Background: Vps30/Atg6 is responsible for both autophagy and vacuolar protein sorting.
Results: Structure of the Vps30 BARA domain was determined and its function was characterized.
Conclusion: BARA domain has a unique fold and is specifically required for autophagy.
Significance: This study will be a basis for elucidating the various functions of Vps30 homologs.
Keywords: Autophagy, Crystal Structure, Crystallography, Phosphatidylinositol 3-Kinase, Structural Biology, Atg6, Beclin 1, Vps30, Vacuolar Protein Sorting
Abstract
Vacuolar protein sorting 30 (Vps30)/autophagy-related protein 6 (Atg6) is a common component of two distinct phosphatidylinositol 3-kinase complexes. In complex I, Atg14 links Vps30 to Vps34 lipid kinase and exerts its specific role in autophagy, whereas in complex II, Vps38 links Vps30 to Vps34 and plays a crucial role in vacuolar protein sorting. However, the molecular role of Vps30 in each pathway remains unclear. Here, we report the crystal structure of the carboxyl-terminal domain of Vps30. The structure is a novel globular fold comprised of three β-sheet-α-helix repeats. Truncation analyses showed that the domain is dispensable for the construction of both complexes, but is specifically required for autophagy through the targeting of complex I to the pre-autophagosomal structure. Thus, the domain is named the β-α repeated, autophagy-specific (BARA) domain. On the other hand, the N-terminal region of Vps30 was shown to be specifically required for vacuolar protein sorting. These structural and functional investigations of Vps30 domains, which are also conserved in the mammalian ortholog, Beclin 1, will form the basis for studying the molecular functions of this protein family in various biological processes.
Introduction
Vacuolar protein sorting 30 (Vps30)4/autophagy-related protein 6 (Atg6) was originally identified by yeast genetic screening as an essential factor for targeting the vacuolar hydrolase, carboxypeptidase Y (CPY), to the vacuole (1). At that time, yeast genetic screening also identified Vps30 as an essential factor for autophagy (2, 3), a bulk degradation pathway, in which a double membrane-bound structure, an autophagosome, sequesters a portion of the cytoplasm and delivers it to the vacuole for degradation (4).
In yeast, two distinct phosphatidylinositol 3-kinase (PI 3-kinase) complexes (complexes I and II) have been reported (5), and share the following three proteins between them: Vps34, the sole PI 3-kinase in yeast, Vps15, a putative protein kinase that activates Vps34 (6, 7), and Vps30. In addition to these three common components, Atg14 and Vps38 are incorporated into complexes I and II, respectively, and contribute to the complex construction by linking the Vps34-Vps15 complex to Vps30 (5). Autophagy, as well as most yeast genes responsible for autophagy, is conserved in higher eukaryotes including mammals. In mammals, Beclin 1, the orthologue of yeast Vps30, is known to be a key regulator of mammalian autophagy (8, 9). Beclin 1 forms a complex with mammalian Vps34 (10), and recent identification of mammalian Atg14 (also called as Atg14L or Barkor) showed that Beclin 1 forms a PI 3-kinase complex whose composition is similar to that of the yeast PI 3-kinase complex I (11–14), although Beclin 1 interacts directly with Vps34 (13, 15). Beclin 1 also forms several distinct PI 3-kinase complexes, some of which contain UVRAG, a putative Vps38 orthologue (11–14), and appears to regulate various membrane trafficking events.
In addition to its complex construction, yeast Atg14 was shown to be crucial for targeting of the PI 3-kinase complex I to the pre-autophagosomal structure (PAS), a perivacuolar structure where most of the Atg proteins colocalize and contribute to autophagosome formation (16–18). Similarly, mammalian Atg14 was shown to target the mammalian PI 3-kinase complex I to a specific site in the endoplasmic reticulum, termed an omegasome, which is a putative site of autophagosome formation in mammals (19, 20). Vps38 was shown to be crucial for targeting of the PI 3-kinase complex II to endosomes in yeast (18). In mammals, UVRAG localizes at endosomes, suggesting that it functions similarly with Vps38 (11). Phosphatidylinositol 3-phosphates produced at autophagic membranes and endosomes by PI 3-kinase complexes recruit effector proteins to regulate each pathway (21–25). Although elucidation of the molecular roles of PI 3-kinase complexes and their components other than Vps30/Beclin 1 is underway, the molecular roles of Vps30/Beclin 1 remain poorly understood.
Thus far, structural study on Vps30/Beclin 1 family proteins has been limited; such research has been restricted to the Bcl-2 homology 3 (BH3) domain (∼20-residue fragment) of Beclin 1 as a complex with Bcl-2 family proteins (26–29). A coiled-coil domain (CCD) has been predicted to be located at the central region of Vps30/Beclin 1 family proteins, and Beclin 1 CCD was shown to mediate interactions with various target proteins including Atg14 and UVRAG (12–15). Residues 244–337 of Beclin 1, which correspond to the C-terminal portion of the CCD and its following ∼70-residue region, are highly evolutionarily conserved; therefore, the region was named the evolutionarily conserved domain (ECD) and its significance in autophagy was clarified (30). Despite the important roles of these conserved domains, their structural information has been totally lacking.
Here, we report the crystal structure of the C-terminal region of Vps30, which overlaps the C-terminal portion of the ECD and the extreme C-terminal region of Vps30. The structure is a novel globular fold comprising three β-sheet-α-helix repeats. In vivo studies have shown that the region is required for autophagy but not for vacuolar protein sorting. Thus, this region is named the β-α repeated, autophagy-specific (BARA) domain. Further analyses demonstrated that BARA is dispensable for the construction of PI 3-kinase complexes, but is crucial for the targeting of complex I to the PAS.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The genes encoding residues 1–557 (full-length), 1–319 (ΔBARA), 187–319 (CCD), and 309–557 of Saccharomyces cerevisiae Vps30 were amplified by polymerase chain reaction and cloned into pGEX6p-1 (GE Healthcare), whereas the gene encoding residues 73–123 of S. cerevisiae Atg14 was cloned into pET-28a(+) (Novagen) into which the glutathione S-transferase (GST) gene had already been inserted. To construct Escherichia coli co-expression plasmids encoding VPS30(187–319) and ATG14(73–123), the VPS30 gene was amplified by PCR and cloned into pET-11a (Novagen). Next, a fragment consisting of a ribosome binding site, GST gene, and the ATG14 gene derived from pET28a(+)-GST-ATG14 was ligated downstream of the VPS30 gene. The constructs were transformed into E. coli BL21(DE3) and expressed as a GST-fused protein. After cell lysis, GST-fused proteins were purified by affinity chromatography using a glutathione-Sepharose 4B (GS4B) column (GE Healthcare). In the case of GST-fused Vps30 proteins, GST was excised from the proteins with PreScission protease (GE Healthcare). Vps30(309–557) was then applied to a Resource S cation-exchange column (GE Healthcare) equilibrated with 20 mm HEPES, pH 6.8, and eluted by a linear gradient from 0 to 1 m NaCl in 20 mm HEPES, pH 6.8. Vps30(1–319) and Vps30(187–319) were again applied to a GS4B column after exchanging the solvent with phosphate-buffered saline using a HiPrep Desalting column (GE Healthcare). In the case of the co-expression experiments of Atg14 and Vps30, GST was excised from GS4B-purified Atg14(73–123) with PreScission protease and the sample was then again applied to a GS4B column after exchanging the solvent with phosphate-buffered saline using the HiPrep desalting column.
