Abstract
A heretofore-unrecognized multigene family encoding diverse immunoglobulin (Ig) domain-containing proteins (DICPs) was identified in the zebrafish genome. Twenty-nine distinct loci mapping to three chromosomal regions encode receptor-type structures possessing two classes of Ig ectodomains (D1 and D2). The sequence and number of Ig domains, transmembrane regions and signaling motifs varies between DICPs. Interindividual polymorphism and alternative RNA processing contribute to DICP diversity. Molecular models indicate that most D1 domains are of the variable (V) type; D2 domains are Ig-like. Sequence differences between D1 domains are concentrated in hypervariable regions on the front sheet strands of the Ig fold. Recombinant DICP Ig domains bind lipids, a property shared by mammalian CD300 and TREM family members. These findings suggest that novel multigene families encoding diversified immune receptors have arisen in different vertebrate lineages and effect parallel patterns of ligand recognition that potentially impact species-specific advantages.
Keywords: zebrafish, innate immunity, lipid binding
1. INTRODUCTION
As the phylogenetically widely divergent species in which immune receptors have been characterized increases, several major trends can be recognized: 1) innate immune receptors have a long evolutionary history with marked similarities in receptor structure and function across wide phylogenetic boundaries [1], 2) primary mediators of adaptive immunity have undergone many changes during the evolution of vertebrates but share remarkable similarities in basic aspects of genetic recombination (rearrangement) and clonal selection [2] and 3) structures of receptors that mediate natural killer (NK)-type function can vary markedly even within members of a single class of vertebrate species (mammals) [3]. It is more difficult to recognize common features of other receptors that are classified as immune-type because of their structural domain composition and signaling properties. Many of these genes are encoded in multigene families and exhibit patterns of structural variation that are predicted to be associated with functional differences. It is likely that at least some receptors encoded by these genes are elements of unrecognized receptor-signaling networks and function through novel mechanisms. The presence of such multigene families in modern representatives of phylogenetically important species emphasizes their significance. Of the various nonmammalian animal models in which these molecules have been identified, the zebrafish (Danio rerio) offers many unique methodological advantages.
We have described variable (V) region-containing transmembrane receptors (novel immune-type receptors [NITRs]) in zebrafish and other bony fish [4]. NITRs are the most complex family of V region-containing immune-type receptors described thus far outside of immunoglobulin (Ig) and T cell antigen receptors (TCRs) [5]. NITRs function in allogeneic recognition in a manner akin to activating/inhibitory NK receptors [6]. A direct cloning strategy [7] identified a distantly related multigene family (modular domain immune-type receptors [MDIRs]) [8]. Through genome scanning utilizing MDIR and NITR Ig domain sequences, an additional multigene family encoding diverse Ig domain-containing proteins (DICPs) was identified. We describe herein the genomic organization, sequence complexity and predicted protein structures of the DICPs in zebrafish, which likely are unique to bony fish. We also demonstrate that recombinant forms of zebrafish DICP Ig domains bind lipids, which is a shared characteristic with members of the mammalian CD300 and TREM families of innate immune receptors [9,10].
2. MATERIALS AND METHODS
2.1. Bioinformatics
Genomic sequences encoding candidate DICP Ig domains were identified on zebrafish chromosomes 3, 14 and 16 with BLAST searches using MDIR and NITR sequences as queries. In silico translation of each Ig domain indicates that several genes encode a frame shift or premature stop codon, permitting their classification as pseudogenes (Supplemental Materials and Methods). Protein sequences were aligned by Clustal W [11]. Phylogenetic trees were constructed from pairwise Poisson correction distances with 2000 bootstrap replications by MEGA5 software [12]. Protein sequence domains were identified with SMART software [13].
2.2. DICP transcripts and genes
A small number of DICP ESTs were identified using BLAST searches of the zebrafish EST database and those appearing to encode full-length proteins were sequenced (Supplemental Materials and Methods). Additional DICP cDNA sequences were obtained by rapid amplification of cDNA ends (RACE) or direct reverse transcriptase-polymerase chain reaction (RT-PCR) with primers complementing predicted exons (Supplemental Materials and Methods).
2.3 DICP D1-D2 cDNA amplicons from chromosome 3
Partial DICP cDNA sequences were generated using primers designed to amplify D1-D2-containing DICP genes on chromosome 3. Forward (CATGTGTTCAGCAGWTMTGGAGAAACTG) and reverse (GATAGACTCCACATCTCCACTGTTTATC) primers were used with Titanium Taq (Clontech) to amplify D1-D2 sequences from pooled kidney and intestine cDNA (zebrafish obtained from EkkWill Waterlife Resources, Ruskin, FL, USA). Amplicons were cloned into pGEM-T Easy (Promega) and sequenced.
2.4 Genomic organization
The genomic organization of DICPs was deduced by comparing cDNA sequences to ZV8 genomic reference sequences: chromosome 3 scaffold 262 (GenBank ID: NW_001878770.2), chromosome 14 scaffold 1719 (GenBank ID: NW_001877436.2) and chromosome 16 scaffold 1952 (GenBank ID: NW_001877662.2). BACs CH73-34H11 (GenBank ID: FP929011) and CH73-322B17 (GenBank ID: FP015862) were used to link two unordered segments within scaffold 1952 that map to chromosome 16.
2.5. Molecular modeling
Theoretical models of DICP D1 domains were generated using the automated protein homology-modeling server SWISS-MODEL [14]. The Structural Classification Of Proteins (SCOP) database was utilized for domain definitions [15]. The Docker program was used to calculate sequence similarity using the Blosum62 matrix. Figures were generated with PyMol (The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC).
2.6. Cloning and expression of hFc chimeras
Recombinant soluble proteins of DICP D1 and D2 ectodomains fused to a human IgG Fc domain were generated by cloning various ectodomains (amplified from pooled hematopoietic tissue cDNA) into the pcDNA3-hsIgG1Fc-Avi fusion vector [16] that introduces a N-terminal start codon, signal peptide and a C-terminal human IgG Fc domain.
DICP D1-hFc and D2-hFc chimeric proteins were expressed and secreted by HEK293T cells. Cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 1 mM sodium pyruvate and 2 mM GlutaMAX (Invitrogen) and transferred to OPTI-MEM I serum-free medium (Invitrogen) for transfection of hFc constructs with Lipofectamine 2000 (Invitrogen). Following transfection, cells were grown for 48 hrs, pooled, and centrifuged at 500 xg for 10 min to clear the supernatant. Recovered supernatants were stored at 4 °C in 0.02% sodium azide. Supernatant harvests were concentrated 10 to 100 fold and the hFc fusion proteins were characterized by Western analyses and quantified using the Easy-Titer Human IgG Assay kit (Thermo Scientific) [16].
2.7. ELISA assay for binding to lipids
Purified lipids (Sigma and Avanti Polar Lipids) were processed as described [9]. Solid phase ELISA assays were conducted as described previously [9]. Either 0.5 μg purified lipid or 50 μl of MBTE/methanol bacterial extract were used to coat plates. Negative control wells were treated in parallel with solvent (100% methanol). Binding efficiency was determined after color development as absorbance at 450 nm. Values were corrected by subtracting the value from negative control wells.
