Abstract
Recent studies questioned whether the PDB contains all compact, single domain protein structures. Here, we show that all quasi-spherical, QS, random protein structures devoid of secondary structure are in the PDB and are excellent templates for all native PDB proteins up to 250 residues. Because QS templates have similar global contour as native, TASSER can refine 98% (90%) of those whose TM-score is 0.4 (0.35) to structures ≥ the 0.5 TM-score threshold (0.74 (0.64) mean TM-score) for CATH/SCOP assignment. Based on this and the fact that at a TM-score of 0.4, 83% (90%) of all (internal) core secondary structure elements are recovered, a 0.40 TM-score is an appropriate fold similarity assignment threshold. Despite claims of Taylor, Trovato and Zhou that many of their structures lack a PDB counterpart, using fr-TM-align, at a 0.45 (0.5) TM-score threshold, essentially all (most) are found in the PDB. Thus, the conclusion that the PDB is likely complete is further supported.
Keywords: Completeness of the PDB, structural alignment, TM-align, TM-score threshold of 0.4, SCOP and CATH fold assignment, protein structure refinement
Introduction
In a series of papers, the likely completeness of the space of single domain protein structures has been explored (1-8); this is one aspect of the protein folding problem that has seen many seminal contributions due to Harold Scheraga(9-12). That is, given a newly determined protein structure, can one find a statistically significant structural match to an already solved protein in the Protein Data Bank (13)? If this is the case, then the PDB is complete; otherwise, it is not. In practice, the traditional way of addressing this issue is to ask if two proteins have the same “fold” or “topology”; for example, whether they have the same SCOP(14) and CATH (15) fold assignment. This inherently discrete view of protein structure is useful in the limit of very high structural similarity. However, if we only focus on the structural, or equivalently geometric, similarity of a pair of proteins without exploiting information regarding their evolutionary relationship, the picture of discrete folds becomes blurred as the structures become evolutionary more distant(5, 8). Consider the idealized case where two proteins have identical core secondary structure arrangements for all but one of their secondary structural elements, whose relative packing angle θ differs by 20°. One would likely assign the pair of proteins to the same fold. Suppose, however, that θ=180°, does the pair share the same fold or not? By many criteria, they would be assigned to a different fold (16). Thus, the issue is the threshold of θ that assigns the pair of proteins to the same topology. Whatever this value may be, at some point, a minor structural fluctuation could shift the fold assignment from the same to a different topology(15-17). This is the fundamental problem with a discrete view of the space of protein structures.
At the other extreme, one can view a protein's structure as a chain contour where local structural fluctuations from the global direction of the chain (as caused by helices, strands and bulges) are averaged out (18, 19). One can then ask if the pair of proteins share a similar global chain contour in that one can readily build the structure of one using the other as the template. This has much in common with protein homology modeling where a structurally related template, which may contain a significant number of gaps, is identified, an alignment to the template made, and then a full-length model is built (20, 21). Consistent with these ideas, protein structure space has been shown to be continuous in that one can morph from one protein structure to another by a transitive series of structurally related intermediates (5). On the other hand, in the limit of high structural similarity, protein space is discrete. This observation has been termed the discrete-continuous duality of protein structure (22).
As indicated above, to ascertain whether a pair of proteins are structurally similar, a comparison metric is needed (23). There are a variety of such metrics including the root mean square deviation from native, RMSD, and the GDT_TS score (24); however, the statistical significance of these measures is length dependent (25). In contrast, the TM-score provided by the TM-align structural alignment algorithm (26) offers the advantage that its statistical significance is length independent (3, 27, 28). The TM-score for a pair of protein structures lies in the range [0,1], with a value of 1.0 for identical structures, and the most probable value for a pair of randomly related proteins is 0.15 (the average best value is 0.30) (28). Previously, we argued that two proteins are structurally related if they share a TM-score ≥ 0.4 (which corresponds to a P-value of 3.4 × 10-5 (28)). Xu and Yang showed that above a TM-score of 0.5, the fold as assessed by CATH (17) and SCOP (29) is likely the same (28). However, based on the ability to yield full-length TASSER models with a TM-score above 0.5 and examining the properties of the smoothed chain contour, we show below that the TM-score threshold value for two structurally related proteins can be reduced to 0.35 (P-value of 2.7 × 10-4). Such structures recapitulate the global chain contour and preserve a significant fraction of the secondary structural elements that enable the construction and subsequent refinement of the model provided by the structural alignment to the template of interest. Finally, we note that Pandit and Skolnick recently developed an improved version of TM-align (26), fr-TM-align (27) that generates alignments with a 9% higher TM-score than TM-align with 7% more residues aligned. The use of fr-TM-align is particularly important in the regime where the original TM-align program gave scores in the range from 0.3-0.5. Here, fr-TM-align generates significantly better alignments, with an improvement of up to 0.15 TM-score units. This is precisely the regime where the P-value of the alignment is the most sensitive (28). Thus, the use of the original TM-score approach may lead one to erroneously conclude that statistically significant structure matches to the PDB are absent, when in reality such matches are detected using the more sensitive fr-TM-align approach.
Over the past several years, we have argued that the structural space of compact, single domain proteins is likely complete (2-5, 8). This conclusion was initially arrived at by comparing proteins of different secondary structure and fold class (2), and then by comparing the structures of proteins in the PDB whose pairwise sequence identity was less than 35%. But one could always argue that the library of solved structures contains proteins that are evolutionarily related and might not cover the space of all possible compact, single domain protein structures (3). By comparing the library of real PDB structures with a set of artificially generated, compact polyalanine structures (4), and subsequently polyvaline structures (chosen because their volume is closer to native structures) containing hydrogen bonded secondary structure elements (30), we demonstrated that at a TM-score threshold of 0.4, 99% of the artificial structures up to 250 residues in length are in the PDB. This number drops to 77% if PDB templates up to 300 residues are used, the PDB300 set; however, this result is actually a significant underestimate as discussed below. The size dependence of the completeness of the PDB partly reflects the TM-score threshold of 0.40; at a TM-score threshold of 0.35, 95% of the artificial proteins have a match to the PDB300. Clearly, the larger size template proteins provide an enriched source of protein structural templates. To explore the length dependence of the library of solved structures, we consider both the full PDB and PDB300 libraries as templates in subsequent analysis.
