Abstract
In mammals, the SET1 family of lysine methyltransferases (KMTs), which includes MLL1-5, SET1A and SET1B, catalyzes the methylation of lysine-4 (Lys-4) on histone H3. Recent reports have demonstrated that a three-subunit complex composed of WD-repeat protein-5 (WDR5), retinoblastoma-binding protein-5 (RbBP5) and absent, small, homeotic disks-2-like (ASH2L) stimulates the methyltransferase activity of MLL1. On the basis of studies showing that this stimulation is in part controlled by an interaction between WDR5 and a small region located in close proximity of the MLL1 catalytic domain [referred to as the WDR5-interacting motif (Win)], it has been suggested that WDR5 might play an analogous role in scaffolding the other SET1 complexes. We herein provide biochemical and structural evidence showing that WDR5 binds the Win motifs of MLL2-4, SET1A and SET1B. Comparative analysis of WDR5-Win complexes reveals that binding of the Win motifs is achieved by the plasticity of WDR5 peptidyl-arginine-binding cleft allowing the C-terminal ends of the Win motifs to be maintained in structurally divergent conformations. Consistently, enzymatic assays reveal that WDR5 plays an important role in the optimal stimulation of MLL2-4, SET1A and SET1B methyltransferase activity by the RbBP5-ASH2L heterodimer. Overall, our findings illustrate the function of WDR5 in scaffolding the SET1 family of KMTs and further emphasize on the important role of WDR5 in regulating global histone H3 Lys-4 methylation.
INTRODUCTION
The landscape of each chromosome emanates from an intricate nucleoprotein complex referred to as the nucleosome. Composed of two copies each of histone H3, H4, H2A and H2B and wrapped by a DNA fragment ∼146 bp, this protein–DNA complex constitutes the first step of DNA compaction and plays an important role in controlling the access of different DNA-binding machineries to various regions of chromosomes. This process is dynamically regulated by ATP-dependent chromatin remodelers along with histone-modifying enzymes and regulates a myriad of biological processes including, but not limited to, chromosome silencing, transcription, DNA replication and DNA damage repair (1).
Several post-translational modifications such as acetylation, phosphorylation, ubiquitylation/sumoylation, ADP ribosylation, PARylation and methylation, have been mapped onto histone proteins and linked to various cellular functions (2). Of interest, methylation of lysine residues by SET domain proteins (Suppressor of Variegation 3–9, Enhancer of Zeste and Trithorax) that frequently exhibits preference towards a precise residue and has the ability to transfer up to three methyl groups to a lysine ε-amine, has been linked to important biological processes. Notably, systematic mapping of methyl–lysine marks on histone H3 demonstrated that histone H3 Lys-4 mono-, di- and tri-methylation (H3K4me1/2/3) decorate the promoter and strong enhancer regions of actively transcribed genes. The same study also showed that H3K4me1 localizes to poised enhancers and chromatin regions undergoing transcriptional transition (3,4). These results further support initial findings linking H3K4 methylation with promoter regions of actively transcribed genes (5–7).
In the budding yeast Saccharomyces cerevisiae, the COMPlex ASsociated with SET1 (COMPASS) (8,9) is the sole protein catalyzing H3K4 methylation whereas in mammals, this reaction is catalyzed by at least six members: MLL1-4, SET1A and SET1B. Each mammalian methyltransferase assembles in a COMPASS-like complex and despite sharing similarities in substrate specificity, the SET1 lysine methyltransferases (KMTs) are not functionally redundant (10–16), and, correlatively, binding studies have demonstrated that members of the SET1 family interact with unique subsets of proteins. The KMTs MLL2 and MLL3 have been shown to interact with activating signal cointegrator 2 (17) while WDR82 and menin associate with SET1A/SET1B (18–20) and MLL1/MLL2 (11,15), respectively. Despite these differences, members of the SET1 family share several features including the complexity of their domain organization and the presence of a catalytic SET domain. In addition, all but MLL5 associate with a three-subunit complex composed of WD-repeat protein-5 (WDR5), retinoblastoma-binding protein-5 (RbBP5) and absent, small, homeotic disks-2-like (ASH2L), collectively referred to as the MLL core complex (21,22), which is required for the stimulation of MLL1 di-/tri-methyltransferase activity both in vitro (23) and in vivo (22,24).
Biochemical and structural studies have recently assigned discrete functions to RbBP5, ASH2L and WDR5 in the assembly of the core complex and stimulation of MLL1 KMT activity. Apart from its DNA binding domain (25,26), ASH2L binds RbBP5 via its SPIa and ryanodine receptor domain and the ASH2L/RbBP5 heterodimer stimulates MLL1 methyltransferase activity (27). In addition to its ASH2L-interacting region, RbBP5 comprises residues that directly bind WDR5, an interaction important for maintaining the integrity of the core complex and indirectly the stimulation of MLL1 KMT activity (28,29). On the opposite side of the WDR5 β-propeller domain, WDR5 binds a small motif preceding the SET domain of MLL1 (23,30). Also referred to as the WDR5-interacting motif (Win), the interaction between the MLL1 Win motif (MLL1Win) and WDR5 is important in the scaffolding of the MLL1–WDR5–RbBP5–ASH2L complex and stimulation of MLL1 KMT activity in vitro (23). Based on these findings and sequence homology between the Win motifs, the same authors surmised that WDR5 would play an analogous role in binding the other members of the SET1 family, namely MLL2-4, SET1A and SET1B (31).
We herein provide biochemical evidence demonstrating that WDR5 binds, with different equilibrium dissociation constants, peptides corresponding to the Win motifs of the other SET1 family members. Correspondingly, the crystal structures of WDR5 in complex with MLL2-4, SET1A and SET1B Win motifs reveal that WDR5 accommodates the different Win motifs using divergent networks of hydrogen bonds and hydrophobic contacts. Finally, we show that WDR5, when in complex with the ASH2L-RbBP5 heterodimer, is important for the full stimulation of MLL1-4, SET1A and SET1B methyltransferase activity. Overall, these findings further support the fundamental role of WDR5 in scaffolding the SET1 complexes and indirect stimulation of H3K4 methylation.