The effect of concentration on lipid binding of hFc fusion proteins in the ELISA assay was evaluated. As a positive control, a hFc-fusion of the Ig domain of murine CLM7, which binds all four purified lipids used in screening [9], was employed. CLM7-hFc was added to ELISA plates at 100 μg/ml (volume 0.10 ml). Dicp3e529-D1-hFc, which exhibits robust lipid binding, was added at 15 μg/ml (volume 0.10 ml). The optimal lipid binding exhibited by CLM7-hFc was obtained at 12-25 μg/ml [9] and assay results were comparable to that of Dicp3e529-D1-hFc at 15 μg/ml. The standard concentration of hFc fusion proteins for assays was 0.10 ml of 10-50 μg/ml.
3. RESULTS AND DISCUSSION
3.1. Identification of DICP Ig domains
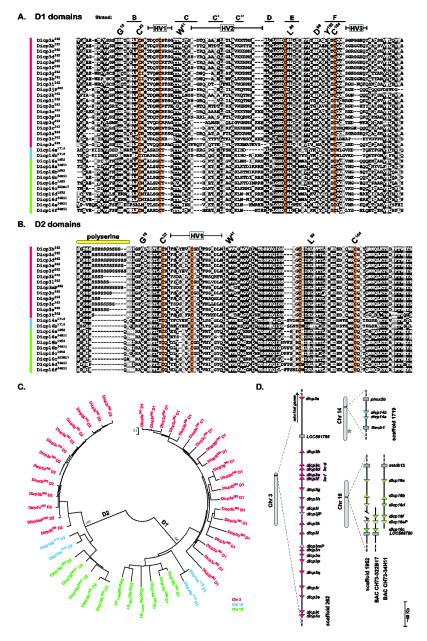
A number of approaches exist for identifying immune receptors in diverse species. We employed a robust series of Ig V-, I- and C2-type motifs from NITRs and MDIRs as queries in tBLASTn searches of the zebrafish genome (version Zv8) to identify unrecognized Ig-region encoding genes and identified the DICP family. The typical DICP consists of two distinct classes of extracellular Ig domains: N-terminal D1 and C-terminal D2 domains, (Figs. 1A-C, Supplemental Figs. S1-S2). DICP D1 domains share more conserved residues with classical V domains (G16, V19, L21, C23, W41, L89, I91, D98, G100, Y102, C104) than do the D2 domains (G16, L21, C23, W41, L89, C104) [17]. Additional pairs of conserved cysteines: C30 and C87 in D1 and C33 and C85 in D2 (Fig 1A-B) are predicted to form intrachain disulfides. Twenty-nine DICP D1 domains were identified on zebrafish chromosomes 3, 14 and 16 (Fig. 1D). The genes corresponding to the D1 domains are designated by: a number that denotes chromosomal location, a letter that denotes the order in which the domains were identified and a superscript that indicates an allele sequence source, e.g., dicp3g262: chromosome 3, scaffold 262 and seventh D1 domain (designated g). (The DICP gene names and symbols appearing in the NIHMS version of the manuscript were changed during the publication process to reflect the nomenclature approved by the Zebrafish Nomenclature Committee. See appendix table for corrected names).
Fig. 1. DICP D1 and D2 domains encoded by genomic sequences.
DICP Ig domains were organized into (A) D1 and (B) D2 domains and aligned. Positions that are 70% or greater identical are shaded in black and those that are structurally related are shaded in gray. Protein symbols are shown on the left with the genomic source (superscript) indicated as an allele designation (scaffold 262, scaffold 1719, scaffold 1952, BAC CH73-34H11 or BAC CH73-322B17). Conserved residues characteristic of immunoglobulin domains are indicated by the IMGT numbering system above the alignments [37]. Hypervariable regions (HV) and predicted Ig strand locations (of D1) also are indicated. A variable length polyserine stretch is evident at the amino terminal region of the D2 domain. Conserved cysteines are shaded with orange. (C) The DICP D1 and D2 domains shown in (A) and (B) were aligned and a neighbor joining tree constructed. (D) The relative chromosomal positions of DICP D1 domains are shown. Triangles represent a single DICP D1 domain; transcriptional orientation is indicated. Predicted pseudogenes are indicated by lighter shades of color and gray rectangles represent linked (non-DICP) genes. The dicp3c and dicp3d D1 domains and the dicp3e and dicp3f D1 domains are predicted to be encoded in single genes (see Supplemental Materials and Methods). BACs were employed to refine the sequence in scaffold 1952. The star on chromosome 14 indicates the location of a NITR gene cluster.
3.2. DICP transcripts
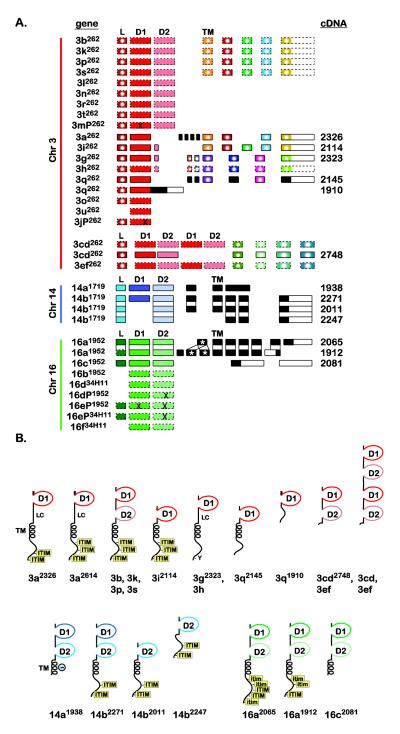
The sequencing of multiple DICP ESTs and cDNAs (Supplemental Materials and Methods and Supplemental Fig. S3) facilitated the characterization of the exon organization and putative translation products from a large number of highly related candidate DICP genes (Fig. 2). Most DICP D1 domain exons are flanked by exons that encode a leader signal sequence and a D2 domain exon; dicp14a and dicp16a are representative. Several genes are comprised of D1 domains that are adjacent to a predicted leader signal sequence, but lack an apparent D2 domain, (e.g. dicp3a and dicp3i). DICP transcripts encoding a single D2 domain can be derived through alternative mRNA splicing, e.g. dicp14b. Two pairs of contiguous D1-D2 sequences are predicted to encode proteins with a D1-D2-D1-D2 configuration (dicp3cd and dicp3ef; see Supplemental Materials and Methods). Based on the genome assemblies (Fig. 1D), which do not reflect the haplotypic and allelic complexity observed in BAC, EST and cDNA analyses, the minimum number of DICP genes and pseudogenes in a zebrafish genome is 27.
Fig. 2. DICP exon organization and predicted protein structures.
(A) DICP genes (Fig. 1) are listed on the left. cDNA identities (Supplemental Materials and Methods and Supplemental Fig. S3) are listed on the right. Exons are represented by rectangles and include: peptide leader (L), D1 domain (D1), D2 domain (D2) and transmembrane domain (TM). Unique exons are black. Similar exons are color coded. Exons that encode peptide sequences that are identical or differ by a single residue are color coded and marked by a white circle. The presence of a stop codon or frameshift within an exon is represented by “X”. Exons predicted from genomic sequence, without a corresponding cDNA, are bound by dashed lines. The dicp16a1912 transcript possesses a duplicated exon (see Supplemental Fig. S7 for details). (B) Schematic representation of DICPs deduced from both cDNAs and predicted genes. Structures directly based on cDNA sequences are indicated by a superscript allele number. D1 = D1 Ig domain; D2 = D2 Ig domain; TM = transmembrane region; LC = region of low sequence complexity; ϴ = negatively charged residue within a TM; ITIM = conventional ITIM and itim = ITIM variant.
