Abstract
Transforming growth factor beta receptor II (TGFβR-II) interacting protein 1 (TRIP-1) is a WD-40 protein that binds to the cytoplasmic domain of the TGF-β type II receptor in a kinase-dependent manner. To investigate the role of TRIP-1 in mineralized tissues, we examined its pattern of expression in cartilage, bone, and teeth and analyzed the relationship between TRIP-1 overexpression and mineralized matrix formation. Results demonstrate that TRIP-1 was predominantly expressed by osteoblasts, odontoblasts, and chondrocytes in these tissues. Interestingly, TRIP-1 was also localized in the extracellular matrix of bone and at the mineralization front in dentin, suggesting that TRIP-1 is secreted by nonclassical secretory mechanisms, as it is devoid of a signal peptide. In vitro nucleation studies demonstrate a role for TRIP-1 in nucleating calcium phosphate polymorphs. Overexpression of TRIP-1 favored osteoblast differentiation of undifferentiated mesenchymal cells with an increase in mineralized matrix formation. These data indicate an unexpected role for TRIP-1 during development of bone, teeth, and cartilage.
Keywords: bone histology, cellular localization, chondrocyte, dental tissues, extracellular matrix
WD-40 repeat containing family of proteins play important roles in cellular functions including signal transduction, mRNA processing, gene regulation, vesicular trafficking, and cell cycle regulation (Neer et al. 1994; Smith et al. 1999; Li and Roberts 2001). WDR5, a member of the WD-40 family of proteins, has been shown to regulate endochondral bone formation (Gori et al. 2006). Recently, mutations in WDR72, a WD repeat protein, caused autosomal recessive hypomaturation amelogenesis imperfecta in humans (El-Sayed et al. 2009). Another study has speculated that premature loss of permanent teeth in patients with Allgrove (4A) syndrome could be related to mutations in AAAS gene, which encodes the protein Aladin, a member of the WD family (Razavi et al. 2010). In recent studies, Gilissen et al. (2010) have reported heterozygous mutations in WDR35, another WD protein, leading to cranioectodermal dysplasia, a disorder characterized by craniosynostosis, and ectodermal and skeletal abnormalities. These studies indicate the importance of the WD-40 family of proteins in the development of bones and teeth.
Transforming growth factor beta receptor II (TGFβR-II) interacting protein-1 (TRIP-1) is a WD protein associated with TGFβR-II and plays an important role in TGF-β signaling. In this pathway, TRIP-1 binds to the cytoplasmic domain of the TGFβR-II in a kinase-dependent manner and is phosphorylated on serine and threonine residues by the receptor kinase (Chen et al. 1995). Studies have also indicated that TRIP-1 acts as a modulator of TGF-β activity in vivo by inhibiting the expression of several genes stimulated by TGF-β (Choy and Derynck 1998). During skeletal development, TRIP-1 functions by regulating osteoblast differentiation. Sheu et al. (2003) have suggested that tartrate-resistant acid phosphatase (TRAP) interacts with TRIP-1 resulting in downstream signaling by TGF-β/Smads. Thus, TRIP-1 might play a functional role in TGF-β signaling through its association with TGFβR-II.
Besides its signaling function, TRIP-1 is an essential subunit of eukaryotic elongation initiation factor 3 (eIF3; Asano et al. 1997). It is a subunit of the eIF3 complex and is also termed as eIF3i, eIF3p36, eIF3S2, or eiF3β. eIF3 is the largest of the eIFs (Behlke et al. 1986) and plays an essential role in the initiation of eukaryotic translation (Kolupaeva et al. 2005; Hinnebusch 2006). Overexpression of TRIP-1 or eIF3i in fibroblasts resulted in improved proliferation, accelerated cell cycle progression, and increased cell size (Ahlemann et al. 2006).
It has long been known that members of the TGF-β super family cytokines regulate a wide variety of biological processes, such as proliferation and cell death (Roberts and Sporn 1993); and defects in TGF-β signal transduction pathways have been known for several developmental defects (Blobe et al. 2000; Massagué et al. 2000). Abundant expression of various TGF-β isoforms is observed in bone during skeletal development, implying their importance in normal hard-tissue development and function. TGF-β and its cognate receptors have also been implicated to play an important role in the regulation of early tooth development (Chai et al. 1999). Studies by Ito et al. (2003), Sasaki et al. (2006), and Seo and Serra (2007) have shown that conditional inactivation of TGFβR-II resulted in a significant reduction in the proliferation of chondrocytes, osteoprogenitor cells, and palatal mesenchyme suggesting that TGFβR-II plays a primary role in regulating mesenchymal cell proliferation during skeletal development. Recent studies by Seo and Serra (2009) show that loss of TGFβR-II caused defects in the development of the skull vault indicating that TGFβR-II plays a critical role in intramembranous bone formation.
Thus, TRIP-1 appears to have several functions during skeletal development. In this study, we report on the developmental expression of TRIP-1 in mineralized tissues. The localization pattern of TRIP-1 would give insight into the complex molecular mechanisms underlying the development of bones and teeth.
Materials and Methods
Cell Culture
Six different cell types were used in this study. T4–4, rat preodontoblast cell line (Hao et al. 2002); MC3T3-E1, mouse preosteoblast cell line (a kind gift from Dr. Franchesi, University of Michigan); C3H10T1/2, mouse embryonic undifferentiated mesenchymal cell line; human bone marrow stem cells (HMSCs; obtained from Tulane Cancer Center); primary preodontoblasts; and primary preosteoblasts obtained from mouse day 3 postnatal pups. T4–4 cells were grown in DMEM/F-12 (Cellgro) containing 10% fetal bovine serum (Cellgro) with 1% antibiotic-antimycotic. MC3T3-E1 cells were grown in aMEM (Cellgro) supplemented with 10% FBS and 1% antibiotics. C3H10T1/2 cells were grown in BME medium (Cellgro) containing 10% FBS and 1% antibiotics. HMSCs, primary preodontoblasts, and preosteoblasts were grown in aMEM medium with 20% fetal bovine serum.
Immunohistochemistry
Embryonic day 13.5 (E13.5) and embryonic day 16.5 (E16.5) whole mouse embryos; postnatal day 1 (P1), day 3 (P3), day 5 (P5), and day 7 (P7) mouse heads; and day 20 (P20) mouse mandibles were collected. The specimens were fixed in 4% paraformaldehyde and embedded in paraffin wax. The day 20 mandibles were demineralized in buffered 10% EDTA for 7 days at 40°C and processed as above. Additionally, long bones from P1, P3, P5, and 4-week-old mice were also fixed and embedded in paraffin wax. Four-week-old long bones were demineralized in buffered 10% EDTA for 7 days at 40°C. Five-µm-thick sections were cut and deparaffinized with xylene and hydrated through graded ethanol. Only the 4-week-old long bone sections were treated with chondroitinase ABC at 0.5 U/ml in PBS for 60 min at room temperature. All the samples were then incubated in 3% H2O2 for 30 min to quench endogenous peroxidase activity and blocked with PBS containing goat serum (Vectastain ABC peroxidase kit) for 60 min at room temperature. Sections were then incubated overnight at 4C with anti-Eif3b antibody (rabbit polyclonal anti-Eif3b [sc-50357] or mouse monoclonal anti-Eif3b [sc-271539]; 1/200, Santa Cruz Biotechnology (Santa Cruz, California) or anti–Type X Collagen antibody (a kind gift from Dr. William A. Horton, Oregon Health Sciences University). Then they were incubated with biotin conjugated secondary antibody, washed 4 times in PBS, incubated with peroxidase-conjugated streptavidin (Vectastain ABC peroxidase kit), and the peroxidase was detected using a DAB kit (Vector Labs, Burlingame, CA). Rabbit secondary antibody with the omission of primary antibody was used as negative control. All experiments were performed in accordance with approved University of Illinois at Chicago animal protocol (Assurance no: A3460.01).
