Abstract
Friedreich's ataxia (FRDA) is a common hereditary degenerative neuro-muscular disorder caused by expansions of the (GAA)n repeat in the first intron of the frataxin gene. The expanded repeats from parents frequently undergo further significant length changes as they are passed on to progeny. Expanded repeats also show an age-dependent instability in somatic cells, albeit on a smaller scale than during intergenerational transmissions. Here we studied the effects of (GAA)n repeats of varying lengths and orientations on the episomal DNA replication in mammalian cells. We have recently shown that the very first round of the transfected DNA replication occurs in the lack of the mature chromatin, does not depend on the episomal replication origin and initiates at multiple single-stranded regions of plasmid DNA. We now found that expanded GAA repeats severely block this first replication round post plasmid transfection, while the subsequent replication cycles are only mildly affected. The fact that GAA repeats affect various replication modes in a different way might shed light on their differential expansions characteristic for FRDA.
INTRODUCTION
Friedreich's ataxia (FRDA) is a common progressive neurodegenerative disorder, resulting from a decrease in the level of frataxin, a protein involved in iron homeostasis in mitochondria (1,2). In most FRDA cases, this loss of function is caused by expansions of the GAA repeat situated in the first intron of the frataxin gene, FXN (3). The expanded GAA repeat interferes with the transcription of the FXN gene, likely by changing its DNA or chromatin structure (4–7). There is a correlation between the length of the GAA repeat and the severity of FRDA symptoms (8,9): normal individuals carry <30 repeated GAA triplets in their FXN alleles, carriers have pre-mutation alleles with 30 to 60 repeats (10); meanwhile, full-mutation alleles with 60 to 1700 repeats are typical of FRDA patients (2,11).
The full- and pre-mutation FXN alleles are quite prone to large-scale changes in the number of repeats during the gametogenesis and first post-zygotic divisions. It is believed that expansions of the GAA repeat occur largely during early post-zygotic divisions. A repeat inherited from the mother could be contracted or expanded with equal probabilities and its propensity to expand increases with mother's age (12). A repeat inherited from the father is always shorter in progeny, as it massively contracts during spermatogenesis (12,13). In fact, it ends up being longer in the progeny than it was in the sperm, since it expands during early embryonic divisions (12,14). The combined probability of expansions and contractions during the intergenerational transmissions is very high, ∼85% (12). Another type of instability, which also contributes to the FRDA pathology, occurs in somatic cells (15,16). The degree of somatic instability differs between human tissues; the highest rate of expansions is observed in dorsal root ganglia, the most affected tissue in FRDA patients (16).
The high instability of the GAA repeat during gametogenesis and early embryogenesis could be due to replication errors as the fork tries to progress through repetitive runs. In a multicopy yeast plasmid, expanded GAA repeats caused replication fork stalling (17). This stalling only occurred in one orientation of the repeat when the GAA run was situated in the lagging strand template. Coincidentally, the repeat instability was more pronounced in this same orientation. Similar observations were also made for repeats in bacterial plasmids (18). When GAA repeats were integrated into a yeast chromosome, fork stalling remained orientation-dependent (19,20), but the rates of repeat expansions became orientation-independent (19,20). At the same time, genetic screening revealed the genes encoding the replication fork components as the key players in repeat expansions in this system, which indicated that replication errors could occur in either orientation of the repeat (19,20). These replication problems are likely due to the unusual structural properties of GAA repeats: they can form DNA triplexes (21), hairpin-like structures (22) and parallel duplexes (23). The orientation dependence of fork stalling at the repeat is highly indicative of triplex formation (24,25). The question remains, however, whether unusual DNA structures of the repeat can be formed prior to or during DNA replication in the context of complex mammalian chromatin.
We have recently shown that the first replication cycle of an episome transfected into mammalian cells goes differently from all the subsequent replication rounds, most likely due to differences in template chromatin structure (26). This first replication cycle appears to initiate at various positions in the episome and depends on the presence of single-stranded DNA regions. Furthermore, it could take place in the late G1 phase of the cell cycle or even in non-dividing cells, implicating this replication mode in DNA repair.
Here we show that carrier-size GAA repeats block the progression of the first episomal replication cycle in mammalian cells. This blockage is much stronger than that previously observed in yeast and bacteria—in fact, it is almost complete. In contrast, subsequent episomal replication cycles were not affected by those repeats and were only mildly inhibited by long, disease-size repeats. This shows that expanded GAA repeats have different effects on different modes of DNA replication.
MATERIALS AND METHODS
Plasmids
The pUCneoGAAn and pUCneoCTTn plasmids were obtained by inserting the repeat-containing EcoRI–EcoRI fragments of pYES-GAA10, pYES-GAA20, pYES-GAA35, pYES-GAA57, pYES-GAA114 and pYES-GAA230 (17) into the blunt-ended AatII site of pUCneo (26) in two orientations. The plasmids were named according to the leading strand template.
Ori(−)GAA57 and Ori(−)CTT57 were obtained by inserting the repeat-containing EcoRI–EcoRI fragment of the pYES-GAA57 (17) into the blunt-ended AatII site of Ori(−) (26) in two orientations.
The pRepControl plasmid was obtained by inserting the HindIII–Eco91I fragment of the pRep4 (Invitrogen), containing both OriP and the EBNA-1 gene, into the ApaI–NdeI-digested pUCneo. RepGAA230 and RepCTT230 were obtained by inserting the repeat-containing EcoRI–EcoRI fragment of pYES-GAA230 into the AatII site of pRep-control.
Detection of alternative DNA conformations of the repeat using S1 nuclease
One microgram of Ori(-)GAA57 plasmid was digested with Nt.BstNBI nicking endonuclease (New England Biolabs) according to the manufacture's protocol, ethanol-precipitated, dissolved in TE (10 mM Tris–HCl, 1 mM EDTA) and then heated at 70° C for 10 min to remove possible alternative DNA structures. Nicked Ori(-)GAA57, as well as a control supercoiled Ori(-)GAA57 plasmids, were digested with 4 units of the S1 nuclease (Fermentas) for 10 min at room temperature. The reactions were stopped with 10 mM EDTA, DNA was precipitated, dissolved in TE and heated again at 70° C for 10 min. At this step, all potential alternative structures were eliminated from the control supercoiled Ori(-)GAA57 plasmid as it was converted into an open circular from by the nuclease. Both samples were then digested with ScaI and NdeI restriction enzymes, and the 957-bp restriction fragments were eluted from agarose, mixed with 1 μg of pUCneo plasmid and digested with 4 units of S1 nuclease for 10 min at room temperature. At this step, S1 nuclease cleaved across the single-stranded breaks that were introduced in the first round of the S1 nuclease digestion. Both samples were then analyzed on 1% agarose gel to reveal double-stranded breaks at expected triplex structures.
Cell cultures and transient transfections
All cell lines (COS-1 monkey fibroblasts, 293A embryonic kidney fibroblasts, 293-EBNA, and HeLa cells) were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum.