Recently, there have been a number of studies that claim that the library of PDB structures is not complete (6, 7, 31). Taylor and coworkers generated a set of structures based on the variation about a known structure by the combinatorial enumeration of all paths connecting different points in a secondary structure lattice and considered five medium sized, three layer βα proteins (6). They compared the resulting structures using DALI (32), TM-align (26) and SAP(33). They concluded that only 6% of the protein like folds they generated are found in nature; however, they conceded that the PDB contains “sufficient components to reconstruct almost any fold”. The issue thus resides in what is a definition of a fold, if the PDB in fact can be used to reconstruct any arbitrary structure. In what follows, we shall examine whether the coverage of fold space is as small as Taylor et al (6) suggest or is in fact much larger.
Another approach aimed at elucidating the coverage of structure space is due Trovato and coworkers (7), who considered 60 residue polyvaline structures generated using the AMBER03 (34) force field by the GROMACS MD simulation package(35). The majority of their simulations were done in vacuum, with a few done in water. Using the TM-score as the structural similarity measure, they find all the folds in the PDB for proteins between 40 to 75 residues, but if a TM-score of 0.45 threshold is considered, then a significant number of their generated structures are absent in the CATH database (15) restricted to templates up to 75 residues in length. However, as shown below, this conclusion is caused by the small size threshold of their allowed PDB templates plus the use of the original, less sensitive TM-align algorithm (26). Recognizing that the average gap length in the template structure is ~26 residues, this means that structures whose maximum length is 75 residues might be too small to use for the assessment of the structural completeness of the PDB; e.g. for a template with 2 gaps, the maximum template TM-score to a 60 residue protein target is ~0.3, well within the structurally insignificant regime.
Another study that examined the completeness of the PDB is due to Dai and Zhou (31), who extended the existing library of PDB structures by permuting loops and considered a maximum of 5 loop permutations on 2936 SCOP domains (14). For proteins between 60 and 200 residues, using the original version of TM-align (26), they conclude that at a TM-score threshold of 0.5, 82% of the loop permuted structures between 180 and 200 residues belong to new fold clusters and are absent in the PDB. We shall explore whether this conclusion holds on further analysis when fr-TM-align is used.
While the above studies are suggestive, perhaps the most rigorous test of the completeness of the PDB is to examine a library of quasi-spherical, QS, random protein structures that are entirely devoid of main chain hydrogen bonds but which possess protein like local geometry and backbone excluded volume (8). If all such QS protein structures were in the PDB, and all PDB templates were in the QS library, this would be the strongest suggestion that the PDB is complete, as they are clearly not related to the structures in the PDB by evolution. Indeed, the QS structures bear the least resemblance to native proteins in that they lack regular secondary structure, backbone hydrogen bonding, protein like binding sites and interfaces (8). In previous work (8), we examined the distribution of TM-scores to the full PDB library and found that 94% of the QS structures up to 250 residues in length are in the PDB. However, it might be argued that such proteins that are entirely devoid of regular secondary structure are poor templates and would result in entirely nonphysical protein structures. To examine this question, we will use the QS structures as templates and examine whether physically realistic models for native protein structures can be built from them. If so, when combined with the fact that almost all the QS structures have a matching template in the PDB, this would strongly argue that the PDB is complete.
The outline of this paper is as follows: First, for smoothed PDB protein structures that define the backbone contour, we examine, as a function of the TM-score of the template to the target, the fraction of secondary structural elements that comprise the core and the fraction of aligned residues in the core and loop regions that are recovered. The goal here is to examine how much global structural information is retained as the TM-score to the native protein structures diminishes. Next, for the quasi-spherical protein structures, we examine their suitability as protein templates and show that for protein structures up to 250 residues in length even in the range of quite low TM-score, the global structural information to the target is encoded. Moreover, the resulting structures can be readily refined by a stripped down version of the structure prediction algorithm, TASSER (36) to give rather good protein models with a mean TM-score of 0.71. By examining the TM-score distribution of smoothed structures that provide the global chain contour, we show that this global chain contour information is present in structures well below a TM-score of 0.4. Here, we further explore the relationship between the TM-score of the QS template and the final TASSER model and show for a QS template score to native of 0.35, 90% of the TASSER models have a TM-score ≥ 0.5. For the Taylor (6), Trovato (7), Zhou (31), QS (8) and PDB200 (a representative set of PDB proteins between 40 and 200 residues) sets, we examine the fraction of structures present in the contemporary PDB library as a function of TM-score. Based on the distribution of TM-scores, we conclude that the PDB contains all the structures in these sets, including the encoding of the global chain contour information needed to build a full-length model with all secondary structural elements present. The composite results provide additional compelling evidence that the PDB is likely complete.
Methods
Fr-TM-align structure comparison algorithm
For each structure under consideration, structural alignments to a reference template library (see below) were done using the fr-TM-align structural alignment algorithm (27), an improved, more sensitive version of the original TM-align approach (26). fr-TM-align compares two structures based on their TM-score defined for a target protein containing N residues as
| (1) |
where is the average distance between a pair of residues in the best structural alignment of a pair of randomly related protein structures. The TM-score lies in the range [0,1], with a TM-score of 1.0 when two structures are identical. The most probable TM-score of randomly related protein structures is 0.15, with the average best alignment score of a pair of randomly related protein structures of 0.30 (26). The advantage of the TM-score is that its statistical significance is protein length independent; thereby allowing the raw TM-score to be used to compare different length protein alignments.
Protein backbone smoothing to detect the global chain contour
While the TM-score is useful for assessing the global structural similarity between two sets of ordered points, one would like to have an approach that more directly detects the similarity in their global chain contours. For each of the compared structures, coordinates are smoothed using the procedure introduced in earlier work to detect surface “U-turns”, regions where the chain reverses global direction between “core” secondary structural elements (19). Consider the replacement of the ith chain position by the average over the ith and i±1 coordinates. This replacement procedure is iterated five times. Then, the relationship between the original coordinates X(i) and the averaged coordinates Y(i) is given by
| (2a) |
where
| (2b) |
Secondary structural elements are defined by the geometric characteristics of the chain-smoothed contour (19). We then define the dimensionless local radius of curvature at the chain location from (Y) as
| (3) |
In practice, to identify the turns, we take the maximum of and denoted by . If , then a U-turn is defined at smoothed residue i. The regions between the U-turns define the set of secondary structural elements, [S]. The resulting smoothed structures are denoted by “sm”.
Figure 1 shows examples of the QS template protein structure (see below for details as to how the QS structures are generated (8)) whose length is the same as the PDB structure 3hkxa along with its structural alignment to the target PDB structure 101m_. The chain averaging procedure results in a smoothed chain contour where local conformational fluctuations are removed.
