Background: Sulforaphane is an important cancer preventative in skin cancer.
Results: SFN induces p21Cip1 via a p53-dependent mechanism that involves p53 stabilization.
Conclusion: p21Cip1 expression is differentially regulated in normal keratinocytes versus cancer cells.
Significance: SFN treatment will slow proliferation of normal epidermal keratinocytes but will cause resident cancer cell apoptosis.
Keywords: Cell Proliferation, Gene Expression, Keratinocytes, p53, Signal Transduction, Sp1
Abstract
Sulforaphane (SFN) is an important cancer preventive agent derived from cruciferous vegetables. We show that SFN treatment suppresses normal human keratinocyte proliferation via a mechanism that involves increased expression of p21Cip1. SFN treatment produces a concentration-dependent increase in p21Cip1 promoter activity via a mechanism that involves stabilization of the p53 protein leading to increased p53 binding to the p21Cip1 promoter p53 response elements. The proximal p21Cip1 promoter GC-rich Sp1 factor binding elements are also required, as the SFN-dependent increase is lost when these sites are mutated. SFN treatment increases Sp1 binding to these elements, and the response is enhanced in the presence of exogenous Sp1 and reduced in the presence of ΔN-Sp3. CpG island methylation alters p21Cip1 promoter activity some systems; however, expression in SFN-treated keratinocytes does not involve changes in proximal promoter methylation. The promoter is minimally methylated, and the methylation level is not altered by SFN treatment. This study indicates that SFN increases p21Cip1 promoter transcription via a mechanism that involves SFN-dependent stabilization of p53 and increased p53 and Sp1 binding to their respective response elements in the p21Cip1 promoter. These results are in marked contrast to the mechanisms observed in skin cancer cell lines and suggest that SFN may protect normal keratinocytes from damage while causing cancer cells to undergo apoptosis.
Introduction
Sulforaphane (SFN)2 is a naturally occurring isothiocyanate derived from cruciferous vegetables. It is an important candidate cancer preventive agent that has activity both in cultured cells and animal models (1–3). SFN is particularly appealing as a preventive agent because it is bioactive in many systems and displays relatively high bioavailability (4). SFN inhibits histone deacetylase activity (2, 5, 6) and regulates xenobiotic metabolism and apoptosis (7–9, 10). SFN stimulates Nrf2-dependent induction of phase I and phase II genes involved in xenobiotic metabolism (9, 11) and increases proteasome subunit level via an Nrf2-dependent mechanism (10).
SFN also influences the cell cycle. Cyclin-dependent kinases (cdk2, cdk4, and cdk6) and cyclin-dependent kinase inhibitors (p27, p21Cip1, etc.) are key enzymes that control cell cycle progression. As cells progress through the cell cycle the cyclins (cyclin A, D, E, etc.) associate with their respective cyclin-dependent kinase subunits (D-type cyclins associate with cdk4 and cdk6, and cyclin E associates with cdk2) (12). The complexes then enter the nucleus where they are activated by phosphorylation (12) and in turn phosphorylate retinoblastoma proteins. Phosphorylation of pRB, for example, leads to release of E2F from the pRB-E2F complex, and free E2F activates expression of genes that regulate entry into S phase (12).
Cyclin-dependent kinase inhibitors suppress cell proliferation by inhibiting activity of cyclin-cdk complexes (12). p21Cip1, for example, interacts with multiple cyclin-cdk complexes, and overexpression of p21Cip1 inhibits proliferation by inhibiting activity of the cyclin D-cdk4/6 and cyclin E-cdk2 complexes (13). Nuclear p21Cip1 is the active form of this protein (12). p21Cip1 level is increased during cell differentiation and in response to oxidative stress and DNA damage (14–17), and transcriptional, posttranscriptional, and epigenetic mechanisms have been described (6, 14–20). In epidermis, p21Cip1 levels increase in response to cell stress, and it is heterogeneously expressed in skin cancer cells (14, 21–24). Of particular interest is the fact that p21Cip1 knock-out results in enhanced tumor formation (23).
Regulation of p21Cip1 transcription is complicated by the fact that it has a key role in cell cycle and so many factors control p21Cip1 expression (15). The p21Cip1 promoter consists of proximal and distal regulatory elements. The proximal elements include GC-rich elements that bind Sp1 and Kruppel-like transcription factors (26–32), whereas the distal promoter (nucleotides −2281/−2261 and −1393/−1374) encodes elements that bind p53.
In cancer cells, SFN regulation of p21Cip1 expression is frequently p53-independent (5, 18, 33). This includes HT-29 colon cancer cells, which express mutant p53 (33), LnCaP prostate cancer cells, which express wild-type p53 (18), and PC-3 prostate cancer cells, which lack p53 (5). In most cases SFN treatment of cancer cells is also associated with increased apoptosis (5, 34). In contrast, we show that SFN-dependent p21Cip1 transcription in normal keratinocytes requires wild-type p53 and that p53 levels are increased via a stabilization mechanism. We further show that Sp1 transcription factors are required and that methylation-related epigenetic regulation (i.e. modulation of methylation of the CpG cluster in the p21Cip1 proximal promoter) is not involved. This is in contrast to our recent findings showing that SFN treatment induces apoptosis in skin cancer cells (34). These findings suggest that topical SFN treatment on the epidermis will halt proliferation of normal epidermal keratinocytes without inducing apoptosis but will cause resident cancer cells to undergo cell death.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Keratinocyte serum-free medium (KSFM) and trypsin was purchased from Invitrogen. Sodium butyrate was from Calbiochem. Dimethyl sulfoxide (DMSO) was purchased from Sigma. R,S-Sulforaphane (SFN, #S8044) was purchased from LKT laboratories (St. Paul, MN). Cycloheximide (239763) was purchased from EMD Chemicals (Gibbstown, NJ).
Mouse monoclonal antibody for β-actin (A5441) was purchased from Sigma. Rabbit polyclonal antibodies for p53 (sc-6243), Sp1 (sc-59x), and Sp3 (sc-644x) were from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal antibodies for p21Cip1 (2947) were from Cell Signaling Technology (Danvers, MA). Rabbit polyclonal antibodies for cdk2 (sc-163), cdk4 (sc-601), cdc25c (sc-327), cyclin A (sc-239), and mouse monoclonal antibody for cyclin B1 (sc-245) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Peroxidase-conjugated anti-mouse IgG (NXA931) and peroxidase-conjugated anti-rabbit IgG (NA934V) were obtained from GE Healthcare. ChIP-grade rabbit anti-Sp1 (ab13370) was from Abcam (Cambridge, MA), and ChIP-grade mouse monoclonal anti-p53 (17–613) was from Millipore (Billerica, MA). Control (sc-37007) and p53 (sc-44218) siRNA were purchased from Santa Cruz.
Plasmids
The human wild-type p21Cip1 promoter-luciferase fusion plasmid, p21-2326, was a gift from Dr. Bert Vogelstein (15). Truncated p21Cip1 plasmids (p21-124, p21-101, and p21-60) were provided by Dr. Toshiyuki Sakai (35). The human Sp1 encoding plasmid was provided by Jon Horowitz (36), and ΔN-Sp3 was previously described (37). The p21Cip1 promoter truncation mutants were constructed using appropriate primers and PCR, and the plasmids are named according to the 5′ most nucleotide from the p21Cip1 promoter sequence. p21-2326 was used as a template to construct Sp1 site deletion mutants including p21-2326 Sp1(Δ1–6), p21-2326 Sp1(Δ1), p21-2326 Sp1(Δ2), p21-2326 Sp1(Δ3), and p21-2326 Sp1(Δ4) in pBluescript II KS(+). These plasmids lack the indicated Sp1 site. p53 site mutants, p21-2326 p53(Δ1), p21-2326 p53(Δ2), and p21-2326 p53(Δ1-2) were also constructed in pBluescript II KS(+). Primary cultures of human epidermal keratinocytes were prepared from human foreskins and maintained in KSFM (38).
Promoter Activity
For p21Cip1 promoter activity analysis, 0.5 μg of p21Cip1 promoter reporter plasmid was mixed with 1 μl of FuGENE 6 reagent diluted with 99 μl of KSFM. The mixture was incubated at 25 °C for 15 min and then added to a 50% confluent culture of primary human epidermal keratinocyte maintained in 2 ml of KSFM in a 9.6-cm2 dish. After 24 h, the cells were treated with the indicated concentrations of SFN. After an additional 24 h, the cells were harvested, and extracts were prepared for assay of luciferase activity.