COS-1 and 293A cells were transfected using lipofectin (Invitrogen), while 293-EBNA cells (Invitrogen) were transfected using turbofect (Fermentas) according to the manufacturer's instructions.
Two-dimensional separation of replication intermediates
Isolation of replication intermediates from mammalian cells and their analysis via two-dimensional (2D) neutral/neutral gel electrophoresis were performed as described in ref. 26.
RESULTS
Replication of an SV40 origin-based plasmid is affected by GAA repeats in a length- dependent and orientation-independent manner
We analyzed the progression of DNA replication through GAA repeats of varying lengths inserted into a pSV2neo-derived plasmid, pUCneo (Figure 1A). The resultant plasmids were introduced into T-antigen-expressing COS-1 monkey fibroblasts by transient transfection. The repeats were positioned on the path of the counterclockwise replication fork initiated at the episomal SV40 origin. We carefully controlled the quality of DNA samples used for transfection to assure that they mainly contained monomeric plasmids. Thirty hours after transfection into mammalian cells, the plasmids and their replication intermediates were isolated. The replication intermediates were digested with the AflIII restriction endonuclease, which generated a fragment with a GAA repeat positioned at approximately one-third of a distance from its end. The replication intermediated were then analyzed by 2D agarose gel electrophoresis followed by Southern hybridization (Figure 2).
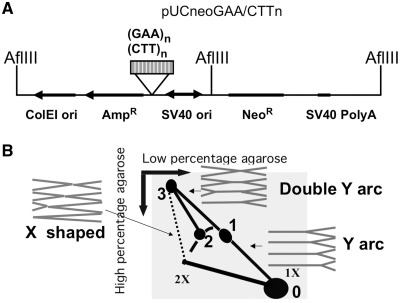
Figure 1.
(A) The scheme of pUCneoGAA/CTTn plasmid. (GAA)n repeat was inserted in pUCneo in two orientations. Replication intermediates were digested by AflIII. The hybridization probe used in 2D gel analysis corresponded to the GAA repeat-containing AflIII fragment. (B) Schematic presentation of the 2D gel electrophoresis of mammalian replication intermediates digested with AflIII. Corresponding shapes of replication intermediates are shown in gray. Spot 0 – unreplicated AflIII fragment; spot 1 – replication stalling of the counterclockwise replication fork at GAA; spot 2 – replication stalling of the clockwise replication fork at GAA; spot 3 – two replication forks from opposite ends of the fragment stalled at the repeat. May also contain intermolecular complexes of two different fragments joined at the GAA repeat. A short stretch of the bubble arc was omitted from this scheme because it is obscured by unreplicated DNA spot in our 2D gel pictures.
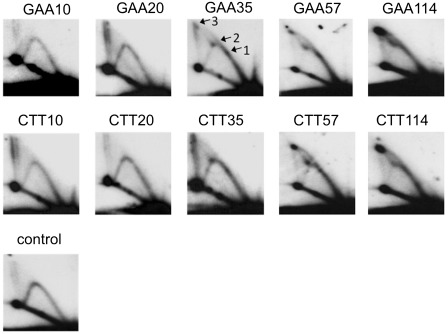
Figure 2.
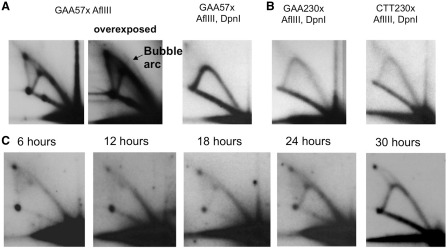
Analysis of pUCneoGAAn/CTTn replication in COS-1 cells by 2D gel electrophoresis. Replication intermediates were digested by AflIII restriction endonuclease as shown in Figure 1. GAAn and CTTn correspond to two orientations of a GAA repeat relative to the SV40 replication origin. Plasmids’ replication was blocked in a length-dependent and orientation-independent manner. The spots 1, 2 and 3, corresponding to Figure 1B, are indicated.
We initially expected that replication stalling at the repeat would result in a 2D pattern consisting of a Y arc with a single bulge (Figure 1B, spot 1), positioned about one-third of the way from the unreplicated DNA spot (Figure 1B, spot 0), as was observed in yeast (17). Unexpectedly, however, the presence of long-normal repeat (GAA)37, pre-mutation length (GAA)57 repeat or mutation length (GAA)114 repeat in the plasmid led to the appearance of two bulges, positioned approximately one-third and two-thirds of the way from the unreplicated spot (Figure 1B, spots 1 and 2). In the presence of the (GAA)37 repeat, there was still a considerable amount of the descending part of the Y arc, which indicates that most of replication forks still progress past the GAA repeat. However, for the (GAA)57 and (GAA)114, the descending part was very faint suggesting that progression of most of the replication forks was blocked by GAA repeat. Since the length of (GAA)114 is more than 300 bp, replication stalling takes the form of a darker region of the arc rather than an isolated spot as was observed for (GAA)37. We believe that most of the forks were blocked by a non-canonical DNA structure formed within the expanded GAA repeat; more stalling was observed at longer repeats where the structure was more likely to form. Differently from our previous observations in yeast, however, we did not see a significant difference in replication stalling between the two orientations of the repeat within the replicon.
Other unexpected elements that were never observed in yeast were two spikes, corresponding to double Y arcs, extending from each bulge. These double Y arcs connected the bulges with spot 3 at the upmost position of the vertical line, corresponding to the X-shaped recombination products. The appearance of a double Y arc extending from the bulge could be explained by the blockage of the replication fork, which reached the GAA repeat first, followed by the completion of the replication by another fork that entered from the opposite end of the fragment. In this case, spot 3 would correspond to the point where two forks coming from the opposite sides meet at the repeat. This pattern is known to occur when the replication fork progression is not just slowed down, but rather completely blocked by the repeat (27). Spot 3 may also contain complexes of two different AflIII fragments from different DNA molecules joined at GAA repeat.
This logic can easily explain the spot 1 as a result of blockage of the counterclockwise replication fork until the clockwise replication fork arrives. The explanation for the second stalling spot at the Y arc (spot 2) is less straightforward. The location of the spot 2 at two-thirds of the way from the unreplicated fragment gave us a hint that it may result from blockage of a replication fork that approached the repeat from the opposite side of the fragment relative to the SV40 origin. In this scenario, the distance that the opposing replication fork has to run to reach the repeat would be exactly two-thirds of the fragment's length.
