformation-specific recognition of carcinogen-DNA adduct in Escherichia coli nucleotide excision repair. Chem. Res. Toxicol. 2007;20:6–10. doi: 10.1021/tx600273h. [DOI] [PubMed] [Google Scholar]
- 17.Seeberg E, Fuchs RP. Acetylaminofluorene bound to different guanines of the sequence -GGCGCC- is excised with different efficiencies by the UvrABC excision nuclease in a pattern not correlated to the potency of mutation induction. Proc. Natl Acad. Sci. USA. 1990;87:191–194. doi: 10.1073/pnas.87.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mu D, Bertrand-Burggraf E, Huang JC, Fuchs RP, Sancar A, Fuchs BP. Human and E.coli excinucleases are affected differently by the sequence context of acetylaminofluorene-guanine adduct. Nucleic Acids Res. 1994;22:4869–4871. doi: 10.1093/nar/22.23.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burnouf D, Koehl P, Fuchs RP. Single adduct mutagenesis: strong effect of the position of a single acetylaminofluorene adduct within a mutation hot spot. Proc. Natl Acad. Sci. USA. 1989;86:4147–4151. doi: 10.1073/pnas.86.11.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain N, Li Y, Zhang L, Meneni SR, Cho BP. Probing the sequence effects on NarI-induced -2 frameshift mutagenesis by dynamic 19F NMR, UV, and CD spectroscopy. Biochemistry. 2007;46:13310–13321. doi: 10.1021/bi701386f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broschard TH, Koffel-Schwartz N, Fuchs RP. Sequence-dependent modulation of frameshift mutagenesis at NarI-derived mutation hot spots. J. Mol. Biol. 1999;288:191–199. doi: 10.1006/jmbi.1999.2667. [DOI] [PubMed] [Google Scholar]
- 22.Koffel-Schwartz N, Fuchs RP. Sequence determinants for -2 frameshift mutagenesis at NarI-derived hot spots. J. Mol. Biol. 1995;252:507–513. doi: 10.1006/jmbi.1995.0515. [DOI] [PubMed] [Google Scholar]
- 23.Mekhovich O, Tang M, Romano LJ. Rate of incision of N-acetyl-2-aminofluorene and N-2-aminofluorene adducts by UvrABC nuclease is adduct- and sequence-specific: comparison of the rates of UvrABC nuclease incision and protein-DNA complex formation. Biochemistry. 1998;37:571–579. doi: 10.1021/bi971544p. [DOI] [PubMed] [Google Scholar]
- 24.Zou Y, Shell SM, Utzat CD, Luo C, Yang Z, Geacintov NE, Basu AK. Effects of DNA adduct structure and sequence context on strand opening of repair intermediates and incision by UvrABC nuclease. Biochemistry. 2003;42:12654–12661. doi: 10.1021/bi034446e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaciuk M, Nowak E, Skowronek K, Tanska A, Nowotny M. Structure of UvrA nucleotide excision repair protein in complex with modified DNA. Nat. Struct. Mol. Biol. 2011;18:191–197. doi: 10.1038/nsmb.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 27.Cho BP, Zhou L. Probing the conformational heterogeneity of the acetylaminofluorene-modified 2'-deoxyguanosine and DNA by 19F NMR spectroscopy. Biochemistry. 1999;38:7572–7583. doi: 10.1021/bi990182d. [DOI] [PubMed] [Google Scholar]
- 28.Gao L, Zhang L, Cho BP, Chiarelli MP. Sequence verification of oligonucleotides containing multiple arylamine modifications by enzymatic digestion and liquid chromatography mass spectrometry (LC/MS) J. Am. Soc. Mass Spectrom. 2008;19:1147–1155. doi: 10.1016/j.jasms.2008.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jain N, Meneni S, Jain V, Cho BP. Influence of flanking sequence context on the conformational flexibility of aminofluorene-modified dG adduct in dA mismatch DNA duplexes. Nucleic Acids Res. 2009;37:1628–1637. doi: 10.1093/nar/gkn1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meneni SR, Shell SM, Gao L, Jurecka P, Lee W, Sponer J, Zou Y, Chiarelli MP, Cho BP. Spectroscopic and theoretical insights into sequence effects of aminofluorene-induced conformational heterogeneity and nucleotide excision repair. Biochemistry. 2007;46:11263–11278. doi: 10.1021/bi700858s. [DOI] [PubMed] [Google Scholar]
- 31.Chakrabarti MC, Schwarz FP. Thermal stability of PNA/DNA and DNA/DNA duplexes by differential scanning calorimetry. Nucleic Acids Res. 1999;27:4801–4806. doi: 10.1093/nar/27.24.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zou Y, Liu TM, Geacintov NE, Van Houten B. Interaction of the UvrABC nuclease system with a DNA duplex containing a single stereoisomer of dG-(+)- or dG-(-)-anti-BPDE. Biochemistry. 1995;34:13582–13593. doi: 10.1021/bi00041a038. [DOI] [PubMed] [Google Scholar]
- 33.Zou Y, Van Houten B. Strand opening by the UvrA(2)B complex allows dynamic recognition of DNA damage. EMBO J. 1999;18:4889–4901. doi: 10.1093/emboj/18.17.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan X, Suzuki N, Grollman AP, Shibutani S. Mutagenic events in Escherichia coli and mammalian cells generated in response to acetylaminofluorene-derived DNA adducts positioned in the Nar I restriction enzyme site. Biochemistry. 2002;41:14255–14262. doi: 10.1021/bi0202878. [DOI] [PubMed] [Google Scholar]
