Background: Ectopically expressed hMSH2 is a tumor biomarker; however, the mechanism of its recognition is unclear.
Results: hMSH2 interacted with both TCRγδ and NKG2D on Vδ2 T cells, resulting in Vδ2 T cell activation. Its expression was up-regulated by EBV infection.
Conclusion: hMSH2 is a ligand for TCRγδ and NKG2D.
Significance: Recognition of hMSH2 by γδ T cells induces innate anti-tumor/virus immunity.
Keywords: Antigen, Innate Immunity, Natural Killer (NK) Cell, T Cell Receptor, Tumor Marker, gamma delta T Cells, Anti-tumor Immunity, Anti-virus Immunity, Endogenous Ligand, Human MutS Homologue 2 (MSH2)
Abstract
Human (h) MutS homologue 2, a nuclear protein, is a critical element of the DNA mismatch repair system. Our previous studies suggest that hMSH2 might be a protein ligand for TCRγδ. Here, we show that hMSH2 is ectopically expressed on a large panel of epithelial tumor cells. We found that hMSH2 interacts with both TCRγδ and NKG2D and contributes to Vδ2 T cell-mediated cytolysis of tumor cells. Moreover, recombinant human MSH2 protein stimulates the proliferation and IFN-γ secretion of Vδ2 T cells in vitro. Finally, hMSH2 expression is induced on the cell surface of Epstein-Barr virus-transformed lymphoblastoid cell lines, and the induction increases the sensitivity of these lymphoblastoid cell lines to γδ T cell-mediated cytolysis. Our data suggest that hMSH2 functions as a tumor-associated or virus infection-related antigen recognized by both Vδ2 TCR and NKG2D, and it plays a role in eliciting the immune responses of γδ T cells against tumor- and virus-infected cells. The recognition of ectopic surface-expressing endogenous antigen by TCRγδ and NKG2D may be an important mechanism of innate immune response to carcinogenesis and viral infection.
Introduction
Human γδ T cells account for a minor fraction of human peripheral T lymphocytes. The innate cell-like feature and MHC-independent antigen recognition define these cells as a special status in tumor surveillance and pathogen defense (1). However, the molecular mechanisms underlying tumor/infected cell recognition by γδ T cells are poorly understood because of the limited number of antigens/ligands recognized by γδ T cells that have been identified so far (2–4).
Substantial evidence indicates that human γδ T cells have the potential to recognize a diverse set of endogenous antigens/ligands, such as MHC class I-related chains A and B (5), members of CD1 family (6), UL16-binding proteins (ULBPs)2 (7, 8), heat shock proteins (HSPs) (9), and F1-ATPase-apolipoprotein A-I complex (10). Most of these molecules are either integral membrane proteins (11) or glycosylphosphatidylinositol-anchored proteins (12), typically expressed poorly or not at all by normal cells (12–14). However, their expression is substantially up-regulated in cancerous or stressed tissues (15–18), serving as a danger signal to alert the immune system.
hMSH2 is a critical element of the highly conserved DNA mismatch repair system, normally located in the nucleus and dimerized with hMSH3 or hMSH6 to form complexes essential for the maintenance of genome integrity (19). Inherited and acquired defects in hMSH2 are closely related to the pathogenesis of hereditary nonpolyposis colon cancer as well as various sporadic cancers (20). Recent findings indicate that hMSH2 also plays important roles in DNA damage signaling, apoptosis, and antibody class switch recombination in B cells (21–23). Previously, with a CDR3δ peptide-based affinity screening system, we identified hMSH2 as a putative tumor-associated protein ligand for human TCRγδ (24).
In this study, we further investigated the recognition of hMSH2 by human γδ T cell receptors and the role of hMSH2 in γδ T cell-mediated immune responses. We found that hMSH2, normally located in the nucleus, was ectopically expressed on the cell surface of different epithelial tumor cells, and its expression was induced by EBV transformation. To the best of our knowledge, our results provide the first evidence that cell surface-expressed hMSH2 is a ligand for both Vδ2 TCR and NKG2D receptors and engages in anti-tumor and anti-virus immunity by enhancing the γδ T cell-mediated cytolysis.
EXPERIMENTAL PROCEDURES
Cell Lines and Culture Medium
HeLa (cervical cancer) was cultured in DMEM supplemented with 10% FBS (Invitrogen). 803 (gastric carcinoma), HO8910 (ovarian cancer), HR8348 (colorectal cancer), and NCI-H520 (lung cancer) cells were maintained in 10% FBS RPMI 1640 medium (Invitrogen). SKOV3 (ovarian cancer), ES-2 (ovarian cancer), and HK-2 (proximal tubular cell line derived from normal kidney) cell lines were cultured in DMEM/F-12 medium (Nuyin, Beijing, China) supplemented with 10% FBS. Cell lines mentioned above were obtained from the Cell Culture Center, Institute of Basic Medicine, Chinese Academy of Medical Sciences. NK-92 cell line, provided by Prof. Zhigang Tian (University of Science and Technology of China), was cultured in α-minimal essential medium (Invitrogen) supplemented with 12.5% FBS, 12.5% horse serum, and 200 IU/ml recombinant IL-2 (Sigma). EBV-transformed B-LCLs 3D5 and LCL1/LCL2 were kind gifts from Prof. Liping Zhu and Dr. Lian Shen (Peking Union Medical College, China); LCL3 was previously established by our laboratory, and LCL4 was freshly generated from a healthy donor as described (7). Briefly, fresh PBMCs separated from healthy donors were transformed with exogenous EBV (B95.8 strain) and cultured in 15% FBS RPMI 1640 medium, 2 mm glutamine, initially supplemented with 0.05 mg/ml cyclosporin A (Novartis Pharma AG, Switzerland) and 0.5% (w/v) PHA (Sigma). All LCLs were maintained in RPMI 1640 medium supplemented with 10% FBS.
Expansion of Human γδ T Cells and Subset Separations
PBMCs from healthy donors were isolated by Ficoll-Hypaque density gradient centrifugation. PBMCs (2 × 106/ml) were cultured for 2–3 weeks in RPMI 1640 medium supplemented with 10% FBS, 2 mm l-glutamine, 10 mm HEPES, and 200 IU/ml recombinant IL-2 in a 24-well culture plate pre-coated with immobilized anti-pan-TCRγδ mAb (Immunotech, Beckman Coulter, Marseille, France). The expanded γδ T cells were used to separate Vδ1 and Vδ2 subsets with FITC-anti-TCRVδ1 (clone TS8.2, Pierce) and FITC anti-TCRVδ2 (clone IMMU389, Immunotech) using the anti-FITC microbeads (Miltenyi Biotec), following the manufacturer's directions.