Several significant features and relationships are observed between DICP proteins: 1) although Dicp3b, Dicp3k, Dicp3p and Dicp3s possess divergent D1 and D2 domains, they, along with Dicp3a and Dicp3i, share transmembrane and cytoplasmic domains that differ by no more than one residue (Supplemental Fig. S4), 2) several DICPs lack D2 domains, 3) Dicp3g and Dicp3h are 99% identical (Supplemental Fig. S5), 4) the leader domains for chromosome 3 DICPs are identical and 5) alternative mRNA splicing produces a variety of different forms of Dicp3q, Dicp14b and Dicp16a (Fig. 2 and Supplemental Fig. S6). It is unclear if dicp16a mRNA variation is a result of alternative splicing or allelic variation as one allele has been identified that encodes one copy of exon 5 and a second allele encodes two copies of exon 5 due to a retrotransposon insertion (Supplemental Fig. S7).
3.3. Predicted functional variation of DICPs
Numerous multigene families of immune receptors include both inhibitory and activating forms. Inhibitory receptors typically are associated with cytoplasmic immunoreceptor tyrosine-based inhibition motifs (ITIMs; S/I/V/LxYxxI/V/L). Activating receptors may possess cytoplasmic immunoreceptor tyrosine-based activation motifs (ITAMs; YxxI/Lx(6-12)YxxI/L) or employ a charged residue within its transmembrane domain that interacts with an ITAM-containing adaptor protein for signaling [18]. DICP transcripts encoding both putative inhibitory and activating receptors have been identified.
Overall, DICPs vary in terms of: 1) number of predicted ectodomains, 2) presence or absence of consensus cytoplasmic ITIMs [19] or variant ITIMs (itims), 3) number of ITIMs/itims, 4) presence or absence of C-terminal tyrosine in the cytoplasmic tail, 5) presence or absence of transmembrane regions, 6) presence or absence of low (sequence) complexity regions and 7) presence or absence of charged residues in the transmembrane domain (Fig. 2B). Most DICPs encode ITIMs/itims and are predicted to be inhibitory. Of the DICPs with defined coding sequence, none possesses a positively charged transmembrane residue, a characteristic of activating function in the KIR, Ly49, and NITR families. However, Dicp14a possesses a transmembrane region with a negatively charged (Glu) residue (GIIIIIEMAALSFPTAILLWIC). This feature is shared with the mammalian activating receptors, CLM-5 and CD300c. It has been reported that CLM-5 partners with and signals via FcRγ [20-22]. Dicp14a may partner and signal via FcRγ or similar adaptor proteins described in zebrafish [23]. Additional DICP transcripts are predicted to encode secreted proteins with unknown function. As observed in other families of innate immune receptors (e.g. NITRs, KIRs, Ly49), putative inhibitory forms of DICPs far outnumber putative activating forms.
3.4. Allelic complexity of DICPs
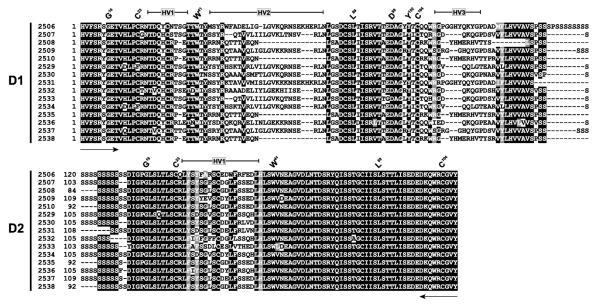
In order to investigate the variability of DICP transcripts, the D1-D2 domains of DICP transcripts encoded on chromosome 3 were amplified from pooled kidney and intestine cDNAs from zebrafish obtained from EkkWill Waterlife Resources and sequenced (Fig. 3). Only two of 15 amplified sequences represent strong matches to the reference genomic sequence which is derived from the Tübingen line of zebrafish (Table 1); specifically, the peptide sequence encoded by cDNA amplicon 2537 matches exactly the predicted Ig domains of Dicp3e262 and the peptide sequence encoded by amplicon 2509 differs from Dicp3f262 by a single residue. D1-D2 domains encoded by two other amplicons, 2530 and 2536, differ from Dicp3l262 and Dicp3p262 by 23 and 33 residues, respectively. Five other amplicons (2507, 2529, 2532, 2533 and 2534) encode D1 and D2 domains that share similarity to two different DICP genes: for example, amplicon 2529 encodes a D1 that is most similar to the D1 domain of Dicp3p262 whereas the D2 domain encoded by this amplicon is most similar to the D2 domain of Dicp3k262. Six amplicons (2506, 2508, 2510, 2531, 2535 and 2538) encode DICP sequences (D1 or D2 or both) that are not present in the reference sequence and corresponding sequences currently are not identifiable by tBLASTn searches; four of these (2508, 2510, 2535 and 2538) may represent new alleles of a single DICP gene. In summary, only one of the fifteen amplicons is predicted to encode a protein that matches exactly the reference sequence; most amplicon sequences encode D1-D2 domains that would be divergent (many with >20 residue differences) from the reference sequence. This allelic complexity exceeds that reported previously for NITRs [24].
Fig. 3. DICP D1-D2 cDNA amplicons.
Partial DICP cDNA sequences were generated using primers designed to amplify D1-D2-containing DICP genes on chromosome 3. The predicted protein sequences encoded by amplicons are aligned (sequence ID number is listed on left) with identical residues shaded in black and structurally similar residues shaded in gray. Primer positions are indicated by arrows below the alignment. Hypervariable (HV) regions and conserved residues characteristic of immunoglobulin domains are indicated by the IMGT numbering system above the alignments [37]. As many of these peptide sequences do not match the reference genomic sequence, they were not assigned a specific gene designation (Table 1).
Table 1. Variation of chromosome 3 DICP D1-D2 cDNA amplicons from reference genomic sequences.
| cDNA Amplicon (GenBank) |
Best Genomic Reference D1a |
Differences with Genomic Reference D1b |
Gaps with Genomic Reference D1c |
Best Genomic Reference D2a |
Differences with Genomic Reference D2b |
Gaps with Genomic Reference D2c |
|---|---|---|---|---|---|---|
| 2506 (JN416864) |
Novel | n.a. | n.a. | Dicp3n262 | 8 | 0 |
| 2507 (JN416865) |
Dicp3l262 | 9 | 0 | Dicp3s262 | 2 | 0 |
| 2508 (JN416866) |
Novel | n.a. | n.a. | Novel | n.a. | n.a. |
| 2590 (JN416867) |
Dicp3f262 | 1 | 0 | Dicp3f262 | 0 | 0 |
| 2510 (JN416868) |
Novel | n.a. | n.a. | Novel | n.a. | n.a. |
| 2529 (JN416869) |
Dicp3p262 | 23 | 0 | Dicp3k262 | 6 | 0 |
| 2530 (JN416870) |
Dicp3l262 | 20 | 1 | Dicp3l262 | 3 | 0 |
| 2531 (JN416871) |
Novel | n.a. | n.a. | Dicp3k262 | 6 | 0 |
| 2532 (JN416872) |
Dicp3l262 | 25 | 1 | Dicp3p262 | 9 | 0 |
| 2533 (JN416873) |
Dicp3l262 | 8 | 0 | Dicp3t262 | 16 | 0 |
| 2534 (JN416874) |
Dicp3p262 | 23 | 0 | Dicp3k262 | 5 | 0 |
| 2535 (JN416875) |
Novel | n.a. | n.a. | Novel | n.a. | n.a. |
| 2536 (JN416876) |
Dicp3p262 | 28 | 0 | Dicp3p262 | 5 | 0 |
| 2537 (JN416877) |
Dicp3e262 | 0 | 0 | Dicp3e262 | 0 | 0 |
| 2538 (JN416878) |
Novel | n.a. | n.a. | Novel | n.a. | n.a. |
DICP ectodomains encoded by the reference genome sequence (scaffold 262) with the highest similarity to DICP cDNA amplicons in Fig 3.