Electroporation and siRNA-mediated Knockdown
Keratinocytes were electroporated with siRNA using the Amaxa electroporator and the VPD-1002 nucleofection kit (Germany). For electroporation, keratinocytes were harvested with trypsin and replated 1 day before use. On the day of electroporation, 1 × 106 of the replated cells were harvested with trypsin and resuspended in KSFM. The cells were collected at 2000 rpm, washed with 1 ml of sterile phosphate buffered saline (pH 7.5), and suspended in 100 μl of keratinocyte nucleofection solution. The cell suspension, which included 3 μg of gene-specific siRNA, was mixed by gentle pipetting and electroporated using the T-018 setting. Warm KSFM (500 μl) was added, and the suspension was transferred a 21.3-cm2 cell culture dish containing 3.5 ml of KSFM.
Immunological Analysis
Equivalent amounts of protein were electrophoresed on a 4–15% denaturing polyacrylamide gradient gels and transferred to nitrocellulose. The membranes were blocked, incubated with a specific primary antibody, washed, and exposed to an appropriate horseradish peroxidase-conjugated secondary antibody. Chemiluminescent detection was used to visualize secondary antibody binding.
Real-time PCR
Total RNA was isolated (Illustra RNAspin Mini kit, GE Healthcare) and reverse-transcribed for quantification by LightCycler 480 PCR system (Roche Applied Science). PCR primers were designed to quantify the abundance p21Cip1 transcript level using LightCycler 480 SYBR Green I, and signals were normalized using cyclophilin A primers. Primers for detection of mRNA levels were as follow p21Cip1: forward (5′-AAGACCATGTGGACCTGTCACTGT-3′) and reverse (5′-AGGGCTTCCTCTTGGAGAAGATCA-3′), p53 forward (5′-TAACAGTTCCTGCATGGGCGGC-3′) and reverse (5′-AGGACAGGCACAAACACGCACC-3′), and cyclophilin A forward (5′-CATCTGCACTGCCAAGACTGA-3′) and reverse (5′-TTCATGCCTTCTTTCACTTTGC-3′).
Chromatin Immunoprecipitation Assay (ChIP)
ChIP assays were conducted as described previously (39). Enrichment of Sp1- and p53-associated DNA sequences in immunoprecipitated samples and input samples was detected by quantitative RT-PCR using sequence-specific primers and LightCycler 480 SYBR Green I Master mix. ChIP primers were as follows. Primers to detect the p21Cip1 proximal promoter Sp1 binding site amplified nucleotides −150/−4 (forward, 5′-GCTGGGCAGCCAGGAGCCTG-3′; reverse, 5′-CTGCTCACACCTCAGCTGGC-3′), p21Cip1 promoter control primers amplified nucleotides −827/−673 (forward, 5′-CTGCTGCAACCACAGGGATTTCTT-3′; reverse, 5′-TGTTGATTGTCACATGCTTC CGGG-3′), and primer to detect the p21Cip1 promoter p53-1 binding site amplified nucleotides −2310/−2205 (forward, 5′-GTGGCTCTGATTGGCTTTCTG; reverse, 5′-CTGAAAACAGGCAGCCCAAG).
Bisulfite-sequencing PCR
Genomic DNA was extracted using the Qiagen DNeasy Blood Tissue kit, and genomic DNA was subjected to bisulfite conversion using the EpiTect Bisulfite kit (Qiagen, Valencia, CA) according to manufacturer's protocol. The promoter sequences of interest were amplified by PCR using Platinum DNA Taq polymerase (Invitrogen). Primer sequences (forward, 5′-AGGAGGGAAGTGTTTTTTTGTAGTA-3′; reverse, 5′-CAACTACTCACACCTCAACTAAC-3′) covering the proximal promoter were designed using MethPrimer software. Bisulfite PCR products were cloned using the TOPO TA cloning kit (Invitrogen). A minimum of 10 clones were prepared and sequenced for each treatment group, and sequencing was performed using the T7 and M13 universal primers.
Protein Turnover
The impact of SFN on p53 turnover was assessed by cycloheximide chase. Keratinocytes were treated with 0 or 20 μm SFN for 24 h before treatment with 50 μg/ml cycloheximide. Cells were harvested and lysed at 0, 15, 30, 45, 90, and 120 min after the addition of cycloheximide. Lysates were analyzed for p53 level by immunoblot.
RESULTS
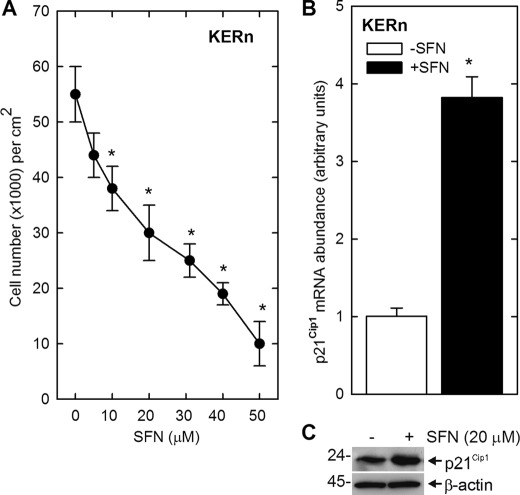
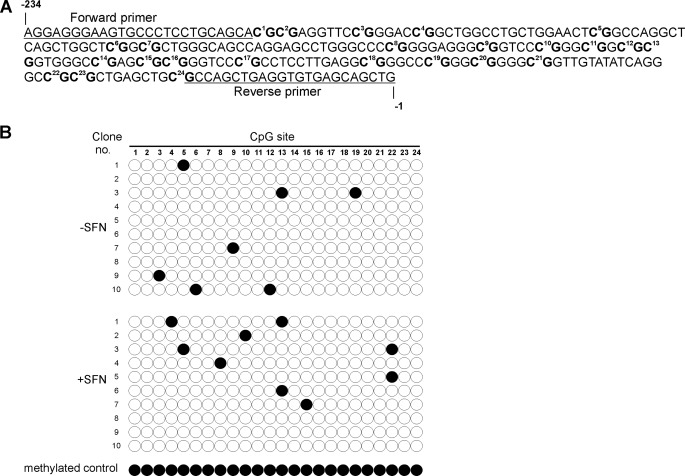
Treatment of keratinocytes with increasing concentrations of SFN produces a concentration-dependent reduction in cell number (Fig. 1A). To understand the molecular basis for this response, we monitored the impact of SFN on p21Cip1 expression. p21Cip1 was selected as a target because it is a key suppressor of keratinocyte proliferation (40–44) and because SFN treatment increases p21Cip1 in other cell types (5, 33, 45). Treatment with 20 μm SFN for 24 h results in a marked increase in p21Cip1 mRNA and protein (Fig. 1, B and C), suggesting that p21Cip1 is involved in suppression of keratinocyte proliferation. We first examined the impact of SFN treatment on p21Cip1 promoter methylation, as SFN is known to function as an epigenetic regulator (46, 47) and because the p21Cip1 promoter CpG island can be hypo- or hypermethylated in various cell types (48). In addition, most information about p21Cip1 promoter methylation status is derived from cancer cells (49, 50). Our results suggest that methylation does not play a major role, as the p21Cip1 promoter is hypomethylated in untreated cells and SFN treatment does not alter the methylation status (Fig. 2B). We were concerned that the low methylation level may be an artifact. To check this, genomic DNA was prepared from untreated cells and then methylated in vitro using CpG methyltransferase, which specifically methylates cytosine residues in CpG motifs (51). As shown in Fig. 2B, this DNA registers as methylated in the bisulfite assay, supporting the conclusion that the p21Cip1 proximal CpG island is hypomethylated in keratinocytes. These findings suggest that DNA methylation is not a key control point in SFN regulation of p21Cip1 expression in normal keratinocytes.
FIGURE 1.
SFN suppresses keratinocyte proliferation and increases p21Cip1 expression. A, human epidermal keratinocytes were seeded in 9.6-cm2 dishes at 20,000 cells/cm2 and permitted to attach overnight. The cultures were then treated with the indicated concentration of SFN and, after 24 h, harvested and counted. Cell counts are the mean ± S.D., and asterisks indicate cells counts that are significantly reduced as compared with the zero SFN treatment group as determined by Student's t test (n = 3, p < 0.005). Similar results were observed in three experiments. B and C, keratinocytes were treated with 20 μm of SFN, and after 24 h extracts were prepared for detection of p21Cip1 mRNA by qPCR and p21Cip1 protein by immunoblot. The mRNA abundance values are the mean ± S.D., n = 3. The asterisk indicates a significant increase over control as determined by Student's t test, p < 0.005. Similar results were observed in each of three experiments.
FIGURE 2.