Normal B lymphocytes were isolated from PBMCs using a human B cell isolated kit II (Miltenyi Biotec). The purity of enriched B cells was evaluated by flow cytometry using a phycoerythrin-conjugated CD19 mAb (Miltenyi Biotec).
Flow Cytometry
For cell surface hMSH2 staining, 1 × 106 cells resuspended in 50 μl of PBS containing 1% BSA were incubated with 2 μg of anti-MSH2 (H-300, Santa Cruz Biotechnology, Santa Cruz, CA) or rabbit IgG for 1 h at 4 °C, followed by staining with FITC-conjugated goat anti-rabbit secondary Ab (The Jackson Laboratory) for 30 min shielded from the light. Suspension cells were pretreated with 2 μl of FcR blocking reagent (Miltenyi Biotec) before adding primary antibody. Cells were fixed with 1% formaldehyde and analyzed on EPICS XL (Beckman Coulter, Bree) or C6 flow cytometer (Accuri Cytometers, Ann Arbor, MI). Data were analyzed with FCS Express Version 3.0 software (De Novo Software, Los Angeles). The percentage positive was calculated by Overton subtraction method. For one- or two-color staining of other cell surface antigens, cells were incubated with FITC/phycoerythrin-conjugated mAbs or isotype-matched control Abs for 30 min at 4 °C.
Synthesis and Reverse Transfection of siRNA Duplexes
Three pairs of siRNA duplexes were synthesized from Invitrogen. Sequences targeting hMSH2 gene (NM 000251.1) were as follows: siRNA I, 5′-TAG GAC TGT GTG AAT TCC CTG ATA A-3′; siRNA II, 5′-CAG CTA GAT GCT GTT GTC AGC TTT G-3′; and siRNA III, 5′-CAA TTG AAA GGA GTC TCC ACG TTC A-3′. StealthTM RNAi universal negative control med GC duplex (Invitrogen) was used as mock control. siRNA duplexes were reverse-transfected into tumor cells at a final concentration of 10 nm in a 6-well format according to the manufacturer's instructions. Briefly, 30 pmol of siRNA or negative control duplex was incubated with 5 μl of LipofectamineTM RNAiMAX (Invitrogen) in 500 μl of Opti-MEM I medium (Invitrogen) for 10–20 min at room temperature. 1.5–2.0 × 105 HeLa, 803, or 3D5 cells were suspended in 2500 μl of complete growth medium without antibiotics and added to each well with siRNA duplex-LipofectamineTM RNAiMAX complexes. The delivery of siRNAs was measured by parallel transfection with BLOCK-iTTM Alexa Fluor® red fluorescent oligonucleotide (Invitrogen) and analyzed by immunofluorescence microscopy at 6–12 h after transfection. Efficiency of gene knockdown was measured by quantitative real time RT-PCR and Western blot as described below; the cell surface expression of hMSH2 was analyzed by flow cytometry and confocal microscopy at 48 or 72 h after the transfection.
Real Time PCR
Total RNA of target cells (2 × 106) was isolated using TRIzol reagent (Promega, Madison, WI). cDNA was synthesized using oligo(dT) (Promega) and Moloney murine leukemia virus reverse transcriptase (Promega) in the reverse transcription reaction. Quantitative real time PCR was performed in a volume of 20 μl containing cDNA template, oligonucleotide primers, and SYBR Green PCR master mix (Applied Biosystems, Warrington, UK) using a 7500 real time PCR system (Applied Biosystems). Primers used for the amplification of hMSH2 and β-actin (endogenous control) were as follows: hMSH2, 5′-TTCATGGCTGAAATGTTGGA (forward) and 5′-ATGCTAACCCAAATCCATCG (reverse); and β-actin, 5′-AGAAAATCTGGCACCACACC(forward) and 5′-TAGCACAGCCTGGATAGCAA (reverse). Cycling conditions were as follows: 95 °C for 10 min, 40 cycles at 95 °C for 15 s and at 55 °C for 45 s. Data were analyzed by Sequence Detector Version 1.2 analysis software (Applied Biosystems).
Western Blot
Total protein from 2 × 106 interference or mock control cells was extracted by Cytobuster protein extraction reagent (Merck) and measured by Micro BCA protein assay kit (Pierce). The protein samples (20 μg) were separated on a 10% SDS-polyacrylamide gel and immunoblotted with mouse anti-hMSH2 mAb (65021-1-Ig, Protein Tech Group, Chicago) for 2 h and incubated with HRP-conjugated goat anti-mouse secondary Ab (The Jackson Laboratory) for 1 h at room temperature. Protein bands were visualized using Supersignal West Pico chemiluminescent substrate (Thermo Scientific). Anti-β-actin mAb (C4, Santa Cruz Biotechnology) served as endogenous control.
Confocal Microscopy
Cells of appropriate number (2–3 × 105) were grown on glass coverslips in 24-well plates overnight, fixed by PBS containing 4% paraformaldehyde for 10–15 min, and blocked in PBS containing 0.5% BSA for 30 min at 4 °C. Cells were then immunolabeled with anti-MSH2 (N-20, Santa Cruz Biotechnology) or rabbit IgG for 1 h and incubated with FITC-conjugated goat anti-rabbit secondary Ab for 30 min at 4 °C. Nuclei were detected by DAPI (Sigma) staining for 5 min. Images were taken with a Leica DMIRE2 inverted microscope fitted with a Leica TCS SP2 SE confocal imager. The objective was ×40 with a numerical aperture of 1.25 (oil objective).
Cytotoxicity and Antibody Blockade Assay
Cell cytotoxicity was determined by CytoTox 96 nonradioactive cytotoxicity assay Kit (Promega) (25). Inhibition of specific lyses by blocking antibodies (20 μg/ml) was performed by preincubating target cells with anti-MSH2 (N-20) and effector cells with anti-TCRγδ (B1.1) (7) and/or anti-NKG2D (149810, R & D Systems) (26) for 1 h at 4 °C. Effector cells of appropriate number were incubated with target cells (HeLa and 3D5, 5 × 104/ml; 803, 1 × 105/ml) in triplicate for 4–5 h at 37 °C. The release of lactate dehydrogenase and percent cytotoxicity were determined by the absorbance of the supernatants at 490 nm.