Number of amino acid differences between the ectodomains encoded by the cDNA amplicon and the reference sequence.
Number of gaps in the alignment between the ectodomains encoded by the cDNA amplicon and the reference sequence.
3.5 DICP D2 domains possess polyserines
Regions of low sequence complexity consisting of variable length triplet nucleotide repeats, which encode two to 16 residue stretches of polyserine, are located N-terminal to G16 in D2 (Fig 1B) and cDNAs encoding these regions have been identified (Fig. 3). This lower sequence complexity of varying lengths in D2 distinguishes DICPs from other multigene families of immune-type receptors. Although the functional relevance of polyserine sequences in DICPs is not yet known, polyserine regions in other proteins have been reported to serve as flexible linker domains [25], affect polypeptide stability [26], and separate distinct functional domains [27]. Polyserine stretches are a conserved feature of vitellogenin in invertebrates and vertebrates [28] and appear to play functional roles in pathogens. Two such examples are ICP4 of Herpes Simplex Virus 1 [29] and gp40 of Cryptosporidium parvum [30-32]. The polyserine stretches in DICPs could function in maintaining cell surface receptor integrity and/or provide steric flexibility in ligand binding or other extracellular interactions.
3.6. Hypervariable regions in DICP ectodomains
Sequence differences between the D1 and D2 domains are illustrated in Figs. 1A-B and 3. The highest degree of intergeneic variation (across all DICP reference sequences) in D1 is observed in three hypervariable regions (HV1-HV3); most variation in D2 is localized to HV1. Notwithstanding the variation in lengths of polyserine stretches, sequence relatedness between the DICPs that map to chromosome 3 is significantly less than that seen for DICP genes that map to chromosomes 14 and 16. The overall sequence differences between genes on chromosome 3 are more regionalized than those on chromosomes 14 and 16. The differences may reflect the lower numbers of sequences being compared for chromosomes 14 and 16 relative to chromosome 3.
3.7. Molecular modeling of DICP D1 domains
Ig domains can be classified as V-, C1-, C2- or I-type based on the characteristic distances between the conserved cysteine residues (C23 C104) that form the B-F disulfide bond, a tryptophan residue (W41) packed against it in the core of the Ig domain fold and overall strand topology. The intercysteine distance in V-type Ig domains ranges from 65 to 75 residues and is appreciably shorter in constant (C) Ig domains (55 to 60 residues) [33]. Intermediate (I-type) Ig domains possess structural features of V domains but exhibit shorter intercysteine distances [34]. All D1 domains from chromosomes 3 and 16 are classified as V domains by InterProScan software (release 30.0) [35]. The D1 domains of Dicp14a and Dicp14b lack one and two amino acids, respectively, that are required for classification as V domains by InterProScan criteria. Although DICP D2 domains possess the Ig framework residues, the distance between the conserved cysteine residues is 62 to 64 residues, which could classify them as I-type Ig domains. However, D2 domains are less than 25% identical to solved Ig structures, which is below the level of similarity that permits homology modeling. InterProScan software classifies DICP D2 domains as Ig-like.
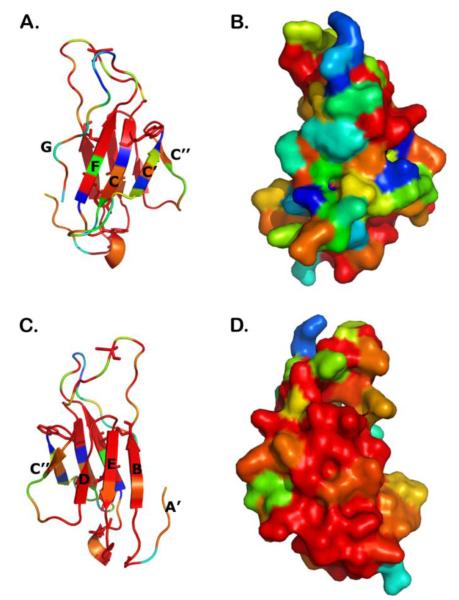
Atomic homology models of D1 domains from all three gene clusters were generated based on the Protein Data Bank. Dicp3a262 D1 is most similar (28% identical) to a V-set domain from the Poliovirus receptor CD155, (PDB ID: 3eowR). Dicp14b1719 D1 is most similar (25% identical) to a shark antibody V region, (PDB ID: 1sq2N); however, Dicp16a1952 D1 is most similar (32% identical) to an I-set Ig domain from the FcγrIII receptor, (PDB ID: 1fnlA). Dicp3f262 D1, which binds phospholipids (see below), is 25% identical to the V domain of an antibody light chain, (PDB ID: 2ghwB), and is 32% identical to the I domain from mouse CNTN4, (PDB ID: 3jxaB). A structural model of Dicp3f262 D1 is shown (Fig. 4).
Fig. 4. Structural modeling of Dicp3f-D1, a predicted V-type Ig domain.
(A) A ribbon diagram of an atomic homology model of Dicp3f-D1 is shown in which color variation indicates sequence differences between Dicp3f-D1 and seven other DICP D1 sequences. Blosum62 similarity values are: <40, blue, 40-50, cyan, 50-60, green, 60-70, yellow, 70-90, orange and 90-100, red. (B) The front sheet (A’GFCC’C”) of Dicp3f is oriented towards the viewer, and depicts the solvent accessible surface. (C and D) The respective ribbon and surface prediction models for Dicp3f-D1, in which the highly conserved back sheet (ABED) is oriented towards the viewer, are shown. Protein sequence variability is confined to a single molecular surface.
The high degree of variation in Dicp3 family members distributes on the front sheet of the Ig-fold (A’GFCC’C” strands, Fig. 4A-B); the back sheet (ABED strands) is predicted to be minimally variant (Fig. 4C-D). In contrast to polymorphic antigen receptors, where sequence variation is clustered on CDR loops, sequence variations in DICPs are distributed over a broader surface encompassing the front sheet and the CDR3-equivalent loop.
Based on the foregoing criteria, nearly all DICP D1 domains are of the V type, which is common to many immune receptors. Joining (J) regions which are conserved features of other V-type receptors such as Igs, TCRs, some NITRs and a few additional IgSF members (e.g. CD8), are absent from DICPs. J regions encode the FGXG peptide motif that facilitates front sheet:front sheet interactions between antigen receptor V domains. V domains that lack the FGXG motif (e.g. CD2 and CD80) do not dimerize using the front sheet:front sheet interface. Notably, the front sheets of the V domains in CD2 and CD80 participate in ligand binding (CD58 for CD2, CTLA-4 for CD80). DICP D1 domains are variable at positions that are clustered to a contiguous solvent exposed surface containing the front sheet F,C,C’ strands and the Ig-TCR CDR3-analogous FG loop, which we propose may influence binding specificities. Taken together, the sequence comparison and modeling data suggest that the front sheet of the DICP D1 domains is used for ligand recognition rather than dimerization.