RNA Isolation and Northern Blot Hybridization Analysis
Total RNA was isolated from the preodontoblast T4–4 cells and preosteoblast MC3T3-E1 cells using Trizol method (Invitrogen) and resolved on 0.8% agarose gel containing formaldehyde and transferred to nylon membrane. 32P labeled rat TRIP-1 cDNA was used as a probe for northern analysis.
Expression and Purification of Recombinant TRIP-1 Protein
Recombinant TRIP-1 protein was expressed in bacteria using pGEX-4T3 system. Briefly, 973-bp fragment corresponding to the coding region of Rat TRIP-1 cDNA was PCR-amplified and cloned into Eco RI/Xho I restriction sites in pGEX4T-3 vector. This plasmid was transformed into BL-21 Escherichia coli cells and the recombinant protein was expressed as a glutathione S-transferase (GST) fusion protein and purified based on manufacturer’s protocol (GE Healthcare, Piscataway, NJ). Thrombin was used to cleave the recombinant protein from GST.
Overexpression of TRIP-1
For stable expression, rat TRIP-1 cDNA was PCR-amplified and cloned into HindIII and ApaI sites of pECFP vector (Clontech Laboratories, Mountain View, CA). Stable transfections with TRIP-1-CFP plasmid DNA were performed with Superfect Transfection reagent (Qiagen, Valencia, CA) according to the manufacturer’s protocol. Selections of the stable colonies were made with G418 sulfate (Sigma, St Louis, MO) at 0.8 mg/ml of medium. Single cell colonies were isolated by plating cells at low density. Control cells were mock transfected with “empty” vector and selected identically.
Generation of Extracellular Matrix from HMSCs
HMSCs were embedded in a collagen scaffold as described previously (Ravindran et al. 2012). The cells were cultured for 48 hr in growth media. Subsequently, differentiation media containing 100 mg/ml ascorbic acid, 10 mM b-glycerophosphate and 10 mM dexamethasone were used for culturing the cells for an additional 2 weeks. At the end of 2 weeks, the cells were treated with buffer 1 (10 mM sodium phosphate, 150 mM sodium chloride, and 0.5% triton x-100) for 30 min at 37°C in a tissue culture incubator. The buffer was then changed to buffer 2 (25mM ammonium hydroxide) and the scaffolds were incubated for 20 min at 37°C. Finally, the scaffolds were washed three times in Hanks balanced salt solution (HBSS) containing no calcium or magnesium. HMSC-derived extracellular matrix (ECM) scaffolds were then subjected to 3 freeze–thaw cycles in liquid nitrogen and 37°C cell culture incubator respectively. The scaffolds were then washed four times in HBSS, fixed in neutral buffered formalin, paraffin embedded, and sectioned (5 mm). The sections were immunostained with anti-Eif3b antibody (rabbit polyclonal anti-Eif3b [sc-50357] or mouse monoclonal anti–Eif3b [sc-271539]; 1/200, Santacruz Biotechnology) and anti-fibronectin antibody (1/200, Sigma) followed by staining with the corresponding fluorescent secondary antibodies. The sections were then imaged using a Zeiss LSM 510 Meta confocal microscope. For cell-derived ECM without the collagen scaffold, the HMSCs were grown to 100% confluency in regular medium without mineralization, and the ECM was isolated as described above.
Immunofluorescence and Confocal Imaging
All cell types used in this study were grown to 60% confluency on cover slips in 6-well plates. Cells were washed three times with PBS, fixed in 4% paraformaldehyde for 1 hr and permeabilized in PBS with 0.5% Triton-x100. The cells were then blocked with 5% bovine serum albumin (BSA) in PBS for 1 hr at room temperature and incubated with anti-Eif3b primary antibody overnight at 40°C (rabbit polyclonal anti-Eif3b [sc-50357] or mouse monoclonal anti–Eif3b [sc-271539]; 1/100, Santacruz Biotechnology). After washing 3 times with PBS, the cells were incubated for 1 hr at room temperature with the goat anti-rabbit TRITC secondary antibody. The cells were then washed extensively in PBS and mounted on glass slides and viewed under a Zeiss 510 LSM Meta confocal microscope.
Protein Isolation and Western Blotting
Total proteins were extracted from C3H10T1/2, MC3T3-E1, and T4–4 cells using M-Per reagent (Pierce, Rockford, IL). Nuclear and cytoplasmic proteins were also extracted from C3H101/2, MC3T3-E1, and T4–4 cells using NE-Per reagent (Pierce). Thirty mg of the total proteins were resolved on a 10% SDS-polyacrylamide gel under reducing conditions. After electrophoresis, the proteins were electro-transferred onto nitrocellulose membrane (Bio-Rad Laboratories, Hercules, CA), blocked with 5% nonfat milk, and probed with rabbit polyclonal anti-Eif3b (sc-50357) or mouse monoclonal anti–Eif3b (sc-271539) antibody (1/500, Santacruz Biotechnology). Tubulin was used as control to ensure equal loading of proteins. Blots were then incubated with HRP-conjugated goat anti-rabbit IgG or anti-mouse IgG secondary antibody (Sigma). After washing three times with PBS containing 0.05% Tween 20 and once with PBS, the bands were visualized by the lightening chemiluminescence reagent (Pierce, Rockford, IL).
To obtain secreted proteins, C3H10T1/2, MC3T3-E1 preosteoblast cells, and T4–4 preodontoblast cells were grown to 80% confluence in their growth medium in 100mm tissue culture dishes. The medium was subsequently changed to serum-free medium for a period of 48 hr. The medium containing the secreted proteins was collected from three 100-mm tissue culture dishes, centrifuged, and dialyzed extensively against double-deionized water. The dialyzed solution was then lyophilized and reconstituted in 500 µl of PBS. Twenty µl of the total proteins were resolved on SDS-PAGE and transferred on to nitrocellulose membrane. Media was also collected from MC3T3-E1 preosteoblast cells and T4–4 preodontoblast cells cultured in the presence of differentiation media. For these experiments, the cells were grown to complete confluence, and the medium was changed to serum-free regular media or differentiation media 48 hr before the desired time point. At the end of the time point, the medium was collected, and the cells were trypsinized and counted to ensure that the cell number did not vary significantly between the time points. Fifteen μl of the reconstituted protein samples obtained from media collected from a single flask at each time point were resolved on SDS-PAGE, and immunoblotting was performed as described previously.