Surface Plasmon Resonance (SPR)
SPR experiments were performed using Biacore 3000 (GE Healthcare). Recombinant human NKG2D-Fc (R & D Systems) and IgG1 Fc (Sino Biological Inc, China) were coupled to the research-grade CM5 sensor chip using the standard amine coupling kit (GE Healthcare). The immobilization level was about 1300 resonance units. After buffer exchange to degassed 0.5% PBST (pH 7.4), affinity-purified soluble rhMSH2 (a six-histidine C-terminal fusion consisting of 323 amino acids and including the MutSI and MutSII domains) or BSA (as a control soluble protein) was injected at 30 μl/min over the immobilized NKG2D-Fc and IgG1 Fc at 25 °C. The binding response at each concentration of rhMSH2 (12.5–200 nm) or BSA (1 μm) was calculated by subtracting the equilibrium response measured in the IgG1 Fc flow cell from the response in the NKG2D flow cell. BIAevaluation version 4.1 software (Biacore AB, Uppsala, Sweden) was used for data analysis. Curve fitting was performed with Sigmaplot 11.0 software (Systat Software, Inc.).
Selective Cell Surface Biotinylation and Immunoblotting
The Pierce cell surface protein isolation kit (Thermo Scientific) was used for selective cell surface biotinylation according to the manufacturer's protocol. Briefly, four T75-cm2 flasks of 90–95% confluent HK-2 (serving as negative control), HeLa, 803, and NCI-H520 cells were washed twice with cold PBS and gently agitated in 0.5 mg/ml EZLink sulfo-NHS-SS-biotin for 30 min at 4 °C. 500 μl of quenching solution was sequentially added to each flask to terminate the biotinylation reaction. Cells were harvested and lysed in 500 μl of Lysis buffer in the presence of protease inhibitor mixture (Thermo Scientific). The cell lysates were clarified by centrifugation at 10,000 × g at 4 °C for 2 min. Labeled proteins were isolated by NeutrAvidin-agarose and eluted with 2× SDS-PAGE sample buffer containing 50 mm DTT. The purified proteins were analyzed by Western blot with anti-hMSH2 mAb (65021-1 Ig) using tumor nuclear proteins as references. The nuclear proteins were prepared by NucBuster protein extraction kit (Novagen) following the manufacturer's protocol.
3H-TdR Incorporation Assay
1 × 105 resting γδ T cells (Vδ2 T cells >85%) were seeded in triplicate in 96-well plates and incubated for 60 h with rhMSH2 (20 μg/well), irrelevant recombinant human His-tagged protein PKM2 (20 μg/well, serving as negative control), or EP5 (100 μg/well, serving as positive control) (24). Cells were cultured for an additional 12 h after adding [3H]thymidine (0.5 μCi/well). The cellular fraction was suspended in scintillation fluid and counted in a beta counter to evaluate 3H-TdR incorporation. The proliferation of Vδ2 T cells was determined by counts/min.
Enzyme-linked Immunosorbent Assay (ELISA)
2 × 106 resting γδ T cells (Vδ2 T cells >85%) were seeded in triplicate in a 24-well plate precoated with rhMSH2 (50 μg/ml), rPKM2 (50 μg/ml, serving as negative control), or anti-pan-TCRγδ mAb (5 μg/ml, serving as positive control). IFN-γ production in cell-free supernatants was determined by ELISA (R & D Systems) after 48 h of incubation.
Statistics
The statistical significance of differences between experimental groups was determined with Student's t test, with p values of less than 0.05 considered significant.
RESULTS
Ectopic Expression of hMSH2 on Epithelial Tumor Cell Lines
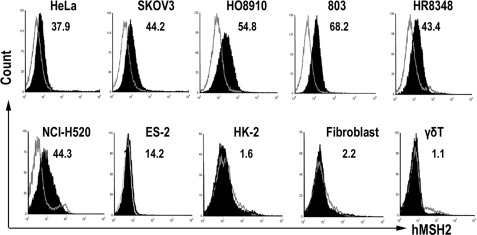
Cell surface expression is a prerequisite for hMSH2 as a ligand for γδ T cells. We examined the surface expression of hMSH2 on seven human epithelial tumor cell lines, including HeLa, SKOV3, HO8910, ES-2, 803, HR8348, and NCI-H520. HK-2, fibroblasts (derived from the normal tissues of an ovarian cancer patient), and γδ T cells were used as normal cell controls. Flow cytometric analysis with specific anti-hMSH2 Ab (H-300) revealed that 10–70% of the cell population among each of the tested tumor lines expressed hMSH2, whereas normal control cells rarely expressed hMSH2 on their surface (<2.2%) (Fig. 1), suggesting broadly ectopic surface expression of hMSH2 during carcinogenesis, which raises the possibility as a ligand for γδ T cells.
FIGURE 1.
Ectopically expressed hMSH2 on human epithelial tumor cell surface. Seven tumor cell lines (HeLa, SKOV3, HO8910, 803, HR8348, NCI-H520, and ES-2) as well as three normal cell controls (HK-2, human fibroblast, and γδ T cells) were stained with H-300 (black full histogram) or isotype-matched control IgG (gray line histogram) and analyzed by flow cytometry. Numbers are percentage of positive cells. Data shown are representative of at least three independent experiments.
Surface Expression of hMSH2 Enhanced γδ T Cell-mediated Tumor Cytolysis in Vitro
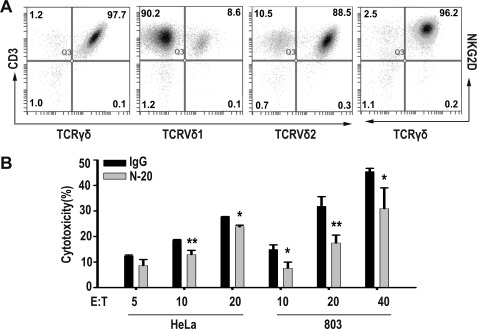
To investigate the significance of ectopically expressed hMSH2 in γδ T cell-mediated anti-tumor immunity, we chose HeLa, 803, and NCI-H520, three tumor cell lines expressing significant surface hMSH2, as targets for specific antibody blockade and target gene knockdown with siRNAs in cytolysis assays. We used γδ T cells that were stimulated and expanded with immobilized anti-pan-TCRγδ mAb as effectors. The expanded γδ T cells were mainly composed of Vδ2 T subset (80–95%) and expressed NKG2D at day 14 (Fig. 2A). Anti-MSH2 Ab (N-20) significantly blocked γδ T cell-mediated cytolysis of HeLa and 803 cells (Fig. 2B).
FIGURE 2.
Anti-hMSH2 Ab (N-20) significantly blocked the γδ T cell-mediated cytolysis of HeLa and 803 cells. A, phenotype of anti-pan-TCRγδ mAb expanded γδ T cells was analyzed by flow cytometry at day 14. Numbers in the picture are percentage of positive cells. B, antibody blockade assay of γδ T cell-mediated cytolysis of HeLa and 803 cells at various E:T ratios. *, p < 0.05; **, p < 0.01, when compared with IgG control.