Methylation status of p21Cip1 in keratinocytes. A, shown is a sequence map of the 24 CpG sites located in the CpG island of the p21Cip1 proximal promoter. Also indicated are the forward and reverse primers used for production of p21Cip1 promoter-specific clones for bisulfite sequence analysis. B, shown is mapping of methylated cytosines in the p21Cip1 promoter using bisulfate-sequencing PCR. Cells were treated with or without 20 μm SFN for 24 h, and DNA extracts were prepared for bisulfite sequencing. A pool of three genomic DNA samples was analyzed. Individual PCR products were cloned, and 10 clones were sequenced. The methylated control sample is genomic DNA that was methylated in vitro using CpG methyltransferase, which specifically methylates cytosine residues in CpG motifs. Filled circles indicate methylated CpG sites; the empty circle indicates unmethylated CpG sites.
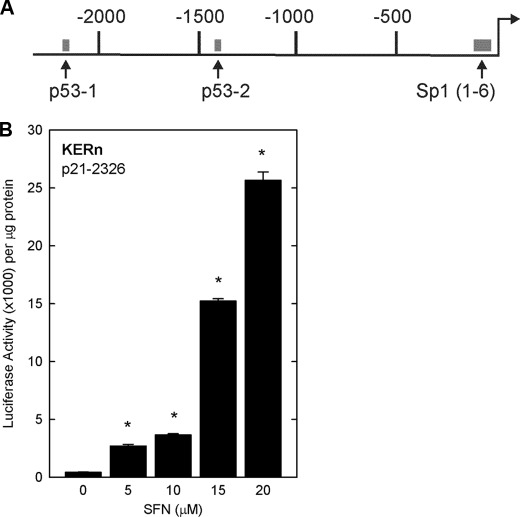
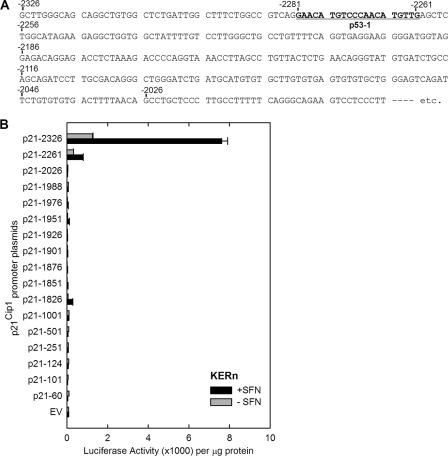
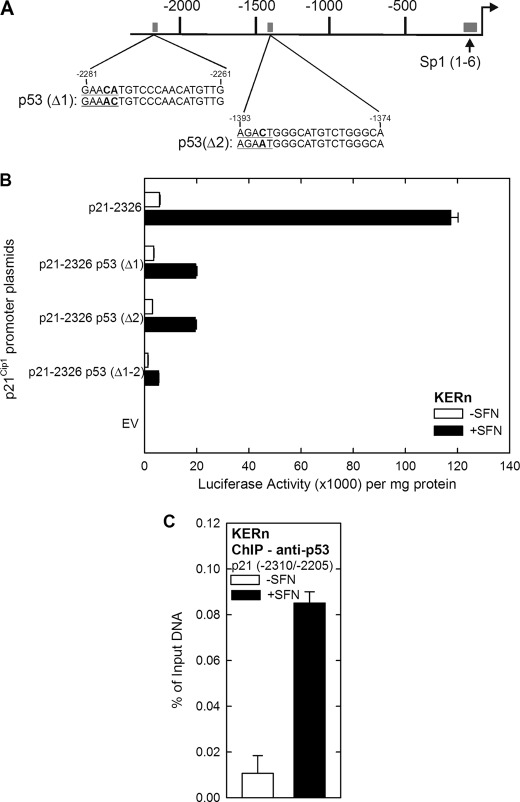
We next determined whether p21Cip1 expression may be regulated by a transcriptional mechanism. To test this, cells were transfected with p21-2326, a plasmid that encodes 2326 nucleotides of the human p21Cip1 promoter linked to luciferase (Fig. 3A) (15), and then treated with or without SFN. Fig. 3B shows that SFN treatment produces a concentration-dependent increase in p21Cip1 promoter activity. Our next goal was to identify the regulatory sites in the p21Cip1 promoter responsible for SFN regulation. We tested response of a series of truncated p21Cip1 promoter-luciferase reporter constructs. These reporters revealed SFN-dependent expression only for promoter lengths equal to or exceeding 2261 nucleotides (Fig. 4), suggesting that an important response element is present in the p21Cip1 distal promoter (nucleotides −2326/−2261). This region contains a previously characterized p53 response element located at nucleotides −2281/−2261 (14). Because the SFN-associated increase in p21Cip1 promoter activity is lost in truncation constructs lacking this segment (Fig. 4), we determined whether p53 has a role in mediating the response to SFN. A second p53 response element is also present in the promoter, and so we individually mutated one or both of these elements (Fig. 5A), and the constructs were transfected into keratinocytes before 24 h of SFN treatment. Fig. 5B shows that mutation of either p53 response element reduces SFN-stimulated promoter activity by 80%, and mutation of both sites results in a complete loss of promoter activity. This is particularly interesting because SFN regulation of p53 has been described as p53-independent in cancer cell lines (5, 18). We next examined p53 interaction on the most distal (p53-1) element using chromatin immunoprecipitation. As shown in Fig. 5C, p53 interaction at this element is markedly increased (8-fold) after treatment with SFN.
FIGURE 3.
SFN increases p21Cip1 promoter activity. A, a schematic of p21Cip1 promoter upstream regulatory region shows the location of the p53 response elements (nucleotides −2281/−2261 and −1393/−1374) and the 6 GC-rich, Sp1(1–6), response elements (nucleotides −120/−50) (71). The distances are in nucleotides relative to the transcription start site, which is indicated by the right-facing arrow. B, keratinocytes were transfected with 0.5 μg of p21-2326 luciferase reporter plasmid and then treated for 24 h with SFN. The cells were harvested, and lysates were assayed for luciferase activity. The values are the mean ± S.D., n = 3. The values indicated by asterisks were significantly increased as measured using the Student's t test, p < 0.005.
FIGURE 4.
SFN activation of the p21Cip1 promoter involves the distal p53 response element. A, shown is a sequence map of the distal p21Cip1 promoter region. Nucleotides are numbered relative to the transcription start site at +1, and the distal (nucleotides −2281/−2261) p53 response element is indicated. B, impact of SFN treatment on p21Cip1 promoter activity is shown. Keratinocytes were transfected with 0.5 μg of each p21Cip1 promoter luciferase reporter plasmid or with empty vector (EV) lacking p21Cip1 promoter sequences. After 24 h, the cells were treated with 20 μm SFN, and after an additional 24 h the cells were harvested for luciferase activity assay. The values are the mean ± S.D. Similar results were observed in each of three experiments.
FIGURE 5.
p53 is required for SFN-dependent p21Cip1 promoter activation. A, shown is a schematic of p21Cip1 promoter distal promoter. The p53-1 (nucleotides −2281/−2261) and p53-2 (nucleotides −1393/−1374) response elements are shown. The top sequence is wild type, and the bottom sequence is mutated. B, keratinocytes were transfected with 0.5 μg of each indicated p21Cip1 luciferase reporter plasmid, and after 24 h cells were incubated with 20 μm SFN for an additional of 24 h. The cells were then harvested, and extracts were prepared for assay of luciferase activity. The values are the mean ± S.D.; n = 3. Promoter activity of the Δ1, Δ2, and Δ1-2 mutants was all significantly reduced compared with wild type, p < 0.005, as assessed using Student's t test. C, a ChIP assay was performed as described under “Experimental Procedures” using p21Cip1 promoter-derived PCR primers encoding the indicated range of nucleotides. The values are the mean ± S.D., and similar results were observed in three experiments.
Regulation of p53 Level
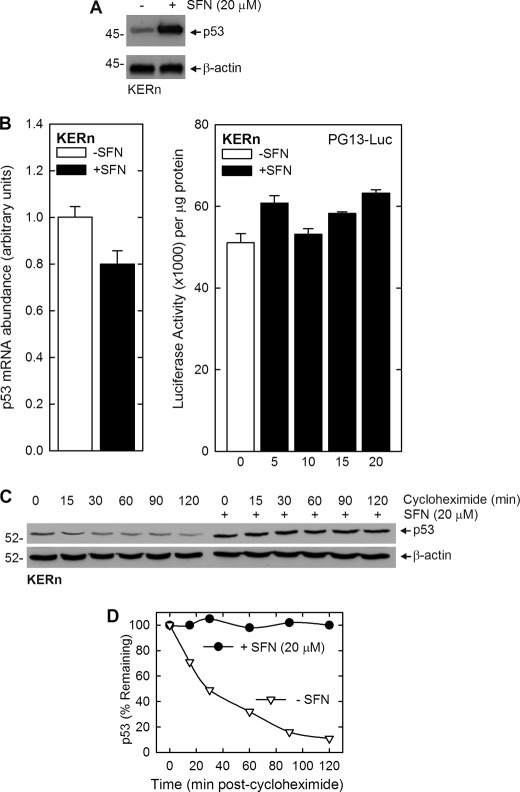
A key question is the mechanism that drives increased p53 binding to the p21Cip1 promoter. To assess this, we treated KERn with SFN and monitored the impact on p53 level. Fig. 6A shows that a 24-h treatment with 20 μm SFN results in a substantial increase in p53 level. We next explored whether this increase is due to changes in p53 mRNA level and gene transcription. Fig. 6B shows that p53 mRNA level and promoter activity is not increased by SFN treatment. We, therefore, assessed the impact of SFN on p53 stability. Cells were treated with 20 μm SFN for 24 h and then treated with protein synthesis inhibitor (cycloheximide) for the indicated times. Fig. 6, C and D, shows that loss of p53 protein is substantial in control cells, but in contrast, p53 levels are stabilized in SFN-treated keratinocytes. Thus, p53 turnover is reduced in SFN-treated keratinocytes.