Figure 1.
The upper panel shows the structures of the QS protein of the same length as 3hkxa and the target protein 101m_; the middle panel shows the smoothed structures generated by the application of eq. 2, with the left hand panel showing the aligned 3hkxa regions to 101m_. The lower panel shows the structural superposition with the target (template) indicated by the thick(thin) tube). The TM-score of the target template alignment of QS template to 101m_ is 0.47; whereas for the smoothed pair of structures, their TM-score is 0.55.
Fraction of aligned core and loop secondary structures
For a target protein containing S secondary structural elements as defined above, we calculate the fraction of aligned secondary elements in a given target protein with respect to a template protein, fsec, as follows: A given target secondary structural element is defined as present if at least a pair of residues in the target are part of the structural alignment to the template. Then, the fraction of matched secondary structural elements is fsec is given by:
| (4a) |
In the lower TM-score range, we have observed that often one or more of the target ends are unaligned; this results from the presence of a non compact dangling tail in the protein structure that may interact with another protein. Of more importance for the generation of the target protein structure from a given template alignment is whether all internal secondary structures are present. To examine this issue, we identify the first and last aligned secondary structure elements in the template's structural alignment to the target, say elements i and j of the target. We then calculate the ratio of the number of internal secondary structural elements aligned from i to j to the total number possible, viz. j-i+1. We define this ratio for the internal secondary structural elements as
| (4b) |
While in principle, we could just have one element aligned which would give an of 1.0, in practice, for non smoothed template structures, given that the average total number of secondary structural elements aligned, fsec ≥ 0.83 for TM-scores ≥ 0.4, this trivial result of a single internal secondary structure aligned will happen rarely. Finally, we also calculate the fraction of aligned target residues in the identified core secondary structural elements, , and the fraction of aligned target residues in the “U-turns” or loops, .
PDB template library
To ensure that the set of PDB structures used for comparison to the Taylor (6), Trovato (7), Zhou (31) sets were available at their time of publication, we employed our PDB template library from October, 2009 comprised of 12,052 monomeric structures with a maximum pairwise sequence identity of 35%. For comparison to the QS structures, we use an updated library of 13,148 template structures. In practice, the results are insensitive to the actual PDB library used, as the space of protein structures is extremely dense. The PDB200, PDB250 and PDB300 are subsets of 4,631, 6,999, and 9,867 proteins of the PDB set that are no more than 200, 250 and 300 residues in length respectively, with a minimum length cutoff of 40 residues. The corresponding set of smoothed structures generated using eq. 2, are termed the PDB200sm, PDB250sm and PDB300sm sets. The list of protein structures, their Cα coordinates, the structural alignments of PDB200 to PDB and PDB200sm to PDBsm may be found at http://cssb.biology.gatech.edu/completeness/PDB/x, x=cafiles, alignments, and alignmentsmooth, respectively.
Quasi-spherical random protein template library
As described in (8), the library of quasi-spherical, QS, random protein structures was constructed as follows: In a sphere whose radius is given by that estimated for a protein of N residues, N points representing the Cαs are randomly placed subject to the constraint that no pair are closer than 3.8 Å. The Cαs are then connected to minimize the overall path length using the solution to the traveling salesman problem provided by the Concorde Traveling Salesman solver given in http://www.tsp.gatech.edu/concorde/ (30). The length distribution is taken from our library of representative PDB proteins between 40 and 300 residues in length, the PDB300 set. This provides the QS300 set of 8,254 proteins whose lengths range from 40 to 300 residues in length. The set of structures corresponding to chain smoothed contours using eq. 2 comprise the QS300sm set.
Structural alignments provided by fr-TM-align with the QS300 set as templates to the PDB250 library as targets were generated, as were the corresponding structural alignments of QS300sm to the PDB250sm library. These provided the initial target template alignments that were then subsequently refined using TASSER. The QS300 library, the structural alignments of QS300 to PDB250 and the corresponding QS300sm alignments to PDB250sm are found at http://cssb.biology.gatech.edu/completeness/QS/x with x=cafiles, alignments, and alignmentsmooth respectively. The alignments using the QS200 and QS200sm sets as the target library are found at x=alignmentsQS200pdb and QS200pdbsm.
Modeling PDB target structures using TASSER
For each PDB250 target, as shown schematically in Figure 1, fr-TM-align provided structural alignments to templates in the QS300 set. We chose proteins up to 250 residues in length as targets to make the test of the utility of the QS300 set as templates a quite difficult one. Up to the top 50 template alignments ranked by their TM-score of the template to the target were selected. Subsequently, contact and distance restraints are derived from these selected template alignments as inputs for TASSER (3) refinement. Other inputs used by TASSER are the predicted secondary structure and solvent accessible surface values for the target PDB250 sequence. Target specific pair potentials that depend on template library sequence profiles are neglected. The final models are the top cluster centroids found by SPICKER (37) clustering from the TASSER low energy trajectory outputs. Possible steric clashes are removed, and main-chain and side-main atoms built using the PULCHRA (38) chain restoration program. The TASSER models may be found at http://cssb.biology.gatech.edu/completeness/QS/TASSER/.
Taylor set
The Taylor set consists of 1,211 protein structures generated using five, three layer β-α proteins that range from 100 to 150 residues in length by varying the number and location of secondary structure elements and then ranking the structures on the basis of a hydrophobic core packing score (6). The resulting proteins, whose lengths range from 108 to 148 residues, were compared to the appropriate PDB300 and PDB template library using fr-TM-align. Similarly, the chain smoothed structures were compared to the PDB300sm and PDBsm template library. The structures and alignments may be found at http://cssb.biology.gatech.edu/completeness/taylor/x, with x=cafiles, alignments and alignmentsmooth, respectively.
Trovato set
Our variant of the Trovato set (7) consists of 28,746 compact, 60 residues, all atom poly VAL protein structures generated by the AMBER03 force field (34) and the GROMACS molecular dynamics simulation package(35). The structures and alignments to PDB300 and PDB, and the smoothed structures may be found at http://cssb.biology.gatech.edu/completeness/trovato/x, with x=cafiles, alignments, and alignmentsmooth respectively.
Zhou set
The Zhou set consists of 2,637 protein domains generated by permuting the loops of native protein structures that range in length from 56 to 200 residues. The structures, distribution of TM-scores and alignments to the PDB300, PDB, and corresponding smoothed structures may be found at http://cssb.biology.gatech.edu/completeness/zhou/x with x=cafiles, alignments, and alignmentsmooth respectively.