Induction of Mineralization and Characterization of the Mineralized Matrices by von Kossa and Alkaline Phosphatase (ALP) Staining
Mineralization was induced with normal growth medium supplemented with 10 mM b-glycerophosphate, 100 mg/ml ascorbic acid, and 10 nM dexamethasone. The C3H10T1/2-CFPTRIP-1 stable transfected cells and C3H10T1/2-CFP mock transfected cells were allowed to undergo differentiation under mineralization conditions (28 days in culture) and fixed with 10% formalin in neutral buffer (Sigma) for 15 min. The slides were washed with distilled water and then treated with 1% AgNO3 for 1 hr, washed again with distilled water, and treated with 2.5% sodium thiosulfate for 5 min. The specimens were then examined under a light microscope and imaged. For ALP staining, the cells were fixed in ice cold methanol for 10 min and incubated with equal volumes of NBT/BCIP reagent (Bio-Rad Laboratories) for 15 to 20 min.
EDX Analysis and SEM
C3H10T1/2-CFP mock and C3H10T1/2-CFPTRIP-1 overexpressing cells were grown on 12-mm cover glass placed in 12-well tissue culture plates under mineralization conditions for 28 days. They were then washed twice with PBS and fixed in 1.5% glutaraldehyde in 0.15 M Sodium cacodylate buffer for 24 hr at 4C. After washing twice with PBS, the samples were dehydrated by passing through a series of graded ethanol solutions of 30%, 50%, 90%, and 100% for 10 min each. The samples were finally dehydrated by immersing them in a solution of Hexamethyldisilazane (HMDS) for 10 min followed by air-drying inside a tissue culture hood. Elemental analysis was directly conducted on a scanning electron microscope (Hitachi S-3000N) equipped with an energy dispersive X-ray device without any further sample processing. They were finally imaged using a Hitachi S-3000N Variable Pressure-SEM (Hitachi High-Technologies Canada, Inc., Canada). JOEL JSM 6320F field emission scanning electron microscope (FESEM) was used to image collagen fibrils and mineralized nodules in the matrix.
In Vitro Nucleation Studies
In vitro nucleation experiments were conducted as previously published (He and George 2004). Briefly, pretreated cover glass was adsorbed with TRIP-1 (20 mg of recombinant TRIP-1 protein in 100 m1 of 25-mm sodium bicarbonate buffer) and incubated overnight at 4°C in a humidified chamber. Twenty mg of BSA-coated cover glass served as controls. After incubation, the samples were rinsed with double-deionized water and placed into a channel connecting two halves of an electrolyte cell, one compartment containing calcium buffer (165 mm NaCl, 10 mm HEPES, 2.5 mm CaCl2, pH 7.4) and the other phosphate buffer (165 mm NaCl, 10 mm HEPES, 1 mm KH2PO4, pH 7.4). A small electric current of 1 mA was passed through the system to facilitate even distribution of the ions on the cover glass coated with the proteins. The buffers were changed regularly to maintain a constant pH. At the end of 14 days, the cover glass was removed, washed with double-deionized water, dehydrated in graded ethanol solutions, and dried with HMDS. The samples were then imaged using a Hitachi S-3000N variable pressure scanning electron microscope. Energy dispersive X-ray (EDX) analysis was also performed for all samples to analyze the presence of calcium and phosphate deposits.
Results
Expression of TRIP-1 During Embryonic Development
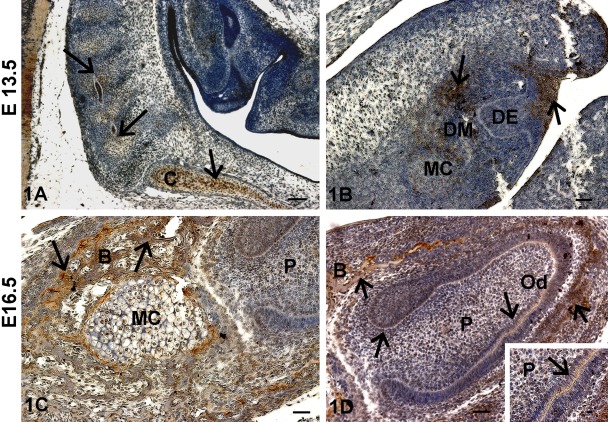
Immunohistochemical analysis at E13.5 showed highly localized expression of TRIP-1 in the chondrocytes of the cartilaginous primordium of the basioccipital bone in the rostral part of the vertebral column with the forming cervical vertebrae (Fig. 1A). Figure 1B shows the bud stage of tooth development and TRIP-1 was detected in the oral epithelium and in the condensing neural-crest-derived ectomesenchymal cells (Higher Magnification, Supplement Fig. 1). In the sagittal section of E16.5 mouse head, TRIP-1 was present in the osteoblasts of the alveolar bone and in the chondrocytes of the Meckel’s cartilage (Fig. 1C). Figure 1D shows predominant localization of TRIP-1 in the dental pulp cells, basement membrane, osteoblasts, and alveolar bone matrix in the developing incisor at E16.5. Expression was also observed in the stem cell niche of the cervical loop at the apical end of the developing incisor.
Figure 1.
Localization of TRIP-1 during embryonic development of bone and teeth. (A) Localization of TRIP-1 in the chondrocytes of the rostral part of the vertebral column with the forming cervical vertebrae. (B) Localization of TRIP-1 in the bud stage of tooth development. (C) Localization of TRIP-1 in Meckel’s cartilage (MC). (D) Localization of TRIP-1 in the developing incisors and alveolar bone at E16.5. Insert shows higher magnification of 1D showing TRIP-1 localization in the basement membrane. Black arrows in all images represent localization of TRIP-1. DM = dental mesenchyme; DE = dental epithelium; B = bone; P = dental pulp cells; C = chondrocytes in the cartilaginous primordium of the basioccipital bone; Od = odontoblasts. A–D bars = 50 µm; inset bar = 10 µm.