FIGURE 6.
SFN increases p53 level by suppressing p53 protein turnover. A, SFN increases the p53 level. Keratinocytes were treated with SFN for 24 h, and extracts were prepared for immunoblot detection of p53 and β-actin. B, SFN does not regulate p53 mRNA level or promoter activity. The values are the mean ± S.D. for three replicates. To measure p53 promoter activity, keratinocytes were transfected with 2 μg of PG13-Luc, which encodes the p53 gene promoter (15), and then treated with 20 μm SFN for 24 h before preparation of extracts and assay of promoter activity. The values are the mean ± S.D. for three separate experiments, and there is no significant change in promoter activity with SFN treatment. C, SFN increases the p53 level by reducing p53 turnover. Keratinocytes were treated with 20 μm SFN for 24 h and then treated with 50 μg/ml cycloheximide for the indicated times (min) before harvest and immunoblot detection of p53 and β-actin. Similar results were observed in three separate experiments. D, shown is a plot of p53 protein levels with time after the addition of cycloheximide. The immunoblot in panel C was scanned, and the level of p53 was plotted. The initial (time = 0) blot intensity point was set at 100% for each curve. These findings suggest a half-life for p53 of 30 min in untreated keratinocytes (open triangles) and stabilization of p53 in SFN-treated cells (closed circles).
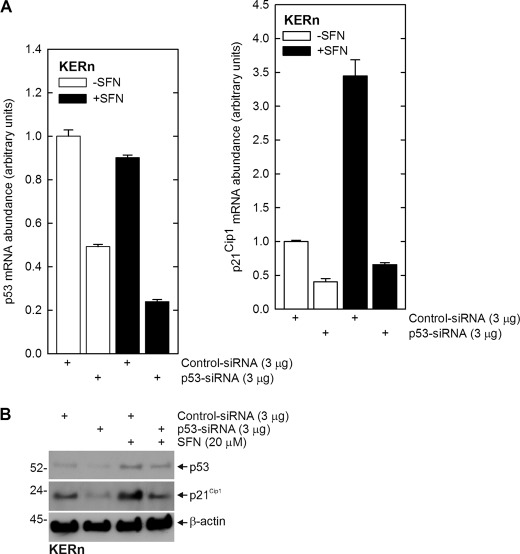
To confirm a role for p53, we next assessed the impact of reduced p53 expression on p21Cip1 mRNA and protein level. Fig. 7, A and B, shows that treatment with p53-siRNA reduces p53 mRNA and protein in both control and SFN-treated cultures and that this is associated with a parallel reduction in p21Cip1 mRNA and protein. These studies suggest that SFN treatment stabilizes p53 protein, and the increased p53 binds to the p21Cip1 promoter to drive gene expression.
FIGURE 7.
Impact of p53 knockdown on SFN-dependent p21Cip1 mRNA and protein level. A, keratinocytes were electroporated with 3 μg of control or p53-siRNA per million cells and after 24 h were treated with or without 20 μm SFN for an additional 24 h. RNA was prepared for detection of p53 and p21Cip1 mRNA. The values are the mean ± S.D., n = 3. Similar results were observed in three experiments. B, reduced p53 expression is associated with reduced p21Cip1 protein level. Keratinocytes were treated with control or p53-siRNA and after 24 h extracts were prepared for detection of p53 and p21Cip1. Similar results were observed in each of three separate experiments.
GC-rich Elements in Proximal p21Cip1 Promoter Are Required for SFN Regulation
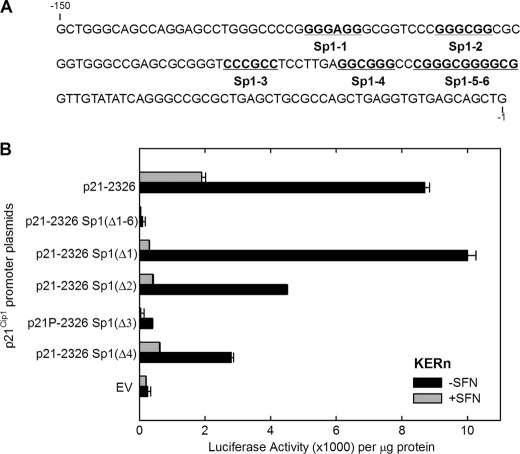
Promoter activity analysis (Fig. 4) suggests that sequences located between −251 and −1 are not involved in the SFN-dependent response; however, this region is known to encode six GC-rich transcription factor binding sites that have an important role in regulating p21Cip1 expression in other systems (52, 53). To assess the role of this region, we selectively mutated the six GC-rich elements (Fig. 8A) and monitored the effect on SFN-dependent transcription. Keratinocytes were transfected with plasmids encoding the indicated constructs, and cells were treated with SFN for 24 h. It is interesting that mutation of these sites impacts SFN-dependent promoter activation. Mutation of single GC-rich elements produces variable outcomes. Fig. 8B suggests that the Sp1–3 site is essential for SFN regulation but in other experiments indicates that other sites are also important. However, a consistent observation is that simultaneous mutation of all six GC-rich elements (Δ1–6) eliminates SFN-dependent promoter activity. Thus, the Sp1 site cluster is required for SFN-dependent induction of p21Cip1.
FIGURE 8.
Effect of deleting GC-rich (Sp1) elements on SFN regulation of p21Cip1 promoter activity in the context of the full-length promoter. A, shown is a sequence map of the proximal region of the p21Cip1 promoter. The six GC-rich (Sp1) sites are identified. The numbers indicate nucleotide position relative to the transcription start site at +1. B, keratinocytes were transfected with 0.5 μg of each p21Cip1 promoter luciferase reporter plasmid or with empty vector (EV) lacking p21Cip1 promoter sequences. After 24 h, the cells were treated with 20 μm SFN. After an additional 24 h the cells were harvested and assayed for luciferase activity. The respective plasmid names identify the deleted Sp1 sites (e.g. Δ1, Δ2, Δ1–6, etc.).
Sp1 Is Required for an SFN-dependent Increase in Promoter Activity
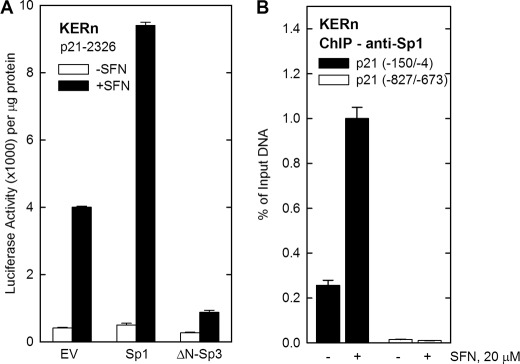
Sp1 interacts with the GC-rich elements in the p21Cip1 promoter (30, 35); therefore, we examined the role of Sp1 factors in mediating the effect of SFN. Keratinocytes were transfected with p21-2326 in the presence of Sp1 or ΔN-Sp3 expression plasmids. Sp1 activates and ΔN-Sp3 inhibits gene expression in keratinocytes (54, 55). In the absence of Sp1 or ΔN-Sp3, SFN treatment produces an 8-fold increase in p21Cip1 promoter activity (Fig. 9A). Treatment with SFN and exogenous Sp1 also produces an 8-fold increase; however, overall basal and stimulated activity increases 2-fold (Fig. 9A). This suggests that Sp1 is required for basal and SFN-stimulated promoter activity. In contrast, ΔN-Sp3 expression suppresses the SFN-dependent increase, further suggesting that Sp1 is required. We next examined Sp1 factor interaction with the proximal promoter GC-rich region. Cells were treated with or without 20 μm SFN for 24 h, and extracts were prepared for ChIP assay using anti-Sp1 antibody. We observe a substantial (5-fold) SFN-dependent increase in Sp1 factor interaction with the proximal promoter GC-rich cluster (nucleotides −150/−4) (Fig. 9B). As a control, we assayed interaction with a region of the promoter (−827/−673) that lacks GC-rich elements. Fig. 9B shows that this region does not interact with Sp1.