3.8. DICPS in bony fish
In order to identify DICP and DICP-related sequences in other (non-zebrafish) vertebrate species, tBLASTn searches were employed with DICP D1 and D2 sequences as queries. A small number of DICP-related sequences were identified in diverse fish species including Cypriniformes (Carp; Cyprinus carpio), Perciformes (tilapia;Oreochromis niloticus), Tetraodontiformes (pufferfish: Tetraodon nigroviridis and Takifugu rubripes), and Salmoniformes (salmon; Salmo salar) (Supplemental Table S1). Structural features of the DICP-related proteins were defined by SMART analyses (Supplemental Fig. S8) and phylogenetic analyses employed to identify non-zebrafish Ig domains most similar to DICP D1 and D2 domains (Supplemental Fig. S9). These results demonstrate that: 1) only one definitive DICP transcript can be currently identified outside of zebrafish and is from the closely related carp (GenBank ID: AB098477), 2) D1-like and D2-like sequences can be identified in secreted and membrane bound proteins in tilapia, salmon and pufferfish, 3) a predicted tilapia transcript (GenBank ID: XM_003458344) possesses two tandem sets of D1-like and transmembrane domains and may represent two transcripts, 4) the four conserved cysteines in both D1 and D2 domains (Fig 1A-B) are present in D1-like and D2-like domains, but their position varies slightly within the Ig scaffold, 5) the high level of sequence diversity between zebrafish D1 and D2 domains and the D1-like and D2-like sequences suggest DICPs have experienced species-specific diversification and 6) no mammalian sequences that are significantly similar to DICPs were identified. In addition, clusters of DICP D1-like sequences can be identified in multiple tilapia genomic scaffolds (not shown). These data suggest that DICPs are encoded by gene clusters in multiple fish species and that the DICPs are restricted to bony fish.
3.9 The chromosome 16 DICP locus shares conserved synteny with human, mouse and chicken chromosomal regions encoding FCR/FCRL
Non-DICP genes that are unequivocal orthologs of mammalian genes and would be useful for evaluating conserved synteny, are absent from the DICP gene cluster on chromosome 3; however, several genes are present at the DICP loci on chromosomes 14 and 16 (Fig 1D) that can be used to identify regions of conserved synteny between the zebrafish DICP loci and mammalian IgSF genes. Specifically, DICP genes on zebrafish chromosome 14 are immediately flanked by phox2b and limch1. Although PHOX2B and LIMCH1 are tightly linked in humans, mice and chicken, no IgSF gene family has been identified near these genes in these species. However, setdb1b, which is adjacent to the DICP gene cluster on zebrafish chromosome 16, is orthologous to SETDB1 on human chromosome 1q21, mouse chromosome 3 (F2) and chicken chromosome 25. All of these chromosomal regions also encode variable numbers of Fc receptor (FCR) and FCR-like (FCRL) molecules as well as the CD1 gene family in mouse and human [36]. SETDB1 and the nearest FCR/FCRL gene are separated by ~1.1 M bp in human, ~0.9 M bp in mouse and ~0.1 M bp in chicken (Supplemental Fig. S10). Given these considerable map distances and large numbers of Ig and adjacent gene loci in vertebrates, the significance of this observation is unclear.
3.10. Lipid binding patterns of DICPs
Based on recent observations that MDIRs and certain CD300 and TREM family members bind lipids [9,10], we investigated the capacity of DICPs to recognize a variety of lipids including those present in bacterial extracts. Twenty DICP D1 domains and four D2 domains were amplified from cDNA and cloned into a hFc expression vector. When transfected into mammalian cells, more than half of these constructs did not produce soluble protein. In our experience, it is not uncommon for constructs expressing certain Ig domains to not produce soluble protein, while other constructs with only small sequence differences produce protein (Cannon and Haire, unpublished). The clones corresponding to Dicp3e-D1, Dicp3f-D1, Dicp3n-D1, Dicp3p-D1, Dicp3s-D1, Dicp14a-D2 and Dicp16a-D1 successfully produced secreted, soluble hFc fusion proteins (Supplemental Fig. S11). Six of the seven D1 and D2 domains in the Fc fusion proteins either matched or differed by two residues from the reference peptide sequence. In contrast, the D1 domain encoded by the Dicp3n-hFc fusion protein (dicp3n505) differs from the D1 domain encoded by the dicp3n262 reference sequence by 10 residues (two of which represent an introduced gap); a highly divergent allele of dicp3n or a new DICP gene may account for the differences (Supplemental Fig. S11). This pattern of sequence diversity is reminiscent of that observed in other DICP cDNA amplicons from chromosome 3 (Fig. 3).
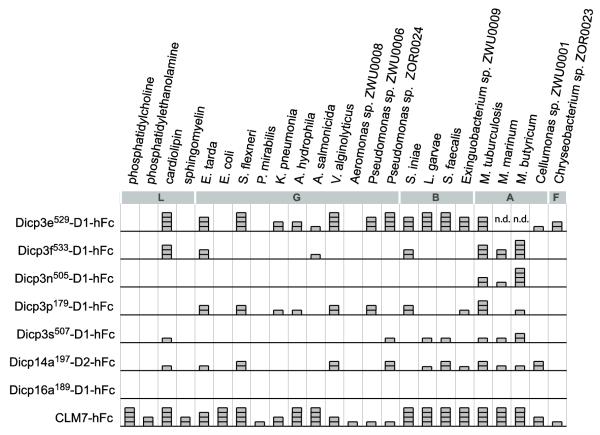
In enzyme-linked immunosorbant assays (ELISAs) for lipid binding, the DICP hFc fusion proteins exhibit a range of lipid binding specificity (Fig. 5). The D1 domain of Dicp3e binds to lipids and bacterial extracts (Supplemental Table S2) and is very robust (ELISA scores of +3 or +4 for 11 of 24 lipid sources). The D1 domains of Dicp3f and Dicp3p along with the D2 domain of Dicp14a exhibit moderate binding (ELISA scores of +2 to +4 for 6 of 24 lipid sources). The D1 domain of Dicp3n displays moderate binding to lipid extracts only from mycobacteria (ELISA scores of +1, +2 and +4). The D1 domain of Dicp3s exhibits weak binding to 7 of 24 lipid sources with only one ELISA score greater than +1. The D1 domain of Dicp16a189 did not bind lipids or bacterial extracts in this assay. Dicp3e529-D1, which binds robustly, and Dicp3f533-D1, which binds moderately, differ by six residues. Although this is a small data set, no clear sequence motif was identified as essential for lipid binding. For example, The D1 domain of Dicp16a, which did not bind lipids in this assay, shares the same core Ig domain residues with the other D1 domains that bound lipids (Supplemental Fig. S11). Identification of residues required for lipid binding is further confounded by the sequence differences in the HV regions between the domains that do and do not bind lipids. In addition, no restriction for DICP binding to extracts from specific classes of bacteria (Gammaproteobacteria, Bacilli, Actinobacteria and Flavobacteria) was observed. The functional implications of lipid binding by DICPs remain to be resolved.
Fig. 5. DICP Ig domains bind phospholipids.
ELISA analysis of interactions of DICP D1-hFc and D2-hFc fusion proteins (Supplemental Fig. S11) with purified lipids (L), along with soluble organic extracts from four different classes of bacteria; Gammaproteobacteria (G), Bacilli (B), Actinobacteria (A) and Flavobacterium (F) (Supplemental Table S2). Mouse CLM7-hFc was included as a positive control [9]. Dicp16a189-D1-hFc failed to score above background in this assay and is considered a negative control. Corrected ELISA values less than 0.03 were scored as zero; 0.03-0.05 as +1; 0.05-0.2 as +2; 0.2-0.5 as +3; >0.5 was scored as +4 and are depicted graphically. n.d. = not determined.