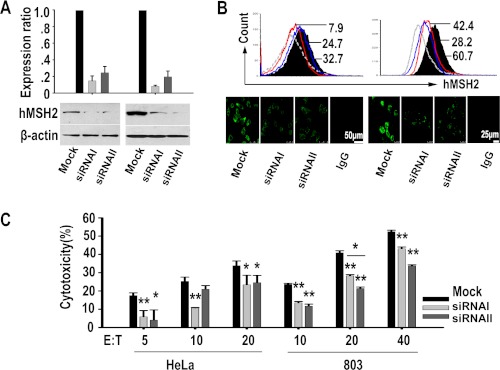
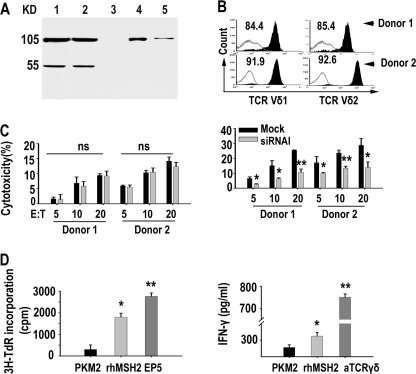
To further validate the hMSH2-mediated specific recognition by γδ T cells, we used small RNA interference to knock down hMSH2 mRNA expression in HeLa and 803 cell lines (Fig. 3). Forty eight hours after specific siRNAs transfection, siRNA duplex I and II resulted in 80 and 66% reduction of hMSH2 mRNA expression in HeLa cells and 92 and 80% reduction in 803 cells, when compared with the mock control (Fig. 3A, top panels). Reduced hMSH2 protein expression was confirmed by Western blot of total protein extracted from siRNA-treated cells or mock control cells (Fig. 3A, bottom panels). Decreased surface expression of hMSH2 on siRNA-treated HeLa and 803 cells was measured by flow cytometry and confocal microscopy (Fig. 3B). The cytotoxicity of human γδ T cells against siRNA-treated HeLa or 803 cells was compared with that of mock controls. At various E:T ratios, the decreased hMSH2 expression with siRNA I or II interference resulted in significantly reduced cytolysis of HeLa and 803 cells by γδ T cells (Fig. 3C). Similar effects were observed in NCI-H520 cells (data not shown). The above results from N-20 Ab blocking or siRNA interference suggest that ectopically expressed hMSH2 mediates the recognition of carcinoma cells by human γδ T cells and contributes to γδ T cell-mediated tumor cytolysis.
FIGURE 3.
hMSH2 knockdown with specific siRNAs decreased the γδ T cell-mediated cytolysis of HeLa and 803 cells. A, top panels, mRNA levels of hMSH2 were determined by quantitative RT-PCR 48 h after siRNA treatments. Data were expressed as means ± S.D. of three independent experiments; bottom panels, 48 h (HeLa) or 72 h (803) after siRNA treatments, hMSH2 protein expression was analyzed by Western blot with anti-hMSH2 mAb. B, surface expression of hMSH2 analyzed by flow cytometry (black full histogram, mock control; gray line, isotype control; red line, siRNA I; blue line, siRNA II) and confocal microscopy, scale bar, 50 μm (HeLa, left panels) or 25 μm (803, right panels). C, γδ T cell-mediated cytotoxicity assays for target cells with hMSH2 knockdown. *, p < 0.05; **, p < 0.01, when compared with mock control. Flow cytometry histograms and images were representative of three independent experiments.
Both TCRγδ and NKG2D Were Involved in Recognition of Surface hMSH2
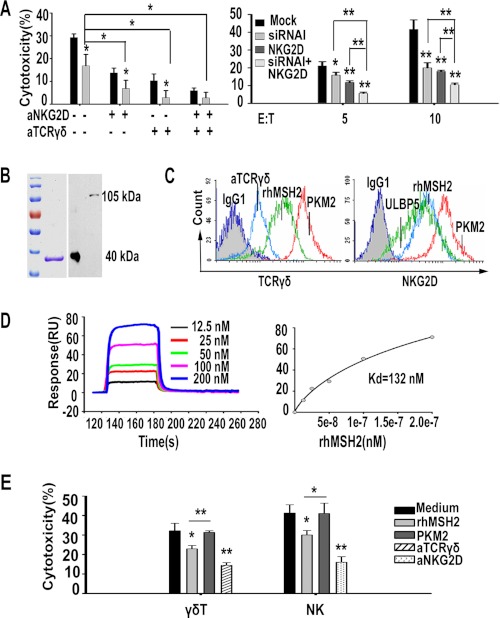
γδ T cells express two main types of recognition receptors, TCRγδ and NKG2D. To determine the functional involvement of TCRγδ and NKG2D in recognizing hMSH2, the expanded γδ T effector cells were pretreated with anti-TCRγδ (B1.1) and/or anti-NKG2D (149810) mAb before incubation with siRNA I-treated HeLa cells or mock control cells. The ectopically expressed hMSH2-mediated recognition by γδ T cells was significantly inhibited by anti-TCRγδ (B1.1), anti-NKG2D (149810), or both mAbs (Fig. 4A, left panel). The combined use of anti-TCRγδ/anti-NKG2D and siRNA I further decreased the cytotoxicity of γδ T effectors when compared with use of anti-TCRγδ/anti-NKG2D or siRNA I alone, suggesting that both TCRγδ and NKG2D may participate in hMSH2-mediated recognition and cytolysis of carcinoma cells by human γδ T cells. Considering the significance of hMSH2 as a NKG2D ligand, we confirmed the recognition of hMSH2 by NKG2D using the NK-92 cell line as effectors. Combination of anti-NKG2D (149810) and siRNA I treatment resulted in a notable decrease in the cytotoxicity of NK-92 cells compared with that using anti-NKG2D or siRNA I alone (Fig. 4A, right panel).
FIGURE 4.