Limited Proteolysis for Crystallization
Purified Vps30(309-557) was incubated with trypsin (enzyme/substrate ratio: 1/300) for 2 h at 25 °C in 20 mm HEPES, pH 6.8, 200 mm NaCl. After addition of phenylmethylsulfonyl fluoride, the trimmed C-terminal domain of Vps30 was purified using a Superdex 75 gel filtration column (GE Healthcare) and concentrated to 4 mg/ml in 20 mm HEPES, pH 6.8, 500 mm NaCl. The concentrated sample was used for crystallization. MALDI-TOF mass analyses revealed that the 11 residues of the N-terminal (309–319) and 18 residues of the C-terminal (540–557) were removed from the sample.
X-ray Crystallography
Crystallization was performed by the sitting drop vapor diffusion method at 20 °C. A 2.0-μl protein solution was mixed with a 0.5 μl of reservoir solution consisting of 0.1 m sodium acetate, pH 4.5, and 2.4 m sodium acetate. For data collection, crystals were soaked in the reservoir solution supplemented with 20% glycerol, flash-cooled, and kept in a stream of nitrogen gas at −178 °C during data collection. Diffraction data of both native and selenomethionine-labeled crystals were collected on an ADSC Quantum 315 charge-coupled device detector using beamline BL-5A (KEK, Japan). Diffraction data were processed using the HKL2000 program suite (31). The initial phasing was performed by the single-wavelength anomalous dispersion method using peak data of the selenomethionine-labeled crystal. After identifying four selenium sites and calculating initial phases using the SOLVE program (32), density modification was performed using the RESOLVE program (33). Model building was performed manually using the COOT program (34), and crystallographic refinement was performed using the Crystallography and NMR system software (35). Data collection, phasing, and refinement statistics are summarized in Table 1.
TABLE 1.
Data collection, phasing, and refinement statistics
| Native | Selenomethionine | |
|---|---|---|
| Data collection statistics | ||
| Wavelength (Å) | 0.9670 | 0.9790 |
| Oscillation range (°) | 180 | 270 |
| Space group | P41212 | P41212 |
| Cell parameters (Å) | a = 59.96, c = 113.98 | a = 60.15, c = 114.57 |
| Resolution range (Å) | 50–2.3 (2.38–2.30) | 50–2.5 (2.59–2.50) |
| Observed reflections | 134,889 | 157,216 |
| Unique reflections | 9,844 | 13,927 |
| Completeness (%) | 99.9 (100.0) | 100.0 (100.0) |
| Rmerge(I)a | 0.074 (0.325) | 0.075 (0.387) |
| Phasing statistics | ||
| Resolution range (Å) | 50–3.2 | |
| No. of selenium sites | 4 | |
| Mean figure of merit | 0.27 | |
| Refinement statistics | ||
| Resolution range (Å) | 50–2.3 | |
| No. of protein atoms | 1,312 | |
| No. of water molecules | 42 | |
| R/Rfree | 0.209/0.243 | |
| R.m.s. deviationb from ideality | ||
| Bond length (Å) | 0.006 | |
| Angles (°) | 1.13 | |
a Rmerge(I) = (ΣΣ|Ii − 〈I〉|)/ΣΣIi, where Ii is the intensity of the ith observation and 〈I〉 is the mean intensity. Values in parentheses refer to the outer shell.
b R.m.s., root mean square.
Cell Strains and Media
We utilized standard methods for yeast manipulation (36, 37). Yeast cells used in this study are listed on Table 2. Yeast cells were incubated in YEPD (1% Bacto-yeast extract, 2% Bacto-peptone, and 2% d-glucose) or S.D. (0.17% yeast nitrogen base without amino acids or ammonium sulfate, 0.5% ammonium sulfate, and 2% d-glucose) + CA (0.5% casamino acid) medium containing appropriate amino acids.
TABLE 2.
Cell strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| SEY6210 | MATα leu2-3,112 ura3-52 his3Δ200 trp1Δ901 lys2-801 suc2-Δ9 | 48 |
| KVY135 | SEY6210 vps30Δ::LEU2 | 49 |
| KOY192 | SEY6210 pho8Δ60::kanMX | Lab stock |
| TKY1290 | SEY6210 VPS38-yEGFP::kanMX | This study |
| TKY1307 | TKY1290 vps30Δ::hphNT1 | This study |
| TKY1308 | KOY192 vps30Δ::hphNT1 | This study |
| TKY1647 | KVY135 mRFP-APE1:HIS3 | This study |
| TKY1675 | TKY1647 ATG14–2xyeGFP:kanMX | This study |
Plasmid Construction for Yeast Experiments
pRS316-based plasmids for Atg14-HA-GFP and Vps38-HA-GFP were generated as reported previously (18). To construct yeast expression plasmids encoding VPS30 tagged with 3× myc (pRS314-VPS30-myc), the 3× myc sequence was amplified by PCR and inserted into pRS314-VPS30 (18). Deletion mutants of VPS30 were produced by inverse PCR of pRS314-VPS30-myc. To construct multicopy plasmids overexpressing the variants, the SacI/SalI fragment derived from these plasmids was ligated into the SacI/SalI site of pRS424. To construct plasmids to express GFP-tagged Vps30 truncates in yeast, a linker sequence containing NheI and NotI sites was inserted into the site between the myc tag sequence and the terminal codon of pRS314-VPS30-myc plasmids. Then, the EGFP fragment was inserted into this site, resulting in pRS314-VPS30-myc-EGFP plasmids. All the constructs were sequenced to confirm their identities. To visualize PAS, mRFP-APE1 was inserted into yeast cells. The mRFP-APE1 fragment from pPS128 (38) was inserted into pRS303 using BamHI, resulting in pRS303-mRFP-APE1 (pTK2672). After digesting pTK2672 using AvrII, it was transformed into KVY135, resulting in TKY1647 yeast cells.