3.11. Summary
With the exception of bony fish NITRs that may function as NK receptors [3], the function of many large families of Ig-containing receptors (with unknown ligands) identified throughout the vertebrate radiations remains unclear. Recently, it has been shown that the differential binding of lipids by members of the mammalian CD300 and TREM gene families is a general feature of this group of molecules and potentially is related to their overall function [9]. Multiple DICP Ig domains described here exhibit a similar capacity to bind free lipid and lipid extracts of different bacteria, including pathogens, suggesting that lipid binding groups the DICPs and CD300/TREM molecules at a functional level.
The predicted inhibitory and activating functions of DICPs and their chromosomal organization in distinct loci along with evidence for retroviral-based transposition, underscores similarities between DICPs with NITRs. Furthermore, nearly all DICPs possess potential O-glycosylation sites in their membrane-proximal extracellular regions, which is characteristic of MDIRs and CD300 molecules. However, the minimum level of overall sequence relatedness does not support a common origin for these gene families.
These findings raise important questions regarding the origins of multigene families encoding Ig domain activating/inhibitory proteins in vertebrates. It appears as if Ig, TCR and FcR (including FcRL) exhibit ubiquitous distribution throughout the bony fish, amphibians, reptiles, birds and mammals. CD300/TREM-like molecules, in which we tentatively have grouped MDIRs [8], are distributed in cartilaginous fish, bony fish and other vertebrates, although the depth of annotation is not comparable to that in mammals. Given the findings reported here, it appears as if far more complex families of Ig domain-encoding cell surface molecules are found in lower vertebrates than in mammals (as has been reported for other Ig encoding families, e.g., avian and amphibian species encode far more putative FcRs than mammals). Given our superior understanding of a large number of vertebrate genomes, it is increasingly more likely that the distributions of DICPs and NITRs may well be restricted to the teleost fish.
The mechanisms whereby multigene families (e.g. DICPs and NITRs) arise and expand are of fundamental interest. Notably, most current reference fish species are egg-laying with ex utero embryonic development. The immunological “needs” of such species may be unique and potentially exceed those of ovoviviparous fish species. Some of the questions raised along these lines likely can be settled with forthcoming genome sequences of representative species of this latter group as well as the resolution of holostean, chondrichthyan as well as the genomes of crosspterygian and sarcopterygian fish species. Regardless of the specific mechanism by which DICPs and NITRs arose and expanded, their wide spread presence in, but simultaneous restriction to, a single large phylogenetic group of vertebrates (the bony fish) emphasizes the highly plastic and dynamic nature of immune molecules.
Supplementary Material
Supplemental Fig S7. Allelic variation of dicp16a includes a retrotransposon insertion and duplication of exon 5. (A) The genomic organization of two dicp16a alleles. Repeat sequences are represented by triangles. (B) Repeat sequences associated with the transposon. (C) Transcript variants of dicp16a. The dicp16a1912-tv2 transcript is a 3′ RACE product and lacks 5′ coding sequence. fs = frameshift.
Supplemental Fig. S8. Vertebrate, non-zebrafish proteins with Ig domains similar to DICP D1 and D2 domains. Sequence domains of bony fish proteins and CNTN5 listed in Supplemental Table S1 are displayed graphically. Ig domains phylogenetically similar to zebrafish DICP D1 and D2 domains (Supplemental Figure S9) are labeled with “D1-like” and “D2-like”, respectively. Sequence identities are indicated by four letter abbreviations of genus-species (e.g. Cyca = Cyprinus carpio) and GenBank accession number or gene symbol.
Supplemental Fig. S9. DICP-like Ig domains. (A) Bony fish Ig domains identified in Supplemental Table S1 (excluding CD4, NCAM2-like and CNTR5 sequences) were aligned and a neighbor joining tree generated. (B-C) Ig domains identified in (A) as phylogenetically similar to DICP D1 and D2 domains were aligned with zebrafish DICP D1 and D2 domains, respectively.
Supplemental Fig. S10. The zebrafish chromosome 16 DICP locus shares conserved synteny with human, mouse and chicken chromosomal regions encoding FCR/FCRL. Figure was modified from UCSC Genome Browser (http://genome.ucsc.edu/). Gene positions were defined by RefSeq except for DICPs and chicken FCR/L which were defined by Ensembl. Sequences shown correspond to zebrafish chromosome 16 (22,533,561 – 24,101,487), chicken chromosome 25 (253,115 – 1,823,755), mouse chromosome 3 (95,127,447 – 96,695,373) and human chromosome 1 (149,369,294 – 150,937,220).
Supplemental Fig. S11. DICP domains employed as Fc fusions. (A) Alignment of DICP Ig domains from hFc fusion proteins with domains encoded by the corresponding genomic sequence. Allele designation is shown as superscript after the gene. (B) Alignment of DICP D1 domains that bound lipid with one (16a) that did not bind lipid in the ELISA-based assay.
Supplemental Fig. S1. DICP D1 domains encoded by genomic sequences. Gene name is followed by genomic DNA source (superscript) as an allele designation (scaffold 262, scaffold 1719, scaffold 1952, BAC CH73-34H11 and BAC CH73-322B17).
Supplemental Fig. S2. DICP D2 domains encoded by genomic sequences. Gene name is followed by genomic DNA source (superscript) as an allele designation (scaffold 262, scaffold 1719, scaffold 1952, BAC CH73-34H11 and BAC CH73-322B17).
Supplemental Fig. S3. DICP proteins encoded by zebrafish cDNAs. Predicted peptide signal sequences and transmembrane domains are boxed. ITIM and ITIM-like sequences are shaded with gray. Negatively charged residues within a TM domain are shaded with black. Cloning of cDNAs is described in Supplemental Materials and Methods.
Supplemental Fig. S4. DICP identity and variability. Transcripts dicp3a2326, dicp3a2614 and dicp3i2114 possess exons that are nearly identical to predicted exons for dicp3b, dicp3k, dicp3p and dicp3s encoded by scaffold 262 (denoted by 262 superscript). Color coding delineates sequences encoded by nearly identical exons.
Supplemental Fig. S5. Dicp3g and Dicp3h. Transcript dicp3g2323 encodes the same peptide sequence predicted to be encoded by dicp3g262 on scaffold 262. The dicp3h262 gene is predicted to encode a nearly identical protein. Color coding delineates sequences encoded by nearly identical exons.
Supplemental Fig S6. Alternative splicing of dicp14b. The proteins encoded by transcript variants of dicp14b are aligned.
Highlights.
The heretofore-unrecognized multigene family of DICPs are described in zebrafish
DICPs include putative inhibitory and activating immune receptors
Interindividual polymorphisms and RNA splicing contribute to DICP diversity
Hypervariable regions of DICP Ig domains may contribute to ligand binding
Recombinant DICP Ig domains bind lipids with varying specificity
Acknowledgements
We thank John Rawls (University of North Carolina), Karen Guillemin and Erika Mittge (University of Oregon), Ed Noga (North Carolina State University), and Carol Kim (University of Maine) for bacteria; Philip Wright for assistance with cDNA synthesis; Wei Chia Lin (Genome Institute of Singapore, Biomedical Sciences Institute) for providing a dicp16c EST clone and Barb Pryor for editorial assistance. The authors are supported the National Institutes of Health (R01 AI057559 to GWL and JAY and R01 AI23337 to GWL).