MATERIALS AND METHODS
Protein purification
Constructs corresponding to the Win and SET domains of MLL1 (3745–3969), MLL2 (5344–5536) and MLL4 (2512–2715) were cloned into a pGST-based vector while those for MLL3 (4714–4911), SET1A (1487–1707) and SET1B (1701–1923) were cloned in a home-made vector (pSMT3). MLL1 and MLL3 were overexpressed as GST and His-SUMO fusion proteins, respectively, in Rosetta cells (Novagen) with 0.1 mM IPTG for 16 h at 18°C. Due to their poor solubility, MLL2, MLL4, SET1A and SET1B were overexpressed in Arctic cells (Stratagene) with 1 mM IPTG for 48 h at 10°C. Cells were harvested in either 50 mM sodium phosphate pH 7.0, 500 mM sodium chloride and 5 mM β-mercaptoethanol (pSMT3) or phosphate buffered saline (pGST), lysed by sonication, clarified by centrifugation and filtration, purified by affinity chromatography and ULP1- (pSMT3) or TEV- (pGST2) cleaved on beads for 16 h at 4°C. Following cleavage, the enzymes were dialyzed into a final buffer of 20 mM Tris–HCl pH 7.5, 300 mM sodium chloride and 5 mM β-mercaptoethanol. The members of the core complex, including ASH2L, RbBP5 and WDR5 (S22–334) were overexpressed and purified as previously described (33). Following gel filtration, the core complex subunits were dialyzed in 20 mM Tris–HCl pH 7.5, 300 mM sodium chloride and 5 mM β-mercaptoethanol. The concentrations of SET1 proteins were quantified either by UV or coomassie-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE) gel.
Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) experiments were performed using a VP-ITC calorimeter (MicroCal) as previously described (29,33). Briefly, a solution of peptide (0.5–1 mM) derived from the Win motif of MLL1, MLL2, MLL3, MLL4, SET1A or SET1B was injected into a solution of WDR5 (0.06 mM). Titrations were performed at 19°C in 20 mM sodium phosphate pH 7.0, 100 mM NaCl and 5 mM β-mercaptoethanol. To decrease the rate of saturation of WDR5 peptidyl-arginine-binding cleft, the volume of injection of the MLL4Win peptide was decreased by half (from 10 to 5 µl) for injections 9–19. Titration curves were analyzed using the Origin software.
Protein crystallization, data collection and structure determination
Equimolar amounts of purified WDR5 and the Win peptides were incubated on ice for 1 h and crystallized using the sitting- or hanging-drop vapor diffusion methods. Crystals of WDR5–Win complexes were obtained in conditions shown in Supplementary Table S1. Briefly, crystallization conditions for each complex vary but the predominant components in the mother liquors are ammonium sulfate and polyethylene glycol 3350. Crystals were sequentially soaked in the mother liquor supplemented with 5, 10, 15 and 20% of glycerol, harvested and flash frozen in liquid nitrogen. Data sets were collected at the Life Science Collaborative Access Team (LS-CAT) beamline at the Advance Photon Source of the Argonne National Laboratory and indexed with HKL2000 (34). The structure of each WDR5–Win complex was solved by molecular replacement using Phaser and apo-WDR5 as a search model (RCSB 2H13.pdb) (35). The structure of each WDR5–Win complex was further improved using iterative cycles of refinement, using either Refmac (36) or Buster (37) and model building with COOT (38). The quality of the structures was validated using Molprobity (39).
In vitro methyltransferase assay
Methyltransferase assays were performed as previously described (29,33). Briefly, each recombinantly purified SET1 KMT was incubated with equimolar amounts of WDR5, RbBP5 and ASH2L in 50 mM Tris pH 8.0, 200 mM NaCl, 3 mM Dithiothreitol (DTT), 5 mM MgCl2, 5% glycerol and 1 mM histone H3 or peptide corresponding to the first N-terminal 15 residues or 10 µM of recombinantly purified H3. Reactions were initiated by adding 1 µCi of radiolabeled S-adenosyl-l-methionine (AdoMet) (0.07 mM of tritiated AdoMet) and stopped by spotting the reactions onto Whatman P-81 filter papers. Free AdoMet was removed by washing the filter papers four times in 250 ml of 50 mM sodium bicarbonate pH 9.0 and methyltransferase activity was quantified by liquid scintillation counting.
RESULTS
Equilibrium dissociation constants of the Win motifs for WDR5
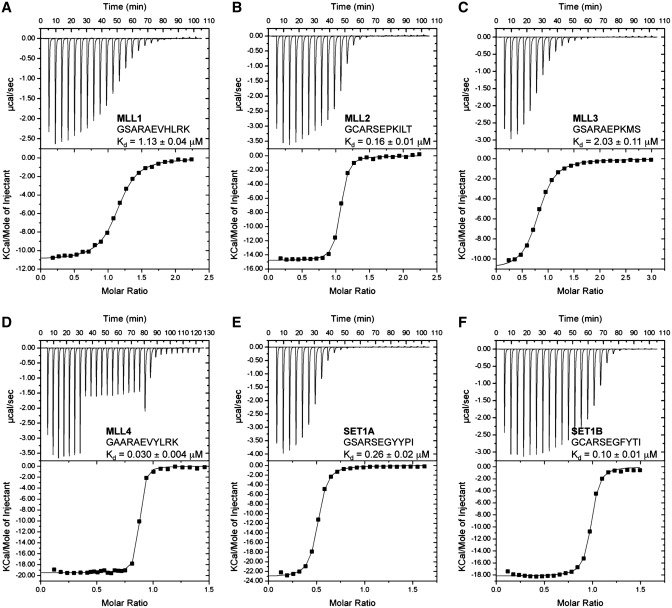
A recent biochemical analysis of WDR5 peptidyl-arginine-binding cleft revealed that this protein binds a 10-residue peptide corresponding to MLL1Win with an equilibrium dissociation constant (KD) of 1.7 µM. The crystal structure of WDR5 in complex with MLL1Win identified that R3765 of MLL1 was critical in conferring binding to WDR5, scaffolding the MLL1–WDR5–RbBP5–ASH2L complex and, correlatively, stimulation of MLL1 methyltransferase activity (23). Accordingly, the same authors posited that a similar region found on other members of the SET1 family would play an analogous role (31) while another study proposed that only MLL1 and MLL4 could bind WDR5 (30). In order to investigate these hypotheses, we first sought to measure the KD values of WDR5 for MLL1-4 and SET1A/BWin motifs. Using ITC, we determined that WDR5 binds MLL1 Win and MLL4Win peptides with a KD of 1.13 µM and 0.03 µM, respectively. Similar titration experiments of WDR5 with MLL2Win and MLL3Win peptides resulted in KD of 0.16 µM and 2.03 µM, respectively, while SET1AWin and SET1BWin peptides bound to WDR5 with a KD of 0.26 µM and 0.10 µM, respectively. Overall, these results demonstrate that WDR5 binds the Win region of all the members of the SET1 family of KMTs. However, given the divergence in the amino acid sequence succeeding the conserved arginine residue and the ∼37-fold range of binding affinities of WDR5 for the Win motifs (Table 1 and Figure 1), our binding analysis suggests that WDR5 interacts with SET1 KMTs using divergent modes.