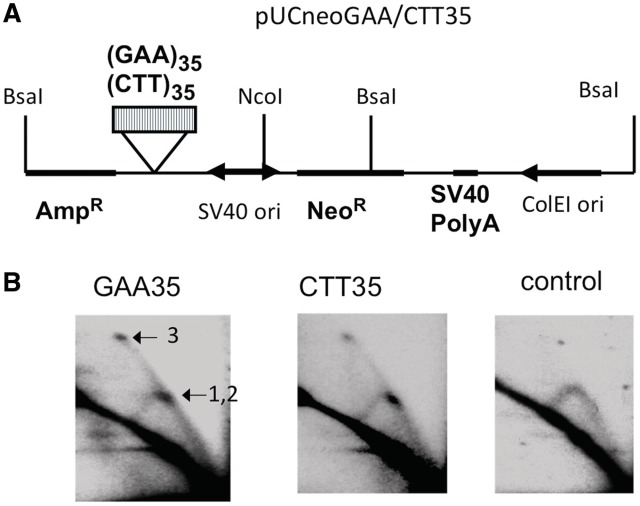
To confirm this idea, we digested the same replication intermediates with the NcoI and BsaI restriction enzymes, positioning the GAA run in the center of the fragment (Figure 3A). In this case, forks entering the fragment from its opposite ends should both reach the repeat at the middle of the fragment, i.e. the two bulges (1 and 2) should now convene at the fragment's center (Figure 3A). This turned out to be the case: one can clearly see a unique bulge in the middle of the arc, with a single spike emanating from it. We believe that the spikes connecting spots 1 and 2 to spot 3 result from replication forks entering the fragment from both ends: one of the forks is stalled, and another approaches from the opposite direction (Figure 3B).
Figure 3.
2D gel electrophoresis of replication intermediates digested so that the (GAA)35 repeat was in the middle of the fragment. (A) The scheme of the digest of pUCneoGAA/CTT35 by BsaI and NcoI. (B) 2D gel electrophoresis of replication intermediates of pUCneo digested with BsaI and NcoI. A single bulge (combined spot 1, and spot 2, Figure 1B) was observed upon BsaI–NcoI digest. A position of the spot 3 is also shown. We concluded that replication approached the GAA repeat from both ends of the fragment.
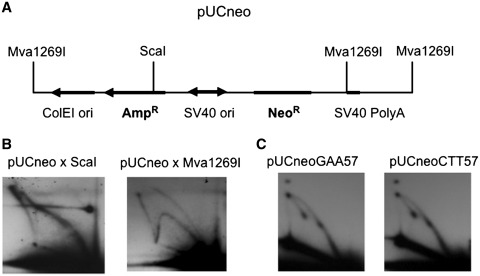
While this was gratifying, we were still left with following question. It was easy to assume that the counterclockwise replication fork could be blocked to the extent that the clockwise replication fork gets enough time to go almost a full circle until it meets its counterpart at the repeat (spot 1 and the spike). For the spot 2, a similar explanation would imply that the clockwise replication fork somehow managed to reach the repeat and get ahead of the counterclockwise fork. This does not seem possible if the two forks start at SV40 origin at the same time, since clockwise fork has almost a full circle to run, while the counterclockwise fork starts very close to the repeat. One possibility could be that the clockwise and counterclockwise replication forks are not synchronized in our episome, and the clockwise replication fork sometimes starts before the counterclockwise. To exclude this, we digested the replication intermediates of pUCneo plasmid at the ScaI site that is located just upstream of the origin (Figure 4A). Under the conditions of this digest, the clockwise replication fork that starts independently from the counterclockwise, and has enough time to go almost a full circle before the counterclockwise fork starts, would generate a bubble arc (Supplementary Figure S1A and S1B). We did not observe a bubble arc, but instead we observed an asymmetric double Y that could account for two replication forks starting simultaneously (Figure 4B, left, Supplementary Figure S1A and S1B). A very similar asymmetric double Y was described in (28). We also observed the Y arc that could potentially originate from dimers. To confirm that the absence of the bubble arc is not an artifact of our analysis, we also digested our replication intermediates with the Mva1269I enzyme cleaving episomal DNA exactly opposite to the origin. Under this digest, the bubble arc was clearly visible (Figure 4B, right).
Figure 4.
(A) The scheme of digest of pUCneo plasmid used to explore whether replication of this plasmid can also occur unidirectionally. (B) Replication intermediates of pUCneo were isolated from COS-1 cells 23 h post transfection and digested by ScaI (left) and Mva1269I (right). The pattern of ScaI digest consists of a Y arc and an asymmetrical double arc. No bubble arc is visible. The control Mva1269I pattern consists of a Y and a bubble arc. The hybridization probe was the same as in all other assays: AflIII-fragment of pUCneo that contained the ampicillin resistant gene. (C) Replication of monomer pUCneoGAA/CTT57 plasmids in COS-1 cells. Replication intermediates were isolated 24 h post-transfection, and digested with AflIII.
Another possibility was that the spot 2 (Figure 1B) originated from the small amount of dimers in the sample. In this case, the replication fork that initiated at the second origin in a dimer can approach the repeat from the other end. To test if this was the case, we purified monomeric pUCneoGAA/CTT57 episomes from an agarose gel, and analyzed their replication in mammalian cells (Figure 4C). We observed the same pattern with two stalling sites and two spikes, thus ruling out that the second spot was the product of dimeric episomes’ replication (Figure 4C).
An inability to explain the appearance of the second stalling site within the framework of the bidirectional, SV40 ori-initiated replication prompted us to analyze the replication mode of the pUCneo episome in more depth. The results of this analysis have been published separately (26). Rather unexpectedly, we found that replication of this episome could be initiated at numerous positions throughout its sequence soon after transfection into mammalian cells. We confirmed that the replication intermediates that we observed are not transferred over from bacteria, but indeed originated from mammalian cells. The alternative replication mode did not rely on the SV40 origin or T-antigen for its initiation but rather initiated in the absence of a regular chromatin structure, and was stimulated by single-stranded DNA regions (26).
It should be noted that for our plasmids, the alternative replication mode apparently co-existed with the conventional replication mode initiated at the SV40 origin. Thus, the replication pattern shown in Figure 1B likely resulted from a superposition of replication intermediates generated in two different replication modes: the SV40-driven and the alternative one. The relative contribution to the episomal replication from those two modes depended on how much DNA was used in transfection, and how long it was inside the cell prior to replication intermediates isolation. For example, analyzing replication of gel-purified monomers (Figure 4C), we observed a decrease in Y arc intensity after spot 1, which may be explained by less efficient SV40-driven replication mode in this particular experiment.
The first replication cycle of the SV40-based episome is blocked by the GAA repeat, while subsequent replication cycles are not affected
Our previous analysis of the alternative replication mode revealed that it is limited to the first replication cycle after episomal transfection into mammalian cells (26). We believe that the pattern of stalling at GAA repeat that we observed (Figures 2–5) was a superposition of the first replication cycle that initiated randomly throughout the sequence, and SV40 origin-initiated replication. Since the first cycle replication initiated everywhere, it mostly produced Y arc, and some of the bubble arc that we could only observe at overexposed pictures (Figure 5A, overexposed panel). During the first replication cycle, replication intermediates consist of the template DNA strands that came from Escherichia coli plasmid and nascent strands synthesized in mammalian cells. In subsequent replication cycles, both nascent and template strands are of mammalian origin. This allows one to distinguish between the first and the subsequent replication cycles using a DpnI restriction digest. This enzyme cleaves GATC sequences when they are methylated or hemimethylated by the bacterial Dam methylase. Thus, the DpnI digest eliminates the non-replicated episomal DNA, as well as the products of its first replication cycle in mammalian cells.