Results/Discussion
The majority of secondary structural elements are aligned for all TM-scores ≥ 0.40
Given the ambiguities in uniquely assigning a protein fold or topology, one means of assessing the equivalence of the global chain contour information is to examine the fraction of target secondary structural elements aligned to the template structure. These can guide the location and orientation of the global chain contour in a low-resolution picture as well as in the original Cα backbone representation. In Figure 2, for the top 100 alignments of the PDB200 set to the PDB set, we plot the fraction of aligned secondary structural elements, fsec given by eq. 4a as a function of TM-score. For a TM-score of 0.40 for the original (non chain smoothed target structures), ~83% of the secondary structural elements defined by the chain smoothing procedure have a template alignment. By a TM-score of 0.5, 90% of the secondary structural elements are aligned. Thus, there is a quite small increase in the number of aligned secondary structural elements as a function of TM-score. Put another way, the majority of secondary structure information is present even at a TM-score of 0.40.
Figure 2.
For the top 100 structural alignments of the PDB200 set to the PDB set, the fraction of aligned target secondary structural elements fsec given by eq. 4a (dashed line) and the fraction of internal aligned secondary structural elements fsecint (solid line) given by eq. 4b (PDB internal) versus the TM-score of the template structure to the native target.
Actually, the cause of these “relatively” low values in the 0.40-0.50 TM-score range is the fact that sometimes the secondary structural elements of one or both ends of the target protein are unaligned. For the construction of accurate models, it is very important that all internal secondary structure elements be present. It should be recognized that the ends of PDB structures might not contact the reminder of the protein (but might contact other proteins in the crystal structure). Since our assertions about the likely completeness of the PDB hold only for compact proteins (2-5, 8), (not dangling tails that can adopt an astronomically large number of conformations), it is not surprising that the tail secondary structural elements might be less well represented than the core of the protein in the structural alignments.
In Figure 2, we also plot the fraction of aligned internal secondary structural elements (given by eq. 4b) versus TM-score. At a TM-score of 0.40, 90% of the internal secondary structural elements are present. This fraction increases to 97% when the TM-score is 0.50, a rather small change. Thus, structures above a TM-score of 0.4 should contain sufficient information to enable the full-length reconstruction of the target structure with the possible exception of the ends; this is explicitly demonstrated in below.
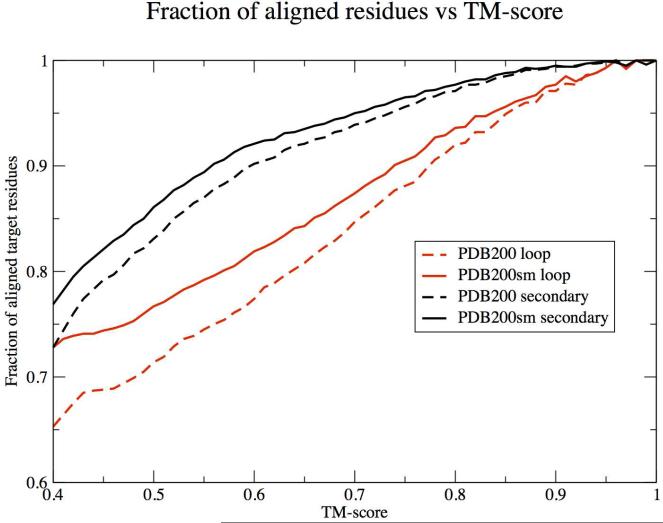
Chain smoothing improves alignment quality in the loop regions
As shown in Table 1 for the PDB200 set where PDB templates whose sequence identity >15% are excluded, chain smoothing improves the average TM-score from 0.65 to 0.70. This is accompanied by an increase in coverage (fraction of aligned residues in the target) from 0.86 to 0.89, if the full PDB template set is used; similar trends are observed for the PDB300 template set. Moreover, on chain smoothing, the number of gaps/target decreases, an effect accompanied by a slight increase in gap length. One might expect that chain smoothing of the target and template increases their structural similarity, with the largest relative improvement in alignment quality coming from the loop regions. This effect is confirmed in Figure 3, where we plot the fraction of aligned residues versus TM-score for the regular secondary structural elements and loops for both the original and smoothed “sm”, structures. For the regular secondary structure (black) regions, their relative differences are smaller than for the loops. However, the fraction of aligned target residues (loop or regular secondary structure) is always larger in the smoothed than in the original structures. Thus, chain smoothing better captures the target-template structural similarity.
Table 1.
Properties of global structural alignments for PDB200, QS, PDB250, Taylor, Trovato, and Zhou sets
| Target | Template Library | Fraction of targets with TM-score ≥0.4 | Average Coveragea,c | Average TM-score | Average Number of Gaps per Targetb,c | Average Gap Length per Targetc | Average Number of Gaps per Templateb,c | Average Gap Length per Templateb,c |
|---|---|---|---|---|---|---|---|---|
| PDB200 | PDBe | 1.00 | 0.86 | 0.65 | 1.19 | 4.87 | 3.98 | 16.7 |
| PDB200sm | PDBsmf | 1.00 | 0.89 | 0.70 | 1.01 | 4.81 | 2.86 | 17.0 |
| PDB200 | PDB300 | 1.00 | 0.84 | 0.64 | 1.27 | 5.19 | 3.05 | 11.2 |
| PDB200sm | PDB300smf | 1.00 | 0.87 | 0.69 | 1.08 | 5.13 | 2.83 | 16.3 |
| QS200 | PDB | 0.99 | 0.78 | 0.44 | 2.97 | 6.63 | 10.6 | 22.6 |
| QS200sm | PDBsm | 0.99 | 0.82 | 0.51 | 2.24 | 7.62 | 6.40 | 28.0 |
| QS200 | PDB300 | 0.85 | 0.76 | 0.43 | 2.96 | 6.84 | 8.20 | 14.3 |
| QS200sm | PDB300sm | 1.00 | 0.80 | 0.49 | 2.46 | 8.15 | 5.43 | 17.9 |
| PDB250 | QS300 | 0.71 | 0.78 | 0.42 | 2.31 | 6.36 | 10.11 | 13.8 |
| PDB250sm | QS300sm | 0.94 | 0.79 | 0.48 | 2.37 | 8.37 | 6.99 | 17.6 |
| PDB250 | TASSER model | 0.98 | 1.00 | 0.71 | - | - | - | - |
| Taylor Set | PDBd | 1.00 | 0.83 | 0.53 | 1.78 | 7.37 | 8.55 | 23.8 |
| Taylor Set | PDBsmf | 1.00 | 0.85 | 0.59 | 1.53 | 7.64 | 5.85 | 27.6 |
| Taylor Set | PDB300 | 1.00 | 0.79 | 0.52 | 2.02 | 8.07 | 6.61 | 15.3 |
| Taylor Set | PDB300smf | 1.00 | 0.83 | 0.57 | 1.67 | 8.65 | 4.94 | 18.7 |
| Trovato Set | PDBd | 1.00 | 0.87 | 0.52 | 0.61 | 3.42 | 5.02 | 26.2 |
| Trovato Set | PDBsmf | 1.00 | 0.90 | 0.59 | 0.44 | 2.84 | 2.69 | 29.4 |
| Trovato set | PDB300 | 1.00 | 0.85 | 0.51 | 0.66 | 3.78 | 4.66 | 18.6 |
| Trovato set | PDB300smf | 1.00 | 0.89 | 0.58 | 0.46 | 3.04 | 2.64 | 21.6 |
| Zhou Set | PDB | 1.00 | 0.81 | 0.54 | 1.44 | 7.37 | 6.18 | 23.9 |
| Zhou Set | PDBsmf | 1.00 | 0.86 | 0.59 | 1.22 | 6.71 | 4.55 | 26.4 |
| Zhou Set | PDB300 | 0.98 | 0.78 | 0.52 | 1.70 | 8.38 | 5.13 | 16.9 |
| Zhou Set | PDB300smf | 1.00 | 0.83 | 0.57 | 1.30 | 7.46 | 3.85 | 19.1 |
Fraction of residues in the target sequence that are part of the best structural alignment.
