Abstract
In vitro assembly of bacteriophage P22 procapsids requires coat protein and sub-stoichiometric concentrations of the internal scaffolding protein. If there is no scaffolding protein, coat protein assembles aberrantly, but only at higher concentrations. Too much scaffolding protein results in partial procapsids. By treating the procapsid as a lattice that can bind and be stabilized by scaffolding protein we dissect procapsid assembly as a function of protein concentration and scaffolding/coat protein ratio. We observe that (i) the coat-coat association is weaker for procapsids than for aberrant polymer formation, (ii) scaffolding protein makes a small but sufficient contribution to stability to favor the procapsid form, and (iii) there are multiple classes of scaffolding protein binding sites. This approach should be applicable to other heterogeneous virus assembly reactions and will facilitate our ability to manipulate such in vitro reactions to probe assembly, and for development of nanoparticles.
Introduction
Bacteriophage P22 has been one of the best documented experimental systems for investigating bacteriophage assembly (Casjens and King, 1974; King et al., 1976; Prevelige and Fane, 2012; Teschke and Parent, 2010) and more recently for the development of virus-derived nanoparticles (Kang et al., 2010; O’Neil et al., 2011). An in vitro assembled procapsid-like particle (PC), which lacks the portal complex and injection proteins, is comprised of 420 coat proteins and a variable number of scaffolding protein molecules (from ~60 to 360), depending on the availability of scaffolding protein in the reaction (King, Hall, and Casjens, 1978; Parent, Zlotnick, and Teschke, 2006). This reaction can be described by:
| 1) |
where Z represents the number of bound scaffolding proteins within a PC (Parent, Suhanovsky, and Teschke, 2007; Parent, Zlotnick, and Teschke, 2006). Kinetics of both extraction of scaffolding protein from procapsids and re-entry of scaffolding protein into empty procapsid shells indicate that there are at least two populations of bound scaffolding proteins (Greene and King, 1994; Parker, Brouillette, and Prevelige, 2001; Teschke and Fong, 1996). There are approximately 60 high affinity scaffolding proteins necessary for assembly of a procapsid (scaffolding/coat protein ratio of 0.14). These bind with a Kd = ~0.3 μM (Parker, Brouillette, and Prevelige, 2001). Higher scaffolding/coat protein ratio results in PCs with additional scaffolding proteins within the procapsid. Thus, bacteriophage P22 procapsids are remarkably nonuniform in scaffolding protein content (Fane and Prevelige, 2003; Parent, Suhanovsky, and Teschke, 2007; Prevelige, Thomas, and King, 1993).
The stability of coat-coat protein interactions, the action of scaffolding protein, and the stability of coat-scaffolding protein interactions all critically affect products of assembly reactions. In the absence of scaffolding protein, coat protein will assemble but into aberrant complexes (Prevelige et al., 1990). While scaffolding protein is required for normal assembly, increasing the affinity of scaffolding protein for coat protein or increasing the scaffolding/coat protein ratio (> 7.0) in an in vitro assembly reaction results in the rapid assembly and entrapment of numerous partial PCs, indicating that scaffolding protein affects kinetics and thermodynamics, and can thus lead to kinetic traps (Parent et al., 2005; Parent, Zlotnick, and Teschke, 2006). Tight regulation of scaffolding protein translation is essential in vivo to ensure a low concentration of scaffolding protein compared to coat protein and to achieve high phage production (King, Hall, and Casjens, 1978).