Figure 5.
(A) Replication stalling was no longer observed upon DpnI digest of replication intermediates. 2D gel electrophoresis of the replication intermediates of pUCneoGAA57 isolated from the COS-1 cells and digested with either AflIII alone, or AflIII and DpnI. To eliminate semimethylated products of the first replication cycle, the digest was performed with 20 units of DpnI, and 10 units of AflIII for about 2 h. Bubble arc can be observed on the overexposed panel of pUCneoGAA57 digested with AflIII (shown by an arrow). (B) An expanded (GAA)230 repeat did not cause replication stalling of the T-antigen-driven replication. 2D gel electrophoresis of the replication intermediates of pUCneoGAA/CTT230 isolated from COS-1 cells and digested with AflIII and DpnI. (C) Replication stalling at the GAA repeat decreased over time. 2D gel electrophoresis of replication intermediates of the pUCneoGAA57 plasmid isolated at 2, 12, 18, 24 and 30 h after COS-1 cells transfection. Only gel-purified monomer plasmid was used for transfection. Replication intermediates were digested with AflIII. The descending part of the Y arc increased, while the bulges and the double Y arc decreased with incubation time.
Figure 5A, right panel, shows the pattern of replication intermediates that were extensively treated by DpnI prior to their electrophoretic separation. Remarkably, after this treatment the replication arc became smooth, while the two bulges, most of the spot 3, and the double Y spike disappeared, indicating that later rounds of replication were not significantly affected by the presence of GAA repeats. Even the longest repeat, (GAA)230, failed to stall the fork during the late, T-antigen driven rounds of the episomal replication (Figure 5B). We concluded, therefore, that only the very first round of episomal replication was strongly inhibited by GAA repeats.
This conclusion was independently confirmed by the analysis of replication through the (GAA)57 repeat at different time points after cell transfection. For these experiments, we used 0.5 µg of the episomal DNA per transfection, i.e. 40-fold less than the amount of DNA used in the experiments shown in Figure 2, in order to promote its rapid utilization by the mammalian replication machinery. Figure 5C shows that there is a gradual decrease in the strength of the fork stalling over time: while the Y arc does not proceed beyond the repeat during the first 12 h post-transfection, its progress, as illustrated by the appearance of the descending arm, becomes evident after 18 h, while the descending part of the arc reaches the same intensity as the ascending part 30 h post-transfection, at which point the spikes and fork stalling drastically decrease.
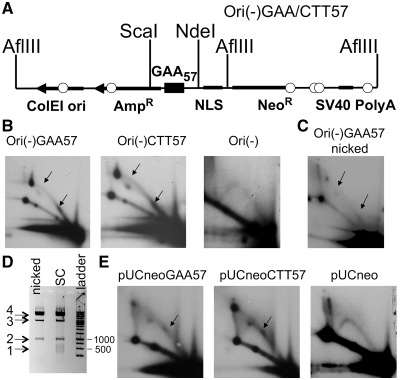
To explore whether replication stalling occurred in the course of the alternative replication, we analyzed the replication of the Ori(-)GAA57 plasmid, which did not contain the SV40 replication origin (Figure 6A). One can see that in COS-1 cells this plasmid was still capable of undergoing some replication, which was characterized by the profound fork stalling at the GAA repeat (Figure 6B). The fact that the descending portion of Y arc beyond the spot 2 was not detectable in the Ori(-) plasmids is consistent with the complete replication blockage at GAA repeat.
Figure 6.
(A) The scheme of the Ori(-)GAA/CTT57 plasmid. In the Ori(-) plasmid, the SV40 replication origin was replaced by the nuclear localization sequence (26). (B) 2D gel electrophoresis analysis of replication intermediates of Ori(-)GAA/CTT57, and Ori(-), isolated from Cos-1 cells. Replication of Ori(-) plasmid was blocked by the (GAA)57 repeat in both orientations. (C) 2D gel electrophoresis analysis of replication intermediates of nicked Ori(-)GAA57 plasmid isolated from Cos-1 cells. Replication of the nicked plasmid that did not contain preformed structures was still affected by GAA repeat. The positions of nicks introduced by Nt. BstNBI are shown by small circles in Figure 6A. We concluded that the GAA repeat forms an alternative structure that stalls replication inside the mammalian cell. (D) S1 nuclease digest of nicked and supercoiled Ori(-)GAA57 plasmids to confirm that alternative DNA structures do not form in the nicked Ori(-)GAA57. 1 – series of bands corresponding to the GAA repeat position that are visible only for the supercoiled sample; 2 – 957 bp ScaI–NdeI fragment; 3 – supercoiled pUCneo, 4 – open circle pUCneo. (E) 2D gel of the replication intermediates of pUCneoGAA57 isolated from 293A cells. Alternative replication of pUCneoGAA57 happens in the absence of T-antigen and is severely stalled by the GAA repeat. The stalling points are indicated by arrows.
We have further confirmed that this first round of alternative replication does not require T-antigen for its initiation. Replication fork stalling at the (GAA)57 repeat was observed upon episome transfection into the human 293A cell line, which does not express viral T-antigen (Figure 6E). We concluded that profound fork stalling at GAA repeats in mammalian cells is only evident during replication in early post-transfection stages (26).
Summarizing, we found that an alternative replication mode for the episome is blocked near completely by the GAA repeat. We cannot, however, deduce from our data whether the very first round of the T-antigen-driven replication is blocked by the repeat as well.