Table 1.
Alignment of SET1 Win motifs
| Amino acid sequence |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Win motifs | P−3 | P−2 | P−1 | P0 | P+1 | P+2 | P+3 | P+4 | P+5 | P+6 | P+7 |
| MLL1Win | G3762 | S | A | R | A | E | V | H | L | R | K |
| MLL2Win | G5062 | C | A | R | S | E | P | K | I | L | T |
| MLL3Win | G4707 | S! | A | R | A | E | P | K | Ma | S | A |
| MLL4Win | G2508 | A | A | R | A | E | V | Y | L | R | K |
| SET1AWin | G1492 | S | A | R | S | E | G | Y | Y | P | I |
| SET1BWin | G1882 | C | A | R | S | E | G | F | Y | T | I |
Underlined residues have not been modeled in the crystal structure. !, C4708S substituted peptide.
aSide chain is missing.
Figure 1.
WDR5 binds MLL1-4, SET1A and SET1B Win motifs. ITC titration experiments with MLL1Win (A), MLL2Win (B), MLL3Win (C), MLL4Win (D), SET1AWin (E) and SET1BWin (F) peptides and WDR5 (upper trace) and the fitted binding curve (lower trace). The sequence of the Win motifs and the KD are indicated as insets. Titration of WDR5 with Win peptides displays binding stoichiometries (N values) between 0.9 and 1.0.
Crystal structures of the WDR5–MLLWin complexes
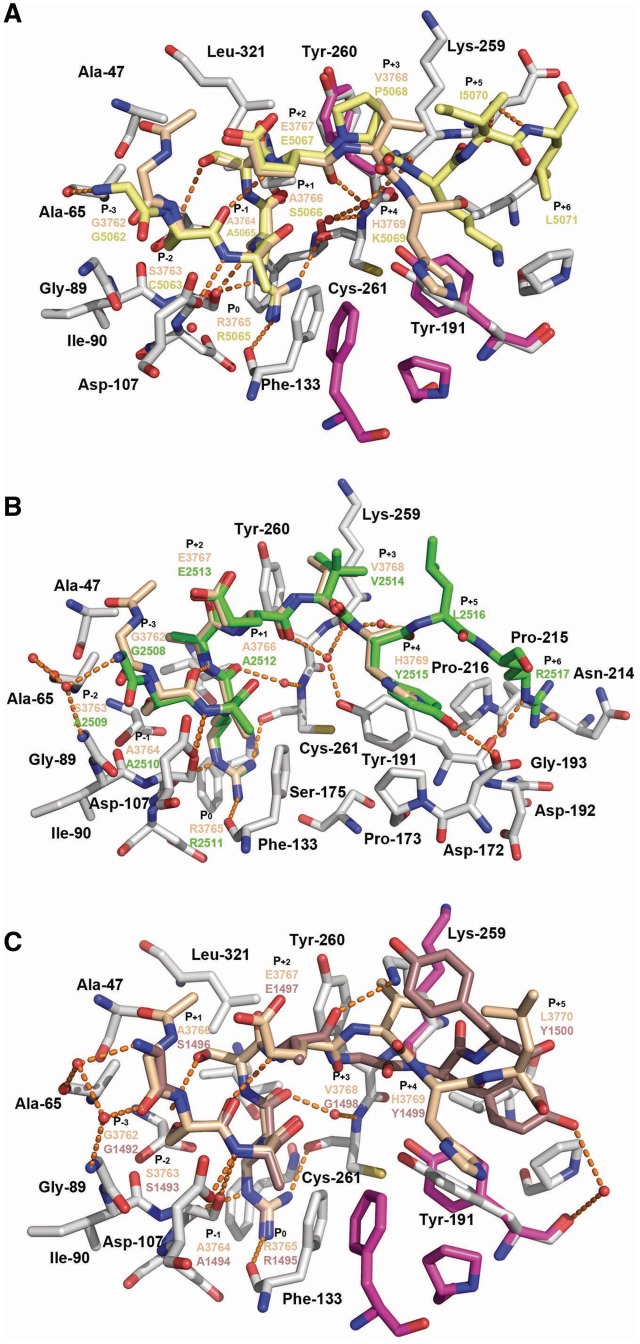
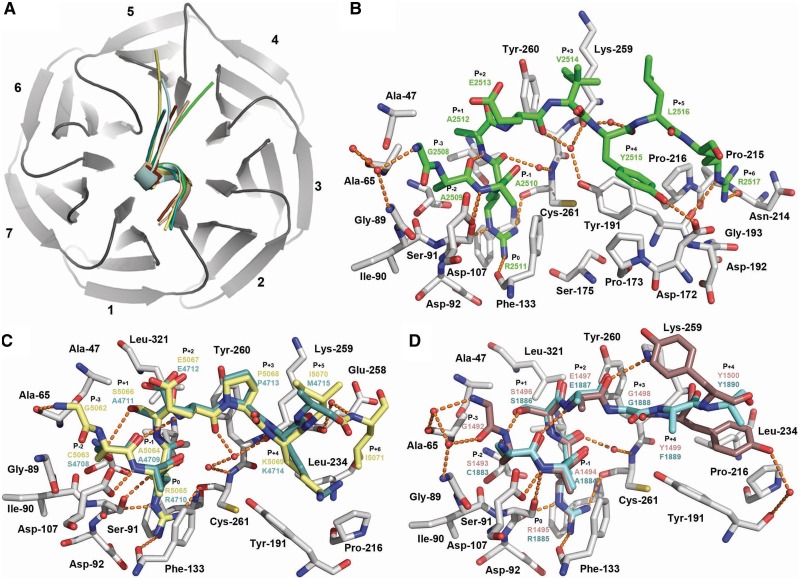
To gain insights into the mechanisms underlying WDR5 ability to bind the structurally divergent C-terminal ends of the Win peptides (Table 1), we determined the crystal structures of WDR5 in complex with peptides corresponding to the Win motifs of MLL2 (1.4 Å resolution), MLL3 (2.2 Å resolution), MLL4 (1.57 Å resolution), SET1A (2.1 Å resolution) and SET1B (2.2 Å resolution) (Supplementary Table S1). The WDR5–Win complexes superimpose with overall r.m.s.d. of < 0.3 Å for all aligned atoms, showing that binding of the Win peptides does not result in noticeable changes in the overall structure of WDR5. Simulated annealing omit maps revealed clear electron density for several residues flanking the central arginine residue of each Win motif (Supplementary Figure S1). However, the distal residues in the peptides, including the M4715 side chain (single letter code refer to residues of the Win motifs), S4716 and A4717 of MLL3Win, K2518 of MLL4Win, P1501 and I1502 of SET1AWin and G1882 and I1892 as well as the side chains of F1889 and Y1890 of SET1BWin lack interpretable electronic density and were not modeled. Close inspection of the WDR5-Win structures reveals that all the peptides bind WDR5 peptidyl-arginine-binding cleft, a crevice located on one side of the WDR5 β-propeller structure (Figure 1), in which each peptide engages in several hydrogen bonds, van der Waals contacts and hydrophobic interactions with WDR5. To facilitate the discussion of the interactions between WDR5 and the Win motifs, we have used a modified version of the Schechter–Berger notation for assigning residues of the Win motifs in which the central arginine residue is denoted as P0 (Table 1). In addition, interactions between WDR5 and the Win motifs have been separated into two categories based on interactions with residues preceding and succeeding the P+2 glutamate residue of the Win motifs (Table 1).
Consistent with the sequence homology of residues flanking the central arginine (P0), including those at P−3, P−2, P−1, P+1 and P+2 positions (Table 1), this region of the peptide adopts similar orientations and engages in several shared direct and water-mediated hydrogen bonds and hydrophobic interactions with WDR5. Similar to MLL1Win, the P−2 to P+2 region folds as a 310 helix in which the side chains of P−3, P−2 and P−1 residues pack tightly within WDR5 peptidyl-arginine-binding cleft and make hydrophobic contacts with Ala-65, Gly-89, Ile-90 and Asp-107 side chains. The same residues in