Coimmunoprecipitation
vps30Δ cells (KVY135) expressing Vps30 mutants (pRS424) and either Atg14-HA-GFP or Vps38-HA-GFP were cultured in S.D. + CA + Ade medium and harvested. After washing the cell pellets with distilled water, cells were suspended in Z buffer (50 mm Tris-HCl, pH 7.5, 1.2 m sorbitol, 1% yeast extract, 2% polypeptone, 1% glucose) containing 0.1 mg/ml of Zymolyase 100T (Seikagaku Corp.) and cultured at 30 °C for 20 min. Spheroplast cells were harvested by centrifugation at 500 × g for 5 min. After washing with Z buffer, the spheroplasts were suspended in lysis buffer (phosphate-buffered saline, pH 7.4, 1 mm EDTA, 1 mm EGTA, 0.5% Tween 20, 1 mm PMSF, 20 μg/ml of leupeptin, 20 μg/ml of benzamidin, 10 μg/ml of pepstatin, and 40 μg/ml of aprotinin) and incubated at 4 °C for 5 min. Samples were then centrifuged at 15,000 × g for 10 min. Supernatants were incubated with anti-c-Myc antibody 9E10 (Covance) at 4 °C for 1 h. After the addition of protein G-Sepharose 4 beads, samples were incubated for an additional hour at 4 °C. Beads were then washed three times with lysis buffer. Bound proteins were eluted with SDS sample buffer and separated by SDS-PAGE. Protein bands were detected by Western blotting using either anti-c-Myc 9E10 (Santa Cruz Biotechnology), anti-HA (Abcam), or anti-Vps34 antibodies.
In Vitro Pulldown Assay
Purified GST-Atg14CCII or GST was incubated with GS4B beads at 4 °C for 10 min. After washing the beads with phosphate-buffered saline, Vps30WT, Vps30ΔBARA, or Vps30CCD were added to the beads and further incubated at 4 °C or 10 min. After washing the beads three times with phosphate-buffered saline, proteins were then eluted with 10 mm glutathione in 50 mm Tris-HCl buffer, pH 8.0.
In Vivo Assays
To quantify the autophagic activity, we employed an Pho8Δ60 alkaline phosphatase assay as described previously (39). A CPY sorting assay was performed with the method modified from a previous report (40). Briefly, mid-log phase yeast cells (A600 = 1) were inoculated in S.D. + CA + Ura + Ade medium containing 50 mm KPO4, pH 5.7, for 2 h. The harvested cells and supernatant media were subjected to trichloroacetic acid (TCA) precipitation. Samples (I: intracellular, corresponding to 0.3 A600 unit; E: extracellular, corresponding to 1.5 A600 unit) were subjected to Western blotting using anti-CPY antibody (1:5000; Lab stock), or anti-Pgk1 antibody (1:10000; Molecular Probes) as a control of cytosolic protein.
Fluorescent Microscopic Observation
The intracellular localization of mRFP-Ape1, Vps30-GFP, Atg14-GFP, or Vps38-GFP fusion proteins were visualized using inverted fluorescent microscopes (IX-71; Olympus, Tokyo, Japan), equipped with an EM-CCD digital camera (ImagEM; Hamamatsu Photonics). Images were acquired using Aquacosmos 2.6 software (Hamamatsu Photonics) and processed using Photoshop software (Adobe Systems). To observe PAS, yeast cells were finally treated with 0.2 μg/ml of rapamycin (Sigma) for 1 h to induce autophagy.
RESULTS
Structure of Vps30BARA
S. cerevisiae Vps30 consists of 557 amino acids, and a coiled-coil motif was predicted in its central region (residues 187–319; supplemental Fig. S1). However, structural information on the C-terminal region of the Vps30 family proteins has not yet been reported. The C-terminal region (residues 320–539) of Vps30 was obtained by limited proteolysis, crystallized, and its structure was determined by x-ray crystallography. The structure was refined against 2.3 Å data to an R-factor of 0.209 and a free R-factor of 0.243 (Table 1). The region corresponding to amino acids 320–538 of Vps30 was modeled, but three loop regions (residues 397–411, 422–432, and 461–487) were omitted from the model because they lacked defined electron density. The structure is comprised of three α-helices (H1, H2, and H3) and three β-sheets (S1, S2, and S3) with an approximate 3-fold symmetry (Fig. 1A). S1, S2, and S3 are three-stranded, anti-parallel β-sheets resembling each other. The helices and the sheets are aligned in the order: S1-H1-S2-H2-S3-H3 from the N terminus to the C terminus (Fig. 1B). Three subdomains (S1-H1, S2-H2, and S3-H3) resemble each other and can be superimposed on each other with root mean square differences of 1.5–2.4 Å for main chain atoms (Fig. 1C); therefore, the subdomains are named repeat 1, repeat 2, and repeat 3, respectively. The hydrophobic residues in H1, H2, and H3 interact with each other (Fig. 1D, left) to form an inter-subdomain helix bundle. In addition, the hydrophobic residues in S1, S2, and S3 interact with the hydrophobic grooves formed between H1-H3, H1-H2, and H2-H3, respectively (Fig. 1D, right), stabilizing the helix bundle. These inter-subdomain interactions make the three subdomains into a stable globular fold.
FIGURE 1.
Structure of Vps30 BARA. A, overall structure of Vps30 BARA. The α-helices and β-strands are indicated with red helical ribbons and cyan arrows, respectively. Secondary structures are labeled, and residues adjacent to the disordered regions are numbered. Amino and carboxy termini are denoted as N and C, respectively. B, topology of Vps30 BARA. The α-helices and β-strands are indicated with boxes and arrows, respectively. Repeats 1, 2, and 3 are colored green, red, and cyan, respectively. The disordered regions are indicated with a broken line. C, superimposition of the secondary structural elements of repeats 2 and 3 on those of repeat 1. Coloring is as in B. D, left, ribbon representation of the three-helix bundle of BARA. Side chains involved in the interaction between helices are indicated with a stick model. Coloring is as in B. Right, ribbon representation of secondary structural elements of BARA. The side chains of conserved hydrophobic residues on the three sheets, which are bound to the grooves formed between helices, are indicated with a stick model. Coloring is as in B. All figures representing molecular structures were generated with PyMOL (47).