Expression of TRIP-1 in Postnatal Developing Tooth
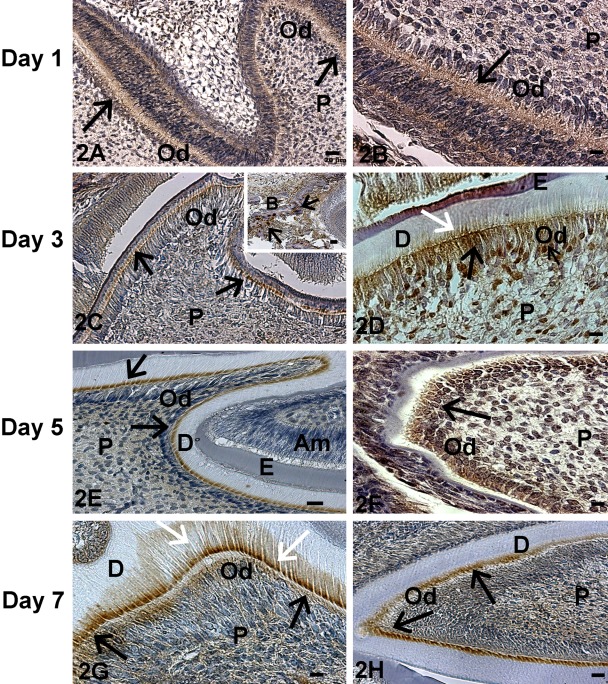
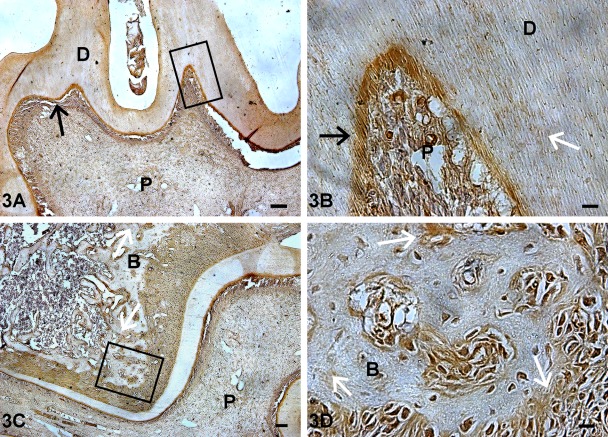
At postnatal day 1, TRIP-1 was localized in the basement membrane of the incisors. In contrast, there was low expression in the molars (Fig. 2A,B). At day 3, localized expression was seen in the odontoblasts of both the developing molar and incisor (Fig. 2C,D). Nuclear localization of TRIP-1 was observed at this developmental stage (Fig. 2C,D). Interestingly, distinct localization of TRIP-1 was observed at the mineralization front during the assembly of the calcified dentin matrix (white arrow in Fig. 2D). TRIP-1 was also localized in the surrounding alveolar bone matrix (Fig. 2C inset). With further development, at day 5 (Fig. 2E,F) and day 7 (Fig. 2G,H) localization of TRIP-1 was observed in the odontoblasts of both the molars and incisors, with predominant expression in the dentin matrix at day 7 (Fig. 2G, white arrow). At day 20, persistent expression of TRIP-1 was observed in the odontoblasts and also localized in the dental pulp cells (Fig. 3A,B). Abundant expression was also observed in the osteoblasts of the alveolar bone (Fig. 3C,D). Thus, TRIP-1 maintains high expression levels in the dental papilla at the bud stage and during its differentiation into fully functional odontoblasts.
Figure 2.
Localization of TRIP-1 in postnatal developing teeth. Localization of TRIP-1 in developing molars of postnatal day 1 (A), day 3 (C), day 5 (E), and day 7 (G) of tooth development. Localization of TRIP-1 in incisors of postnatal day 1 (B), day 3 (D), day 5 (F), and day 7 (H) of tooth development. Black arrows point to expression in the odontoblasts. White arrows represent expression in the ECM. P = dental pulp cells; B = bone; Od = odontoblasts; Am = ameloblasts; D = dentin; E = enamel. A,E,H bars = 20 µm; B,D,F,G bars = 10 µm; inset bar = 20 µm.
Figure 3.
Localization of TRIP-1 in postnatal day 20 mouse incisor and surrounding alveolar bone. (A,C) Localization of TRIP-1 in the dentin matrix of the molars and the alveolar bone of postnatal day 20 mouse mandible. (B,D) Enlarged images show the presence of TRIP-1 in the mineralizing dentin matrix and in the alveolar bone at day 20. Black arrows point to expression in the odontoblasts. White arrows represent expression in the ECM. P = dental pulp cells; B = bone; D = dentin. A bar = 100 µm; B,D bars = 10 µm; C bar = 50 µm.
Expression of TRIP-1 in Developing Mouse Femurs
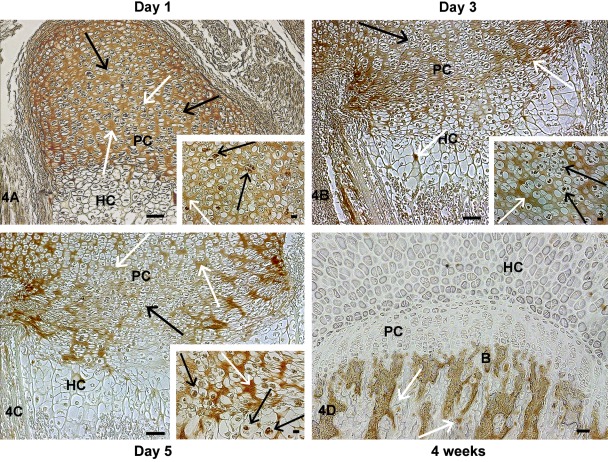
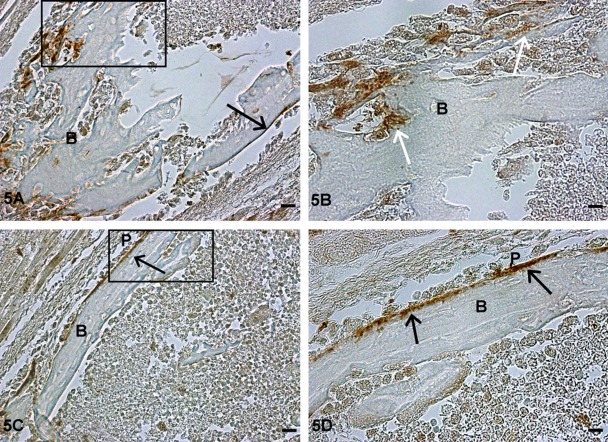
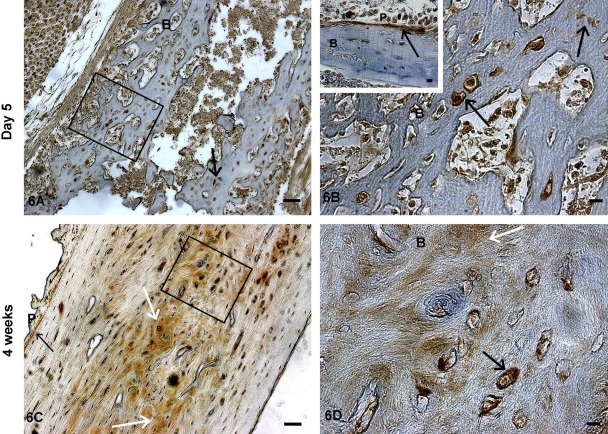
We then examined the localization of TRIP-1 during endochondral ossification by analysis of various skeletal elements at different developmental stages. At postnatal day 1, TRIP-1 expression was detected in the extracellular matrix of the proliferating chondrocytes and also localized in the nucleus of some proliferating cells, but no expression was observed in the hypertrophic chondrocytes of the growth plate (Fig. 4A). Increased expression of TRIP-1 in the cartilage matrix was observed at day 3 with some localization in the periosteum (Fig. 4B, 5A–D). The level of expression was more widespread in the chondrocytic matrix at postnatal day 5 with localization in prehypertrophic extracellular matrix and in the periosteum (Fig. 4C, 6A,B). Interestingly, at 4 weeks of development, no expression of TRIP-1 was seen in the proliferating cells and in the cartilage matrix (Fig. 4D). However, predominant expression was seen in the bone spicules of the primary spongiosa, osteocytes, and mineralized matrix of cortical bone (Fig. 6C,D; Supplement Figs 2-4).