Blockage of the first replication cycle could be caused by non-B DNA structures pre-existing in episomal DNA, as well as the structures that formed inside mammalian cells
The first replication cycle of a transfected plasmid could be blocked by a DNA structure that either was formed by the repeat inside a mammalian cell or pre-existed in the plasmid used for transfection. Magnesium ions, which are normally present in the transfection media, could promote the formation of triplex by the GAA repeat (21). To determine whether the alternative replication can be affected by DNA structures that form inside the cell, we eliminated the pre-existing structures and tested whether stalling at the GAA repeat would still occur. We capitalized on the fact that triplex or sticky DNA structures are readily formed in superhelical plasmids in presence of magnesium ions or under acidic pH, but are extremely unfavorable in open circular or linear DNA (29,30). First, we used the plasmid sample isolated by alkali lysis from bacteria and dissolved in TE pH 8.5, the conditions in which triplex structure does not form. Second, we introduced single-stranded breaks by Nt.BstNBI nicking enzyme, and additionally heated the sample for 20 min at 70° C in the presence of EDTA to get rid of all potential alternative DNA structures. Nt.BstNBI introduced six single-stranded breaks scattered around the plasmid (Figure 6A, shown by circles). To confirm that our nicked plasmid sample does not contain alternative DNA conformations, we employed the S1 nuclease, which cleaves DNA at single-stranded regions existing in intramolecular triplexes or other alternative DNA structures. The supercoiled GAA-containing plasmid was used as a control, since it is known to form triplex structure under acidic pH in S1 nuclease buffer (25). We treated nicked and supercoiled Ori(-)GAA57 with the S1 nuclease, followed by the digestion with the ScaI and NdeI restriction enzymes and purification of the 957-bp fragment containing (GAA)57-repeat from an agarose gel (Figure 6D, band 2). This protocol allowed us to analyze only the single-stranded breaks generated by the S1 nuclease in the repeat-containing DNA fragment. (We deliberately limited our analysis to this ScaI–NdeI fragment to avoid interference with DNA fragments generated upon the S1 digestion opposite to the single-stranded breaks made by Nt.BstNBI.) We then subjected the eluted fragments to yet another round of the S1 digest to convert single-stranded breaks into the double-stranded ones. A supercoiled pUCneo plasmid was added to the samples to confirm the S1 nuclease activity that converted the supercoiled pUCneo (Figure 6D, band 3) into the open circle (Figure 6D, band 4). Subsequent gel-electrophoretic analysis showed multiple double-stranded breaks at the position of GAA repeat in the supercoiled but not in the nicked plasmid (Figure 6D, band 1). These data confirm that nicked plasmid in our transfection experiments did not contain pre-formed alternative structures.
When we transfected Cos-1 cells with Ori(-)GAA57 plasmid, in which the alternative DNA structures of the GAA repeat were eliminated by nicking and heating, fork stalling at the repeat was still observed. Note, however, that both the replication arc and stall sites became less pronounced than in the case of superhelical DNA (Figure 6C). We believe that this decrease in replication efficiency was due to the fact that initiation of the alternative replication depends on DNA supercoiling (26). Thus, a nick should be repaired and supercoiling introduced in order for the transfected plasmid to start replicating. No supercoiled plasmid was visible in the initial sample that was used in transfection (Supplementary Figure S2), so the arc that we observed could not be a result of a preformed structure in the molecules that escaped nicking. Fork stalling observed in the last experiment must, therefore, be associated with a DNA structure that was formed inside a mammalian cell. Decreased efficiency of the replication stalling might be due to the fact that some of the plasmids got chromatinized even before the nicks were sealed, making triplex formation impossible.
We conclude that while a significant fraction of repeat-mediated stalling observed during the first replication round is likely due to pre-existing non-canonical DNA structures formed in the transfection media, structures formed inside the cell impede the replication fork progression as well.
The initiation of alternative replication is stimulated by the presence of GAA repeats
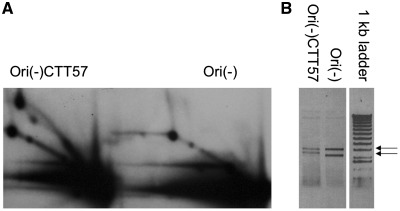
As discussed above, the elongation stage of the alternative replication is severely inhibited by the presence of even relatively short GAA repeats. Remarkably, however, the same repeats seem to increase the rate of alternative replication initiation. This is evident, for example, from the fact that the intensity of the replication arc approaching the repeat is much higher than that for the control, repeat-free plasmid (Figure 7A). Note that about the same amount of the plasmid was loaded for GAA-containing and control plasmids (Figure 7B). Similar increases in the alternative replication efficiency of the repeat-containing plasmids were consistently observed in all our experiments, which required us to load significantly more replication intermediates for the control plasmid to equalize them with the intermediates of the repeat-containing plasmids (Figure 6B).
Figure 7.
(A) The first round of replication of the plasmid containing GAA repeat is more pronounced than that of a control plasmid. The same amount of plasmids Ori(-)CTT57 and Ori(-) were used for 293A transfections. The cells were lysed 6 h after transfection, replication intermediates were isolated and digested with AflIII. The samples were intentionally run together to allow direct comparison. (B) The samples Ori(-)CTT57 and Ori(-) contain about the same amount of plasmid.
Replication of Epstein–Barr virus-derived episomes is mildly affected by the GAA repeats
One explanation for why only the first replication cycle of the pSV-derived episome was inhibited by the GAA repeats could be that T-antigen serves as a replicative helicase during all subsequent replication cycles. T-antigen is a very potent helicase that was shown to pass through alternative DNA structures (31,32). To evaluate this hypothesis, we decided to analyze the replication of GAA repeats in another episomal system driven by the Epstein–Barr virus (EBV) replication origin (33). In this case, replication is initiated at the OriP origin upon binding of the EBNA protein (34), followed by the loading of other components of the mammalian replication fork (35). Consequently, the MCM2-7 complex, rather than T-antigen, serves as the replication helicase here.
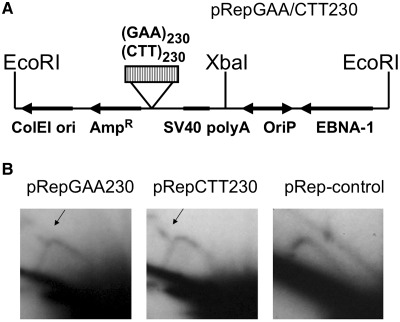
293-EBNA cells were transfected with the pRepGAA230 or pRepCTT230 plasmids, which harbored the (GAA)230 repeat in two orientations (Figure 8A). Replication intermediates were isolated in 6 days after transfection, and digested by EcoRI and XbaI enzymes, which positioned the repeat at one-third of the length of the fragment from an OriP-proximal end. Since replication intermediates were isolated 6 days post-transfection, they contained only a small portion of initial bacterial sample (no more than 10% of episomal replication intermediates, 20 ng of which were loaded on the 2D gel in Figure 8B). An extensive DpnI digest was then performed to eliminate this residual amount of initial bacterial DNA. The activity of DpnI enzyme was controlled in Supplementary Figure S3 (Supplementary Data). This digest would also eliminate the residual products of the first replication cycle, although we never observed them 2 days post-transfection anyway (26). An extensive DpnI digest was then performed to make sure that we were only analyzing plasmids that replicated in mammalian cells more than once. Electrophoretic analysis of the replication intermediates revealed the presence of a characteristic double Y arc with a bulge (Figure 8B, shown by arrows), similar to what was observed during the first round of replication of the pUCneoGAA35 plasmid. The appearance of the double Y arc was indicative of replication being blocked by the GAA repeat at least in a fraction of plasmids. It should be noted, however, that this blockage was observed in only a fraction of molecules in contrast to a near-complete blockage of the first replication round by the same repeat (Figure 2). The observed pattern of fork stalling was not an artifact of large-scale deletions at the GAA230 repeat: it stayed intact throughout the experiment, and there were no significant changes in the length of the restriction fragment in question as is evident from the lack of shift in the position of the descending part of the Y arc.
Figure 8.