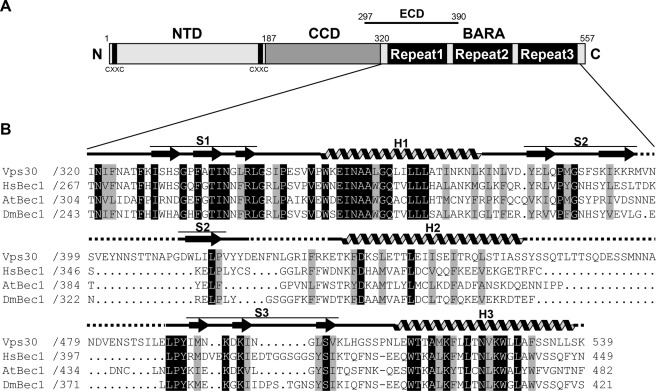
Sequence alignment of Vps30 with its higher eukaryote homologues (Beclin 1) shows that the C-terminal region is highly conserved among them (Fig. 2B). The Beclin 1 ECD corresponds to residues 297–390 of Vps30, which comprise the C-terminal portion of the predicted coiled-coil, repeat 1, and the N-terminal portion of repeat 2. The region C-terminal to the ECD is also conserved, especially the residues constituting the secondary structures of repeat 3 (Fig. 2B). Most of the hydrophobic residues contributing to the interaction among the three subdomains in Fig. 1D are conserved (Fig. 2B). These observations suggest that the three-repeat architecture of the C-terminal region of Vps30 is conserved among Vps30/Beclin 1 homologues. On the basis of its unique β-sheet-α-helix repeat and its functional significance in autophagy described below, the C-terminal region is named the BARA domain. Comparison of the BARA structure with the PDB data base using the DALI search engine (41) revealed that the three-repeat architecture of BARA, namely a three-helix bundle surrounded by three β-sheets, is novel.
FIGURE 2.
Domain architecture of Vps30 and sequence alignment with homologues. A, domain architecture of Vps30. B, sequence alignment of the C-terminal region of Vps30/Beclin 1 homologues. Perfectly conserved residues are shaded black, and residues conserved as hydrophobic are shaded gray. The secondary structural elements of BARA are shown above the alignment. HsBec1, Homo sapiens Beclin 1; At, Arabidopsis thaliana; Dm, Drosophila melanogaster.
Previous study showed that the ECD of Beclin 1 is crucial for mammalian autophagy (30). The ECD corresponds to the C-terminal portion of CCD and the N-terminal portion of BARA so that deletion of the ECD could influence both CCD and BARA structures. Therefore, we propose that the residues constituting BARA should be treated as a structural and functional unit when studying the function of Vps30/Beclin 1 domains. Consequently, Vps30 can be structurally divided into three regions: N-terminal domain that is structurally uncharacterized (NTD, residues 1–186), CCD (residues 187–319), and BARA (residues 320–557) (Fig. 2A).
Vps30BARA Is Required for Autophagy
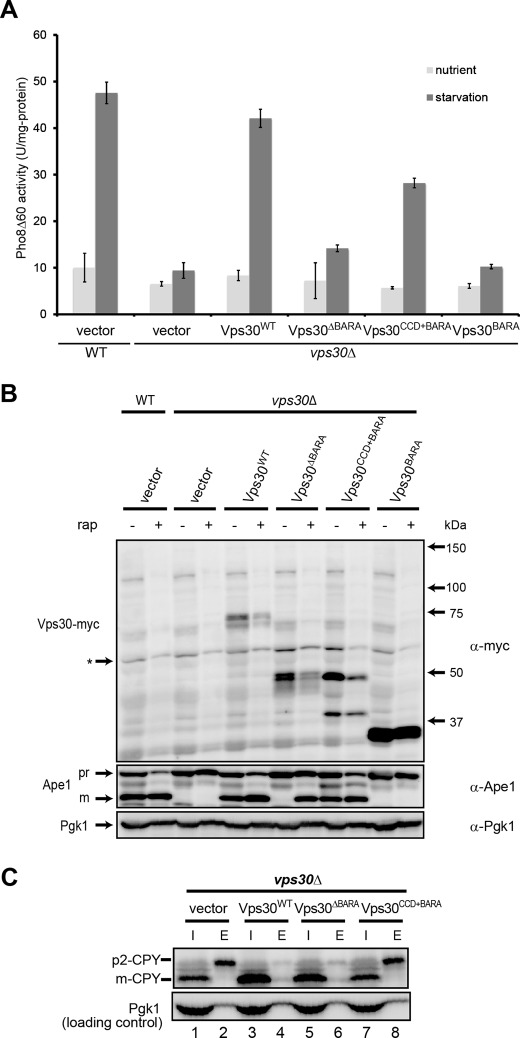
Because Vps30 BARA (Vps30BARA) is conserved among Vps30/Beclin 1 homologues, it is speculated that Vps30BARA plays important roles in the functioning of Vps30. To investigate the contribution of Vps30BARA on autophagy, cells expressing truncated forms of Vps30 were subjected to the Pho8Δ60 assay, a method commonly used for the assessment of autophagic activity (42, 43). This method utilizes a genetically engineered cytosolic form of an alkaline phosphatase, Pho8 (Pho8Δ60), which is delivered into the vacuole exclusively by autophagy, and activated. Thus, the autophagic activity correlates well with the phosphatase activity. As shown in Fig. 3A, wild-type cells as well as vps30Δ cells expressing wild-type Vps30 (Vps30WT cells) showed increased phosphatase activity in response to starvation conditions for 4 h, whereas vps30Δ cells showed no increase in the activity. Compared with these cells, Vps30ΔBARA or Vps30BARA cells showed only a slight increase in activity.
FIGURE 3.