FIGURE 9.
Sp1 is required for the SFN-dependent increase in p21Cip1 promoter activity. A, keratinocytes were transfected with 0.5 μg of p21-2326 luciferase reporter plasmid in the presence of 0.5 μg of Sp1 or ΔN-Sp3 expression vector. After 24 h the cells were treated with 20 μm SFN for 24 h, cells were harvested, and extracts were assayed for luciferase activity. B, ChIP assay was performed as described under “Experimental Procedures” using p21Cip1 promoter-derived PCR primers encoding the indicated range of nucleotides. The values are the mean ± S.D. (n = 3), and similar results were observed in three experiments.
The findings in Figs. 8 and 9 suggest that the GC-rich elements may not selectively mediate SFN induction of p21Cip1 expression. It is more likely that they are required for both basal and stimulated promoter activity, and so when these sites are mutated, the promoter cannot respond to SFN. This is consistent with the face that elimination of all six GC-rich elements reduces both basal and stimulated promoter activity (Fig. 8).
Impact of SFN on Cell Cycle
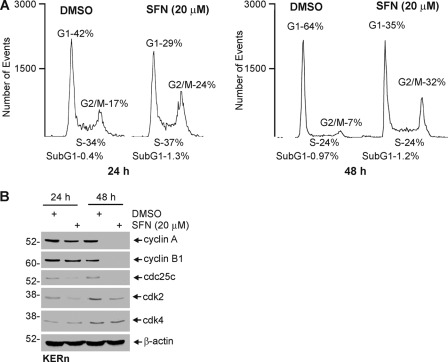
In some systems SFN causes cells to accumulate in G2/M (18). To assess the impact on cell cycle in keratinocytes, we treated cells for 24 and 48 h and examined the cell cycle stage. These studies show a prominent reduction in G1 phase cells and accumulation in G2/M (Fig. 10A). The effect is more dramatic at 48 h. To analyze the mechanism responsible for these changes, we studied the impact on cell cycle regulatory protein expression. Fig. 10B shows a reduction in cyclin A, cyclin B1, cdc25c, and cdk2 levels in SFN-treated cells. In contrast, cdk4 did not markedly change in level. These findings are consistent with accumulation of cells at the G2/M stage of the cell cycle and general cessation of cell proliferation. SFN is also known to induce apoptosis in cancer cell models (8, 56–59); however, we observed no change in apoptosis as evidenced by a lack of increase in sub-G1 cells (Fig. 10A) and by an absence of procaspase or PARP cleavage in SFN-treated keratinocytes. Moreover, Bax and Bcl2 levels are below the limit of detection in these cells, and none were detected (not shown).
FIGURE 10.
SFN treatment alters cell cycle progression but does not induce apoptosis. A, SFN treatment causes loss of cells in G1 and accumulation of cells in G2/M. Keratinocytes were treated with 0 or 20 μm SFN for 24 or 48 h before cell sorting. Similar results were observed in each of three experiments. B, keratinocytes were treated for 24 and 48 h, and extracts were prepared for detection of the indicated cell cycle regulators. Similar results were observed in each of three experiments.
DISCUSSION
SFN Impact on Epidermis and Keratinocytes
SFN is a naturally occurring dietary agent found in cruciferous vegetables that is an important candidate dietary cancer prevention agent. SFN impacts a number of cellular processes. For example, SFN inhibits histone deacetylase activity (2, 5, 6), stimulates apoptosis, and induces G2/M arrest (8, 18, 60–63). The impact of SFN has been monitored in epidermis. Treatment with broccoli extract or SFN increases the phase I and phase II enzyme response in mouse and human epidermis (9, 11), suppresses UVB-induced skin carcinogenesis in SKH-1 mice (64), and reduces 7,12-dimethyl benz[a]anthracene (DMBA)-induced skin cancer (65). SFN also reduces UVB-dependent inflammation in HR-1 hairless mice (66).
The mechanism of SFN action in keratinocytes has also been studied. These studies used a line of immortal human keratinocytes, HaCaT, that retain some characteristics of normal cells. SFN treatment induces phase I and II enzymes (7, 67), inhibits AP1 transcription factor function (68), and reduces inflammatory response (66) in these cells. However, HaCaT cells harbor mutant p53 (69), and so they are not a good model for studying regulation of p53-dependent genes such as p21Cip1. Additional studies have been completed in SCC-13 cells, but these also harbor a p53 mutation (34). For this reason, in the present study, we examine the impact of SFN on primary cultures of normal human keratinocytes. It is important to study normal keratinocytes, as the impact of SFN on these cells has not been studied and because normal keratinocytes are a primary epidermal cell type challenged by external carcinogenic stress and treated by chemopreventive agents.
Our studies show that SFN suppresses normal human keratinocytes proliferation and that this is associated with increased p21Cip1 expression. p21Cip1 is a cyclin-dependent kinase inhibitor that blocks cell cycle progression (20) and is a key regulator of keratinocyte proliferation (40–44, 70). Our studies show that p21Cip1 levels increase in SFN-treated cells. A key issue is the mechanism responsible for this increase. SFN is known to influence epigenetic regulatory proteins, including the DNA methyltransferases (DNMT1 and DNMT3a), which methylate cytosine residues in CpG motifs to suppress gene expression (46). SFN also increases histone acetylation at the p21Cip1 promoter in prostate cancer cells (8, 72).
The p21Cip1 promoter encodes a large CpG island in the proximal region of the promoter (nucleotides −234/−1), and so reduced methylation in this region could increase p21Cip1 expression. p21Cip1 promoter methylation status has been extensively studied in cancer cells and tumors (48, 49, 73). The p21Cip1 promoter, for example, is hypermethylated in low grade mucosa-associated lymphoid tissue lymphoma (73). However, p21Cip1 promoter methylation status has not been widely studied in primary cultures of normal cells. We, therefore, determined whether the p21Cip1 promoter is methylated in keratinocytes and whether methylation status is regulated by SFN. Interestingly, the p21Cip1 promoter is hypomethylated in keratinocytes, and treatment with SFN does not alter methylation status. These studies are the first to rule out a role for DNA methylation in regulation of p21Cip1 expression in normal keratinocytes.
SFN Regulation of p21Cip1 Is p53-dependent
The SFN-dependent increase in p21Cip1 protein is associated with a parallel increase in level of the corresponding mRNA. To assess the mechanism of regulation, we examined the impact of SFN treatment on p21Cip1 promoter activity. The p21Cip1 distal promoter encodes two p53 binding sites, p53-1 (nucleotides −2281/−2261) and p53-2 (nucleotides −1393/−1374) (14), that drive p21Cip1 expression in some systems (35, 74). Our promoter truncation and point mutagenesis studies indicate that the p53-1 and p53-2 are required for SFN-dependent induction of p21Cip1 gene expression. ChIP studies reveal that p53 interacts at these p53 binding sites at an 8-fold higher level after SFN treatment and that the SFN-dependent p21Cip1 increase is attenuated by p53 knockdown. These studies strongly suggest that SFN treatment increases p53 interaction with the p21Cip1 promoter to drive increased gene expression (Fig. 11).
FIGURE 11.
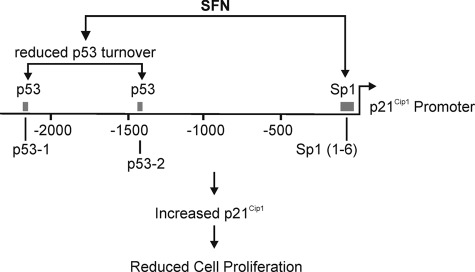
SFN regulation of p21Cip1 gene expression. Our studies indicate that SFN treatment stimulates p53 interaction at the p53 response elements in the distal promoter and Sp1 interaction at the GC-rich (Sp1) elements in the proximal promoter to increase gene expression. Nucleotide number is indicated relative to the transcription start site. The p53 and Sp1 response elements are indicated by boxes, and the transcription start site is the right-facing arrow.
These findings are unique in that SFN-mediated regulation of p21Cip1 promoter activity is p53-dependent. This finding in normal cells is in contrast to findings in tumor cells where regulation of p21Cip1 expression is p53-independent. Examples include HT-29 colon cancer cells, which express mutant p53 (33), LnCaP prostate cancer cells, which express wild-type p53 (18), and PC-3 prostate cancer cells, which lack p53 (5). Oxidative stress and MAPK signaling play key roles in inducing p21Cip1 in HT-29 cells (33). PC-3 prostate cancer cells lack p53, and in these cells SFN induction of p21Cip1 is associated with reduced histone acetylation on the p21Cip1 promoter (5). Our ChIP studies show that that p53 and Sp1 transcription factors accumulate at the p21Cip1 promoter after SFN treatment and that methylation of the CpG islands is not involved in making this chromatin accessible. However, in future studies we will need to determine whether chromatin acetylation is required for expression as has been reported in prostate cancer cells (5).