(A) The scheme of the pRepGAA/CTT230 plasmid, digested with EcoRI and XbaI. (B) Replication of pRepGAA/CTT230 is only mildly affected by the repeat. The plasmid replication intermediates were isolated from 293-EBNA cells 6 days after transfection, digested with EcoR1, XbaI and DpnI, and analyzed by 2D gel electrophoresis. The hybridization probe corresponded to the XbaI–EcoRI fragment containing the GAA repeat. The presence of the double Y arcs is indicative of a partial replication blockage.
DISCUSSION
This study demonstrates, for the first time, that GAA repeats can block the replication fork progression in mammalian cells likely due to the triplex formation. Remarkably, different modes of episomal replication in mammalian cells were affected differently by GAA repeats. We have previously demonstrated that the first round of replication of the SV40-derived plasmids in mammalian cells could be carried out by the alternative replication mode, which does not require the presence of the viral replication origin and is initiated at various sites in the plasmid (26). Here we report that this replication mode is almost completely blocked by the presence of GAA repeats. On the other hand, replication of an EBV-derived plasmid, initiated by the EBNA protein at the OriP origin, is only mildly inhibited by long GAA repeats. Finally, T-antigen-driven replication of the pSV2-derived plasmids was not affected by the GAA repeats after the second replication round.
Thus, GAA repeats impede the progression of the mammalian replication fork, which is involved in both alternative and OriP-initiated replication in mammalian cells, but have no effect on the progression of a fork where viral T-antigen serves as a replication helicase. This is not surprising, given that T-antigen is an exceptionally potent DNA helicase, and was shown to unravel various unusual DNA structures (31,32). Note, also, that another expandable repeat, (GGC)n, stalled the progression of T-antigen-driven forks (36); however, this stalling was less pronounced than the stalling observed in yeast (37). We believe that the latter stalling resulted from the formation of a hairpin-like structure in the lagging strand template DNA, i.e. when the replicative helicase has already passed the repeat. The formation of such hairpin structures in mammalian cells has been recently demonstrated (38).
A particularly potent stalling of the first episomal replication cycle by the GAA repeat is most likely caused by its unusual DNA conformation(s). In naked superhelical DNA, this repeat can form a triplex (21) or a more complex sticky DNA structure, a triplex formed between the two separate GAA runs (29). Since alternative replication operates on superhelical DNA that just started to assemble in chromatin, appearance of these structures ahead of the replication fork would inevitably block its progression. In the EBV-derived plasmid, the native replication fork replicates chromatinized DNA templates. The presence of a regular nucleosome structure would limit the ability of a GAA repeat to form unusual DNA conformations, explaining much more modest replication inhibition.
We believe that the first replication round is stalled so profoundly, compared to the subsequent rounds, because the inhibitory DNA triplexes pre-exist in plasmids used for the transfection. DNA triplexes can easily form in superhelical DNA isolated from bacteria in the presence of magnesium ions in the transfection mix. To address the question whether the GAA repeat can affect replication in the lack of the pre-formed triplex, we introduced nicks into our plasmid DNA followed by heating it in the absence of bivalent cations, which unraveled all pre-formed DNA structures (30,39). After transfecting mammalian cells with this nicked DNA, we still detected replication stalling, although somewhat less pronounced. This result suggests that a triplex or some other non-canonical structure can form in DNA inside the mammalian cells. The latter observation is also consistent with the data on the modest inhibition of the fork progression by GAA repeats in EBV-derived plasmids.
While the GAA repeat did not serve as a replication origin during alternative DNA replication, as it was evident from the lack of a pronounced bubble arc in our 2D gels, it did increase the efficiency of the first replication round, somehow making a plasmid more competent in terms of replication initiation. This characteristic of an alternative initiation mode was also observed in other systems, when a particular sequence made a DNA template more prone to replication without being an origin (40,41).
The increase in the efficiency of alternative replication could be due to the formation of a non-canonical DNA structure by the GAA repeat. Indeed, non-canonical DNA structures such as hairpins and intramolecular triplexes are accompanied by single-stranded regions. We have previously shown that the presence of single-stranded DNA induced by superhelicity can promote alternative replication initiation (26). However, the subsequent electrophoretic analysis of replication through the BsaI–NcoI DNA fragment containing the GAA repeat in its center did not reveal a trace of the bubble arc, which rules out that the repeat per se serves as a replication initiation point. We hypothesize, therefore, that a single-stranded DNA part of the intramolecular triplex might serve as a loading dock for the replication fork components that would then initiate replication elsewhere.
What are the implications of our findings for the FRDA pathogenesis? It is generally believed that expansions of GAA repeats occur in early embryonic divisions during intergenerational transmissions in FRDA families (12). Recently, it was also demonstrated that these repeats expand with an almost 100% probability upon transcription factor-mediated reprogramming of fibroblasts from FRDA patients (42,43). Pluripotent cells’ chromatin configuration is more open than that of differentiated cells (44). The state of chromatin in mammalian zygote is understudied; however, the major portion of chromatin in sea urchin zygote is in unfolded nucleosome structure, which mostly consists of non-regular positioned nucleosomes (45). These data hint that open chromatin structure may facilitate GAA repeat expansions. In our case, replication stalling is much more pronounced in the lack of regular chromatin in episomal DNA, as it makes triplex formation more feasible. This consideration makes it tempting to speculate that abnormal replication of long (GAA)n repeats in the context of open chromatin could contribute to their expansions. Note, however, that the analogy between the chromatin structure of our episomes and the chromatin of the mammalian zygote is not substantiated by the experimental data and remains purely speculative.
Another possibility comes from recent observations that the instability of the FRDA repeat in mammalian chromosomes was strongly enhanced by transcription through the repetitive region (46,47). It is possible that single-stranded DNA segments accumulating upon the formation of stable DNA–RNA complexes in the repeat could promote alternative replication initiation, which would also lead to the repeat's instability. Supporting this notion, the replication efficiency was much higher for pUCneoGAA57 compared to Ori(-)GAA57 in 293 cells. Since these cells lack T-antigen, transcription from a promoter adjacent to the SV40 origin could contribute to the initiation of alternative DNA replication.
Finally, we have previously shown that the alternative replication mode was also detected outside of the S-phase of the cell cycle, synchronized at the G1/S border (26). This indicates that the alternative replication mode could be associated with cellular DNA repair. In fact, repair can be stimulated by the presence of alternative DNA conformations including those formed by GAA repeats (48). Instability of the GAA repeat was also observed in post-mitotic cells (16), which could be attributed to alternative replication/repair process.
In summary, we found, for the first time, that different modes of replication in mammalian cells are affected differently by the expandable GAA repeats causative of FRDA. This result might help to explain the differences in the rates and scales of repeat instability between somatic and embryonic cell divisions characteristic for this disease.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3.