Assessment of autophagy and CPY-sorting activities. A, autophagic activity of pho8Δ60 vps30Δ yeast cells (TKY1308) expressing Vps30 mutants. Autophagic activity was measured using Pho8Δ60 alkaline phosphatase (ALP) assay as described under “Experimental Procedures.” Error bars indicate the standard deviation of three independent experiments. B, Ape1 maturation in vps30Δ cells expressing Vps30 mutants. Lysates of the vps30Δ cells (KVY135) carrying pRS314-based VPS30 mutants were subjected to Western blotting and Ape1 and Vps30 bands were detected using anti-Ape1 and anti-Myc antibodies, respectively. As a loading control, Pgk1 was detected using anti-Pgk1 antibody. C, CPY sorting in vps30Δ cells expressing Vps30 mutants. The vps30Δ cells (KVY135) carrying pRS314-based VPS30 mutants were subjected to CPY sorting assay as described under “Experimental Procedures.”
Next, we examined autophagic activity of cells expressing truncated forms of Vps30 by monitoring aminopeptidase 1 (Ape1) maturation. The proform of Ape1 (prApe1) is transported to the vacuole via the cytoplasm-to-vacuole targeting pathway under nutrient-rich conditions and by autophagy in response to starvation conditions or rapamycin treatment. In the vacuole, prApe1 is processed into a mature form (mApe1), which can be monitored by Western blotting for Ape1. Therefore, the mApe1:prApe1 ratio observed in this assay reflects the progress of autophagy. As shown in Fig. 3B, Vps30WT and Vps30CCD+BARA cells showed a strong mApe1 band and a weak prApe1 band in response to rapamycin treatment, whereas vps30Δ cells and Vps30BARA cells showed no mApe1 band and a strong prApe1 band on the blot. These results are well consistent with those obtained by the Pho8Δ60 assay, showing that Vps30NTD is dispensable and Vps30BARA is insufficient for autophagy. Vps30ΔBARA cells showed strong mApe1 and prApe1 bands. Because monitoring Ape1 maturation is a much more sensitive method to detect autophagic activity compared with the Pho8Δ60 assay, this partial progression of Ape1 maturation may reflect the weak but remaining autophagic activity in these cells and suggest that Vps30BARA is important for efficient autophagy.
Vps30BARA Is Dispensable for Vacuolar Protein Sorting
In addition to autophagy, Vps30 is also involved in the vacuolar protein sorting pathway. The contribution of Vps30BARA on vacuolar protein sorting was studied by a carboxypeptidase Y (CPY) sorting assay, which observes the secretion of a vacuolar hydrolase CPY to the extracellular space from cells defective in Golgi apparatus-endosome transport, a critical step in CPY sorting that requires PI 3-kinase complex II. It has been reported that vps30Δ and vps38Δ cells but not atg14Δ cells show missorting of CPY, resulting in the secretion of the Golgi-derived p2 form of CPY (p2-CPY) (2, 22). We first confirmed that p2-CPY was secreted in vps30Δ cells, whereas in Vps30WT cells, p2-CPY was scarcely secreted and mCPY was accumulated in the vacuole (Fig. 3C, lanes 1–4). In Vps30ΔBARA cells, most CPY were detected as mCPY in the intracellular extracts, and p2-CPY was scarcely detected in the extracellular extracts as in Vps30WT cells (Fig. 3C, lanes 3–6). In contrast, in Vps30CCD+BARA cells, significant amounts of p2-CPY were detected in the extracellular extracts and mCPY was only weakly detected in the intracellular extracts as in vps30Δ cells (Fig. 3C, lanes 1, 2, 7, and 8). These results suggest that Vps30NTD but not Vps30BARA is required for vacuolar protein sorting.
Vps30BARA Is Not Required for Interaction with Atg14 or Vps38
Vps30 interacts directly with Atg14 and forms the autophagy-specific PI 3-kinase complex I. Because Vps30BARA has been shown as indispensible for autophagy, we next examined whether Vps30BARA is responsible for the interaction with Atg14 by coimmunoprecipitation experiments. Myc-tagged Vps30WT and three truncated forms of Vps30, Vps30ΔBARA, Vps30BARA, and Vps30CCD+BARA, were co-expressed with Atg14 fused to a 3× hemagglutinin (HA) tag and green fluorescent protein (GFP; Atg14-HA-GFP) in vps30Δ cells, and Myc-tagged Vps30s were pulled down with anti-Myc antibody and protein G-Sepharose beads. As shown in Fig. 4, Vps30WT, Vps30ΔBARA, and Vps30CCD+BARA but not Vps30BARA interacted with Atg14. These results suggest that Vps30BARA is dispensable for the interaction with Atg14. Similarly, immunoprecipitation experiments using vps30Δ cells co-expressing truncated mutants of Vps30 and Vps38-HA-GFP showed that Vps30BARA is dispensable for the interaction with Vps38. Vps34 was coimmunoprecipitated with Vps30WT and truncated forms of Vps30 except for Vps30BARA, which is consistent with the previous report that Vps30 interacts with Vps34 through either Atg14 or Vps38 (5).
FIGURE 4.
Analysis of Vps30 interaction with Atg14 and Vps38. Coimmunoprecipitation experiments were performed as described under “Experimental Procedures.” Protein bands for Vps30, Atg14, Vps34, and Vps38 were detected using anti-Myc, anti-HA, anti-Vps34, and anti-HA antibodies, respectively. The samples used for the upper three panels were prepared from vps30Δ yeast cells (KVY135) expressing Vps30 mutants and Atg14-HA-GFP, whereas those used for the bottom panel were prepared from KVY135 cells expressing Vps30 mutants and Vps38-HA-GFP. Asterisks indicate degraded products of Vps30.
Vps30BARA Is Required for Targeting of PI 3-Kinase Complex I to PAS
Vps30BARA is crucial for autophagy; nevertheless, Vps30BARA was shown to be dispensable for the construction of PI 3-kinase complex I. What then is the function of Vps30BARA in autophagy? To examine the role of Vps30BARA in PAS targeting of PI 3-kinase complex I, we first monitored the localization of GFP-tagged Vps30 truncates in cells expressing mRFP-tagged Ape1 as a PAS marker (Fig. 5A).
FIGURE 5.
Vps30BARA is required for efficient PAS targeting of Vps30 and Atg14. A, the vps30Δ mRFP-APE1 yeast cells (TKY1647) expressing Vps30 mutants fused to GFP were subjected to microscopic observation. Vps30-GFP and mRFP-Ape1 in cells with rapamycin treatment for 1 h were observed using fluorescent microscopy. B, the vps30Δ mRFP-APE1 ATG14-GFP yeast cells (TKY1675) expressing Vps30 mutants were subjected to microscopic observation. Atg14-GFP and mRFP-Ape1 in cells with rapamycin treatment for 1 h were observed using fluorescent microscopy. Arrows indicate the PAS. A bar indicates 2 μm.