SFN Regulates p53 Protein Stability
We also studied the impact of SFN on p53 expression. p53 is known to be regulated by both transcriptional and post-transcriptional mechanisms (75). Our studies show that SFN does not increase p53 mRNA level or promoter activity. In contrast, SFN inhibits p53 protein turnover, and this accounts for the substantial increase in p53 level. The half-life of p53 is 30 min in untreated cells, but p53 is highly stable in SFN-treated keratinocytes. The fact that this is associated with an increase in p21Cip1 mRNA and promoter activity suggests that the p53 is transcriptionally active. p53 stabilization has not been previously described in SFN-treated skin-derived cells, but Pelling and co-workers (76) showed that apigenin increases p53 stability in mouse keratinocytes. Thus, it may be that stabilization is a common mechanism of regulating p53 levels by chemopreventive agents in keratinocytes.
Further studies will be necessary to understand the details of this regulation. For example, SFN treatment causes a general increase in proteasome activity in SCC-13 skin cancer cells (34). Because a major mode of p53 down-regulation is via the proteasome (25), it is interesting that SFN stabilizes p53 in normal keratinocytes. Thus, it will be important in future studies to monitor the impact of SFN on mechanisms that control p53 turnover, including impact on the E3 ubiquitin ligase, Hdm2, which ubiquitinates p53 leading to its degradation in the proteasome (25), and impact on proteasome activity. It is also worth noting that not all p53-regulated genes are impacted by this rise in p53 level. Bax is generally not detected in keratinocytes, except under extreme conditions, and did not observe Bax after SFN treatment (not shown).
SFN Regulation of p21Cip1; Role for GC-rich Response Elements
An important cluster of transcription regulatory sites is present in the p21Cip1 proximal promoter between nucleotides −150 and −1. This is a cluster of six GC-rich transcription factor binding sites (35). Various signaling pathways regulate p21Cip1 gene expression by stimulating Sp1, Sp3, and Kruppel-like transcription factor interactions at these sites (26–32). We show that mutation of these six GC-rich eliminates both basal and SFN-stimulated promoter activity. We propose that these sites are necessary for both basal and SFN-stimulated transcription and that in their absence SFN has no impact on transcription. In support of an essential role for these sites, we observed an increase in Sp1 factor binding at these locations after SFN treatment. In addition, combining treatment with SFN and Sp1 overexpression increases promoter activation compared with either treatment. Moreover, expression of ΔN-Sp3, an inhibitory form of Sp3 (37, 54, 55), reduces the SFN-dependent increase. Taken together, these findings suggest that SFN increases p21Cip1 expression by increasing p53 interaction at the p53 response elements in the distal promoter and Sp1 interaction at the GC-rich elements in the proximal promoter but that SFN acts mainly through a p53-dependent mechanism.
In summary, our findings indicate that SFN increases p53 levels by inhibiting p53 turnover (Fig. 11). The increase in p53 is associated with increased p53 binding to the p21Cip1 promoter. In addition, SFN treatment increases Sp1 factor interaction with the p21Cip1 promoter. These events are both required for SFN activation of p21Cip1 expression, which ultimately leads to cessation of cell proliferation. At the biological level, these findings suggest that topical SFN treatment on the epidermis will halt proliferation of normal epidermal keratinocytes via a p53- and p21Cip1-dependent mechanism but will not cause these cells to undergo apoptosis. In contrast, our previous report indicates SFN causes SCC-13 skin cancer cells to undergo apoptosis (34). These results suggest that topical treatment with SFN may cause cancer cells to undergo cell death while only slowing growth of normal keratinocytes. Such an outcome would provide a clear therapeutic advantage of SFN as a skin cancer-preventive agent.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AR046494 (to R. E.).
- SFN
- sulforaphane
- p21Cip1
- p21 cyclin-dependent kinase inhibitor
- cdk
- cyclin-dependent kinase
- Nrf2
- nuclear factor E2-related factor 2
- KSFM
- keratinocyte serum-free medium.
REFERENCES
- 1. Myzak M. C., Tong P., Dashwood W. M., Dashwood R. H., Ho E. (2007) Sulforaphane retards the growth of human PC-3 xenografts and inhibits HDAC activity in human subjects. Exp. Biol. Med. (Maywood) 232, 227–234 [PMC free article] [PubMed] [Google Scholar]
- 2. Myzak M. C., Dashwood W. M., Orner G. A., Ho E., Dashwood R. H. (2006) Sulforaphane inhibits histone deacetylase in vivo and suppresses tumorigenesis in Apc− mice. FASEB J. 20, 506–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Singh A. V., Xiao D., Lew K. L., Dhir R., Singh S. V. (2004) Sulforaphane induces caspase-mediated apoptosis in cultured PC-3 human prostate cancer cells and retards growth of PC-3 xenografts in vivo. Carcinogenesis 25, 83–90 [DOI] [PubMed] [Google Scholar]
- 4. Dashwood R. H., Ho E. (2007) Dietary histone deacetylase inhibitors. From cells to mice to man. Semin. Cancer Biol. 17, 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Myzak M. C., Hardin K., Wang R., Dashwood R. H., Ho E. (2006) Sulforaphane inhibits histone deacetylase activity in BPH-1, LnCaP and PC-3 prostate epithelial cells. Carcinogenesis 27, 811–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Myzak M. C., Karplus P. A., Chung F. L., Dashwood R. H. (2004) A novel mechanism of chemoprotection by sulforaphane. Inhibition of histone deacetylase. Cancer Res. 64, 5767–5774 [DOI] [PubMed] [Google Scholar]
- 7. Wagner A. E., Ernst I., Iori R., Desel C., Rimbach G. (2010) Sulforaphane but not ascorbigen, indole-3-carbinole, and ascorbic acid activates the transcription factor Nrf2 and induces phase-2 and antioxidant enzymes in human keratinocytes in culture. Exp. Dermatol. 19, 137–144 [DOI] [PubMed] [Google Scholar]
- 8. Clarke J. D., Hsu A., Yu Z., Dashwood R. H., Ho E. (2011) Differential effects of sulforaphane on histone deacetylases, cell cycle arrest, and apoptosis in normal prostate cells versus hyperplastic and cancerous prostate cells. Mol. Nutr. Food Res. 55, 999–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kwak M. K., Kensler T. W. (2010) Targeting NRF2 signaling for cancer chemoprevention. Toxicol. Appl. Pharmacol. 244, 66–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kwak M. K., Wakabayashi N., Greenlaw J. L., Yamamoto M., Kensler T. W. (2003) Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol. Cell. Biol. 23, 8786–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kwak M. K., Egner P. A., Dolan P. M., Ramos-Gomez M., Groopman J. D., Itoh K., Yamamoto M., Kensler T. W. (2001) Role of phase 2 enzyme induction in chemoprotection by dithiolethiones. Mutat. Res. 480, 305–315 [DOI] [PubMed] [Google Scholar]
- 12. Abukhdeir A. M., Park B. H. (2008) P21 and p27. Roles in carcinogenesis and drug resistance. Expert. Rev. Mol. Med. 10, e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xiong Y., Hannon G. J., Zhang H., Casso D., Kobayashi R., Beach D. (1993) p21 is a universal inhibitor of cyclin kinases. Nature 366, 701–704 [DOI] [PubMed] [Google Scholar]
- 14. el-Deiry W. S., Tokino T., Waldman T., Oliner J. D., Velculescu V. E., Burrell M., Hill D. E., Healy E., Rees J. L., Hamilton S. R. (1995) Topological control of p21WAF1/CIP1 expression in normal and neoplastic tissues. Cancer Res. 55, 2910–2919 [PubMed] [Google Scholar]
- 15. el-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. (1993) WAF1, a potential mediator of p53 tumor suppression. Cell 75, 817–825 [DOI] [PubMed] [Google Scholar]