Figure 4.
Localization of TRIP-1 in the femoral growth plate region during early endochondral ossification. Localization of TRIP-1 in the growth plate region in the long bone sections of postnatal day 1 (A), day 3 (B), day 5 (C), and 4-week-old (D) mice. Black arrows represent expression in chondrocytes. White arrows represent expression in the ECM. Inset shows higher magnification images of TRIP-1 expression in nucleus of the chondrocytes. HC = hypertrophic chondrocytes; PC = proliferating chondrocytes; B = bone. A–C bars = 50 µm; D bar = 100 µm; inset bar = 10 µm.
Figure 5.
Localization of TRIP-1 in the long bone of postnatal day 3 developing mice. (A,C) Localization of TRIP-1 in the preosteoblasts and periosteun of postnatal day 3 mouse long bone. (B,D) Higher magnification of the boxed region showing the presence of TRIP-1 in the mineralizing bone matrix and in the periosteum at day 3. Black arrows represent expression of TRIP-1 in periosteum. White arrows represent expression of TRIP-1 in preosteoblasts. B = bone; P = periosteum. A,C bars = 20 µm; B,D bars = 10 µm.
Figure 6.
Localization of TRIP-1 in postnatal day 5 and 4-week-old long bones of developing mice. Localization of TRIP-1 in the long bone sections of postnatal day 5 (A) and 4-week-old (C) mice. (B, D) Enlarged images show the presence of TRIP-1 in the mineralizing matrix and in osteocytes, osteoblasts, and periosteum. Insert in Fig. 6B shows the expression of TRIP-1 in the peroisteum. Black arrows point to expression of TRIP-1 in the bone osteocytes and periosteum. White arrows represent expression of TRIP-1 in the ECM. B = bone; P = periosteum. A,C bars = 50 µm; B,D, and inset bars = 10 µm.
Subcellular Localization of TRIP-1
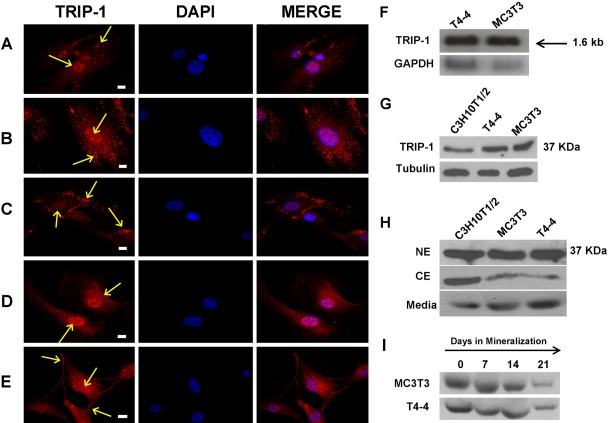
Subcellular localization of TRIP-1 was analyzed by immunocytochemical analysis in different cell types, namely primary pulp cells and calvarial cells from 3-day pups, HMSCs, T4–4, and MC3T3 cells. In all cell types studied, TRIP-1 was predominantly expressed in the cytoplasm, although some expression was observed in the nucleus and on the plasma membrane (Fig. 7A–E). Northern blot analysis was performed to identify TRIP-1 transcript in T4–4 preodontoblasts and MC3T3-E1 preosteoblasts (Fig. 7F). A single transcript of 1.6 kb was detected in both cell types.
Figure 7.
Immunolocalization of TRIP-1 in different cell types. Figures 7A–E represent confocal images of the immunolocalization of TRIP-1 in different cell types: (A) primary odontoblasts; (B) primary osteoblasts; (C) human marrow stromal cells; (D) T4–4 preodontoblasts; and (E) MC3T3-E1 preosteoblast cells. Localization of TRIP-1 is represented by yellow arrows. Scale bars = 10 µm. (F) Northern blotting analysis of T4–4 preodontoblasts and MC3T3-E1 preosteoblast cells showing expression of TRIP-1. The size of TRIP-1 transcript obtained was 1.6 kb. (G) Western blot analysis of total protein lysates from C3H10T1/2 undifferentiated mesenchymal cells, T4–4 preodontoblasts, MC3T3-E1 preosteoblasts with TRIP-1 antibody showing expression of TRIP-1 at 37 KDa. (H) Western blot analysis of nuclear, cytoplasmic, and the secretome of the above 3 cell types shows distinct presence of TRIP-1 at 37 KDa. (I) Western blot analysis of proteins isolated from the secretory pool before (day 0) and after inducing differentiation (days 7, 14, and 21) for both T4–4 and MC3T3 cells showing the presence of TRIP-1. A-E TRIP-1, DAPI, and MERGE bars = 10 µm.
Protein expression was confirmed by western blotting of the total proteins isolated from T4–4, MC3T3-E1, and C3H10T1/2 cells. A distinct band at 37 kDa was observed (Fig. 7G). Tubulin was used as a control to demonstrate equal loading of total protein. To further confirm the subcellular localization of TRIP-1, nuclear and cytoplasmic proteins were extracted from the above-mentioned three cell types. TRIP-1 expression was observed more in the nuclear fractions when compared to the cytoplasmic fractions (Fig. 7H). Since expression of TRIP-1 was observed in the matrix during tooth development, we performed Western blot analysis on the proteins isolated from the secretory pool before and after differentiation for both T4–4 and MC3T3 cells. Results in Fig. 7I confirmed that TRIP-1 is indeed secreted.
Expression of TRIP-1 in the Extracellular Matrix
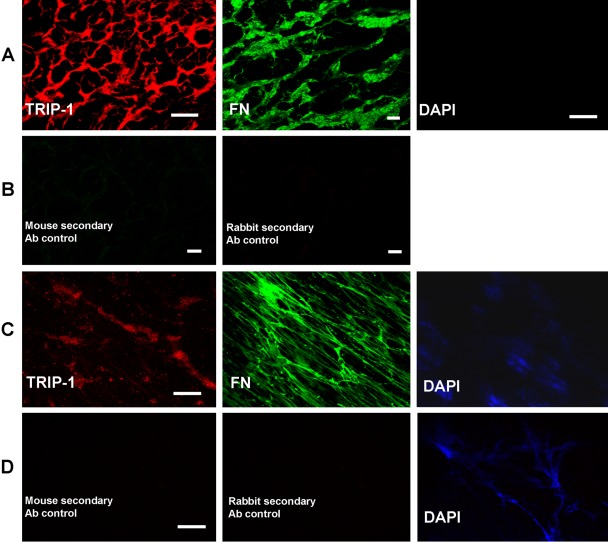
A signal peptide is absent in TRIP-1. However, expression was observed in the matrix and in the secretome of various cell lines studied. To further confirm that TRIP-1 is indeed a secretory protein, HMSCs were differentiated on a 3-D collagen scaffold, and the extracellular matrix secreted by the cells were immunostained with TRIP-1 antibody. Confocal images of the ECM showed TRIP-1 localized in the HMSC matrix (Fig. 8A). Fibronectin, a predominant ECM protein was used as a positive control. Negative staining for DAPI shows the absence of intact cells or any nuclear material. Also both rabbit and mouse secondary antibodies with the omission of primary antibodies were used as negative controls (Fig. 8B). These results were further confirmed by performing the same experiments on the ECM isolated directly from HMSCs. Results in Fig. 8C show the presence of TRIP-1 in the ECM secreted by HMSCs. Absence of intact cells was confirmed by staining for the cell nuclei with DAPI. Fibronectin was used as a positive control and secondary antibodies with the omission of primary antibodies were used as negative control (Fig. 8D). Results from the above experiments suggest that TRIP-1 is indeed a secretory protein and is probably secreted by nonclassical secretory method in the absence of a signal peptide.