Appendix
Table S3.
Official names for zebrafish DICP genes.*
| Zebrafish chromosome 3 DICPs | |
|---|---|
| Name in manuscript | Official name |
| dicp3a | dicp1.1 |
| dicp3b | dicp1.2 |
| dicp3c-d | dicp1.3-4 |
| dicp3e-f | dicp1.5-6 |
| dicp3g | dicp1.7 |
| dicp3h | dicp1.8 |
| dicp3i | dicp1.9 |
| dicp3jP | dicp1.10P |
| dicp3k | dicp1.11 |
| dicp3l | dicp1.12 |
| dicp3mP | dicp1.13P |
| dicp3n | dicp1.14 |
| dicp3o | dicp1.15 |
| dicp3p | dicp1.16 |
| dicp3q | dicp1.17 |
| dicp3r | dicp1.18 |
| dicp3s | dicp1.19 |
| dicp3t | dicp1.20 |
| dicp3u | dicp1.21 |
| Zebrafish chromosome 14 DICPs | |
|---|---|
| Name in manuscript | Official name |
| dicp14a | dicp2.1 |
| dicp14b | dicp2.2 |
| Zebrafish chromosome 16 DICPs | |
|---|---|
| Name in manuscript | Official name |
| dicp16a | dicp3.1 |
| dicp16b | dicp3.2 |
| dicp16c | dicp3.3 |
| dicp16d | dicp3.4 |
| dicp16eP | dicp3.5P |
| dicp16f | dicp3.6 |
Official gene names are approved by the Zebrafish Nomenclature Committee (www.zfin.org)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DATA DEPOSITION: Sequence data have been deposited with GenBank under accession numbers: JN416849 - JN416885.
Reference List
- [1].Litman GW, Cooper MD. Commentary: Why study the evolution of immunity? Nat Immunol. 2007;8:547–548. doi: 10.1038/ni0607-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Du PL, Zucchetti I, De SR. Immunoglobulin superfamily receptors in protochordates: before RAG time. Immunol Rev. 2004;198:233–248. doi: 10.1111/j.0105-2896.2004.00122.x. [DOI] [PubMed] [Google Scholar]
- [3].Yoder JA, Litman GW. The phylogenetic origins of natural killer receptors and recognition: relationships, possibilities, and realities. Immunogenetics. 2011;63:123–141. doi: 10.1007/s00251-010-0506-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Yoder JA. Form, function and phylogenetics of NITRs in bony fish. Dev Comp Immunol. 2009;33:135–144. doi: 10.1016/j.dci.2008.09.004. [DOI] [PubMed] [Google Scholar]
- [5].Litman GW, Hawke NA, Yoder JA. Novel immune-type receptor genes. Immunol Rev. 2001;181:250–259. doi: 10.1034/j.1600-065x.2001.1810121.x. [DOI] [PubMed] [Google Scholar]
- [6].Cannon JP, Haire RN, Magis AT, Eason DD, Winfrey KN, Hernandez Prada JA, Bailey KM, Jakoncic J, Litman GW, Ostrov DA. A bony fish immunological receptor of the NITR multigene family mediates allogeneic recognition. Immunity. 2008;29:228–237. doi: 10.1016/j.immuni.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cannon JP, Haire RN, Litman GW. Identification of diversified genes that contain immunoglobulin-like variable regions in a protochordate. Nat Immunol. 2002;3:1200–1207. doi: 10.1038/ni849. [DOI] [PubMed] [Google Scholar]
- [8].Cannon JP, Haire RN, Mueller MG, Litman RT, Eason DD, Tinnemore D, Amemiya CT, Ota T, Litman GW. Ancient divergence of a complex family of immune-type receptor genes. Immunogenet. 2006;58:362–373. doi: 10.1007/s00251-006-0112-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cannon JP, O’Driscoll ML, Litman GW. Specific lipid recognition is a general feature of CD300 and TREM molecules. Immunogenet. 2011 doi: 10.1007/s00251-011-0562-4. in press. [DOI] [PubMed] [Google Scholar]
- [10].Choi SC, Simhadri VR, Tian L, Gil-Krzewska A, Krzewski K, Borrego F, Coligan JE. Cutting Edge: Mouse CD300f (CMRF-35-Like Molecule-1) Recognizes Outer Membrane-Exposed Phosphatidylserine and Can Promote Phagocytosis. J Immunol. 2011;187:3483–3487. doi: 10.4049/jimmunol.1101549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- [12].Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011 doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nuc Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lo CL, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. SCOP database in 2002: refinements accommodate structural genomics. Nucleic Acids Res. 2002;30:264–267. doi: 10.1093/nar/30.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cannon JP, O’Driscoll ML, Litman GW. Construction, expression and purification of chimeric protein reagents based on immunoglobulin Fc regions. Methods Mol Biol. 2011;748:51–67. doi: 10.1007/978-1-61779-139-0_4. [DOI] [PubMed] [Google Scholar]
- [17].Barclay AN. Membrane proteins with immunoglobulin-like domains--a master superfamily of interaction molecules. Sem Immunol. 2003;15:215–223. doi: 10.1016/s1044-5323(03)00047-2. [DOI] [PubMed] [Google Scholar]
- [18].Barrow AD, Trowsdale J. You say ITAM and I say ITIM, let’s call the whole thing off: the ambiguity of immunoreceptor signalling. Eur J Immunol. 2006;36:1646–1653. doi: 10.1002/eji.200636195. [DOI] [PubMed] [Google Scholar]
- [19].Daeron M, Jaeger S, Du PL, Vivier E. Immunoreceptor tyrosine-based inhibition motifs: a quest in the past and future. Immunol Rev. 2008;224:11–43. doi: 10.1111/j.1600-065X.2008.00666.x. [DOI] [PubMed] [Google Scholar]
- [20].Fujimoto M, Takatsu H, Ohno H. CMRF-35-like molecule-5 constitutes novel paired receptors, with CMRF-35-like molecule-1, to transduce activation signal upon association with FcRgamma. Int Immunol. 2006;18:1499–1508. doi: 10.1093/intimm/dxl083. [DOI] [PubMed] [Google Scholar]
- [21].Nakano T, Tahara-Hanaoka S, Nakahashi C, Can I, Totsuka N, Honda S, Shibuya K, Shibuya A. Activation of neutrophils by a novel triggering immunoglobulin-like receptor MAIR-IV. Mol Immunol. 2008;45:289–294. doi: 10.1016/j.molimm.2007.04.011. [DOI] [PubMed] [Google Scholar]
- [22].Clark GJ, Ju X, Tate C, Hart DN. The CD300 family of molecules are evolutionarily significant regulators of leukocyte functions. Trends Immunol. 2009;30:209–217. doi: 10.1016/j.it.2009.02.003. [DOI] [PubMed] [Google Scholar]
- [23].Yoder JA, Orcutt TM, Traver D, Litman GW. Structural characteristics of zebrafish orthologs of adaptor molecules that associate with transmembrane immune receptors. Gene. 2007;401:154–164. doi: 10.1016/j.gene.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yoder JA, Mueller MG, Wei S, Corliss BC, Prather DM, Willis T, Litman RT, Djeu JY, Litman GW. Immune-type receptor genes in zebrafish share genetic and functional properties with genes encoded by the mammalian lymphocyte receptor cluster. Proc Natl Acad Sci USA. 2001;98:6771–6776. doi: 10.1073/pnas.121101598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Anderson TA, Levitt DG, Banaszak LJ. The structural basis of lipid interactions in lipovitellin, a soluble lipoprotein. Structure. 1998;6:895–909. doi: 10.1016/s0969-2126(98)00091-4. [DOI] [PubMed] [Google Scholar]