Only gaps >3 residues are considered.
Only templates with a TM-score ≥ 0.4 are considered.
Structural alignments to the entire PDB library without chain length restrictions.
All template structures with a sequence identity >15% to the target are excluded.
Figure 3.
Comparison of the fraction of structurally aligned residues in the regular core secondary structure regions (black) and loop regions (red) for the PDB200 (dashed lines) and smoothed PDB200sm (solid lines) sets to the top 100 structures in the PDBsm set.
All Quasi-spherical protein structures are contained in the PDB and vice versa
To further demonstrate that the library of QS structures contains all the protein structures in the PDB and vice versa (8), we consider the most extreme case of quasi-spherical proteins packed into the same spherical volume as a native protein of the same length but which are essentially devoid of any regular secondary structure and backbone hydrogen bonds. We now demonstrate that the QS300 proteins are excellent templates for every compact protein in the PDB and can be used to build rather good quality structures. As shown in Table 1, 99% of the QS200 proteins as the target have a match to the PDB template set, with a TM-score ≥0.40; their mean TM-score is 0.44. Comparing the chain contours as in the QS200sm set, the mean TM-score improves to 0.51. As above, the number of target and template gaps diminish on chain contour smoothing but now the gap lap increases rather than decreases. Since we have shown in Figure 2 that above a TM-score of 0.5, 97% of internal secondary structure elements are preserved, this is a more than adequate threshold. However, as we next show, even at much lower TM-scores, the QS300 structures are excellent templates. If this holds for compact structures that are entirely devoid of secondary structure elements, then we would expect it to be even more true when more protein like structures are produced such as was done in the Taylor (6), Trovato (7), Zhou (31) sets.
The quasi-spherical structures are excellent templates for PDB structures
For the native PDB250 set as targets, Table 1 seems to show the problematic result that if the QS300 set is used as the template library, then only 71% of PDB targets have a TM-score ≥0.40. In fact, if we compare the smoothed PDB250sm set to the QS300sm set, then 94% ≥ of the QS300 templates have a TM-score to the smoothed native structure ≥0.40. Using the QS300 set as templates, a total of 8,254 PDB targets (PDB250 set) were modeled using TASSER. As shown in Table 1, the mean TM-score of the resulting TASSER model is 0.71, with 98% of the targets having a TM-score≥ 0.40. (The few that are below this threshold reflect errors in top cluster selection rather than the incompleteness of the template library).
In Figure 4, we plot the cumulative fraction of targets whose TM-score of the best QS template, best smoothed QS template and first ranked TASSER model exceed the TM-score on the abscissa. The effect of chain smoothing is dramatic and shows that the global chain contour information of the target is strongly encoded in the template. This is why the TM-score of the first ranked TASSER model (ranked based on the cluster centroid density by SPICKER (37)) generally dramatically improves on refinement.
Figure 4.
Cumulative fraction of target structures whose best QS template (dot-dot-dashed), smoothed (dashed) and first ranked TASSER model has a TM score ≥ value on the abscissa.
In Figure 5, we further demonstrate that this conservation of the global chain contour information is what underlies the dramatic TM-score improvement on TASSER refinement. The top panel shows that on chain smoothing the TM-score to native uniformly improves (the dashed line corresponds to the same TM-score values for the TM-scores of QS and QS300sm templates, abbreviated QS-sm in the figure). The middle (bottom) panel compares the TM-score of the TASSER model to native to that of the QS300 template (QS300sm template) to native. The few cases in the middle panel that show a diminution in TASSER model TM-score relative to the top ranked QS template, as noted above, are due to poor model selection. The improvement in structure quality due to TASSER refinement is dramatic. The reason is clearly delineated in the lower panel where the TM-score of the QS300sm templates to native covers a higher range of TM-scores. In other words, the chain contour provided by the QS300 template contains a significant amount of native chain contour information. This is captured by TASSER on model building and subsequent refinement.
Figure 5.
For the PDB250 target set and the QS300 template set: Upper panel: Comparison of the TM-score to native of the QS-sm template to that of the corresponding QS template to native. Middle panel: Comparison of the TM-score to native of the TASSER model to that of corresponding QS template to native. Lower Panel: Comparison of the TM-score to native of the TASSER model to that of corresponding QS-sm template to native.