WDR5 form a wall that maintains the side chain of the P−2 residue in an orientation permissive to make a hydrogen bond with the backbone amide of the P+1 residue. The alanine residue in the P−1 position binds analogously in all WDR5–Win complexes. Indeed, the P−1 amide group engages in a hydrogen bond with the Asp-107 carboxylate group and its side chain makes hydrophobic contacts with the side chains of Phe-133 and Phe-149. Similar to histone H3 R2 (35,40–42) and MLL1 R3765 (30,31), the central arginine (P0) of the Win motifs extends inside the central toroid cavity of WDR5 in which its guanidinium group interacts with the side chains of Phe-133 and Phe-263. The P0 residue also makes several direct hydrogen bonds with the main chains of Ser-91, Phe-133 and Cys-261 and a water-mediated hydrogen bond with the Ser-175 backbone carbonyl group. Following the central arginine residue, the P+1 position is occupied by a small residue such as serine (MLL2, SET1A and SET1B) or alanine (MLL3 and MLL4) while the P+2 position is occupied by a glutamate residue. Consistent with the sequence homology between the P+1 and P+2 residues, this portion of the Win motifs shares several interactions with WDR5. The P+1 residue makes van der Waals contacts with the side chains of Ala-47 and Tyr-260 while in MLL4 the P+2 carboxylate moiety makes two additional water-mediated hydrogen bonds with the backbone amides of the P+3 and P−3 residues. In addition, the hydroxyl group of the MLL2Win and SET1BWin P+1 serine residue makes a hydrogen bond with the sulfhydryl group of the P−2 cysteine residue.
Despite high sequence homology and analogous binding modes of the P−3, P−2, P−1 and P0 residues, structural alignment of the WDR5–Win complexes reveals notable structural differences. In comparison with MLL1Win, the backbone of the MLL2Win and MLL4Win P−3 residue is rotate 180°, which results in additional water-mediated and direct hydrogen bonds with WDR5 (Figure 3). This rotation leads to a small structural displacement of the P−2 backbone with negligible consequences on the positioning of C5063 (MLL2), S4708 (MLL3), A2509 (MLL4), S1493 (SET1A) and C1883 (SET1B) side chains and their interactions with WDR5. However, in clear contrast to the P−2, P−1, P0, P+1 and P+2 residues, the P+3 to P+5 residues adopt divergent orientations. MLL2Win and MLL3Win peptides, which bind analogously to WDR5, interact in such a way that their respective C-termini point towards blade 5 of WDR5 while MLL4Win and MLL1Win adopt a conformation placing their C-terminal ends towards blade 4 of the β-propeller. SET1AWin and SET1BWin peptides bind in an intermediate orientation such that their C-termini are maintained on the surface of blades 4 and 5 of WDR5 (Figure 2A). Given the structural differences adopted by residues succeeding the P+2 residue (Figure 3), we purported that each peptide would engage in distinct interactions with WDR5. The backbone of P+3 Y2514 of MLL4Win, which binds similarly to H3769 of MLL1Win, forms direct and water-mediated hydrogen bonds with Asp-172 and Lys-259 carboxylate and carbonyl groups, respectively. In addition, its aromatic ring makes hydrophobic contacts with the side chains of Tyr-191, Pro-173 and Phe-149. In contrast to the solvent exposed P+4 L2516, the P+5 R2517 side chain is nestled in a small cleft formed by the surface of blade 4 and the loop connecting blades 4 and 5 of WDR5 (Figure 3B). This crevice, which is composed of Tyr-191, Asp-192, Gly-193, Asn-214, Pro-215 and Pro-216, likely orients the R2517 side chain in a permissive orientation to engage in direct hydrogen bonds with the backbone carbonyl groups of Tyr-191 and Asn-214 and van der Waals contacts with Pro-216.