Figure 8.
Expression of TRIP-1 in the ECM of HMSCs. (A) Expression of TRIP-1 and Fibronectin in the ECM secreted by differentiating HMSCs grown on scaffolds. (B) Secondary antibody negative controls. (C) Expression of TRIP-1 and Fibronectin in the ECM secreted by HMSCs. (D) Secondary antibody negative controls. A TRIP-1 and DAPI bars = 20 µm; A FN bars = 10 µm; B bars = 10 µm; C and D bars = 20 µm.
Overexpression of TRIP-1 Induces Cellular Differentiation and Formation of Mineralized Nodules
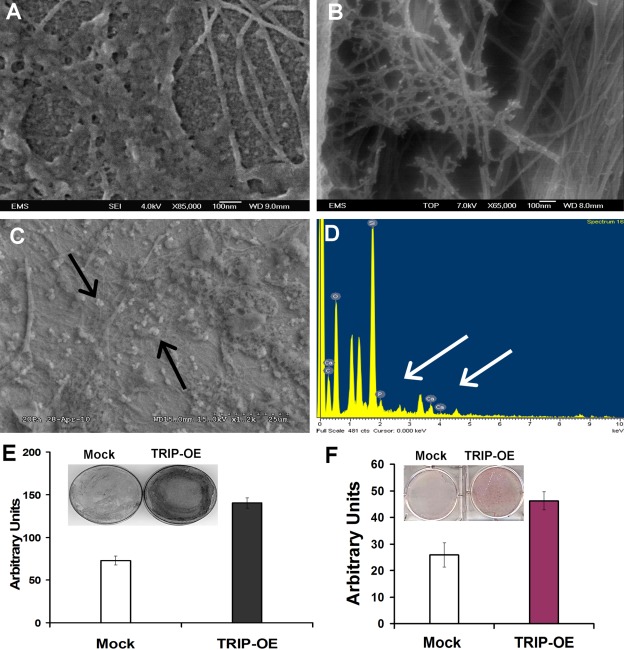
To gain insight into the role of TRIP-1 in the ECM, a stable cell line expressing TRIP-1 as a CFP-fusion protein was generated. The C3H10T1/2-TRIP-1 overexpressing cells and mock cells (with the empty vector) were cultured in the presence of osteogenic differentiation medium. FESEM imaging of the TRIP-1 overexpressed cells showed the presence of abundant collagen fibrils with mineralized nodules (Fig. 9B). SEM analysis in Fig. 9C depicts the calcified matrix deposited by TRIP-1 overexpressing cells, and Fig. 9D is the corresponding EDX analysis confirming the presence of calcium and phosphate in the deposits (white arrows). Figure 9A represents the matrix deposited by mock cells. To further confirm the differentiation and formation of mineralized matrix by TRIP-1 overexpressing cells, we performed von Kossa and ALP staining. Results in Fig. 9E and 9F demonstrate that von Kossa and ALP staining were significantly higher in TRIP-1 overexpressing cells when compared with mock cells. Taken together, these data suggest that TRIP-1 might have a functional role during matrix mineralization.
Figure 9.
Analysis of the extracellular matrix secreted by TRIP-1 overexpressing cells. (A) Scanning electron micrograph of control cells showing deposition of proteinaceous ECM with no mineral deposits. (B,C) Scanning electron micrograph of the matrix secreted by C3H10T1/2-TRIP-1 overexpressing cells subjected to in vitro mineralization for 28 days. Note deposition of collagen fibrils containing numerous mineral deposits (black arrows). (D) EDX analysis of the mineral deposits formed by C3H10T1/2-TRIP-1 overexpressing cells grown in differentiation media. Calcium and phosphate peaks are marked by white arrows. (E and F) von Kossa and ALP staining of C3H10T1/2-mock and C3H10T1/2-TRIP-1 overexpressing cells and their corresponding quantification.
rTRIP-1 Induces Nucleation of Calcium Phosphate Polymorphs
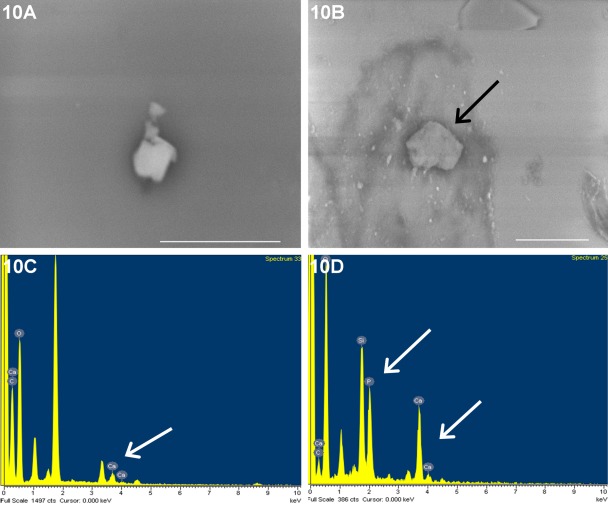
To demonstrate the involvement of TRIP-1 in mineralization, in vitro nucleation studies were performed with rTRIP-1. Figure 10B represents SEM image of mineral deposits obtained from in vitro nucleation experiments performed for 14 days on TRIP-1 coated glass plates, and their corresponding EDX analysis is shown in Fig. 10D. The ratio of calcium and phosphate was determined to be 0.83 (SEM = 0.19, n=4). This suggests that the calcium phosphate polymorphs were amorphous. BSA-coated cover glass was used as a control (Fig. 10A), and the EDX analysis performed on the deposits is shown in Fig. 10C.
Figure 10.
In vitro nucleation of calcium phosphate polymorph by TRIP-1. (B,D) Electron micrograph of calcium phosphate deposits on rTRIP-1 coated cover glass subjected to in vitro nucleation and its corresponding EDX analysis respectively. B scale bar = 10 µm. White arrows point to calcium and phosphate peaks. 10A and 10C are the images of deposits obtained from BSA coated cover glass used as a control and its EDX analysis respectively. Bars = 5μm.