The next question to be explored is the relationship between an initial TM-score value of the QS template and the final TM-score obtained on TASSER refinement. In Figure 6, for the set of initial TM-scores provided by the best QS300 alignment to the PDB250 target, we examine the cumulative fraction of TASSER models whose TM-score meets or exceeds a given value. At a TM-score of 0.32 (upper left panel), only 9.5% of the TASSER models have a TM-score ≥ 0.50; their mean TM-score is 0.33. The critical region where the refinement dramatically improves is for initial TM-scores around 0.34-0.35. For an initial TM-score of 0.34, 72% of the TASSER models have a TM-score ≥ 0.50, with a mean TM-score of 0.55. When the initial TM-score is 0.35, now 90% of the TASSER models exceed a TM-score of 0.50, and have a mean TM-score of 0.64. Using our standard TM-score threshold of 0.40, (next to bottom, right hand lower panel), now 98% of the TASSER models have a TM-score ≥ 0.50, with a mean TM-score of 0.74. For completeness, we also consider the case of the small subset (21) of targets whose best QS300 TM-score is 0.49. Now, 100% of the targets have a TASSER TM-score ≥ 0.61, with a mean TM-score of 0.75. Given that the quasi-spherical protein structures can be refined to give very high TM-score structures, this shows that in the current situation, a raw TM-score above 0.4 and even below (to a TM-score of ~0.35) contains sufficient global contour information that high quality full length models can be built starting from templates that are devoid of secondary structure. This analysis for the QS structures plus the results for the PDB200 set (see Figures 2 and 3) clearly demonstrate that a template with a TM-score ≥ 0.40 retains essentially all the global fold information necessary for building structures whose TM-score ≥ 0.50, the threshold by which Dai and Zhang conclude one can confidently assign a protein to a CATH fold. (31). So even by the more restrictive definition implicit in a discrete view of structure space, the PDB is quite likely complete.
Figure 6.
For a given initial TM-score of the best QS300 template to the PDB250 structure (indicated by both the figure legend and the dashed line), the cumulative fraction of targets whose top (first ranked) TASSER model's TM score ≥ the value on the abscissa. The TM score threshold of 0.40 is indicated by the dotted line. In the bottom right hand panel, for an initial TM score of 0.45, we employ the same convention as above, but now in addition, for an initial QS best template TM-score of 0.49 in the dot dashed line, we show the cumulative fraction of targets whose first ranked TASSER model has a TM score ≥ value on the abscissa.
The Taylor, Trovato and Zhou structures are all found in the PDB library
Using fr-TM-align, we now examine the fraction of structures in the Taylor (6), Trovato (7), and Zhou (31) sets present in the full PDB as well as in the PDB300 set, where we restrict the library to proteins ≤ 300 residues in length. As shown in Figures 7 A and B, where we compare the cumulative fraction of targets with a TM-score ≥ value on the abscissa, all three sets have virtually identical behavior as a function of TM-score. Thus, despite their very disparate means of preparation, we find a similar distribution of PDB template matches. Their cumulative TM-score curve lies between the QS200 structures and the PDB200 structures. The former lack secondary structure and the latter, even at a 15% identity threshold to the PDB, probably still detect a significant number of evolutionarily related proteins.
Figure 7.
A. Cumulative fraction of QS200, QS200sm, Taylor, Trovato, Zhou PDB200, PDB200sm targets whose TM-score ≥ abscissa for the templates in the PDB library. B. Same target sets as in A but using the PDB300 library as templates.
As shown in Table 1, with the full PDB as the template library, the mean TM-scores of the Taylor, Trovato and Zhou sets are 0.53, 0.52 and 0.54 respectively. Similar behavior is seen when the PDB300 set is used as the template library, with a small diminution in template quality. The mean number of gaps/target is small, with the smallest value of 0.61 for the Trovato set, comprised of 60 residue proteins. 96% of the Trovato structures have a TM-score ≥ 0.45, while 63% have a TM-score ≥ 0.50. However as shown in Figure 1, a TM-score of 0.40 is sufficient for recovery of above 90% of the core secondary structural elements and even for the QS proteins is more than adequate to generate quite high TM-score structures, (see Figure 6). If we consider the smoothed Trovato structures, the mean TM-score to the full PDB (PDB300) is 0.59 (0.57). For chain smoothed structures, 99.8% of targets have a TM-score≥ 0.45, and 95% have a TM-score ≥0.5. Thus, essentially every representative of the Trovato set is clearly contained in the PDB structural library.
We further analyze the behavior of Taylor and Zhou sets of target structures in Figure 8, where we compare the PDB300 and PDB template library for both the original and smoothed structures. For comparison, the QS target set is shown as well. Chain smoothing clearly increases the value of the best TM-score. Now, the mean value of the TM-score for the Taylor and Zhou sets is 0.59. As was found previously for the PDB200 and QS200 sets, the number of gaps/target decreases as the structural similarity of the target and the template increases due to chain smoothing.
Figure 8.
In the top, middle and lower panel for the Taylor, Zhou and QS200 sets, the cumulative fraction of targets that have a match to the PDB and PDB300 template library as a function of TM-score. Also shown are the cumulative fractions of targets that have a best TM-score template for the contour smoothed targets and templates as indicated by PDBsm and PDB300sm.
In Figure 9, we consider for the Taylor, Zhou and QS200 target structure libraries, (shown in the dashed lines), the fraction of unmatched targets in the PDB set at the given TM-score threshold of 0.45 (red) and 0.50 (black) as a function of target protein length. For both the Taylor and Zhou sets, using a TM-score threshold of 0.45, essentially all targets have a TM-score match (98.5% and 99% respectively), independent of chain length. For a TM-score threshold of 0.5, for the Taylor set, roughly 22-33% of targets lack a PDB template. Similarly, for the Zhou set, the fraction of unmatched templates monotonically increases from 11% to 23% as a function of chain length. However, this is misleading, as shown when we consider the smoothed templates in the solid lines. For the Taylor set, even at a TM-score of 0.50, the fraction of unmatched targets does not exceed 1.7%. Similarly, for the Zhou set, the fraction of unmatched targets does not exceed 5.0%.
Figure 9.
Fraction of unmatched targets in the Taylor, Zhou and QS sets to the PDB library as a function of the number of target protein residues. Red (black) indicates a TM-score threshold of 0.45 (0.50). Dashed (solid) lines are for the original (smoothed) structure.