- 35.Meneni S, Liang F, Cho BP. Examination of the long-range effects of aminofluorene-induced conformational heterogeneity and its relevance to the mechanism of translesional DNA synthesis. J. Mol. Biol. 2007;366:1387–1400. doi: 10.1016/j.jmb.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang F, Meneni S, Cho BP. Induced circular dichroism characteristics as conformational probes for carcinogenic aminofluorene-DNA adducts. Chem. Res. Toxicol. 2006;19:1040–1043. doi: 10.1021/tx0601253. [DOI] [PubMed] [Google Scholar]
- 37.Liang F, Cho BP. Enthalpy-entropy contribution to carcinogen-induced DNA conformational heterogeneity. Biochemistry. 2010;49:259–266. doi: 10.1021/bi901629p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elmquist CE, Wang F, Stover JS, Stone MP, Rizzo CJ. Conformational differences of the C8-deoxyguanosine adduct of 2-amino-3-methylimidazo[4,5-f]quinoline (IQ) within the NarI recognition sequence. Chem. Res. Toxicol. 2007;20:445–454. doi: 10.1021/tx060229d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang F, DeMuro NE, Elmquist CE, Stover JS, Rizzo CJ, Stone MP. Base-displaced intercalated structure of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme, a hotspot for -2 bp deletions. J. Am. Chem. Soc. 2006;128:10085–10095. doi: 10.1021/ja062004v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang F, Elmquist CE, Stover JS, Rizzo CJ, Stone MP. DNA sequence modulates the conformation of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme. Biochemistry. 2007;46:8498–8516. doi: 10.1021/bi700361u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown K, Hingerty BE, Guenther EA, Krishnan VV, Broyde S, Turteltaub KW, Cosman M. Solution structure of the 2-amino-1- methyl-6-phenylimidazo[4,5-b]pyridine C8-deoxyguanosine adduct in duplex DNA. Proc. Natl Acad. Sci. USA. 2001;98:8507–8512. doi: 10.1073/pnas.151251898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koehl P, Valladier P, Lefevre JF, Fuchs RP. Strong structural effect of the position of a single acetylaminofluorene adduct within a mutation hot spot. Nucleic Acids Res. 1989;17:9531–9541. doi: 10.1093/nar/17.23.9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veaute X, Fuchs RP. Polymorphism in N-2-acetylaminofluorene induced DNA structure as revealed by DNase I footprinting. Nucleic Acids Res. 1991;19:5603–5606. doi: 10.1093/nar/19.20.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gillet LC, Alzeer J, Scharer OD. Site-specific incorporation of N-(deoxyguanosin-8-yl)-2-acetylaminofluorene (dG-AAF) into oligonucleotides using modified 'ultra-mild' DNA synthesis. Nucleic Acids Res. 2005;33:1961–1969. doi: 10.1093/nar/gki335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shapiro R, Hingerty BE, Broyde S. Minor-groove binding models for acetylaminofluorene modified DNA. J. Biomol. Struct. Dyn. 1989;7:493–513. doi: 10.1080/07391102.1989.10508506. [DOI] [PubMed] [Google Scholar]
- 46.Zou Y, Luo C, Geacintov NE. Hierarchy of DNA damage recognition in Escherichia coli nucleotide excision repair. Biochemistry. 2001;40:2923–2931. doi: 10.1021/bi001504c. [DOI] [PubMed] [Google Scholar]
- 47.Malta E, Verhagen CP, Moolenaar GF, Filippov DV, van der Marel GA, Goosen N. Functions of base flipping in E. coli nucleotide excision repair. DNA Repair. 2008;7:1647–1658. doi: 10.1016/j.dnarep.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 48.Malta E, Moolenaar GF, Goosen N. Base flipping in nucleotide excision repair. J. Biol. Chem. 2006;281:2184–2194. doi: 10.1074/jbc.M508901200. [DOI] [PubMed] [Google Scholar]
- 49.Scharer OD. Multistep damage recognition, pathway coordination and connections to transcription, damage signaling, chromatin structure, cancer and aging: current perspectives on the nucleotide excision repair pathway. DNA Repair. 2011;10:667. doi: 10.1016/j.dnarep.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 50.Liu Y, Reeves D, Kropachev K, Cai Y, Ding S, Kolbanovskiy M, Kolbanovskiy A, Bolton JL, Broyde S, Van Houten B, et al. Probing for DNA damage with beta-hairpins: similarities in incision efficiencies of bulky DNA adducts by prokaryotic and human nucleotide excision repair systems in vitro. DNA Repair. 2011;10:684–696. doi: 10.1016/j.dnarep.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruan Q, Liu T, Kolbanovskiy A, Liu Y, Ren J, Skorvaga M, Zou Y, Lader J, Malkani B, Amin S, et al. Sequence context- and temperature-dependent nucleotide excision repair of a benzo[a]pyrene diol epoxide-guanine DNA adduct catalyzed by thermophilic UvrABC proteins. Biochemistry. 2007;46:7006–7015. doi: 10.1021/bi700294k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.