Recognition and binding of hMSH2 by both TCRγδ and NKG2D. A, hMSH2-mediated recognition of HeLa cells by human γδ T cells (left panel) or NK-92 cells (right panel) was blocked by anti-TCRγδ and/or anti-NKG2D mAbs. *, p < 0.05; **, p < 0.01, compared with mock control. B, affinity-purified rhMSH2 (around 40 kDa) was verified by reducing SDS-PAGE following Coomassie Blue staining (left panel) and was blotted with anti-MSH2 mAb (65021-1 Ig) (right panel). Full-length hMSH2 protein in HeLa cell lysates showed a mass of 105 kDa. C, flow cytometric analyses of specific binding by TCRγδ (left panel) and NKG2D (right panel) to rhMSH2. D, SPR analysis of NKG2D interacting with rhMSH2. Left panel, BIAcore sensorgram measuring binding between recombinant hMSH2 at gradient concentrations and immobilized NKG2D; right panel, plots of the equilibrium binding responses from the sensorgrams as a function of hMSH2 concentration. E, preincubation with rhMSH2 (20 μg/well) significantly reduced the cytolysis of γδ T cells (E/T 20:1) and NK cells (E/T 10:1) toward HeLa cells. *, p < 0.05; **, p < 0.01, compared with nontreated or rPKM2 (20 μg/well)-treated groups.
We used affinity-purified rhMSH2, which was judged homogeneous by a single band on 10% SDS-polyacrylamide gel following Coomassie Blue staining (Fig. 4B), to confirm the specific binding of hMSH2 by TCRγδ or NKG2D receptors. Expanded γδ T cells were incubated with rhMSH2, rPKM2 (negative control for both TCRγδ and NKG2D), anti-TCRγδ (positive control for TCRγδ), or ULBP5 (positive control for NKG2D) before staining with FITC-conjugated anti-pan-TCRγδ or anti-NKG2D (1D11). As shown in Fig. 4C, decreased fluorescence intensity of TCRγδ and NKG2D was observed in the rhMSH2 preincubated group and the positive control groups, suggesting a specific binding of hMSH2 to both receptors. Furthermore, SPR analysis suggested a direct binding of rhMSH2 to NKG2D-Fc. The Kd value derived from nonlinear curve fitting of the standard Langmuir binding isotherm was 132 nm for rhMSH2 (Fig. 4D). Preincubation with soluble rhMSH2 protein for 1 h at 4 °C resulted in a significant decrease of killing activity of γδ T cells and NK cells against HeLa cells when compared with rPKM2 blocking group and nontreated group (Fig. 4E). These results, together with our previous data (24, 27), support that γδ T cells recognize hMSH2 via both TCRγδ and NKG2D.
Surface hMSH2 Expressed on Epithelial Tumor Cells Was a Full-length Protein
To characterize the cell surface-expressed hMSH2, the membrane proteins of HeLa, 803, NCI-H520, and HK-2 cells were biotinylated and purified. Western blot analysis with anti-hMSH2 mAb (65021-1 Ig) demonstrated that the membrane-bound hMSH2 was a full-length protein with a mass of 105 kDa (Fig. 5A). No truncated form of hMSH2 protein was detected.
FIGURE 5.
hMSH2-stimulated activation of Vδ2 T subset in vitro. A, ectopically expressed hMSH2 on HeLa (lane 4) and 803 (lane 5) cell surface is a full-length protein with a mass of 105 kDa. HK-2 cell surface protein (lane 3) and HeLa, 803 nuclear protein (lanes 1 and 2) were used as negative and positive controls for Western blot. The nuclear protein extracted from HeLa and 803 cells displayed an additional 55-kDa band. B, purity evaluation of sorted Vδ1 and Vδ2 cells with flow cytometry. Numbers shown are percentage of positive cells. C, Vδ1 and Vδ2 T cell-mediated cytolysis of HeLa cells. siRNA I-treated HeLa cells exhibited decreased sensitivity to Vδ2-mediated cytolysis. Data shown are representative of three independent experiments from two donors. *, p < 0.05; **, p < 0.01, compared with mock control. ns, no significance, D, rhMSH2 activates expanded γδ T cells (Vδ2 >85%) by triggering proliferative (left panel) and IFN-γ (right panel) responses. Data shown are means ± S.D. (error bar) of three independent experiments. *, p < 0.05; **, p < 0.01, compared with irrelevant protein control PKM2.
hMSH2 Preferentially Activates Vδ2 Subset in Vitro
To determine the specific subset that recognizes the surface-expressed hMSH2 on tumor cells, the purified Vδ1 and Vδ2 subsets (purity 84–95%) were used as effector cells in cytotoxicity assays (Fig. 5B). The Vδ1 subset exhibited lower cytotoxicity against HeLa cells compared with the Vδ2 subset. The cytotoxicity was not inhibited by siRNA I treatment (Fig. 5C, left panel). In contrast, Vδ2 T cell-mediated cytotoxicity was significantly reduced in siRNA I-treated groups (Fig. 5C, right panel). These results indicate that hMSH2 is preferentially recognized by Vδ2 T cells. This is consistent with the fact that hMSH2 was identified using a Vδ2 CDR3 peptide (OT3) as the probe (24).
rhMSH2 was used to stimulate proliferation and IFN-γ production of Vδ2 T cells in vitro. Incubation of expanded Vδ2 T cells (purity >85%) with immobilized rhMSH2 resulted in marked proliferation (p < 0.05) (Fig. 5D, left panel) and a significant increase in IFN-γ concentration (p < 0.05) (Fig. 5D, right panel) compared with irrelevant recombinant control protein PKM2, demonstrating the ability of hMSH2 to activate human Vδ2 T cells in vitro.
Inducible Expression of hMSH2 on EBV-transformed Human PBMCs
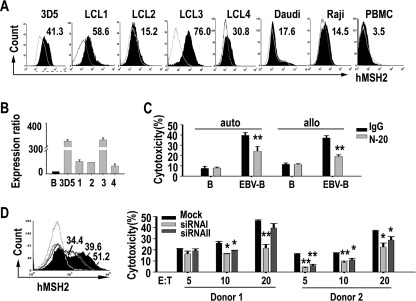
γδ T cells have been implicated in immune responses against EBV infection (28). Some γδ T cells ligands can be induced on EBV-infected B cells and facilitate γδ T cell-mediated killing (7, 16). To determine the role of hMSH2 in γδ T cell-mediated anti-viral immunity, we infected PBMCs isolated from healthy donor with EBV and analyzed hMSH2 expression. We found that surface hMSH2 appeared on EBV-transformed LCLs but not on untreated PBMCs (Fig. 6A). hMSH2 mRNA expression was also elevated in LCLs compared with normal B cells, indicating EBV infection up-regulates hMSH2 expression (Fig. 6B). The sensitivity of freshly generated EBV-transformed B cells to autologous or allogenic γδ T cell-mediated cytolysis was 2–3-fold greater than that of normal B cells (Fig. 6C). Treatment with N-20 Ab significantly reduced the specific cytolysis of these EBV-infected cells (Fig. 6C).