As an example of a structure where a suitable PDB template was not found in the PDB by Dai and Zhou (31) but which is found in the current analysis, we present results for 1a8la1_46, shown in their Figure 3 as an example of a “new” fold. In Figure 10, we show the structural alignment of 1rw8A to 1a8la1_46. This is the third best template identified by fr-TM-align in the PDB template library. The top two templates (3ig3A and 1wer) are unaligned to the C-terminus of 1a8la1_46, but have essentially the same TM-score. To avoid issues of the structural completeness when some secondary structural elements are absent on a chain terminus, we focus on 1r8wA. In the left hand side of Figure 10, we show the structural alignments of the 1r8wA to 1a8la1_46. A total of 97/113 residues are aligned with a TM-score of 0.50 and a RMSD of 4.68 Å. As shown for the structural alignment of the chain smoothed 1r8wA to 1a8la1_46, the TM-score increases to 0.52, with 109/113 residues aligned with a RMSD of 4.65 Å. Thus, as indicated by the failure rate in Figure 9, most of the structures in the Zhou set have highly significant matches to structures in the PDB.
Figure 10.
Example of a significant structural match to permuted 1a8la1 structure (1a8la1_46) identified in Dai and Zhou (31) as lacking a match in the PDB. Left hand side: the TM-score of 1rw8A to 1al8a1_46 is 0.50. Right hand side, structural alignment of the smoothed 1al8a1_46 to 1rw8A; the corresponding TM-score is 0.52.
By comparison, for the QS200 set, while comparison of smoothed contours dramatically reduces the fraction of unmatched targets at a TM-score threshold of 0.45 to no more than 2.7%, depending on target chain length, up to 44% of targets are unmatched at a TM-score threshold of 0.5. This is consistent with Figure 8. Similar results as above are recovered if the PDB300 library is used (See Supporting Information, Figure 1).
Conclusions
Why did the Taylor (6), Trovato (7) and Zhou (31) groups conclude that a significant fraction of their structures are not in the PDB? There are three causes for their disagreement with our current results: First, they used the original TM-align (26) algorithm that in the TM-score regime below 0.5 often fails to detect structurally similar templates. As demonstrated elsewhere, the improved fr-TM-align algorithm (27) does not suffer from this limitation and detects more subtle structural similarities. A second issue is the size distribution of the proteins in the template library. Trovato, (7) in particular, restricted the size of templates to no more than 75 residues. Given that the average gap length of a template alignment is roughly 26 residues when the full PDB is used as the template library, this is too small a template size range to detect all structurally similar proteins. A third issue is the question of what TM-score threshold is relevant. As shown when the QS300 structures are used as templates for the PDB250 library, as well as by analysis of the fraction of core secondary structural elements matched as a function of TM-score, a TM-score of 0.4 is more than sufficient to provide a template whose full length model will exceed a TM-score of 0.5. Moreover, the similarity of global chain contours outside the conserved core implicit in a structure whose TM-score is 0.40 is sufficient that the chain smoothed models will often have a TM-score above 0.4. Thus, if one adopts the practical definition that a structure is present in the PDB if it can provide a geometrically suitable template that contains more than 90% of the core secondary structure and can be converted into a full length model that can be readily refined to a TM-score above 0.5, then our original conclusion that the library of compact single domain proteins is likely complete is further strengthened by the present study. This conclusion obviates any necessity of arbitrary fold classification, but if one wishes to classify proteins into folds, since a TM-score of 0.5 is all that is needed to make a confident SCOP and CATH classification (28), then the library of all folds is present in the Taylor, Trovato and Zhou sets. Moreover, it is implicitly found in the most distant class of structures from native proteins, the QS set.
One of the more surprising results of this study is that the QS templates, despite their lack of regular secondary structure, contain all the global chain contour information needed to recapitulate the PDB structural library of single domain proteins. The only assumptions used in the construction of the QS proteins are that they adopt the same volume as a native protein of the same length and that the excluded volume of the residues is preserved. This strongly suggests that global chain contour information, (as can be readily recovered on chain smoothing), of real proteins is simply the result of these two effects, packing and excluded volume. As shown elsewhere (8), packing defects and chain excursions resulting from the inclusion of hydrogen bonded secondary structures introduce protein like cavities and binding interfaces, essential factors for intermolecular interactions 8. However, from the point of view of global structure alone, there is nothing special or unique about the library of structures found in the PDB. The full space of compact, single domain protein structures likely arises from purely geometric effects. As concluded some time ago, one need not invoke evolution to rationalize most of the structural features of proteins (4).
With regards to the practical solution of the protein folding problem, we also argued that the protein structure prediction problem, at least for single domain proteins, could be solved by matching to the library of PDB templates (3). The key issue is to identify templates for the 30% of targets where contemporary structure prediction methods fail (39, 40). To date, all algorithms employ native structures as templates. However, the present results on the QS templates and the other sets suggest that chain smoothing might be a useful way of improving initial template model quality in threading. This idea will be pursued in the near future for template selection, initial alignment generation and side chain contact prediction. As suggested by a reviewer, we will also explore the ramifications of chain smoothing in fold classification, e.g. how many structures significantly align to a chain smoothed structure as well as the utility of this approach in defining fold families.
In conclusion, this work further argues that the library of solved PDB structures is likely complete and clearly demonstrates that the majority of secondary structural elements and global chain contour similarity is retained for structures with a TM-score to native of 0.4. Such structures are a rich source of information that needs to be better exploited, both in the design of improved structure prediction algorithms and approaches to model refinement. Moreover, while the notion of discrete folds is convenient as a classification tool, it may perhaps be more productive to focus on the key structural features that a pair proteins have in common without resorting to arbitrary assignments of fold similarity. If such similarity is detected, then as demonstrated for the QS structures, quite high quality models can be built, regardless of whether the templates have any local secondary structure in common, nor whether they are evolutionarily related or not. The key challenge is to develop methods that can routinely identify this similarity in the limit when their evolutionary relationship, if any, cannot be detected.
Supplementary Material
Supporting information available:
Figure S.1 Fraction of unmatched targets in the Taylor, Zhou and QS sets to the PDB300 library as a function of the number of target protein residues. Red (black) indicates a TM-score threshold of 0.45 (0.50). Dashed (black) lines are for the original (smoothed) structures. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgements/Dedication
This paper is dedicated with deep affection and admiration to Professor Harold Scheraga on the occasion of his 90th birthday. This research was supported in part by grant No. GM-48835 of the Division of General Medical Sciences of the National Institutes of Health. We thank the Taylor, Trovato and Zhou groups for generously providing us with their structural libraries.