- 16. Schwaller J., Koeffler H. P., Niklaus G., Loetscher P., Nagel S., Fey M. F., Tobler A. (1995) Posttranscriptional stabilization underlies p53-independent induction of p21WAF1/CIP1/SDI1 in differentiating human leukemic cells. J. Clin. Invest. 95, 973–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Esposito F., Cuccovillo F., Vanoni M., Cimino F., Anderson C. W., Appella E., Russo T. (1997) Redox-mediated regulation of p21(waf1/cip1) expression involves a post-transcriptional mechanism and activation of the mitogen-activated protein kinase pathway. Eur. J. Biochem. 245, 730–737 [DOI] [PubMed] [Google Scholar]
- 18. Herman-Antosiewicz A., Xiao H., Lew K. L., Singh S. V. (2007) Induction of p21 protein protects against sulforaphane-induced mitotic arrest in LNCaP human prostate cancer cell line. Mol. Cancer Ther. 6, 1673–1681 [DOI] [PubMed] [Google Scholar]
- 19. Park J. W., Jang M. A., Lee Y. H., Passaniti A., Kwon T. K. (2001) p53-independent elevation of p21 expression by PMA results from PKC-mediated mRNA stabilization. Biochem. Biophys. Res. Commun. 280, 244–248 [DOI] [PubMed] [Google Scholar]
- 20. Gorospe M., Wang X., Holbrook N. J. (1999) Functional role of p21 during the cellular response to stress. Gene Expr. 7, 377–385 [PMC free article] [PubMed] [Google Scholar]
- 21. Matsuta M., Kon S., Sasaki K., Matsuta M. (1997) Immunohistochemical detection of p21WAF1/CIP1 and p53 proteins in formalin-fixed paraffin-embedded tissue sections of squamous cell carcinoma of the skin. J. Dermatol. Sci. 14, 233–239 [DOI] [PubMed] [Google Scholar]
- 22. Ahmed N. U., Ueda M., Ichihashi M. (1997) p21WAF1/CIP1 expression in non-melanoma skin tumors. J. Cutan. Pathol. 24, 223–227 [DOI] [PubMed] [Google Scholar]
- 23. Weinberg W. C., Fernandez-Salas E., Morgan D. L., Shalizi A., Mirosh E., Stanulis E., Deng C., Hennings H., Yuspa S. H. (1999) Genetic deletion of p21WAF1 enhances papilloma formation but not malignant conversion in experimental mouse skin carcinogenesis. Cancer Res. 59, 2050–2054 [PubMed] [Google Scholar]
- 24. Inohara S., Kitagawa K., Kitano Y. (1996) Coexpression of p21Waf1/Cip1 and p53 in sun-exposed normal epidermis but not in neoplastic epidermis. Br. J. Dermatol. 135, 717–720 [PubMed] [Google Scholar]
- 25. Helton E. S., Chen X. (2007) p53 modulation of the DNA damage response. J. Cell. Biochem. 100, 883–896 [DOI] [PubMed] [Google Scholar]
- 26. Fried R. M., Voelkel E. F., Rice R. H., Levine L., Tashjian A. H., Jr. (1988) Evidence for multiple bone resorption-stimulating factors produced by normal human keratinocytes in culture. Endocrinology 122, 2467–2475 [DOI] [PubMed] [Google Scholar]
- 27. Mandal S., Davie J. R. (2010) Estrogen regulated expression of the p21 Waf1/Cip1 gene in estrogen receptor-positive human breast cancer cells. J. Cell. Physiol. 224, 28–32 [DOI] [PubMed] [Google Scholar]
- 28. Wu J., Lingrel J. B. (2004) KLF2 inhibits Jurkat T leukemia cell growth via up-regulation of cyclin-dependent kinase inhibitor p21WAF1/CIP1. Oncogene 23, 8088–8096 [DOI] [PubMed] [Google Scholar]
- 29. Yokota T., Matsuzaki Y., Miyazawa K., Zindy F., Roussel M. F., Sakai T. (2004) Histone deacetylase inhibitors activate INK4d gene through Sp1 site in its promoter. Oncogene 23, 5340–5349 [DOI] [PubMed] [Google Scholar]
- 30. Koutsodontis G., Moustakas A., Kardassis D. (2002) The role of Sp1 family members, the proximal GC-rich motifs, and the upstream enhancer region in the regulation of the human cell cycle inhibitor p21WAF-1/Cip1 gene promoter. Biochemistry 41, 12771–12784 [DOI] [PubMed] [Google Scholar]
- 31. Lee T. H., Chuang L. Y., Hung W. C. (2000) Induction of p21WAF1 expression via Sp1-binding sites by tamoxifen in estrogen receptor-negative lung cancer cells. Oncogene 19, 3766–3773 [DOI] [PubMed] [Google Scholar]
- 32. Prowse D. M., Bolgan L., Molnár A., Dotto G. P. (1997) Involvement of the Sp3 transcription factor in induction of p21Cip1/WAF1 in keratinocyte differentiation. J. Biol. Chem. 272, 1308–1314 [DOI] [PubMed] [Google Scholar]
- 33. Shen G., Xu C., Chen C., Hebbar V., Kong A. N. (2006) p53-independent G1 cell cycle arrest of human colon carcinoma cells HT-29 by sulforaphane is associated with induction of p21Cip1 and inhibition of expression of cyclin D1. Cancer Chemother. Pharmacol. 57, 317–327 [DOI] [PubMed] [Google Scholar]
- 34. Balasubramanian S., Chew Y. C., Eckert R. L. (2011) Sulforaphane suppresses polycomb group protein level via a proteasome-dependent mechanism in skin cancer cells. Mol. Pharmacol. 80, 870–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nakano K., Mizuno T., Sowa Y., Orita T., Yoshino T., Okuyama Y., Fujita T., Ohtani-Fujita N., Matsukawa Y., Tokino T., Yamagishi H., Oka T., Nomura H., Sakai T. (1997) Butyrate activates the WAF1/Cip1 gene promoter through Sp1 sites in a p53-negative human colon cancer cell line. J. Biol. Chem. 272, 22199–22206 [DOI] [PubMed] [Google Scholar]
- 36. Udvadia A. J., Templeton D. J., Horowitz J. M. (1995) Functional interactions between the retinoblastoma (Rb) protein and Sp-family members. Superactivation by Rb requires amino acids necessary for growth suppression. Proc. Natl. Acad. Sci. U.S.A. 92, 3953–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Adhikary G., Crish J. F., Gopalakrishnan R., Bone F., Eckert R. L. (2005) Involucrin expression in the corneal epithelium. An essential role for Sp1 transcription factors. Invest. Ophthalmol. Vis. Sci. 46, 3109–3120 [DOI] [PubMed] [Google Scholar]
- 38. Efimova T., Deucher A., Kuroki T., Ohba M., Eckert R. L. (2002) Novel protein kinase C isoforms regulate human keratinocyte differentiation by activating a p38 δ mitogen-activated protein kinase cascade that targets CCAAT/enhancer-binding protein α. J. Biol. Chem. 277, 31753–31760 [DOI] [PubMed] [Google Scholar]
- 39. Chew Y. C., Adhikary G., Wilson G. M., Reece E. A., Eckert R. L. (2011) Protein kinase C (PKC) δ suppresses keratinocyte proliferation by increasing p21(Cip1) level by a KLF4 transcription factor-dependent mechanism. J. Biol. Chem. 286, 28772–28782 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 40. Wong P. P., Pickard A., McCance D. J. (2010) p300 alters keratinocyte cell growth and differentiation through regulation of p21(Waf1/CIP1). PLoS. ONE 5, e8369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cheng F., McLaughlin P. J., Verderame M. F., Zagon I. S. (2009) The OGF-OGFr axis utilizes the p16INK4a and p21WAF1/CIP1 pathways to restrict normal cell proliferation. Mol. Biol. Cell 20, 319–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Aliouat-Denis C. M., Dendouga N., Van den Wyngaert I., Goehlmann H., Steller U., van de Weyer I., Van Slycken N., Andries L., Kass S., Luyten W., Janicot M., Vialard J. E. (2005) p53-independent regulation of p21Waf1/Cip1 expression and senescence by Chk2. Mol. Cancer Res. 3, 627–634 [DOI] [PubMed] [Google Scholar]
- 43. Devgan V., Mammucari C., Millar S. E., Brisken C., Dotto G. P. (2005) p21WAF1/Cip1 is a negative transcriptional regulator of Wnt4 expression downstream of Notch1 activation. Genes Dev. 19, 1485–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hauser P., Ma L., Agrawal D., Haura E., Cress W. D., Pledger W. J. (2004) Efficient down-regulation of cyclin A-associated activity and expression in suspended primary keratinocytes requires p21(Cip1). Mol. Cancer Res. 2, 96–104 [PubMed] [Google Scholar]