In vps30Δ cells expressing Vps30WT-GFP (Vps30WT-GFP cells), GFP signals were observed as dots, some of which were merged with those of mRFP-Ape1 in response to rapamycin treatment. This result suggests that some populations of Vps30 localize to the PAS and others to endosomes as reported previously (18). Similar co-localization with mRFP-Ape1 was observed for Vps30CCD+BARA-GFP although the number of total dots was decreased. This result suggests that Vps30CCD+BARA localizes to the PAS, whereas its localization to endosomes is impaired. In Vps30ΔBARA-GFP cells, the GFP signals were observed as dots, which were seldom merged with those of mRFP-Ape1, suggesting that Vps30ΔBARA can localize to endosomes but not to the PAS. In Vps30BARA-GFP cells, the signals were diffused in the cytoplasm and not merged with the dots of mRFP-Ape1. In the latter two cells, the signals of mRFP-Ape1 were strong, suggesting that autophagy was inactive and mRFP-Ape1 accumulated in the cytosol. All these results are consistent with the activities of Vps30 truncates in autophagy and CPY sorting (Fig. 3).
We next monitored the localization of GFP-tagged Atg14 in cells expressing Vps30 truncates and mRFP-tagged Ape1 (Fig. 5B). In Vps30WT cells, the signals of Atg14-GFP were merged with those of mRFP-Ape1 in response to rapamycin treatment. In Vps30CCD+BARA cells, the signals of Atg14-GFP were observed as dots, some of which were merged with those of mRFP-Ape1. In Vps30ΔBARA cells, the signals of Atg14-GFP were also observed as dots, but less frequently, and they were seldom merged with those of mRFP-Ape1. In Vps30BARA cells, the signals of Atg14-GFP were diffused in the cytosol and not merged with those of mRFP-Ape1 as in the case in vps30Δ cells containing a control vector. These results are well consistent with the localization of Vps30 truncates, strongly suggesting that Vps30BARA is required for efficient PAS targeting of PI 3-kinase complex I.
Characterization of Functions of Vps30CCD and Vps30NTD
At this point, the autophagy-specific function of Vps30BARA has been established. But what then is the function of the other two domains, Vps30CCD and Vps30NTD? PAS targeting of PI 3-kinase complex I requires not only Vps30BARA but also Vps30CCD (Fig. 5) and Atg14 (18). Immunoprecipitation experiments suggest that Vps30CCD is crucial for the interaction with Atg14 (Fig. 4). These data suggest that one function of Vps30CCD is to link Vps30BARA and Atg14 together for PAS targeting of complex I. We subsequently studied the direct interaction between Vps30 and Atg14 using recombinant proteins. Three coiled-coil motifs (I, II, and III) were predicted in the N-terminal region of Atg14, among which the coiled-coil II (CCII) was shown to be sufficient for the interaction with Vps30 (18). GST-fused Atg14CCII (residues 73–123), Vps30WT, Vps30ΔBARA, and Vps30CCD were expressed in E. coli, purified, and used for pulldown assays. Although GST-Atg14CCII was unstable and the purified sample contained many degradation products, Vps30WT, Vps30ΔBARA, and Vps30CCD all interacted with GST-Atg14CCII (Fig. 6A). We also co-expressed GST-Atg14CCII with Vps30CCD in E. coil, and purified GST-Atg14CCII was co-eluted with Vps30CCD from glutathione-Sepharose 4B beads without degradation. These two proteins behaved as a complex during further purification (Fig. 6B and data not shown). These results suggest that Vps30CCD is sufficient for interaction and stabilization of Atg14.
FIGURE 6.
Vps30CCD directly interacts with Atg14CCII. A, in vitro pulldown assay between GST-Atg14CCII and Vps30 mutants. Asterisks indicate degraded products of GST-Atg14CCII and Vps30CCD. B, purification of GST-Atg14CCII coexpressed with Vps30CCD in E. coli. After purification with GS4B beads (lane 1), GST was excised from Atg14CCII with PreScission protease (lane 2) and free GST was removed from the sample using GS4B beads (lane 3). The asterisk indicates a degradation product of Vps30CCD. Protein bands in A and B were stained with Coomassie Brilliant Blue.
Vps30NTD was shown to be required for vacuolar protein sorting, but not autophagy (Fig. 3), which can be attributed to Vps30NTD for interaction with Vps38, but not with Atg14 (Fig. 4). To study the significance of the Vps30-Vps38 interaction through Vps30NTD on endosome targeting of Vps38, we monitored the intracellular localization of Vps38-GFP in cells expressing truncated forms of Vps30. As previously reported (18), Vps38 was localized to the punctate structures corresponding to endosomes in Vps30WT cells (Fig. 7). In Vps30ΔBARA cells, Vps38 showed a localization pattern similar to that in cells expressing Vps30WT, although the signal intensity was much lower (Fig. 7). In Vps30BARA or Vps30CCD+BARA cells, Vps38 was diffused in the cytosol and the signal intensity was decreased to an undetectable level as in the case of vps30Δ cells containing a control vector (Fig. 7). These results, together with those of the immunoprecipitation experiments, suggest that the Vps30-Vps38 interaction through Vps30NTD (and possibly Vps30CCD) is crucial for the interaction and stabilization of Vps38 and its targeting to endosomes.
FIGURE 7.
Vps30NTD is required for endosome-targeting of Vps38. The VPS38-GFP vps30Δ yeast cells (TKY1307) expressing Vps30 mutants in logarithmic phase were subjected to microscopic observation. Vps38-yEGFP was observed using fluorescent microscopy. A bar indicates 2 μm.
DISCUSSION
Thus far, structural information of the Vps30/Beclin 1 family proteins has been limited. Here, we succeeded in determining the structure of the C-terminal region of Vps30 by x-ray crystallography. The structure shows a unique β-sheet-α-helix repeat architecture, so it is named BARA (Fig. 1). Because the residues constituting the secondary structural elements are highly conserved among Vps30/Beclin 1 family proteins (Fig. 2B), the domain appears to be structurally and functionally conserved among them. Based on structural information and sequence alignment, we divided Vps30/Beclin1 into three domains: NTD, CCD, and BARA.