The structure of scaffolding protein in the context of a procapsid is poorly defined. A recent procapsid cryo-EM reconstruction suggests that every coat protein may have an scaffolding protein bound (Chen et al., 2011). This would be more scaffolding protein per PC than has been observed by biochemical assays, which indicate between 60 and 360 scaffolding proteins per PC (Greene and King, 1994; Parent, Zlotnick, and Teschke, 2006; Parker, Brouillette, and Prevelige, 2001; Prevelige, Thomas, and King, 1993). The C-terminal helix-turn-helix domain of scaffolding protein is known to interact with coat protein via elcrostatic interactions (Cortines et al., 2011; Parent et al., 2005; Parker and Prevelige, 1998). Only parts of scaffolding protein that are directly in contact with the contiguous procapsid are visible in reconstructions, suggesting that most of scaffolding protein is in varied orientations, likely interacting with other scaffolding protein molecules (Chen et al., 2011; Thuman-Commike et al., 2000). There are several explanations to rectify the difference between biochemical observations and the recent image reconstruction. Perhaps some of the observed density in the reconstruction is due to the interaction of the N-terminus of scaffolding protein with the interior of PC (Padilla-Meier and Teschke, 2011; Suhanovsky and Teschke, 2011). Another explanation is that the 415 sites in a native procapsid are filled randomly, but molecular crowding prevents more than 300 to 360 scaffolding proteins from fitting into any one procapsid; thus, the average of many particles would show scaffolding protein density at each possible binding site.
During assembly scaffolding protein is required to catalyze, stabilize and direct the geometry of PC formation (Prevelige, Thomas, and King, 1988). In its simplest case, PC stability can be described as the sum of contributions from coat protein (C) and scaffolding protein (S):
| 2) |
where Z is the number of bound scaffolding proteins within PCs (Parent, Zlotnick, and Teschke, 2006). Since PCs are the final reaction product formed under a given set of conditions (Parent, Zlotnick, and Teschke, 2006), the law of mass action for the reaction dictates the following:
| 3) |
When the concentration of each free species and the number of bound scaffolding protein molecules was measured experimentally, the results of this analysis indicated that under specified experimental conditions each coat protein contributes −7.2 kcal/mol to PC stability and each scaffolding protein contributes −6.1 kcal/mol (Parent, Zlotnick, and Teschke, 2006).
There are three critical asumptions inherent in this approach. i) Scaffolding protein contributes 100% of its binding energy to PC stability. ii) As indicated in equation 3, a PC with Z scaffolding proteins is “complete”, and that iii) all reaction products have exactly Z scaffolding proteins. However as described above, PCs are not uniform with respect to the amount of scaffolding protein incorporated and generally assemble with substoichiometric quantities of scaffolding protein. Taking a different perspective, here we develop an analysis of PC stability where we consider scaffolding protein binding to a lattice of available sites. This analysis yields new insights into the contribution of scaffolding protein to PC stability.
Results
Theory of scaffolding protein contribution to PC stability
In the approach described above, we assumed that scaffolding protein is an essential component with fixed amounts of scaffolding protein per PC. We reanalyzed the data of Parent et al., 2006, with additional data to cover a wider range of scaffolding/coat protein ratios (described in the Materials and Methods). By considering scaffolding protein as an optional component in assembly we can describe association of a hypothetical PC in the absence of scaffolding protein (PC_w/out_S):
| 4) |
To emphasize the concentration dependence of assembly, equation 4 is put in terms of μ°, the equilibrium chemical potential of PC, coat protein (C), and scaffolding protein (S) (e.g. μ°PC = −RT ln[PC] under standard conditions) to generate equation 5.
| 5) |
Equation 6 presents a more realistic description of PC stability, as in vitro assembled PCs necessarily contain scaffolding protein.
| 6) |
However, equation 6 suggests that scaffolding protein binding is uniform. In fact, Z represents a distribution (different PC particles have different amounts of bound scaffolding protein) and, as will be shown later, not all sites make the same energetic contribution.
To evaluate PC assembly as a function of scaffolding protein we compare equations 5 and 6 to generate equation 6. For simplicity, and to place energies on a scale more typical for protein-protein interactions, instead of using the energy for the entire PC without scaffolding protein (ΔG°PC_w/out_S) we use ΔGPC,C, S=0 for the association energy per coat protein in the PC in the absence of scaffolding protein.
| 7) |
Extrapolation of ΔGPC_w/out_S to [S] = 0 will lead to the energy of coat protein-coat protein interaction for PC formation. Equation 7 indicates that PC stability can be enhanced by incorporation of an arbitrary number of scaffolding protein molecules, Z, to available sites.