FUNDING
A Summer Discovery grant, and the Penn State Institute of Diabetes and Obesity Undergraduate research grant (to G.S.C.); Merck Company Foundation grant (to M.P.); NIH grant GM60987 (to S.M.M.); NIH grant GM087472; and research grant from Friedreich’s Ataxia Research Alliance (to M.M.K.). Funding for open access charge: NIH grant GM60987, NIH grant GM087472, and Undergraduate Research Support Funding.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Campuzano V, Montermini L, Lutz Y, Cova L, Hindelang C, Jiralerspong S, Trottier Y, Kish S, Faucheux B, Trouillas P, et al. Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes. Hum. Mol. Genet. 1997;6:1771–1780. doi: 10.1093/hmg/6.11.1771. [DOI] [PubMed] [Google Scholar]
- 2.Durr A, Cossee M, Agid Y, Campuzano V, Mignard C, Penet C, Mandel JL, Brice A, Koenig M. Clinical and genetic abnormalities in patients with Friedreich's ataxia. N. Engl. J. Med. 1996;335:1169–1175. doi: 10.1056/NEJM199610173351601. [DOI] [PubMed] [Google Scholar]
- 3.Campuzano V, Montermini L, Molto M, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, et al. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 4.Gottesfeld JM, Pandolfo M. Development of histone deacetylase inhibitors as therapeutics for neurological disease. Future Neurol. 2009;4:775–784. doi: 10.2217/fnl.09.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greene E, Mahishi L, Entezam A, Kumari D, Usdin K. Repeat-induced epigenetic changes in intron 1 of the frataxin gene and its consequences in Friedreich ataxia. Nucleic Acids Res. 2007;35:3383–3390. doi: 10.1093/nar/gkm271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saveliev A, Everett C, Sharpe T, Webster Z, Festenstein R. DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature. 2003;422:909–913. doi: 10.1038/nature01596. [DOI] [PubMed] [Google Scholar]
- 7.Krasilnikova MM, Kireeva ML, Petrovic V, Knijnikova N, Kashlev M, Mirkin SM. Effects of Friedreich's ataxia (GAA)n*(TTC)n repeats on RNA synthesis and stability. Nucleic Acids Res. 2007;35:1075–1084. doi: 10.1093/nar/gkl1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montermini L, Richter A, Morgan K, Justice CM, Julien D, Castellotti B, Mercier J, Poirier J, Capozzoli F, Bouchard JP, et al. Phenotypic variability in Friedreich ataxia: role of the associated GAA triplet repeat expansion. Ann. Neurol. 1997;41:675–682. doi: 10.1002/ana.410410518. [DOI] [PubMed] [Google Scholar]
- 9.Filla A, De Michele G, Cavalcanti F, Pianese L, Monticelli A, Campanella G, Cocozza S. The relationship between trinucleotide (GAA) repeat length and clinical features in Friedreich ataxia. Am. J. Hum. Genet. 1996;59:554–560. [PMC free article] [PubMed] [Google Scholar]
- 10.Montermini L, Andermann E, Labuda M, Richter A, Pandolfo M, Cavalcanti F, Pianese L, Iodice L, Farina G, Monticelli A, et al. The Friedreich ataxia GAA triplet repeat: premutation and normal alleles. Hum. Mol. Genet. 1997;6:1261–1266. doi: 10.1093/hmg/6.8.1261. [DOI] [PubMed] [Google Scholar]
- 11.Pandolfo M. The molecular basis of Friedreich ataxia. Adv. Exp. Med. Biol. 2002;516:99–118. doi: 10.1007/978-1-4615-0117-6_5. [DOI] [PubMed] [Google Scholar]
- 12.De Michele G, Cavalcanti F, Criscuolo C, Pianese L, Monticelli A, Filla A, Cocozza S. Parental gender, age at birth and expansion length influence GAA repeat intergenerational instability in the X25 gene: pedigree studies and analysis of sperm from patients with Friedreich's ataxia. Hum. Mol. Genet. 1998;7:1901–1906. doi: 10.1093/hmg/7.12.1901. [DOI] [PubMed] [Google Scholar]
- 13.Pianese L, Cavalcanti F, De Michele G, Filla A, Campanella G, Calabrese O, Castaldo I, Monticelli A, Cocozza S. The effect of parental gender on the GAA dynamic mutation in the FRDA gene. Am. J. Hum. Genet. 1997;60:460–463. [PMC free article] [PubMed] [Google Scholar]
- 14.Delatycki MB, Paris D, Gardner RJ, Forshaw K, Nicholson GA, Nassif N, Williamson R, Forrest SM. Sperm DNA analysis in a Friedreich ataxia premutation carrier suggests both meiotic and mitotic expansion in the FRDA gene. J. Med. Genet. 1998;35:713–716. doi: 10.1136/jmg.35.9.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma R, Bhatti S, Gomez M, Clark RM, Murray C, Ashizawa T, Bidichandani SI. The GAA triplet-repeat sequence in Friedreich ataxia shows a high level of somatic instability in vivo, with a significant predilection for large contractions. Hum. Mol. Genet. 2002;11:2175–2187. doi: 10.1093/hmg/11.18.2175. [DOI] [PubMed] [Google Scholar]
- 16.De Biase I, Rasmussen A, Monticelli A, Al-Mahdawi S, Pook M, Cocozza S, Bidichandani S. Somatic instability of the expanded GAA triplet-repeat sequence in Friedreich ataxia progresses throughout life. Genomics. 2007;90:1–5. doi: 10.1016/j.ygeno.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Krasilnikova MM, Mirkin SM. Replication stalling at Friedreich's ataxia (GAA)n repeats in vivo. Mol. Cell. Biol. 2004;24:2286–2295. doi: 10.1128/MCB.24.6.2286-2295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pollard L, Sharma R, Gomez M, Shah S, Delatycki M, Pianese L, Monticelli A, Keats B, Bidichandani S. Replication-mediated instability of the GAA triplet repeat mutation in Friedreich ataxia. Nucleic Acids Res. 2004;32:5962–5971. doi: 10.1093/nar/gkh933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim H, Narayanan V, Mieczkowski P, Petes T, Krasilnikova M, Mirkin S, Lobachev K. Chromosome fragility at GAA tracts in yeast depends on repeat orientation and requires mismatch repair. EMBO J. 2008;27:2896–2906. doi: 10.1038/emboj.2008.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shishkin AA, Voineagu I, Matera R, Cherng N, Chernet BT, Krasilnikova MM, Narayanan V, Lobachev KS, Mirkin SM. Large-scale expansions of Friedreich's ataxia GAA repeats in yeast. Mol. Cell. 2009;35:82–92. doi: 10.1016/j.molcel.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gacy A, Goellner G, Spiro C, Chen X, Gupta G, Bradbury E, Dyer R, Mikesell M, Yao J, Johnson A, et al. GAA instability in Friedreich's ataxia shares a common, DNA-directed and intraallelic mechanism with other trinucleotide diseases. Mol. Cell. 1998;1:583–593. doi: 10.1016/s1097-2765(00)80058-1. [DOI] [PubMed] [Google Scholar]
- 22.Heidenfelder B, Makhov A, Topal M. Hairpin formation in Friedreich's ataxia triplet repeat expansion. J. Biol. Chem. 2003;278:2425–2431. doi: 10.1074/jbc.M210643200. [DOI] [PubMed] [Google Scholar]