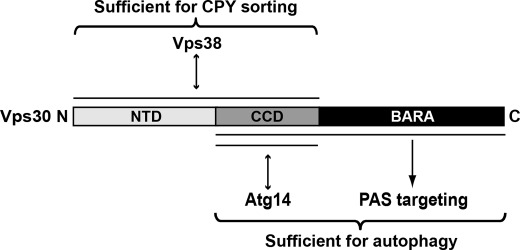
Based on this domain definition, we established the functions of NTD, CCD, and BARA of Vps30. They are summarized in Fig. 8. Vps30BARA is dispensable for the interaction with both Atg14 and Vps38 and for CPY sorting, but is crucial for autophagy through the targeting of the PI 3-kinase complex I to the PAS. On the other hand, Vps30NTD is necessary for the interaction with Vps38 but not with Atg14, and is crucial for CPY sorting but not for autophagy. Vps30CCD is crucial for the interaction with Atg14 and presumably with Vps38 too, and thus appears to be essential for both autophagy and CPY sorting. During truncation analysis, we succeeded in obtaining Vps30 mutants whose activity is restricted to either autophagy (Vps30CCD+BARA) or CPY sorting (Vps30ΔBARA). These Vps30 mutants would be helpful for studying the Vps30 functions in autophagy and CPY sorting separately. Furthermore, such truncation analyses would also be beneficial to study mammalian Beclin 1, which interacts with various factors and appears to play crucial roles in various membrane trafficking events besides autophagy (9, 11).
FIGURE 8.
Summary of the functions of Vps30 domains revealed in this study.
Because targeting of Vps30 to the PAS requires Atg14 (18), it is suggested that the Vps30-Atg14 complex is targeted to the PAS interdependently, for which Vps30CCD+BARA plays a crucial role. How then is the Vps30-Atg14 complex targeted to the PAS? Systematic and quantitative analysis of the PAS localization of Atg proteins using fluorescence microscopy showed that the Vps30-Atg14 complex localizes to the PAS depending on the presence of Atg9, Atg13, or Atg17 (44). Therefore, the simplest idea is that the Vps30-Atg14 complex localizes to the PAS through direct interaction with these proteins. Alternatively, it is also possible that the Vps30-Atg14 complex interacts with some unidentified factor(s) on the PAS. In either case, Vps30BARA may be directly involved in such interactions together with Atg14. It is not clear whether Vps30CCD is also directly involved in such interactions or whether its role is restricted to linking Vps30BARA and Atg14. Conserved residues are clustered on one surface of Vps30BARA (supplemental Fig. S2, left). Because this surface contains the N terminus, it may form a continuous surface with the Vps30CCD-Atg14 complex. We speculate that such a conserved continuous surface might be responsible for recognizing conserved factor(s) such as Atg9 and Atg13, through which the PI 3-kinase complex I might be targeted to the PAS. Likewise, PI 3-kinase complex II might be targeted to endosomes through the interaction between the Vps30ΔBARA-Vps38 complex and some factor(s) on endosomes. Very recently, crystal structures of the CCD and the C-terminal regions of Beclin 1 have been reported (45, 46), which show that the C-terminal region has a globular fold similar to Vps30 BARA and Beclin 1 ECD is a part of CCD and BARA. It was also reported that Beclin 1 BARA possesses an aromatic finger (Phe-Phe-Trp sequence) in the loop region, which mediates direct association with lipids, especially cardiolipin, and this lipid-binding ability is important for mammalian autophagy (45). However, sequence alignment shows that the aromatic finger is not conserved in Vps30 (Fig. 2B), and thus the PAS-targeting function of Vps30 BARA could not be attributed to lipid binding. Structural studies on the Vps30-Atg14 and Vps30-Vps38 complexes and identification of their interacting partner(s) are required for revealing the molecular mechanism of proper targeting of PI 3-kinase complexes.
Supplementary Material
Acknowledgments
We are grateful to Dr. Hitoshi Nakatogawa for providing helpful comments on the manuscript and Miou Matsunami and Manami Akioka for technical support. The synchrotron radiation experiments were performed at the beamline BL-5A at KEK, Japan.
This work was supported in part by Grants-in-aid for Scientific Research on Priority Areas (to N. N. N.) and Targeted Proteins Research Program (to F. I. and Y. O.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, by Grant for Basic Science Research Projects from The Sumitomo Foundation (to N. N. N.), and by a Leave a Nest grant BioGarage award (to T. K.).

This article contains supplemental Figs. S1 and S2.
- Vps
- vacuolar protein sorting
- Atg
- autophagy-related
- BARA
- β-α-repeated, autophagy-specific
- CCD
- coiled-coil domain
- CPY
- carboxypeptidase Y
- GS4B
- glutathione-Sepharose 4B
- NTD
- N-terminal domain
- PAS
- pre-autophagosomal structure
- ECD
- evolutionarily conserved domain
- prApe1
- proform of Ape1
- mApe1
- mature Ape1.