Figure 3.
Comparative analysis Win motif binding modes. (A) Superimposition of WDR5-bound MLL1 (beige) and MLL2 (yellow) peptides. (B) Overlay of MLL1 (beige) and MLL4 (green) peptides in complex with WDR5 (grey). (C) Structural alignment of WDR5–MLL1Win and WDR5–SET1AWin complex in which carbons of the peptides are highlighted as in Figure 2. Residues of WDR5 peptidyl-arginine-binding adopting divergent conformation between the complexes are rendered as pink carbon atoms. Hydrogen bonds and water molecules are rendered as in Figure 2.
Figure 2.
Crystal structure of WDR5 in complex with MLL2-4, SET1A and SET1B Win motifs. (A) Overall structure of WDR5 (dark gray) showing the seven blades and the relative orientations of MLL2 (yellow), MLL3 (turquoise), MLL4 (green), SET1A (firebrick) and SET1B (cyan) Win motifs. (B) Crystal structure of WDR5–MLL4Win complex. Zoomed view of WDR5 peptidyl-arginine-binding cleft in which WDR5 and MLL4 carbon atoms are rendered in gray and green, respectively. (C) Crystal structures of WDR5–MLL2Win and WDR5–MLL3Win complexes. MLL2 and MLL3 carbon atoms are highlighted as in (A). (D) Crystal structures of WDR5–SET1AWin and WDR5–SET1BWin complexes. SET1A and SET1B carbon atoms are colored as in (A). Water molecules and key hydrogen bonds are depicted as red spheres and orange dash lines, respectively.
Titration analysis revealed that MLL4Win binds ∼37-fold more tightly to WDR5 than the highly homologous MLL1Win peptide (Figure 1 and Table 1). Owing to the high sequence homology between these two motifs and structural homology between the WDR5-bound MLL1Win and MLL4Win complexes, the differences in binding affinities are seemingly at odds. However, while a comparative analysis of WDR5–MLL1Win and WDR5–MLL4Win complexes reveals that both MLL1Win and MLL4Win P+4 residues share extensive interactions with WDR5, it also shows that a water-mediated interaction of the MLL1Win P+4 H3769 side chain with the WDR5 Asp-172 carboxylate group is replaced by a direct hydrogen bond between the same residue in WDR5 and the side chain hydroxyl group of MLL4Win P+4 Y2514; a difference that presumably explain the dissimilarity between MLL1Win and MLL4Win binding affinities for WDR5.
Owing to the presence of a proline residue in the P+3 position of MLL2Win and MLL3Win, we postulated that these peptides would bind WDR5 differently than MLL1Win and MLL4Win. Consistent with this hypothesis, structural alignment of WDR5–MLL2Win and WDR5–MLL1Win complexes reveals that the C-terminal end of MLL2Win shifts towards the loop preceding β18, resulting in several novel interactions between MLL2Win and WDR5 (Figure 3B). In MLL2Win, the P+3 P5068 residue makes close van der Waals contacts with the side chains of Lys-259 and Tyr-260. The WDR5 peptidyl-arginine-binding cleft also undergoes notable structural changes. The phenolic side chain of Tyr-191 rotates of 90° positioning its aromatic ring in a near parallel orientation with the P+4 lysine residue of MLL2Win/MLL3Win. In addition, the same lysine residue in MLL2Win/MLL3Win forms a novel hydrogen bond with the Lys-259 carbonyl group and hydrophobic contacts with Leu-234. Finally, in contrast to all other Win motifs, the MLL2Win P+5 L5071 residue lies within a hydrophobic cleft composed of Pro-216 and Leu-234. Overall, the crystal structure of MLL2Win bound to WDR5 suggests that the P+3 residue acts as a structural determinant underlying the binding mode of a peptide within WDR5 peptidyl-arginine-binding cleft.
After observing that the P+3 position in MLL2Win/MLL3Win peptides is a key point of structural divergence between the Win peptides, we postulated that the P+3 glycine residue of SET1AWin/ SET1BWin motifs would also exhibit structural differences compared to other Win motifs. Accordingly, in contrast to MLL1-4 Win peptides, the backbone carbonyl of the SET1AWin P+2 glutamate residue undergoes a 180° rotation that results in novel hydrogen bonds between the amide groups of the P+3 and P+4 residues and the backbone carbonyls of the P−1 residue and Lys-259 of WDR5, respectively. Notably, in comparison with the WDR5–MLL1Win complex, the side chains of Lys-259 and Tyr-191 shift toward the Win peptides, which results in the formation of additional hydrophobic contacts with the SET1AWin P+3 glycine and P+5 tyrosine residues (Figure 3C). In addition, the same tyrosine residue in SET1A engages in several hydrophobic contacts with the lateral chain of Leu-234 and Pro-216.
Overall, the crystal structures of the WDR5–Win complexes highlight WDR5's ability in binding the Win motifs using alternative structural determinants. More importantly, our structural studies suggest that the plasticity of WDR5 peptidyl-arginine-binding cleft stems from its ability to operate subtle but key structural changes that accommodate the binding of structurally divergent residues in position P+3. Finally, crystal structures of the WDR5–Win complexes suggest that the type of residue at the P+3 position will likely play an important role in the binding mode adopted by the peptide within the WDR5 peptidyl-arginine-binding cleft.