- 23.LeProust E, Pearson C, Sinden R, Gao X. Unexpected formation of parallel duplex in GAA and TTC trinucleotide repeats of Friedreich's ataxia. J. Mol. Biol. 2000;302:1063–1080. doi: 10.1006/jmbi.2000.4073. [DOI] [PubMed] [Google Scholar]
- 24.Wells R. DNA triplexes and Friedreich ataxia. FASEB J. 2008;22:1625–1634. doi: 10.1096/fj.07-097857. [DOI] [PubMed] [Google Scholar]
- 25.Potaman V, Oussatcheva E, Lyubchenko Y, Shlyakhtenko L, Bidichandani S, Ashizawa T, Sinden R. Length-dependent structure formation in Friedreich ataxia (GAA)n*(TTC)n repeats at neutral pH. Nucleic Acids Res. 2004;32:1224–1231. doi: 10.1093/nar/gkh274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandok GS, Kapoor KK, Brick RM, Sidorova JM, Krasilnikova MM. A distinct first replication cycle of DNA introduced in mammalian cells. Nucleic Acids Res. 2011;39:2103–2115. doi: 10.1093/nar/gkq903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward TR, Hoang ML, Prusty R, Lau CK, Keil RL, Fangman WL, Brewer BJ. Ribosomal DNA replication fork barrier and HOT1 recombination hot spot: shared sequences but independent activities. Mol. Cell. Biol. 2000;20:4948–4957. doi: 10.1128/mcb.20.13.4948-4957.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santamaria D, Viguera E, Martinez-Robles ML, Hyrien O, Hernandez P, Krimer DB, Schvartzman JB. Bi-directional replication and random termination. Nucleic Acids Res. 2000;28:2099–2107. doi: 10.1093/nar/28.10.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakamoto N, Chastain P, Parniewski P, Ohshima K, Pandolfo M, Griffith J, Wells R. Sticky DNA: self-association properties of long GAA.TTC repeats in R.R.Y triplex structures from Friedreich's ataxia. Mol. Cell. 1999;3:465–475. doi: 10.1016/s1097-2765(00)80474-8. [DOI] [PubMed] [Google Scholar]
- 30.Samadashwily GM, Dayn A, Mirkin SM. Suicidal nucleotide sequences for DNA polymerization. EMBO J. 1993;12:4975–4983. doi: 10.1002/j.1460-2075.1993.tb06191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baran N, Pucshansky L, Marco Y, Benjamin S, Manor H. The SV40 large T-antigen helicase can unwind four stranded DNA structures linked by G-quartets. Nucleic Acids Res. 1997;25:297–303. doi: 10.1093/nar/25.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kopel V, Pozner A, Baran N, Manor H. Unwinding of the third strand of a DNA triple helix, a novel activity of the SV40 large T-antigen helicase. Nucleic Acids Res. 1996;24:330–335. doi: 10.1093/nar/24.2.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirchmaier AL, Sugden B. Plasmid maintenance of derivatives of oriP of Epstein–Barr virus. J. Virol. 1995;69:1280–1283. doi: 10.1128/jvi.69.2.1280-1283.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gahn TA, Schildkraut CL. The Epstein–Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989;58:527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- 35.Dhar SK, Yoshida K, Machida Y, Khaira P, Chaudhuri B, Wohlschlegel JA, Leffak M, Yates J, Dutta A. Replication from oriP of Epstein–Barr virus requires human ORC and is inhibited by geminin. Cell. 2001;106:287–296. doi: 10.1016/s0092-8674(01)00458-5. [DOI] [PubMed] [Google Scholar]
- 36.Voineagu I, Surka CF, Shishkin AA, Krasilnikova MM, Mirkin SM. Replisome stalling and stabilization at CGG repeats, which are responsible for chromosomal fragility. Nat. Struct. Mol. Biol. 2009;16:226–228. doi: 10.1038/nsmb.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pelletier R, Krasilnikova M, Samadashwily G, Lahue R, Mirkin S. Replication and expansion of trinucleotide repeats in yeast. Mol. Cell. Biol. 2003;23:1349–1357. doi: 10.1128/MCB.23.4.1349-1357.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu G, Chen X, Bissler JJ, Sinden RR, Leffak M. Replication-dependent instability at (CTG) x (CAG) repeat hairpins in human cells. Nat. Chem. Biol. 2010;6:652–659. doi: 10.1038/nchembio.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakamoto N, Ohshima K, Montermini L, Pandolfo M, Wells R. Sticky DNA, a self-associated complex formed at long GAA*TTC repeats in intron 1 of the frataxin gene, inhibits transcription. J. Biol. Chem. 2001;276:27171–27177. doi: 10.1074/jbc.M101879200. [DOI] [PubMed] [Google Scholar]
- 40.Krysan P, Calos M. Replication initiates at multiple locations on an autonomously replicating plasmid in human cells. Mol. Cell. Biol. 1991;11:1464–1472. doi: 10.1128/mcb.11.3.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vashee S, Cvetic C, Lu W, Simancek P, Kelly T, Walter J. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 2003;17:1894–1908. doi: 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ku S, Soragni E, Campau E, Thomas EA, Altun G, Laurent LC, Loring JF, Napierala M, Gottesfeld JM. Friedreich's ataxia induced pluripotent stem cells model intergenerational GAATTC triplet repeat instability. Cell Stem Cell. 2010;7:631–637. doi: 10.1016/j.stem.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mirkin SM. Getting to the core of repeat expansions by cell reprogramming. Cell Stem Cell. 2010;7:545–546. doi: 10.1016/j.stem.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sha K, Boyer LA. The chromatin signature of pluripotent cells. 2009 Stem Book, ed. The Stem Cell Research Community, StemBook, doi:10.3824/stembook.1.45.1, http://www.stembook.org. [PubMed] [Google Scholar]
- 45.Imschenetzky M, Puchi M, Gutierrez S, Montecino M. Sea urchin zygote chromatin exhibit an unfolded nucleosomal array during the first S phase. J. Cell Biochem. 1995;59:161–167. doi: 10.1002/jcb.240590205. [DOI] [PubMed] [Google Scholar]
- 46.Soragni E, Herman D, Dent SY, Gottesfeld JM, Wells RD, Napierala M. Long intronic GAA*TTC repeats induce epigenetic changes and reporter gene silencing in a molecular model of Friedreich ataxia. Nucleic Acids Res. 2008;36:6056–6065. doi: 10.1093/nar/gkn604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ditch S, Sammarco MC, Banerjee A, Grabczyk E. Progressive GAA.TTC repeat expansion in human cell lines. PLoS Genet. 2009;5:e1000704. doi: 10.1371/journal.pgen.1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao J, Bacolla A, Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability and evolution. Cell. Mol. Life Sci. 2010;67:43–62. doi: 10.1007/s00018-009-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.