FIGURE 6.
Inducible expression of hMSH2 on EBV-transformed human PBMCs. A, flow cytometric analysis of surface expression of hMSH2 on EBV-transformed LCLs. Numbers shown are percentage of positive cells. B, relative quantification of hMSH2 mRNA expression in 3D5 and four other LCLs by quantitative RT-PCR. 1–4, LCL1–4. Data presented as N-fold differences in hMSH2 mRNA expression in the five LCL strains relative to normal B cells. Data presented were means ± S.D. of three independent experiments. C, cytotoxicity assays of autologous (auto) or allogenic (allo) γδ T cells against normal B cells or freshly generated EBV-transformed B cell line LCL4. The cytotoxicity of autologous/allogenic γδ T cells was blocked by N-20. The data shown represent one of three independent experiments with consistent results. *, p < 0.05; **, p < 0.01, compared with rabbit IgG control group. D, hMSH2 knockdown in 3D5 with siRNAs I or II resulted in a decreased cytotoxicity of γδ T cells. Left panel, 60 h after siRNAi (black line) or II (black line) transfection, surface expression of hMSH2 was examined by flow cytometry and compared with mock control (black full histogram). One representative result from three independent experiments was shown. Right panel, cytotoxicity assay of expanded γδ T cells against siRNA I or II treated-3D5 cells and mock control. Data shown are means ± S.D. of two independent experiments from two donors. *, p < 0.05; **, p < 0.01, compared with mock control.
To further confirm that the induced hMSH2 facilitates the killing of EBV-transformed cells by γδ T cells, specific siRNAs interference was done in LCL 3D5 (Fig. 6D). Consistent with that observed in carcinoma cells, specific cytolysis of siRNAs-treated 3D5 cells by γδ T cells was significantly reduced, demonstrating that the surface expression of hMSH2 mediated the recognition of LCLs by γδ T cells. Taken together, these results suggest that the cell surface-expressed hMSH2 is inducible and may mediate the specific clearance of EBV-infected cells by human γδ T cells.
DISCUSSION
Most γδ T cells have innate cell-like features and can be activated upon recognition of a diverse set of conserved yet poorly defined endogenous tumor-associated antigens/ligands through their TCR and/or NKG2D receptors (1). Understanding these natural ligands will be helpful not only for the elucidation of antigen recognition mechanism by which γδ T cells discriminate healthy from malignant cells but also for the development of efficient tumor immunotherapeutic strategies (29). In this study, we provide evidence for the surface-expressed hMSH2 on carcinoma cells to serve as a tumor-derived recognition antigen for human peripheral γδ T cells via both TCRγδ and NKG2D receptors. In addition, we show for the first time that hMSH2 is up-regulated on EBV-transformed B cells and involved in γδ T cell-mediated anti-viral cytotoxicity.
Based on our previous study, we hypothesized that hMSH2 may be a tumor-associated ligand for TCRγδ. To test this possibility, we first examined the cell surface expression of hMSH2 on cervical, gastric, ovarian, colorectal, and lung carcinoma cell lines. We observed a ubiquitous ectopic expression of hMSH2 at the surface of the tested tumor cell lines. Aberrant cell surface expression of hMSH2 provided a ligand for γδ T cell receptors. The ectopically expressed hMSH2 was correlated with an enhanced γδ T cell-cytolytic activity against carcinoma cells, suggesting an hMSH2-mediated recognition of carcinoma cells by γδ T cells. Such ectopic expression of hMSH2 on the cell surface may reflect an alert mechanism to the innate immune system when they suffer stress or are in a process of carcinogenesis.
Despite the central role of TCR-dependent recognition in γδ T cell activation, NKG2D takes a critical part in γδ T cytolytic response to tumor cells by binding to stress-inducible ligands MHC class I-related chains and ULBPs (30, 31). NKG2D has been described to express on essentially all Vδ2 T cells and on the immobilized mAb expanded γδ T cells. TCR- and NKG2D-mediated recognition of hMSH2 was supported by the results that hMSH2 silenced target cells became less sensitive to the cytotoxicity of γδ T cells. Specific binding analyses with flow cytometry and SPR provided further evidence for the dual recognition. Taking into account the constitutive expression of NKG2D on Vδ2 T cells, NK cells, and CD8+ T cells (32), the NKG2D-hMSH2 interaction could be important in hMSH2-associated tumor immune surveillance.
Vδ2 T cells are the main subset of human peripheral γδ T cells that recognize phospho-Ags and produce Th1 cytokines such as IFN-γ. They preferentially kill hematopoietic tumors (33, 34). More recent studies reveal their ability to kill carcinoma cells pretreated by phospho-Ags (35), suggesting that tumor recognition of these cells strongly depends on isopentenyl pyrophosphate production. However, it is likely that some other molecules are involved in the recognition process but are as yet undefined. hMSH2 as appears to be such a molecule. Indeed, the Vδ2 TCR-mediated recognition of hMSH2 was suggested by the specific binding of OT3-grafted Vδ2 TCR to rhMSH2 proteins in our previous study (24, 27). The functional significance of hMSH2 recognition by γδ T cells is supported by our observation that the surface expression of hMSH2 on HeLa cells is correlated with the cytotoxicity of Vδ2 T cells. hMSH2 readily activates Vδ2 T cells to proliferate and produce IFN-γ. Thus, hMSH2-mediated recognition of carcinoma cells by Vδ2 T cells opens new perspectives for Vδ2 T cell-based tumor immunotherapy.
The human herpesvirus EBV is thought to act as a triggering agent for autoimmune diseases and cancer in genetically susceptible individuals (36). Proliferation of γδ T cells and overexpression of HSPs and ULBPs in acute or chronic EBV infections have been reported previously (7, 16, 28, 37). The mRNA transcription and cell-surface expression of hMSH2 were up-regulated by EBV infection, leading to a substantial enhancement of γδ T cell cytotoxicity toward EBV-transformed cells. These results suggest that hMSH2 may be stress-inducible, a common feature of most γδ T cell ligands, and potentially important for immunity in EBV infections. Interestingly, although previous reports studied Vδ1 T cell response to EBV infection, our study suggests a role of Vδ2 T cells in anti-EBV immunity.