In vitro PC formation as a function of scaffolding protein
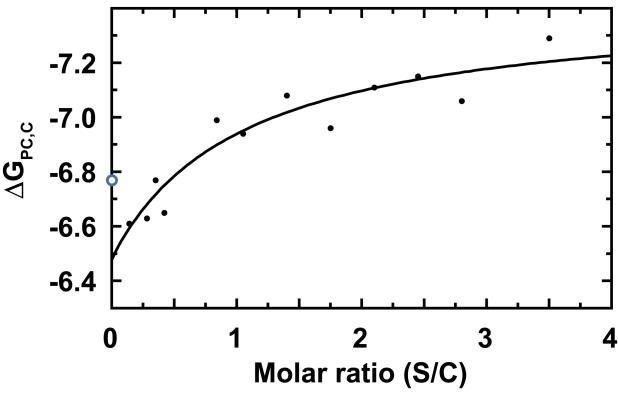
Equation 7 is consistent with our observation that increasing the scaffolding protein concentration in an assembly reaction leads to a greater yield of PC (Parent, Zlotnick, and Teschke, 2006). Figure 1, a plot of ΔGPC,C versus the input ratio of scaffolding/coat protein (S/C) in the in vitro assembly reaction, shows the hyperbolic shape expected for a saturable binding curve, i.e., the energy from scaffolding protein is additive, but depends on scaffolding protein binding to limited number of sites within PCs. The hyperbolic curve is a function of the intrinsic coat protein-coat protein interaction energy in the context of a PC (ΔGPC,C,S=0), the number of specific binding sites for scaffolding protein (Snum), the energy contribution per scaffolding protein (ΔGPC,S), and the midpoint of binding (K’s) in terms of the input S/C ratio:
| 8) |
Figure 1. Scaffolding protein increases PC stability.
ΔGPC_w/out_S becomes stronger at higher relative concentrations of scaffolding protein. The hyperbolic curve fit indicates each scaffolding protein contributes −1.30 ± 0.24 kcal/mol to a base PC stability of −6.45 ± 0.11 kcal/mol per coat protein. The midpoint of the binding fit is S/C = 0.88. Note that the ΔGPC_w/out_S for 0 scaffolding protein is −6.77 kcal/mol, (blue, open circle) calculated from the critical concentration for uncontrolled coat protein assembly (Figure 2) and is not included in the curve fit. This graph includes new data, and data from (Parent, Zlotnick, and Teschke, 2006).
Since the number of scaffolding protein sites and the scaffolding protein contribution to PC stability (Snum and ΔGPC,S) are correlated, we considered several possibilities. Snum was determined independently by fitting a plot of bound scaffolding protein per PC against the initial ratio of scaffolding/coat protein; the resulting hyperbolic fit indicated a maximum of 294 ± 33 scaffolding proteins per PC. Alternatively, approximately 360 scaffolding protein sites were estimated from a curve fit where an assumption of a minimum number of bound scaffolding protein molecules for PC formation was used (Materials and Methods, data not shown). This is similar to the maximum number of 350 scaffolding protein per PC determined previously (Parent, Zlotnick, and Teschke, 2006). A maximum of 420 sites is theoretically possible if every coat protein in a PC binds a scaffolding protein. These values are integral multiples of the 60 asymmetric units in an icosahedron. ΔGPC,C,S=0 and ΔGPC,S for each Snum value were thus determined with equation 8 (Figure 1, Table 1). For an Snum of 300, each scaffolding protein contributes ΔGPC,S of −1.30 ± 0.24 kcal/mol to PC stability (Table 1). The different values of Snum lead to the same qualitative result that each scaffolding protein contributes only a small amount of energy to assembly. By extrapolation to [S] = 0, each coat protein contributes −6.45 ± 0.11 kcal/mol to PC stability, which is similar to that determined previuosly (Parent, Zlotnick, and Teschke, 2006).
Table 1. Assembly of PC and the binding of S to PC.