References
- 1.Chothia C, Finkelstein AV. Annu Rev Biochem. 1990;59:1007–1039. doi: 10.1146/annurev.bi.59.070190.005043. [DOI] [PubMed] [Google Scholar]
- 2.Kihara D, Skolnick J. J Mol Biol. 2003;334:793–802. doi: 10.1016/j.jmb.2003.10.027. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Skolnick J. Proc Natl Acad Sci U S A. 2005;102:1029–1034. doi: 10.1073/pnas.0407152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Hubner IA, Arakaki AK, Shakhnovich E, Skolnick J. Proc Natl Acad Sci U S A. 2006;103:2605–2610. doi: 10.1073/pnas.0509379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skolnick J, Arakaki AK, Lee SY, Brylinski M. Proc Natl Acad Sci U S A. 2009;106:15690–15695. doi: 10.1073/pnas.0907683106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor WR, Chelliah V, Hollup SM, MacDonald JT, Jonassen I. Structure. 2009;17:1244–1252. doi: 10.1016/j.str.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 7.Cossio P, Trovato A, Pietrucci F, Seno F, Maritan A, Laio A. PLoS computational biology. 2010;6:e1000957. doi: 10.1371/journal.pcbi.1000957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brylinski M, Gao M, Skolnick J. Why not consider a spherical protein? Implications of backbone hydrogen bonding for protein structure and function. Phys Chem Chem Phys. 2011 doi: 10.1039/c1cp21140d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnautova YA, Vorobjev YN, Vila JA, Scheraga HA. Proteins. 2009;77:38–51. doi: 10.1002/prot.22414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maisuradze GG, Senet P, Czaplewski C, Liwo A, Scheraga HA. J Phys Chem A. 2010;114:4471–4485. doi: 10.1021/jp9117776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pillardy J, Arnautova YA, Czaplewski C, Gibson KD, Scheraga HA. Proc Natl Acad Sci U S A. 2001;98:12351–12356. doi: 10.1073/pnas.231479298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu G, Narayan M, Kurinov I, Ripoll DR, Welker E, Khalili M, Ealick SE, Scheraga HA. J Am Chem Soc. 2006;128:1204–1213. doi: 10.1021/ja055313e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, Young J, Yukich B, Zardecki C, Berman HM, Bourne PE. Nucleic Acids Res. 2011;39:D392–401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Nucleic Acids Res. 2008;36:D419–425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuff AL, Sillitoe I, Lewis T, Clegg AB, Rentzsch R, Furnham N, Pellegrini-Calace M, Jones D, Thornton J, Orengo CA. Nucleic Acids Res. 2011;39:D420–426. doi: 10.1093/nar/gkq1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuff A, Redfern OC, Greene L, Sillitoe I, Lewis T, Dibley M, Reid A, Pearl F, Dallman T, Todd A, Garratt R, Thornton J, Orengo C. Structure. 2009;17:1051–1062. doi: 10.1016/j.str.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuff AL, Sillitoe I, Lewis T, Redfern OC, Garratt R, Thornton J, Orengo CA. Nucleic Acids Res. 2009;37:D310–314. doi: 10.1093/nar/gkn877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu WP, Kolinski A, Skolnick J. Proteins. 1997;29:443–460. doi: 10.1002/(sici)1097-0134(199712)29:4<443::aid-prot5>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 19.Kolinski A, Skolnick J, Godzik A, Hu WP. Proteins. 1997;27:290–308. [PubMed] [Google Scholar]
- 20.Yang Z, Lasker K, Schneidman-Duhovny D, Webb B, Huang CC, Pettersen EF, Goddard TD, Meng EC, Sali A, Ferrin TE. UCSF Chimera, MODELLER, and IMP: An integrated modeling system. J Struct Biol. 2011 doi: 10.1016/j.jsb.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mariani V, Kiefer F, Schmidt T, Haas J, Schwede T. Assessment of template based protein structure predictions in CASP9. Proteins. 2011 doi: 10.1002/prot.23177. 2011. [DOI] [PubMed] [Google Scholar]
- 22.Sadreyev RI, Kim BH, Grishin NV. Current opinion in structural biology. 2009;19:321–328. doi: 10.1016/j.sbi.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wrabl JO, Grishin NV. Journal of computational biology : a journal of computational molecular cell biology. 2008;15:317–355. doi: 10.1089/cmb.2007.0161. [DOI] [PubMed] [Google Scholar]
- 24.Zemla A, Venclovas C, Moult J, Fidelis K. Proteins Suppl. 1999;3:22–29. doi: 10.1002/(sici)1097-0134(1999)37:3+<22::aid-prot5>3.3.co;2-n. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Skolnick J. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Skolnick J. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pandit SB, Skolnick J. BMC bioinformatics. 2008;9:531. doi: 10.1186/1471-2105-9-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu J, Zhang Y. Bioinformatics. 2010;26:889–895. doi: 10.1093/bioinformatics/btq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andreeva A, Howorth D, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Nucleic Acids Res. 2004;32:D226–229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Applegate DL, Bixby RE, Chvatal V, Cook W, Espinoza DG, Goycoolea M, Helsgaun K. Operations Res Lett. 2009;37:11–15. [Google Scholar]
- 31.Dai L, Zhou Y. J Mol Biol. 2011;408:585–595. doi: 10.1016/j.jmb.2011.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holm L, Rosenstrom P. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor WR. Methods Mol Biol. 2000;143:19–32. doi: 10.1385/1-59259-368-2:19. [DOI] [PubMed] [Google Scholar]
- 34.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P. Journal of computational chemistry. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 35.Lindahl E, Hess B, van der Spoel D. J Mol Mod. 2001;7:306–317. [Google Scholar]
- 36.Zhang Y, Skolnick J. Proc Natl Acad Sci U S A. 2004;101:7594–7599. doi: 10.1073/pnas.0305695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Skolnick J. Journal of computational chemistry. 2004;25:865–871. doi: 10.1002/jcc.20011. [DOI] [PubMed] [Google Scholar]
- 38.Rotkiewicz P, Skolnick J. Journal of computational chemistry. 2008;29:1460–1465. doi: 10.1002/jcc.20906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee SY, Skolnick J. Biophysical journal. 2010;99:3066–3075. doi: 10.1016/j.bpj.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bazzoli A, Tettamanzi AG, Zhang Y. J Mol Biol. 2011;407:764–776. doi: 10.1016/j.jmb.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information available:
Figure S.1 Fraction of unmatched targets in the Taylor, Zhou and QS sets to the PDB300 library as a function of the number of target protein residues. Red (black) indicates a TM-score threshold of 0.45 (0.50). Dashed (black) lines are for the original (smoothed) structures. This material is available free of charge via the Internet at http://pubs.acs.org.