- 45. Kim J. H., Han Kwon K., Jung J. Y., Han H. S., Hyun Shim J., Oh S., Choi K. H., Choi E. S., Shin J. A., Leem D. H., Soh Y., Cho N. P., Cho S. D. (2010) Sulforaphane increases cyclin-dependent kinase inhibitor, p21 protein in human oral carcinoma cells, and nude mouse animal model to induce G2/M cell cycle arrest. J. Clin. Biochem. Nutr. 46, 60–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Meeran S. M., Patel S. N., Tollefsbol T. O. (2010) Sulforaphane causes epigenetic repression of hTERT expression in human breast cancer cell lines. PLoS. ONE 5, e11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dashwood R. H., Myzak M. C., Ho E. (2006) Dietary HDAC inhibitors. Time to rethink weak ligands in cancer chemoprevention? Carcinogenesis 27, 344–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kawamata N., Inagaki N., Mizumura S., Sugimoto K. J., Sakajiri S., Ohyanagi-Hara M., Oshimi K. (2005) Methylation status analysis of cell cycle regulatory genes (p16INK4A, p15INK4B, p21Waf1/Cip1, p27Kip1, and p73) in natural killer cell disorders. Eur. J. Haematol. 74, 424–429 [DOI] [PubMed] [Google Scholar]
- 49. Ying J., Srivastava G., Gao Z., Zhang X., Murray P., Ambinder R., Tao Q. (2004) Blood 103, 743–746 [DOI] [PubMed] [Google Scholar]
- 50. Majid S., Kikuno N., Nelles J., Noonan E., Tanaka Y., Kawamoto K., Hirata H., Li L. C., Zhao H., Okino S. T., Place R. F., Pookot D., Dahiya R. (2008) Genistein induces the p21WAF1/CIP1 and p16INK4a tumor suppressor genes in prostate cancer cells by epigenetic mechanisms involving active chromatin modification. Cancer Res. 68, 2736–2744 [DOI] [PubMed] [Google Scholar]
- 51. Renbaum P., Abrahamove D., Fainsod A., Wilson G. G., Rottem S., Razin A. (1990) Cloning, characterization, and expression in Escherichia coli of the gene coding for the CpG DNA methylase from Spiroplasma sp. strain MQ1(M.SssI). Nucleic Acids Res. 18, 1145–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Datto M. B., Yu Y., Wang X. F. (1995) Functional analysis of the transforming growth factor β-responsive elements in the WAF1/Cip1/p21 promoter. J. Biol. Chem. 270, 28623–28628 [DOI] [PubMed] [Google Scholar]
- 53. Zeng Y. X., Somasundaram K., el-Deiry W. S. (1997) AP2 inhibits cancer cell growth and activates p21WAF1/CIP1 expression. Nat. Genet. 15, 78–82 [DOI] [PubMed] [Google Scholar]
- 54. Banks E. B., Crish J. F., Eckert R. L. (1999) Transcription factor Sp1 activates involucrin promoter activity in non-epithelial cell types. Biochem. J. 337, 507–512 [PMC free article] [PubMed] [Google Scholar]
- 55. Banks E. B., Crish J. F., Welter J. F., Eckert R. L. (1998) Characterization of human involucrin promoter distal regulatory region transcriptional activator elements. A role for Sp1 and AP1 binding sites. Biochem. J. 331, 61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen M. J., Tang W. Y., Hsu C. W., Tsai Y. T., Wu J. F., Lin C. W., Cheng Y. M., Hsu Y. C. (2012) Apoptosis induction in primary human colorectal cancer cell lines and retarded tumor growth in SCID mice by sulforaphane. Evid. Based. Complement Alternat. Med. 2012, 415231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sakao K., Singh S. V. (2012) d,l-Sulforaphane-induced apoptosis in human breast cancer cells is regulated by the adapter protein p66Shc. J. Cell. Biochem. 113, 599–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hamsa T. P., Thejass P., Kuttan G. (2011) Induction of apoptosis by sulforaphane in highly metastatic B16F-10 melanoma cells. Drug Chem. Toxicol. 34, 332–340 [DOI] [PubMed] [Google Scholar]
- 59. Jeon Y. K., Yoo D. R., Jang Y. H., Jang S. Y., Nam M. J. (2011) Sulforaphane induces apoptosis in human hepatic cancer cells through inhibition of 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4, mediated by hypoxia inducible factor-1-dependent pathway. Biochim. Biophys. Acta 1814, 1340–1348 [DOI] [PubMed] [Google Scholar]
- 60. Jakubíková J., Sedlák J., Mithen R., Bao Y. (2005) Role of PI3K/Akt and MEK/ERK signaling pathways in sulforaphane- and erucin-induced phase II enzymes and MRP2 transcription, G2/M arrest, and cell death in Caco-2 cells. Biochem. Pharmacol. 69, 1543–1552 [DOI] [PubMed] [Google Scholar]
- 61. Cho S. D., Li G., Hu H., Jiang C., Kang K. S., Lee Y. S., Kim S. H., Lu J. (2005) Involvement of c-Jun N-terminal kinase in G2/M arrest and caspase-mediated apoptosis induced by sulforaphane in DU145 prostate cancer cells. Nutr. Cancer 52, 213–224 [DOI] [PubMed] [Google Scholar]
- 62. Traka M., Gasper A. V., Smith J. A., Hawkey C. J., Bao Y., Mithen R. F. (2005) Transcriptome analysis of human colon Caco-2 cells exposed to sulforaphane. J. Nutr. 135, 1865–1872 [DOI] [PubMed] [Google Scholar]
- 63. Bhamre S., Sahoo D., Tibshirani R., Dill D. L., Brooks J. D. (2009) Temporal changes in gene expression induced by sulforaphane in human prostate cancer cells. Prostate 69, 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dinkova-Kostova A. T., Jenkins S. N., Fahey J. W., Ye L., Wehage S. L., Liby K. T., Stephenson K. K., Wade K. L., Talalay P. (2006) Protection against UV-light-induced skin carcinogenesis in SKH-1 high-risk mice by sulforaphane-containing broccoli sprout extracts. Cancer Lett. 240, 243–252 [DOI] [PubMed] [Google Scholar]
- 65. Xu C., Huang M. T., Shen G., Yuan X., Lin W., Khor T. O., Conney A. H., Kong A. N. (2006) Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res. 66, 8293–8296 [DOI] [PubMed] [Google Scholar]
- 66. Shibata A., Nakagawa K., Yamanoi H., Tsuduki T., Sookwong P., Higuchi O., Kimura F., Miyazawa T. (2010) Sulforaphane suppresses ultraviolet B-induced inflammation in HaCaT keratinocytes and HR-1 hairless mice. J. Nutr. Biochem. 21, 702–709 [DOI] [PubMed] [Google Scholar]
- 67. Gao X., Dinkova-Kostova A. T., Talalay P. (2001) Powerful and prolonged protection of human retinal pigment epithelial cells, keratinocytes, and mouse leukemia cells against oxidative damage. The indirect antioxidant effects of sulforaphane. Proc. Natl. Acad. Sci. U.S.A. 98, 15221–15226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhu M., Zhang Y., Cooper S., Sikorski E., Rohwer J., Bowden G. T. (2004) Phase II enzyme inducer, sulforaphane, inhibits UVB-induced AP-1 activation in human keratinocytes by a novel mechanism. Mol. Carcinog. 41, 179–186 [DOI] [PubMed] [Google Scholar]
- 69. Charlot J. F., Nicolier M., Prétet J. L., Mougin C. (2006) Modulation of p53 transcriptional activity by PRIMA-1 and Pifithrin-α on staurosporine-induced apoptosis of wild-type and mutated p53 epithelial cells. Apoptosis. 11, 813–827 [DOI] [PubMed] [Google Scholar]
- 70. Okuyama R., LeFort K., Dotto G. P. (2004) A dynamic model of keratinocyte stem cell renewal and differentiation. Role of the p21WAF1/Cip1 and Notch1 signaling pathways. J. Investig. Dermatol. Symp. Proc. 9, 248–252 [DOI] [PubMed] [Google Scholar]
- 71. Schavinsky-Khrapunsky Y., Huleihel M., Aboud M., Torgeman A. (2003) Role of protein kinase C and the Sp1-p53 complex in activation of p21(WAF-1) expression by 12-O-tetradecanoylphorbol-13-acetate in human T cells. Oncogene 22, 5315–5324 [DOI] [PubMed] [Google Scholar]
- 72. Ho E., Clarke J. D., Dashwood R. H. (2009) Dietary sulforaphane, a histone deacetylase inhibitor for cancer prevention. J. Nutr. 139, 2393–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Go J. H. (2003) Methylation analysis of cyclin-dependent kinase inhibitor genes in primary gastrointestinal lymphomas. Mod. Pathol. 16, 752–755 [DOI] [PubMed] [Google Scholar]
- 74. Ocker M., Schneider-Stock R. (2007) Histone deacetylase inhibitors. Signaling toward p21Cip1/waf1. Int. J. Biochem. Cell Biol. 39, 1367–1374 [DOI] [PubMed] [Google Scholar]
- 75. Brooks C. L., Gu W. (2011) The impact of acetylation and deacetylation on the p53 pathway. Protein Cell 2, 456–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. McVean M., Xiao H., Isobe K., Pelling J. C. (2000) Increase in wild-type p53 stability and transactivational activity by the chemopreventive agent apigenin in keratinocytes. Carcinogenesis 21, 633–639 [DOI] [PubMed] [Google Scholar]