REFERENCES
- 1. Seaman M. N., Marcusson E. G., Cereghino J. L., Emr S. D. (1997) Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35 gene products. J. Cell Biol. 137, 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kametaka S., Okano T., Ohsumi M., Ohsumi Y. (1998) Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 273, 22284–22291 [DOI] [PubMed] [Google Scholar]
- 3. Tsukada M., Ohsumi Y. (1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169–174 [DOI] [PubMed] [Google Scholar]
- 4. Mizushima N., Yoshimori T., Ohsumi Y. (2011) The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132 [DOI] [PubMed] [Google Scholar]
- 5. Kihara A., Noda T., Ishihara N., Ohsumi Y. (2001) Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152, 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stack J. H., DeWald D. B., Takegawa K., Emr S. D. (1995) Vesicle-mediated protein transport. Regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J. Cell Biol. 129, 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stack J. H., Herman P. K., Schu P. V., Emr S. D. (1993) A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J. 12, 2195–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676 [DOI] [PubMed] [Google Scholar]
- 9. Cao Y., Klionsky D. J. (2007) Physiological functions of Atg6/Beclin 1. A unique autophagy-related protein. Cell Res. 17, 839–849 [DOI] [PubMed] [Google Scholar]
- 10. Kihara A., Kabeya Y., Ohsumi Y., Yoshimori T. (2001) Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2, 330–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Itakura E., Kishi C., Inoue K., Mizushima N. (2008) Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell 19, 5360–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matsunaga K., Saitoh T., Tabata K., Omori H., Satoh T., Kurotori N., Maejima I., Shirahama-Noda K., Ichimura T., Isobe T., Akira S., Noda T., Yoshimori T. (2009) Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 11, 385–396 [DOI] [PubMed] [Google Scholar]
- 13. Sun Q., Fan W., Chen K., Ding X., Chen S., Zhong Q. (2008) Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. U.S.A. 105, 19211–19216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhong Y., Wang Q. J., Li X., Yan Y., Backer J. M., Chait B. T., Heintz N., Yue Z. (2009) Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 11, 468–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liang C., Feng P., Ku B., Dotan I., Canaani D., Oh B. H., Jung J. U. (2006) Autophagic and tumor suppressor activity of a novel Beclin1-binding protein UVRAG. Nat. Cell Biol. 8, 688–699 [DOI] [PubMed] [Google Scholar]
- 16. Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. (2001) The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20, 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Suzuki K., Ohsumi Y. (2010) Current knowledge of the pre-autophagosomal structure (PAS). FEBS Lett. 584, 1280–1286 [DOI] [PubMed] [Google Scholar]
- 18. Obara K., Sekito T., Ohsumi Y. (2006) Assortment of phosphatidylinositol 3-kinase complexes. Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol. Biol. Cell 17, 1527–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Matsunaga K., Morita E., Saitoh T., Akira S., Ktistakis N. T., Izumi T., Noda T., Yoshimori T. (2010) Autophagy requires endoplasmic reticulum targeting of the PI 3-kinase complex via Atg14L. J. Cell Biol. 190, 511–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Axe E. L., Walker S. A., Manifava M., Chandra P., Roderick H. L., Habermann A., Griffiths G., Ktistakis N. T. (2008) Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182, 685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Obara K., Sekito T., Niimi K., Ohsumi Y. (2008) The ATG18-ATG2 complex is recruited to autophagic membranes via PtdIns(3)P and exerts an essential function. J. Biol. Chem. 283, 23972–23980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burda P., Padilla S. M., Sarkar S., Emr S. D. (2002) Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase. J. Cell Sci. 115, 3889–3900 [DOI] [PubMed] [Google Scholar]
- 23. Vanhaesebroeck B., Guillermet-Guibert J., Graupera M., Bilanges B. (2010) The emerging mechanisms of isoform-specific PI3K signaling. Nat. Rev. Mol. Cell Biol. 11, 329–341 [DOI] [PubMed] [Google Scholar]
- 24. Simonsen A., Tooze S. A. (2009) Coordination of membrane events during autophagy by multiple class III PI 3-kinase complexes. J. Cell Biol. 186, 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Obara K., Noda T., Niimi K., Ohsumi Y. (2008) Transport of phosphatidylinositol 3-phosphate into the vacuole via autophagic membranes in Saccharomyces cerevisiae. Genes Cells 13, 537–547 [DOI] [PubMed] [Google Scholar]
- 26. Feng W., Huang S., Wu H., Zhang M. (2007) Molecular basis of Bcl-xL's target recognition versatility revealed by the structure of Bcl-xL in complex with the BH3 domain of Beclin-1. J. Mol. Biol. 372, 223–235 [DOI] [PubMed] [Google Scholar]
- 27. Ku B., Woo J. S., Liang C., Lee K. H., Hong H. S., E X., Kim K. S., Jung J. U., Oh B. H. (2008) Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine γ-herpesvirus 68. PLoS pathogens 4, e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oberstein A., Jeffrey P. D., Shi Y. (2007) Crystal structure of the Bcl-XL-Beclin 1 peptide complex. Beclin 1 is a novel BH3-only protein. J. Biol. Chem. 282, 13123–13132 [DOI] [PubMed] [Google Scholar]
- 29. Sinha S., Colbert C. L., Becker N., Wei Y., Levine B. (2008) Molecular basis of the regulation of Beclin 1-dependent autophagy by the γ-herpesvirus 68 Bcl-2 homolog M11. Autophagy 4, 989–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Furuya N., Yu J., Byfield M., Pattingre S., Levine B. (2005) The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy, and tumor suppressor function. Autophagy 1, 46–52 [DOI] [PubMed] [Google Scholar]
- 31. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 32. Terwilliger T. C., Berendzen J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Terwilliger T. C. (2000) Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system. A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 36. Amberg D. C., Burke D., Strathern J. N. (2005) Methods in Yeast Genetics 2005: A Cold Spring Harbor Laboratory Course Manual, Cold Spring Harbor, NY [Google Scholar]
- 37. Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. (2004) A versatile toolbox for PCR-based tagging of yeast genes. New fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 [DOI] [PubMed] [Google Scholar]
- 38. Strømhaug P. E., Reggiori F., Guan J., Wang C. W., Klionsky D. J. (2004) Atg21 is a phosphoinositide-binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol. Biol. Cell 15, 3553–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Noda T., Ohsumi Y. (1998) Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273, 3963–3966 [DOI] [PubMed] [Google Scholar]
- 40. Gabriely G., Kama R., Gerst J. E. (2007) Involvement of specific COPI subunits in protein sorting from the late endosome to the vacuole in yeast. Mol. Cell Biol. 27, 526–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Searching protein structure databases with DaliLite version 3. Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Noda T., Matsuura A., Wada Y., Ohsumi Y. (1995) Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 210, 126–132 [DOI] [PubMed] [Google Scholar]
- 43. Noda T., Klionsky D. J. (2008) The quantitative Pho8Δ60 assay of nonspecific autophagy. Methods Enzymol. 451, 33–42 [DOI] [PubMed] [Google Scholar]
- 44. Suzuki K., Kubota Y., Sekito T., Ohsumi Y. (2007) Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12, 209–218 [DOI] [PubMed] [Google Scholar]
- 45. Huang W., Choi W., Hu W., Mi N., Guo Q., Ma M., Liu M., Tian Y., Lu P., Wang F. L., Deng H., Liu L., Gao N., Yu L., Shi Y. (2012) Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane-binding protein. Cell Res. 22, 473–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li X., He L., Che K. H., Funderburk S. F., Pan L., Pan N., Zhang M., Yue Z., Zhao Y. (2012) Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat. Commun. 3, 662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. DeLano W. L. (2002). The PyMOL Molecular Graphics System, DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
- 48. Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. (1988) Protein sorting in Saccharomyces cerevisiae. Isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8, 4936–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kirisako T., Ichimura Y., Okada H., Kabeya Y., Mizushima N., Yoshimori T., Ohsumi M., Takao T., Noda T., Ohsumi Y. (2000) The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 151, 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.