Discussion
Type II TGF-β receptor interacting protein-1 (TRIP-1) is a member of a family of structurally conserved proteins, the WD-40 repeat proteins (Chen et al. 1995). The WD-40 proteins contain 4 or more copies of a conserved Trp-Asp motif, the so-called WD-40 repeat, which forms a scaffold for binding other proteins (Neer and Smith 1996). TRIP-1 has been identified as a phosphorylation target of the TGFβR-II kinase during in vitro studies (Chen et al. 1995) and also as a functional component of eukaryotic translation initiator factor 3 (eiF3) multiprotein complexes (Asano et al. 1997). Choy and Derynck (1998) have reported modulation of Smad pathway by TRIP-1, and Sheu et al. (2003) have demonstrated the same effect in osteoblasts. Recent work has shown that loss of TRIP-1 in lung epithelial cells dramatically increases the ability of these cells to undergo epithelial-mesenchymal transformation in response to TGF-β1 treatment (Perez et al. 2011). In this study, we characterized the physiological expression of TRIP-1 during bone and teeth development to determine if TRIP-1 plays an important role in a biological process such as matrix mineralization.
Immunohistochemical analysis shows the localization of TRIP-1 in several cell types responsible for mineralized matrix formation in the developing mouse embryo. Tooth development takes place by the reciprocal interactions and controlled programming between the oral epithelium and the neural-crest-derived mesenchyme. Signals transmitted by the epithelial cells promote condensation of the mesenchymal cells called the dental papilla. At the bud stage of tooth development, TRIP-1 was expressed in both the epithelial cells of the oral ectoderm and specifically in the dental papilla. With further development, the dental papilla differentiates to form the odontoblast cells that will initially produce the uncalcified predentin matrix and subsequently mineralized dentin, while the dental lamina differentiates to form the ameloblasts that will produce enamel. Expression of TRIP-1 was observed in both ameloblasts and odontoblasts until day 3 in the molars. At later developmental stages, TRIP-1 is expressed only by the odontoblasts. As the incisors are continuously erupting, expression of TRIP-1 was seen in both odontoblasts and ameloblasts. Undifferentiated pulp cells show lower expression levels of TRIP-1 during early stages and higher expression with further development. These observations suggest that TRIP-1 might play a regulatory role in the differentiation of both ameloblasts and odontoblasts and perhaps might participate in the crosstalk between these two cell types.
An interesting observation was the localization of TRIP-1 in the mineralizing dentin matrix. At day 3, the odontoblasts are fully polarized and secrete type I collagen and several noncollagenous proteins necessary for matrix mineralization. During this process, TRIP-1 was localized at the mineralization front where the first crystals of calcium phosphate are deposited. This suggests that TRIP-1 might play a role in matrix mineralization. This function was demonstrated by in vitro nucleation assay, which demonstrated that TRIP-1 could bind calcium.
Skeletal development in vivo occurs via two major processes, namely intramembranous and endochondral ossification. Intramembranous ossification occurs by direct differentiation of mesenchymal cells into osteoblasts. TRIP-1 was expressed in the osteoblasts of the intramembranous bone as early as E16.5. Predominant localization was seen even at day 20 in preosteoblasts and differentiated osteoblasts. Endochondral bone formation is a complex and tightly regulated process that involves programmed proliferation and maturation of chondrocytes followed by terminal differentiation, hypertrophy, and replacement of cartilage by bone. During endochondral bone formation, TRIP-1 was localized in the periosteum, osteoblasts, and bone marrow at day 3 and 5; while at 4 weeks of development, increased expression was seen in the bone matrix and in osteocytes. Based on the physiological expression pattern, it is possible that TRIP-1 might play a role in both intramembranous and endochondral bone formation.
During embryogenesis, condensations of mesenchymal cells form, within which chondrocytes develop, proliferate, and differentiate to form a cartilage template that contains distinct zones of resting, proliferative, and hypertrophic chondrocytes; and these cells are spatially arranged (Seo and Serra 2007). Chondrogenesis is the earliest step in endochondral bone formation. It begins with the condensation of undifferentiated mesenchyme, leading to increase in cell density. These undifferentiated progenitor cells differentiate into chondrocytes. TRIP-1 was localized at E13.5 specifically in the chondrocytes of the vertebral body and in the chondrocytes of the cartilaginous primordium of the basioccipital bone. No staining was present in the intervertebral disc. During endochondral ossification, TRIP-1 expression was controlled spatially and temporally. In the epiphyseal growth plate, proliferating chondrocytes undergo maturation and hypertrophic differentiation. At day 1 and 3, localization of TRIP-1 was seen in the extracellular matrix of the proliferating chondrocytes. With further development, TRIP-1 signal disappears from the cartilage matrices of proliferating and prehypertrophic chondrocytes and was specifically seen in the bone spicules of the ossification zone. In the craniofacial region, chondrocytes of the Meckel’s cartilage also expressed TRIP-1. Based on the localization pattern, it could be concluded that TRIP-1 is required for chondrocyte proliferation and differentiation during the formation of axial and appendicular skeleton.
Subcellular localization of TRIP-1 in primary osteoblasts, odontoblasts, and cell lines showed specific localization patterns. Localization at the plasma membrane suggests its probable participation in the TGF-β signaling pathway. Localization of TRIP-1 in the cytoplasm suggests its involvement in the protein translational machinery, whereas a functional role for nuclear TRIP-1 is yet to be determined.
In this study, an interesting observation is the localization of TRIP-1 in the extracellular matrix. A classical signal peptide is absent in TRIP-1; however, Western blot analysis performed on total proteins isolated from the secretome of C3H10T1/2, T4–4, and MC3T3-E1 showed the presence of TRIP-1. TRIP-1 in the ECM was also confirmed by immunohistochemical analysis of the HMSC-derived ECM. In a recent study, several proteins have been identified in the secretome of human mesenchymal stem cells that do not have a signal peptide (Chiellini et al. 2008). It was suggested that cells secrete exosomes as a mechanism to extrude some proteins. Thus, TRIP-1 could be secreted to the ECM by such nonclassical secretory mechanisms.
Presence of TRIP-1 in the ECM and at the mineralization front led us to investigate its function as a calcium-binding protein. SEM analysis of the matrix deposited by C3H10T1/2 cells overexpressing TRIP-1 grown under differentiation conditions, showed deposition of mineralized deposits on fibrillar collagen. Intense von Kossa and alkaline phosphatase staining confirmed that TRIP-1 might play a functional role in osteogenic differentiation and mineralization of the extracellular matrix. In vitro nucleation experiments confirmed that immobilized TRIP-1 had the potential to nucleate calcium phosphate polymorphs.
In conclusion, based on the temporal and spatial expression pattern of TRIP-1 during bone and teeth development, it is possible that TRIP-1 might reveal a previously unknown function. We envisage TRIP-1 to function in osteoblast differentiation, perhaps through the canonical TGF-β signaling pathway, whereas its presence in the ECM suggests that it is a secreted protein and can be involved in the mineralization of the extracellular matrix.