Thus, we envision that when the host cells are in a cancerous or distressed condition, ectopic expression of hMSH2 on the cell surface will occur to serve as a danger signal to alert the innate immune system. The signal is sensed by TCR and NKG2D expressed on Vδ2 T cells. TCRδ2 and NKG2D will work synergistically to initiate effective immune responses against tumor or virus-infected cells. However, it remains enigmatic how hMSH2 transports to the cell surface. As a nuclear protein, hMSH2 contains neither signal peptide sequence for membrane transport nor classic C-terminal KDEL sequence for endoplasmic reticulum retention. The re-location of this molecule to the membrane may be due to the change of membrane composition or the common alteration of other mismatch repair proteins (such as hMSH6) in malignant cells (38, 39). Besides, it is not known whether cell surface-expressed hMSH2 molecules themselves act as immunogenic determinants for human γδ T cells and NK cells, or whether they work as an Ag presentation molecule, just like CD1c and F1-ATPase do (40, 41). A better understanding of these underlying mechanisms will help us to determine the precise role of hMSH2 in γδ T cell-mediated immune surveillance and to optimize hMSH2-based therapeutic strategies for malignancy.
Acknowledgments
We thank Prof. Liping Zhu and Dr. Lian Shen for cell lines. Special thanks are extended to Dr. Ning Kang for insightful comments and suggestions.
This work was supported by Grant 30930083 from the National Natural Science Foundation.
- ULBP
- UL16-binding protein
- h
- human
- rh
- recombinant human
- EBV
- Epstein-Barr virus
- LCL
- lymphoblastoid cell line
- PBMC
- peripheral blood mononuclear cell
- SPR
- surface plasmon resonance
- TCR
- T cell receptor.
REFERENCES
- 1. Bonneville M., O'Brien R. L., Born W. K. (2010) γδ T cell effector functions. A blend of innate programming and acquired plasticity. Nat. Rev. Immunol. 10, 467–478 [DOI] [PubMed] [Google Scholar]
- 2. Wilson I. A., Stanfield R. L. (2001) Unraveling the mysteries of γδ T cell recognition. Nat. Immunol. 2, 579–581 [DOI] [PubMed] [Google Scholar]
- 3. Kreslavsky T., von Boehmer H. (2010) γδTCR ligands and lineage commitment. Semin. Immunol. 22, 214–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gomes A. Q., Martins D. S., Silva-Santos B. (2010) Targeting γδ T lymphocytes for cancer immunotherapy. From novel mechanistic insight to clinical application. Cancer Res. 70, 10024–10027 [DOI] [PubMed] [Google Scholar]
- 5. Groh V., Steinle A., Bauer S., Spies T. (1998) Recognition of stress-induced MHC molecules by intestinal epithelial γδ T cells. Science 279, 1737–1740 [DOI] [PubMed] [Google Scholar]
- 6. Spada F. M., Grant E. P., Peters P. J., Sugita M., Melián A., Leslie D. S., Lee H. K., van Donselaar E., Hanson D. A., Krensky A. M., Majdic O., Porcelli S. A., Morita C. T., Brenner M. B. (2000) Self-recognition of CD1 by γδ T cells. Implications for innate immunity. J. Exp. Med. 191, 937–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kong Y., Cao W., Xi X., Ma C., Cui L., He W. (2009) The NKG2D ligand ULBP4 binds to TCRγ9/γδ2 and induces cytotoxicity to tumor cells through both TCRγδ and NKG2D. Blood 114, 310–317 [DOI] [PubMed] [Google Scholar]
- 8. Lança T., Correia D. V., Moita C. F., Raquel H., Neves-Costa A., Ferreira C., Ramalho J. S., Barata J. T., Moita L. F., Gomes A. Q., Silva-Santos B. (2010) The MHC class Ib protein ULBP1 is a nonredundant determinant of leukemia/lymphoma susceptibility to γδ T-cell cytotoxicity. Blood 115, 2407–2411 [DOI] [PubMed] [Google Scholar]
- 9. Hirsh M. I., Junger W. G. (2008) Roles of heat shock proteins and γδ T cells in inflammation. Am. J. Respir. Cell Mol. Biol. 39, 509–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scotet E., Martinez L. O., Grant E., Barbaras R., Jenö P., Guiraud M., Monsarrat B., Saulquin X., Maillet S., Estève J. P., Lopez F., Perret B., Collet X., Bonneville M., Champagne E. (2005) Tumor recognition following Vγ9Vδ2 T cell receptor interactions with a surface F1-ATPase-related structure and apolipoprotein A-I. Immunity 22, 71–80 [DOI] [PubMed] [Google Scholar]
- 11. Botzler C., Li G., Issels R. D., Multhoff G. (1998) Definition of extracellular localized epitopes of Hsp70 involved in an NK immune response. Cell Stress Chaperones 3, 6–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Groh V., Bahram S., Bauer S., Herman A., Beauchamp M., Spies T. (1996) Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc. Natl. Acad. Sci. U.S.A. 93, 12445–12450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Multhoff G., Botzler C., Wiesnet M., Müller E., Meier T., Wilmanns W., Issels R. D. (1995) A stress-inducible 72-kDa heat-shock protein (HSP72) is expressed on the surface of human tumor cells but not on normal cells. Int. J. Cancer 61, 272–279 [DOI] [PubMed] [Google Scholar]
- 14. Poggi A., Venturino C., Catellani S., Clavio M., Miglino M., Gobbi M., Steinle A., Ghia P., Stella S., Caligaris-Cappio F., Zocchi M. R. (2004) Vδ1 T lymphocytes from B-CLL patients recognize ULBP3 expressed on leukemic B cells and up-regulated by trans-retinoic acid. Cancer Res. 64, 9172–9179 [DOI] [PubMed] [Google Scholar]
- 15. Groh V., Rhinehart R., Secrist H., Bauer S., Grabstein K. H., Spies T. (1999) Broad tumor-associated expression and recognition by tumor-derived γδ T cells of MICA and MICB. Proc. Natl. Acad. Sci. U.S.A. 96, 6879–6884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kotsiopriftis M., Tanner J. E., Alfieri C. (2005) Heat shock protein 90 expression in Epstein-Barr virus-infected B cells promotes γδ T-cell proliferation in vitro. J. Virol. 79, 7255–7261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vetter C. S., Groh V., thor Straten P., Spies T., Bröcker E. B., Becker J. C. (2002) Expression of stress-induced MHC class I-related chain molecules on human melanoma. J. Invest. Dermatol. 118, 600–605 [DOI] [PubMed] [Google Scholar]