Role of WDR5 in stimulating the KMT activity of the SET1 family of methyltransferases
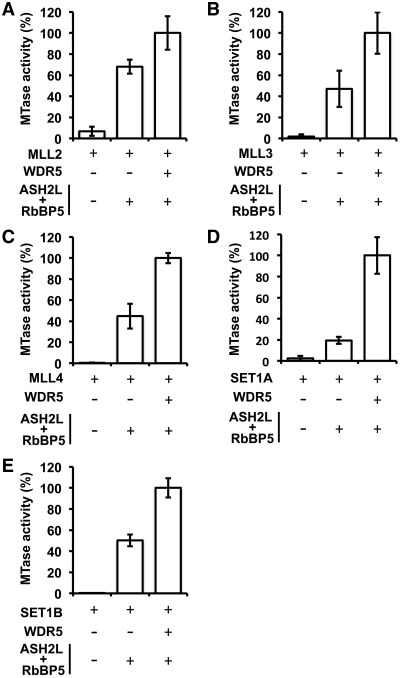
The Win motif of MLL1 is important for its interaction with the WDR5–RbBP5–ASH2L complex and thereby stimulation of its methyltransferase activity (23). After showing that WDR5 binds the Win motifs of MLL1-4 and SET1A/B, we examined the role of the WDR5–RbBP5–ASH2L complex in stimulating the other members of the SET1 family of methyltransferases. A comparison of SET1 KMT activity in the absence or presence of the WDR5–RbBP5–ASH2L complex confirmed that the core complex enhances the methyltransferase activity of all members, with the exception of MLL5, but with differences in the fold of stimulation. Specifically, the WDR5–RbBP5–ASH2L complex stimulated MLL2/MLL3 and MLL4 activity by ∼52/256- and ∼15-fold, respectively, while co-incubation of SET1A and SET1B with the core complex subunits increased methylation of histone H3 by ∼40- and ∼1290-fold (Figure 4). The stimulatory roles of the core complex on SET1B can be rationalized by the nearly undetectable activity of these KMTases on histone H3 in the absence of the WDR5–RbBP5–ASH2L complex. Overall, these observations support initial in vivo data showing that the members of the core complex are critical for global levels of histone H3K4 methylation.
Figure 4.
Stimulation of the SET1 family of KMTs activity on histone H3 by the core complex. Radiometric methyltransferase assays performed with MLL2 (A), MLL3 (B), MLL4 (C), SET1A (D) and SET1B (E) either in the absence or presence of WDR5 and the RbBP5–ASH2L complex. Methyltransferase assays were performed at a concentration of enzyme in the linear range of activity of MLL2 (1 µM), MLL3 (0.5 µM), MLL4 (1 µM), SET1A (1 µM) and SET1B (0.5 µM) complexes. Activity is represented as the average of three independent experiments performed in triplicate. Error bars indicate the standard deviation between the assays.
After showing that the core complex subunits stimulate the methyltransferase activity of MLL2-4, SET1A and SET1B, we sought to measure the contribution of WDR5 in the enzymatic activity of the complex (Figure 4). Interestingly, incubation of the ASH2L/RbBP5 heterodimer with each KMT failed to fully stimulate, to various extent, the enzymatic activity of each member of the SET1 family. Specifically, the absence of WDR5 resulted in a 2-fold decrease of activity for SET1A, MLL3 and MLL4, while co-incubation of SET1B and MLL2 with the ASH2L-RbBP5 heterodimer decreased the methylation of histone H3 by 5-fold and 1.5-fold, respectively (Figure 4). Overall, these results clearly demonstrate the importance of WDR5 in stimulating the methyltransferase activity of MLL2-4, SET1A and SET1B and support recent studies suggesting that WDR5 is key for the activity for all members of the SET1 family of KMTs.
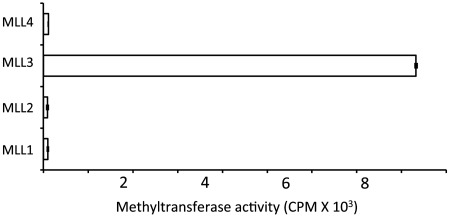
Following the identification that the WDR5–RbBP5–ASH2L complex stimulates all members of the SET1 family, we sought to evaluate the methylation of histone H3 by MLL1-4 in the absence of the core complex subunits. To examine the catalytic activity of each MLL, we incubated each KMT with a histone H3 peptide and a tritiated cofactor. As shown in Figure 5, we failed to detect histone H3 methylation by MLL1, MLL2 and MLL4 while a similar assay performed with MLL3 resulted in a notable accumulation of the methylated peptide. These observations suggest that MLL3 enzymatic activity may play a role independent of the WDR5–RbBP5–ASH2L complex.
Figure 5.
MLL3 methylates histone H3 in the absence of the core complex subunits. Comparative analysis of MLL's enzymatic activity in the absence of the core complex subunits. Assays were performed in the presence of 5.0 µM of enzymes. Activity is represented as in Figure 4.
DISCUSSION
The results presented herein provides biochemical and structural evidence that the WDR5 peptidyl arginine-binding cleft is important in the binding of all the members of the SET1 family of methyltransferases stimulated by the core complex.
Of interest, we observed that MLL2 and MLL3 Win motifs bind similarly to WDR5. Owing to the presence of a proline residue in P+3, Song et al. (30) had previously proposed that MLL2/MLL3 might interact with WDR5 using a different mode of binding. Comparative analyses of WDR5–MLL2Win and WDR5–MLL1Win complexes support this hypothesis as the C-terminal ends of each peptide are maintained by specific sets of hydrogen bonds, hydrophobic contacts and van der Waals interactions. Similarly, we also observed noticeable differences between WDR5–SET1AWin and WDR5–MLL1Win complexes that include rotation of the SET1AWin P+2 backbone, a change that could be attributed to the presence of a glycine residue in P+3, which results in reorientation of the P+4 and P+5 residues. These structural differences are accommodated by the reorganization of residues forming the WDR5 peptidyl-arginine-binding cleft including Lys-259 and Tyr-191. Although, it was evidenced by several mutational studies (23,28,30,33,35,41) that the N-terminal end of the Win motifs is important for binding to WDR5, our comparative analysis suggest that the propensity of WDR5 in binding the members of the SET1 family of KMTs is achieved by the plasticity of its peptidyl-arginine-binding cleft and more specifically its ability to accommodate the divergent C-terminal region of the Win motifs.