Acknowledgments
We are grateful to Dr. Qi Gao for providing assistance with isolation of primary osteoblast and odontoblast cells.
Footnotes
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by National Institutes of Health Grant DE 11657 and the Brodie Endowment Fund.
Supplementary material for this article is available on the Journal of Histochemistry & Cytochemistry Web site at http://jhc.sagepub.com/supplemental.
References
- Ahlemann M, Zeidler R, Lang S, Mack B, Munz M, Gires G. 2006. Carcinoma-associated eIF3i overexpression facilitates mTOR-dependent growth transformation. Mol Carcinog. 45:957–967 [DOI] [PubMed] [Google Scholar]
- Asano K, Kinzy TG, Merrick WC, Hershey JW. 1997. Conservation and diversity of eukaryotic translation initiation factor eIF3. J Biol Chem. 272:1101–1109 [DOI] [PubMed] [Google Scholar]
- Behlke J, Bommer UA, Lutsch G, Henske A, Bielka H. 1986. Structure of initiation factor eIF-3 from rat liver: hydrodynamic and electron microscopic investigations. Eur J Biochem. 157:523–530 [DOI] [PubMed] [Google Scholar]
- Blobe GC, Schiemann WP, Lodish HF. 2000. Role of transforming growth factor β in human disease. N Engl J Med. 342:1350–1358 [DOI] [PubMed] [Google Scholar]
- Chai Y, Zhao J, Mogharei A, Xu B, Bringas P, Jr, Shuler C, Warburton D. 1999. Inhibition of transforming growth factor-β type II receptor signaling accelerates tooth formation in mouse first branchial arch explants. Mech Dev. 86:63–74 [DOI] [PubMed] [Google Scholar]
- Chen RH, Miettinen PJ, Maruoka EM, Choy L, Derynck R. 1995. A WD-domain protein that is associated with and phosphorylated by the type II TGF-beta receptor. Nature. 377:548–552 [DOI] [PubMed] [Google Scholar]
- Chiellini C, Cochet O, Negroni L, Samson M, Poggi M, Ailhaud G, Alessi MC, Dani C, Amri EZ. 2008. Feb. Characterization of human mesenchymal stem cell secretome at early steps of adipocyte and osteoblast differentiation. BMC Mol Biol. 9:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy L, Derynck R. 1998. The type II transforming growth factor (TGF)-beta receptor-interacting protein TRIP-1 acts as a modulator of the TGF-beta response. J Biol Chem. 273:31455–31462 [DOI] [PubMed] [Google Scholar]
- El-Sayed W, Parry DA, Shore RC, Ahmed M, Jafri H, Rashid Y, Al-Bahlani S, Al Harasi S, Kirkham J, Inglehearn CF, et al. 2009. Mutations in the beta propeller WDR72 cause autosomal-recessive hypomaturation amelogenesis imperfecta. Am J Hum Genet. 85:699–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilissen G, Arts H, Hoischen A, Spruijt L, Mans DA, Arts P, Van Lier B, Steehouwer M, Van Reeuwijk J, Kant SG, et al. 2010. Exome sequencing identifies WDR35 variants involved in Sensenbrenner syndrome. Am J Hum Genet. 87:418–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gori F, Friedman LG, Demay MB. 2006. Wdr5, a WD-40 protein, regulates osteoblast differentiation during embryonic bone development. Dev Biol. 295:498–506 [DOI] [PubMed] [Google Scholar]
- Hao J, Narayanan K, Ramachandran A, He G, Almushayt A, Evans C, George A. 2002. Odontoblast cells immortalized by telomerase produce mineralized dentin-like tissue both in vitro and in vivo. J Biol Chem. 277:19976–19981 [DOI] [PubMed] [Google Scholar]
- He G, George A. 2004. Dentin matrix protein 1 immobilized on type I collagen fibrils facilitates apatite deposition in vitro. J Biol Chem. 279:11649–11656 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AW. 2006. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 31:553–560 [DOI] [PubMed] [Google Scholar]
- Ito Y, Yeo JY, Chytil A, Han J, Bringas P, Jr, Nakajima A, Shuler CF, Moses HL, Chai Y. (2003). Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development.130:5269–5280 [DOI] [PubMed] [Google Scholar]
- Kolupaeva VG, Unbehaun A, Lomakin IB, Hellen CU, Pestova TV. 2005. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA. 11:470–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Roberts R. 2001. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell. Mol. Life Sci. 58:2085–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J, Blain SW, Roger SL. (2000). TGF-β signaling in growth control, cancer, and heritable disorders. Cell. 103:295–309 [DOI] [PubMed] [Google Scholar]
- Neer EJ, Schmidt CJ, Nambudripad R, Smith TF. 1994. The ancient regulatory-protein family of WD-repeat proteins. Nature. 371:297–300 [DOI] [PubMed] [Google Scholar]
- Neer EJ, Smith TF. 1996. G protein heterodimers: New structures propel new questions. Cell. 84:175–178 [DOI] [PubMed] [Google Scholar]
- Perez RE, Navarro A, Rezaiekhaligh MH, Mabry SM, Ekekezie II. 2011. TRIP-1 regulates TGF-β1-induced epithelial-mesenchymal transition of human lung epithelial cell line A549. Am J Physiol Lung Cell Mol Physiol. 300:L799–L807 [DOI] [PubMed] [Google Scholar]
- Ravindran S, Gao Q, Kotecha M, Magin RL, Karol S, Bedran-Russo A, George A. 2012. Biomimetic extracellular matrix-incorporated scaffold induces osteogenic gene expression in human marrow stromal cells. Tissue Eng. 18:295–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razavi Z, Mehdi Taghdiri M, Eghbalian F, Bazzazi N. 2010. Premature loss of permanent teeth in Allgrove (4A) syndrome in two related families. Iran J Pediatr. 20:101–106 [PMC free article] [PubMed] [Google Scholar]
- Roberts AB, Sporn MB. 1993. Physiological actions and clinical applications of transforming growth factor-beta (TGF-beta). Growth Factors. 8:1–9 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Ito Y, Bringas P, Jr, Chou S, Urata MM, Slavkin H, Chai Y. 2006. TGFbeta-mediated FGF signaling is crucial for regulating cranial neural crest cell proliferation during frontal bone development. Development. 133:371–381 [DOI] [PubMed] [Google Scholar]
- Seo HS, Serra R. 2007. Deletion of Tgfbr2 in Prx1-cre expressing mesenchyme results in defects in development of the long bones and joints. Dev Biol. 310:304–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Serra R. 2009. Tgfbr2 is required for development of the skull vault. Dev Biol. 334:481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu T, Schwarz EM, Martinez DA, O’Keefe RJ, Rosier RN, Zuscik MJ, Puzas JE. 2003. A phage display technique identifies a novel regulator of cell differentiation. J Biol. Chem. 278:438–443 [DOI] [PubMed] [Google Scholar]
- Smith TF, Gaitatzes C, Saxena K, Neer EJ. 1999. The WD repeat: a common architecture for diverse functions. Trends Biochem Sci. 24:181–185 [DOI] [PubMed] [Google Scholar]