- 18. Rölle A., Mousavi-Jazi M., Eriksson M., Odeberg J., Söderberg-Nauclér C., Cosman D., Kärre K., Cerboni C. (2003) Effects of human cytomegalovirus infection on ligands for the activating NKG2D receptor of NK cells: Up-regulation of UL16-binding protein (ULBP) 1 and ULBP2 is counteracted by the viral UL16 protein. J. Immunol. 171, 902–908 [DOI] [PubMed] [Google Scholar]
- 19. Acharya S., Wilson T., Gradia S., Kane M. F., Guerrette S., Marsischky G. T., Kolodner R., Fishel R. (1996) hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc. Natl. Acad. Sci. U.S.A. 93, 13629–13634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chung D. C., Rustgi A. K. (1995) DNA mismatch repair and cancer. Gastroenterology 109, 1685–1699 [DOI] [PubMed] [Google Scholar]
- 21. Peters A. C., Young L. C., Maeda T., Tron V. A., Andrew S. E. (2003) Mammalian DNA mismatch repair protects cells from UVB-induced DNA damage by facilitating apoptosis and p53 activation. DNA Repair 2, 427–435 [DOI] [PubMed] [Google Scholar]
- 22. Seifert M., Scherer S. J., Edelmann W., Böhm M., Meineke V., Löbrich M., Tilgen W., Reichrath J. (2008) The DNA-mismatch repair enzyme hMSH2 modulates UV-B-induced cell cycle arrest and apoptosis in melanoma cells. J. Invest. Dermatol. 128, 203–213 [DOI] [PubMed] [Google Scholar]
- 23. Eccleston J., Yan C., Yuan K., Alt F. W., Selsing E. (2011) Mismatch repair proteins MSH2, MLH1, and EXO1 are important for class-switch recombination events occurring in B cells that lack nonhomologous end joining. J. Immunol. 186, 2336–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen H., He X., Wang Z., Wu D., Zhang H., Xu C., He H., Cui L., Ba D., He W. (2008) Identification of human T cell receptor γδ-recognized epitopes/proteins via CDR3δ peptide-based immunobiochemical strategy. J. Biol. Chem. 283, 12528–12537 [DOI] [PubMed] [Google Scholar]
- 25. Jenkins M. R., Tsun A., Stinchcombe J. C., Griffiths G. M. (2009) The strength of T cell receptor signal controls the polarization of cytotoxic machinery to the immunological synapse. Immunity 31, 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Halary F., Pitard V., Dlubek D., Krzysiek R., de la Salle H., Merville P., Dromer C., Emilie D., Moreau J. F., Déchanet-Merville J. (2005) Shared reactivity of Vδ2(neg) γδ T cells against cytomegalovirus-infected cells and tumor intestinal epithelial cells. J. Exp. Med. 201, 1567–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xi X., Guo Y., Chen H., Xu C., Zhang H., Hu H., Cui L., Ba D., He W. (2009) Antigen specificity of γδ T cells depends primarily on the flanking sequences of CDR3δ. J. Biol. Chem. 284, 27449–27455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. De Paoli P., Gennari D., Martelli P., Cavarzerani V., Comoretto R., Santini G. (1990) γδ T cell receptor-bearing lymphocytes during Epstein-Barr virus infection. J. Infect. Dis. 161, 1013–1016 [DOI] [PubMed] [Google Scholar]
- 29. Thedrez A., Sabourin C., Gertner J., Devilder M. C., Allain-Maillet S., Fournié J. J., Scotet E., Bonneville M. (2007) Self/non-self discrimination by human γδ T cells. Simple solutions for a complex issue? Immunol. Rev. 215, 123–135 [DOI] [PubMed] [Google Scholar]
- 30. Rincon-Orozco B., Kunzmann V., Wrobel P., Kabelitz D., Steinle A., Herrmann T. (2005) Activation of Vγ 9Vδ 2 T cells by NKG2D. J. Immunol. 175, 2144–2151 [DOI] [PubMed] [Google Scholar]
- 31. Nedellec S., Sabourin C., Bonneville M., Scotet E. (2010) NKG2D costimulates human V γ 9V δ 2 T cell antitumor cytotoxicity through protein kinase Cθ-dependent modulation of early TCR-induced calcium and transduction signals. J. Immunol. 185, 55–63 [DOI] [PubMed] [Google Scholar]
- 32. Groh V., Wu J., Yee C., Spies T. (2002) Tumor-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature 419, 734–738 [DOI] [PubMed] [Google Scholar]
- 33. Sturm E., Braakman E., Fisch P., Vreugdenhil R. J., Sondel P., Bolhuis R. L. (1990) Human V γ 9-V δ 2 T cell receptor-γδ lymphocytes show specificity to Daudi Burkitt's lymphoma cells. J. Immunol. 145, 3202–3208 [PubMed] [Google Scholar]
- 34. Gomes A. Q., Correia D. V., Grosso A. R., Lança T., Ferreira C., Lacerda J. F., Barata J. T., Silva M. G., Silva-Santos B. (2010) Identification of a panel of 10 cell surface protein antigens associated with immunotargeting of leukemias and lymphomas by peripheral blood γδ T cells. Haematologica 95, 1397–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Viey E., Fromont G., Escudier B., Morel Y., Da Rocha S., Chouaib S., Caignard A. (2005) Phosphostim-activated γδ T cells kill autologous metastatic renal cell carcinoma. J. Immunol. 174, 1338–1347 [DOI] [PubMed] [Google Scholar]
- 36. Rouse B. T., Sehrawat S. (2010) Immunity and immunopathology to viruses: What decides the outcome? Nat. Rev. Immunol. 10, 514–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cheung R. K., Dosch H. M. (1993) The growth transformation of human B cells involves superinduction of hsp70 and hsp90. Virology 193, 700–708 [DOI] [PubMed] [Google Scholar]
- 38. Knudsen N. Ø., Andersen S. D., Lützen A., Nielsen F. C., Rasmussen L. J. (2009) Nuclear translocation contributes to regulation of DNA excision repair activities. DNA Repair 8, 682–689 [DOI] [PubMed] [Google Scholar]
- 39. Meng X., Riordan N. H., Riordan H. D., Mikirova N., Jackson J., González M. J., Miranda-Massari J. R., Mora E., Trinidad Castillo W. (2004) Cell membrane fatty acid composition differs between normal and malignant cell lines. P R Health Sci. J. 23, 103–106 [PubMed] [Google Scholar]
- 40. Barral D. C., Brenner M. B. (2007) CD1 antigen presentation: How it works. Nat. Rev. Immunol. 7, 929–941 [DOI] [PubMed] [Google Scholar]
- 41. Mookerjee-Basu J., Vantourout P., Martinez L. O., Perret B., Collet X., Périgaud C., Peyrottes S., Champagne E. (2010) F1-adenosine triphosphatase displays properties characteristic of an antigen presentation molecule for Vγ9Vδ2 T cells. J. Immunol. 184, 6920–6928 [DOI] [PubMed] [Google Scholar]