- [26].Hasper A, Soteropoulos P, Perlin DS. Modification of the N-terminal polyserine cluster alters stability of the plasma membrane H(+)-ATPase from Saccharomyces cerevisiae. Biochim Biophys Acta. 1999;1420:214–222. doi: 10.1016/s0005-2736(99)00100-5. [DOI] [PubMed] [Google Scholar]
- [27].Howard MB, Ekborg NA, Taylor LE, Hutcheson SW, Weiner RM. Identification and analysis of polyserine linker domains in prokaryotic proteins with emphasis on the marine bacterium Microbulbifer degradans. Protein Sci. 2004;13:1422–1425. doi: 10.1110/ps.03511604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Smolenaars MM, Madsen O, Rodenburg KW, Van der Horst DJ. Molecular diversity and evolution of the large lipid transfer protein superfamily. J Lipid Res. 2007;48:489–502. doi: 10.1194/jlr.R600028-JLR200. [DOI] [PubMed] [Google Scholar]
- [29].Bates PA, DeLuca NA. The polyserine tract of herpes simplex virus ICP4 is required for normal viral gene expression and growth in murine trigeminal ganglia. J Virol. 1998;72:7115–7124. doi: 10.1128/jvi.72.9.7115-7124.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cevallos AM, Zhang X, Waldor MK, Jaison S, Zhou X, Tzipori S, Neutra MR, Ward HD. Molecular cloning and expression of a gene encoding Cryptosporidium parvum glycoproteins gp40 and gp15. Infect Immun. 2000;68:4108–4116. doi: 10.1128/iai.68.7.4108-4116.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cevallos AM, Bhat N, Verdon R, Hamer DH, Stein B, Tzipori S, Pereira ME, Keusch GT, Ward HD. Mediation of Cryptosporidium parvum infection in vitro by mucin-like glycoproteins defined by a neutralizing monoclonal antibody. Infect Immun. 2000;68:5167–5175. doi: 10.1128/iai.68.9.5167-5175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Strong WB, Gut J, Nelson RG. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infect Immun. 2000;68:4117–4134. doi: 10.1128/iai.68.7.4117-4134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Williams AF, Barclay AN. The immunoglobulin superfamily-domains for cell surface recognition. Ann Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- [34].Harpaz Y, Chothia C. Many of the immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set which is close to that containing variable domains. J Mol Biol. 1994;238:528–539. doi: 10.1006/jmbi.1994.1312. [DOI] [PubMed] [Google Scholar]
- [35].Zdobnov EM, Apweiler R. InterProScan--an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- [36].Davis RS. Fc receptor-like molecules. Ann Rev Immunol. 2007;25:525–560. doi: 10.1146/annurev.immunol.25.022106.141541. [DOI] [PubMed] [Google Scholar]
- [37].Giudicelli V, Duroux P, Ginestoux C, Folch G, Jabado-Michaloud J, Chaume D, Lefranc MP. IMGT/LIGM-DB, the IMGT comprehensive database of immunoglobulin and T cell receptor nucleotide sequences. Nucleic Acids Res. 2006;34:D781–D784. doi: 10.1093/nar/gkj088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig S7. Allelic variation of dicp16a includes a retrotransposon insertion and duplication of exon 5. (A) The genomic organization of two dicp16a alleles. Repeat sequences are represented by triangles. (B) Repeat sequences associated with the transposon. (C) Transcript variants of dicp16a. The dicp16a1912-tv2 transcript is a 3′ RACE product and lacks 5′ coding sequence. fs = frameshift.
Supplemental Fig. S8. Vertebrate, non-zebrafish proteins with Ig domains similar to DICP D1 and D2 domains. Sequence domains of bony fish proteins and CNTN5 listed in Supplemental Table S1 are displayed graphically. Ig domains phylogenetically similar to zebrafish DICP D1 and D2 domains (Supplemental Figure S9) are labeled with “D1-like” and “D2-like”, respectively. Sequence identities are indicated by four letter abbreviations of genus-species (e.g. Cyca = Cyprinus carpio) and GenBank accession number or gene symbol.
Supplemental Fig. S9. DICP-like Ig domains. (A) Bony fish Ig domains identified in Supplemental Table S1 (excluding CD4, NCAM2-like and CNTR5 sequences) were aligned and a neighbor joining tree generated. (B-C) Ig domains identified in (A) as phylogenetically similar to DICP D1 and D2 domains were aligned with zebrafish DICP D1 and D2 domains, respectively.
Supplemental Fig. S10. The zebrafish chromosome 16 DICP locus shares conserved synteny with human, mouse and chicken chromosomal regions encoding FCR/FCRL. Figure was modified from UCSC Genome Browser (http://genome.ucsc.edu/). Gene positions were defined by RefSeq except for DICPs and chicken FCR/L which were defined by Ensembl. Sequences shown correspond to zebrafish chromosome 16 (22,533,561 – 24,101,487), chicken chromosome 25 (253,115 – 1,823,755), mouse chromosome 3 (95,127,447 – 96,695,373) and human chromosome 1 (149,369,294 – 150,937,220).
Supplemental Fig. S11. DICP domains employed as Fc fusions. (A) Alignment of DICP Ig domains from hFc fusion proteins with domains encoded by the corresponding genomic sequence. Allele designation is shown as superscript after the gene. (B) Alignment of DICP D1 domains that bound lipid with one (16a) that did not bind lipid in the ELISA-based assay.
Supplemental Fig. S1. DICP D1 domains encoded by genomic sequences. Gene name is followed by genomic DNA source (superscript) as an allele designation (scaffold 262, scaffold 1719, scaffold 1952, BAC CH73-34H11 and BAC CH73-322B17).
Supplemental Fig. S2. DICP D2 domains encoded by genomic sequences. Gene name is followed by genomic DNA source (superscript) as an allele designation (scaffold 262, scaffold 1719, scaffold 1952, BAC CH73-34H11 and BAC CH73-322B17).
Supplemental Fig. S3. DICP proteins encoded by zebrafish cDNAs. Predicted peptide signal sequences and transmembrane domains are boxed. ITIM and ITIM-like sequences are shaded with gray. Negatively charged residues within a TM domain are shaded with black. Cloning of cDNAs is described in Supplemental Materials and Methods.
Supplemental Fig. S4. DICP identity and variability. Transcripts dicp3a2326, dicp3a2614 and dicp3i2114 possess exons that are nearly identical to predicted exons for dicp3b, dicp3k, dicp3p and dicp3s encoded by scaffold 262 (denoted by 262 superscript). Color coding delineates sequences encoded by nearly identical exons.
Supplemental Fig. S5. Dicp3g and Dicp3h. Transcript dicp3g2323 encodes the same peptide sequence predicted to be encoded by dicp3g262 on scaffold 262. The dicp3h262 gene is predicted to encode a nearly identical protein. Color coding delineates sequences encoded by nearly identical exons.
Supplemental Fig S6. Alternative splicing of dicp14b. The proteins encoded by transcript variants of dicp14b are aligned.