In metazoans, WDR5 plays a critical role in regulating/maintaining proper levels of histone H3K4 methylation (21,22,24) while in Drosophila, deletion of the will die slowly protein (WDR5 homolog) leads to global loss of methylation and ultimately to lethality (43). Similarly, morpholino knockdown of WDR5 in Xenopus leads to severe developmental deficiencies including gut, hematopoietic and somatic defects (21). We propose that the plasticity of the WDR5 peptidyl-arginine-binding cleft plays an important role in scaffolding all of the SET1 complexes and underlies the fundamental roles of WDR5 in regulating various developmental programs (21,44).
MLL3, a unique member of the SET1 family of HKMTs
The characterization of SET1 enzymatic activity on histone H3 presented herein provides the first comparative analysis for the SET1 family of methyltransferases. While the SET domains of MLL1, MLL2, and MLL4 displayed negligible enzymatic activity in the absence of the WDR5–RbBP5–ASH2L complex, we observed detectable histone H3 methyltransferase activity for MLL3 independent of the core complex. This observation that MLL3 efficiently methylates histone H3 in the absence of the core complex while MLL2 (MLL4 in rodents) strictly depends on the presence of the core complex subunits is interesting. Indeed, while initial studies had suggested that MLL3 and MLL4 are functionally redundant in retinoic acid receptor transactivation in mouse embryonic fibroblasts (45), targeted inactivation of MLL3 resulted in several defects including lower body mass, hypofertility and increased formation of ureter epithelial tumors (45,46). Based on our findings, we propose that the ability of MLL3 to methylate histone H3 in the absence of the core complex subunits may underlie some of the non-overlapping functions between MLL3 and MLL4.
The expanding role of WDR5
The recent findings that WDR5 binds to proteins unrelated to the trithorax group proteins shed new light on the ever-expanding role of WDR5. It has been shown recently that Oct4 forms a complex with WDR5 independent of the presence of RbBP5, ASH2L or the SET1 family members (44). By identifying an interaction between WDR5 and the embryonic stem cell core transcriptional network, the authors of this study established that WDR5 expression is necessary for the efficient formation of induced pluripotent stem cells (44). In addition, Gan et al. (47), recently demonstrated that WDR5 regulates smooth muscle cell-selective gene activation by interacting with the homeodomain of pituitary homeobox 2. Similarly, WDR5 has been shown to interact with the VISA-associated signaling complex during virus infection-triggered IRF3 and NF-κB activation (48) and with the NRC/NCoA6 interacting factor 1 (49). Finally, recent MudPit analysis identified WDR5 as a subunit of the non-specific lethal complex (50). These examples further emphasize the important role of WDR5 in scaffolding these complexes and we propose that the plasticity of WDR5 peptidyl-arginine-binding cleft, or its recently characterized V-shape cleft (29), plays an important role in these biological processes.
ACCESSION NUMBER
Coordinates and structure factors for the WDR5-MLL2Win, WDR5-MLL3Win, WDR5-MLL4Win, WDR5-SET1AWin, WDR5-SET1BWin complexes have been deposited in the protein data bank (rcsb.org) with the accession number 3UVK.pdb, 3UVL.pdb, 3UVM.pdb, 3UVN.pdb and 3UVO.pdb respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figure 1.
FUNDING
Canadian Institute of Health Research grant (to J.-F.C.); an early research award from the Ministry of Research and Innovation (province of Ontario). Dr Couture holds a Canada Research Chair in Structural Biology and Epigenetics. Natural Sciences and Engineering Research Council of Canada (CREATE) (scholarship to P.Z.). Funding for open access charge: Canadian Institute of Health Research grant (to J.-F.C.); an early research award from the Ministry of Research and Innovation (province of Ontario).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr Alain Doucet, Sylvain Lanouette and Vanessa Mongeon for providing comments on the manuscript.
REFERENCES
- 1.Allis CD, Jenuwein T, Reinberg D. Epigenetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2007. [Google Scholar]
- 2.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 5.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 7.Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 8.Tenney K, Shilatifard A. A COMPASS in the voyage of defining the role of trithorax/MLL-containing complexes: linking leukemogensis to covalent modifications of chromatin. J. Cell Biochem. 2005;95:429–436. doi: 10.1002/jcb.20421. [DOI] [PubMed] [Google Scholar]
- 9.Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl Acad. Sci. USA. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 2007;282:20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Hayes DN, Shanmugam KS, Bhattacharjee A, Biondi CA, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol. Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- 12.Patel SR, Kim D, Levitan I, Dressler GR. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev. Cell. 2007;13:580––592. doi: 10.1016/j.devcel.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T, Mazo A, Eisenbach L, Canaani E. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol. Cell Biol. 2007;27:1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho EA, Prindle MJ, Dressler GR. BRCT domain-containing protein PTIP is essential for progression through mitosis. Mol. Cell Biol. 2003;23:1666–1673. doi: 10.1128/MCB.23.5.1666-1673.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol. Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JH, Skalnik DG. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J. Biol. Chem. 2005;280:41725–41731. doi: 10.1074/jbc.M508312200. [DOI] [PubMed] [Google Scholar]
- 17.Goo YH, Sohn YC, Kim DH, Kim SW, Kang MJ, Jung DJ, Kwak E, Barlev NA, Berger SL, Chow VT, et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol. Cell Biol. 2003;23:140–149. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol. Cell Biol. 2008;28:7337–7344. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JH, Skalnik DG. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A Histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol. Cell Biol. 2008;28:609–618. doi: 10.1128/MCB.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JH, Tate CM, You JS, Skalnik DG. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J. Biol. Chem. 2007;282:13419–13428. doi: 10.1074/jbc.M609809200. [DOI] [PubMed] [Google Scholar]
- 21.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 22.Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 23.Patel A, Vought VE, Dharmarajan V, Cosgrove MS. A conserved arginine-containing motif crucial for the assembly and enzymatic activity of the mixed lineage leukemia protein-1 core complex. J. Biol. Chem. 2008;283:32162–32175. doi: 10.1074/jbc.M806317200. [DOI] [PubMed] [Google Scholar]
- 24.Steward MM, Lee JS, O'Donovan A, Wyatt M, Bernstein BE, Shilatifard A. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat. Struct. Mol. Biol. 2006;13:852–854. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Wan B, Wang KC, Cao F, Yang Y, Protacio A, Dou Y, Chang HY, Lei M. Crystal structure of the N-terminal region of human Ash2L shows a winged-helix motif involved in DNA binding. EMBO Rep. 2011;12:797–803. doi: 10.1038/embor.2011.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarvan S, Avdic V, Tremblay V, Chaturvedi CP, Zhang P, Lanouette S, Blais A, Brunzelle JS, Brand M, Couture JF. Crystal structure of the trithorax group protein ASH2L reveals a forkhead-like DNA binding domain. Nat. Struct. Mol. Biol. 2011;18:857–859. doi: 10.1038/nsmb.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao F, Chen Y, Cierpicki T, Liu Y, Basrur V, Lei M, Dou Y. An Ash2L/RbBP5 heterodimer stimulates the MLL1 methyltransferase activity through coordinated substrate interactions with the MLL1 SET domain. PLoS One. 2010;5:e14102. doi: 10.1371/journal.pone.0014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Odho Z, Southall SM, Wilson JR. Characterization of a novel WDR5-binding site that recruits RbBP5 through a conserved motif to enhance methylation of histone H3 lysine 4 by mixed lineage leukemia protein-1. J. Biol. Chem. 2010;285:32967–32976. doi: 10.1074/jbc.M110.159921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Avdic V, Zhang P, Lanouette S, Groulx A, Tremblay V, Brunzelle J, Couture JF. Structural and biochemical insights into MLL1 core complex assembly. Structure. 2011;19:101–108. doi: 10.1016/j.str.2010.09.022. [DOI] [PubMed] [Google Scholar]
- 30.Song JJ, Kingston RE. WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J. Biol. Chem. 2008;283:35258–35264. doi: 10.1074/jbc.M806900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patel A, Dharmarajan V, Cosgrove MS. Structure of WDR5 bound to mixed lineage leukemia protein-1 peptide. J. Biol. Chem. 2008;283:32158–32161. doi: 10.1074/jbc.C800164200. [DOI] [PubMed] [Google Scholar]
- 32.Couture JF, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat. Struct. Mol. Biol. 2007;14:689–695. doi: 10.1038/nsmb1273. [DOI] [PubMed] [Google Scholar]
- 33.Avdic V, Zhang P, Lanouette S, Voronova A, Skerjanc I, Couture JF. Fine-tuning the stimulation of MLL1 methyltransferase activity by a histone H3-based peptide mimetic. FASEB J. 2011;25:960–967. doi: 10.1096/fj.10-171959. [DOI] [PubMed] [Google Scholar]
- 34.Minor ZOaW. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Macromol. Crystallogr. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Couture JF, Collazo E, Trievel RC. Molecular recognition of histone H3 by the WD40 protein WDR5. Nat. Struc.t Mol. Biol. 2006;13:698–703. doi: 10.1038/nsmb1116. [DOI] [PubMed] [Google Scholar]
- 36.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta. Crystallogr. D Biol. Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 37.Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han Z, Guo L, Wang H, Shen Y, Deng XW, Chai J. Structural basis for the specific recognition of methylated histone H3 lysine 4 by the WD-40 protein WDR5. Mol. Cell. 2006;22:137–144. doi: 10.1016/j.molcel.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 41.Ruthenburg AJ, Wang W, Graybosch DM, Li H, Allis CD, Patel DJ, Verdine GL. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat. Struct. Mol. Biol. 2006;13:704–712. doi: 10.1038/nsmb1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuetz A, Allali-Hassani A, Martin F, Loppnau P, Vedadi M, Bochkarev A, Plotnikov AN, Arrowsmith CH, Min J. Structural basis for molecular recognition and presentation of histone H3 by WDR5. EMBO J. 2006;25:4245–4252. doi: 10.1038/sj.emboj.7601316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hollmann M, Simmerl E, Schafer U, Schafer MA. The essential Drosophila melanogaster gene wds (will die slowly) codes for a WD-repeat protein with seven repeats. Mol. Genet. Genomics. 2002;268:425–433. doi: 10.1007/s00438-002-0768-0. [DOI] [PubMed] [Google Scholar]
- 44.Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee S, Lee DK, Dou Y, Lee J, Lee B, Kwak E, Kong YY, Lee SK, Roeder RG, Lee JW. Coactivator as a target gene specificity determinant for histone H3 lysine 4 methyltransferases. Proc. Natl Acad. Sci. USA. 2006;103:15392–15397. doi: 10.1073/pnas.0607313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J, Kim DH, Lee S, Yang QH, Lee DK, Lee SK, Roeder RG, Lee JW. A tumor suppressive coactivator complex of p53 containing ASC-2 and histone H3-lysine-4 methyltransferase MLL3 or its paralogue MLL4. Proc. Natl Acad. Sci. USA. 2009;106:8513–8518. doi: 10.1073/pnas.0902873106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gan Q, Thiebaud P, Theze N, Jin L, Xu G, Grant P, Owens GK. WD repeat-containing protein 5, a ubiquitously expressed histone methyltransferase adaptor protein, regulates smooth muscle cell-selective gene activation through interaction with pituitary homeobox 2. J. Biol. Chem. 2011;286:21853–21864. doi: 10.1074/jbc.M111.233098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang YY, Liu LJ, Zhong B, Liu TT, Li Y, Yang Y, Ran Y, Li S, Tien P, Shu HB. WDR5 is essential for assembly of the VISA-associated signaling complex and virus-triggered IRF3 and NF-kappaB activation. Proc. Natl Acad. Sci. USA. 2010;107:815–820. doi: 10.1073/pnas.0908967107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garapaty S, Xu CF, Trojer P, Mahajan MA, Neubert TA, Samuels HH. Identification and characterization of a novel nuclear protein complex involved in nuclear hormone receptor-mediated gene regulation. J. Biol. Chem. 2009;284:7542–7552. doi: 10.1074/jbc.M805872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cai Y, Jin J, Swanson SK, Cole MD, Choi SH, Florens L, Washburn MP, Conaway JW, Conaway RC. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J. Biol. Chem. 2010;285:4268–4272. doi: 10.1074/jbc.C109.087981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.