Results are from curve fits of Equation 8 to Figure 1 and extrapolation of titrations in Figure 3 to infinite dilution. The table shows coat-coat protein association energy (ΔGPC_w/out_S), the effect of scaffolding protein on the coat-coat protein association (ΔGPC,S), and the dissociation constant of scaffolding protein for PC (1/KS). Note that the product of Snum and ΔGPC,S is constant; the curve fit to Figure 1 is identical for all values of Snum.
| Snum | ΔGPC_w/out_S (kcal/mol) |
ΔGPC,S (kcal/mol) |
1/KS (μM) |
|---|---|---|---|
| 300 | −6.45 | −1.30 | 0.77 ± 0.70 |
| 360 | −6.45 | −1.08 | 1.29 ± 0.95 |
| 420 | −6.45 | −0.93 | 1.88 ± 1.25 |
Critical concentration for spontaneous assembly
For viral capsids, the mimimal concentration of coat protein for efficient assembly is a ‘pseudo-critical’ concentration. The prefix ‘pseudo-‘ is required because the concentration of free coat protein is not truly constant over a broad range of coat protein input concentrations when the assembly products are particles of defined size (this explicitly arises from equation 3); for open-ended non-capsid polymer (like tubulin) there is a true critical concentration where free protein is constant and can freely equilibrate with protein forming the polymeric complex (Tanford, 1980; Zlotnick, 1994). The pseudo-critical concentration for assembly of coat protein into PCs, in the absence of scaffolding protein, was 16 μM, determined by extrapolation of the data in Figure 1 to S = 0 (ΔGPC,C,S=0). We compared this value for PC formation in the absence of scaffolding protein to an experimentally measured value for spontaneous assembly of coat protein, which can be induced by concentrating coat protein monomers.
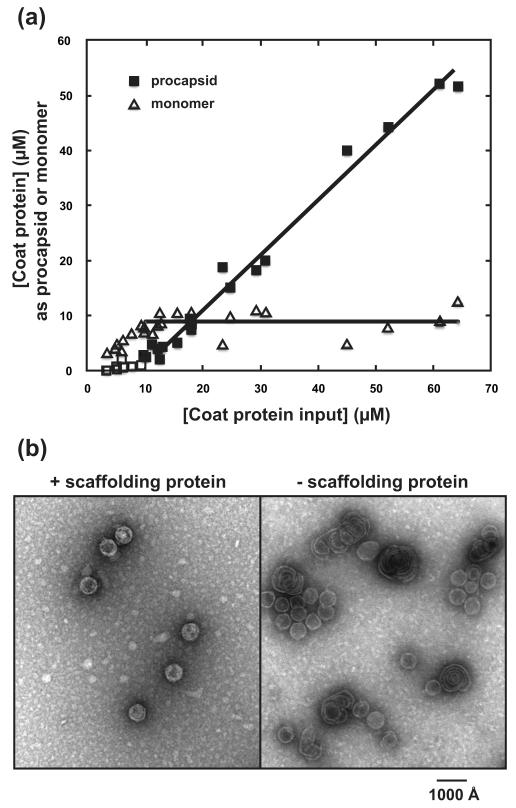
We determined the critical concentration for spontaneous assembly of coat protein by quantifying the concentrations of coat protein monomer and non-PC polymer from reactions with different initial concentrations of coat protein. Figure 2A shows a classic critical concentration isotherm, expected for open-ended polymers (Katen and Zlotnick, 2009). The average concentration for unassembled coat protein (for reactions where input [C] > 10 μM) was 8.9 ± 2.2 μM. Above [C] = 10 μM, all additional coat protein was incorporated into polymer, indicated by the slope of the polymer line of 0.99. The concentration where non-PC polymer appeared, estimated from the x-intercept of the polymer line (for reactions where total [C] > 10 μM) was 8.5 ± 2.1 μM. Many, if not all of the resulting particles, had aberrant morphology (Figure 2B).
Figure 2. coat protein polymerization in the absence of scaffolding protein.
(A) Assembly at different concentrations of coat protein demonstrates a critical concentration at ≤10μM coat protein (open squares and triangles), therefore straight lines were fit to data for total [C] > 10 μM (closed squares and triangles). For coat protein monomer, the average concentration was 8.9 ± 2.2 μM. For assembled non-PC polymer, the x-intercept was 8.5 ± 2.1 μM coat protein. Assembly reactions of concentrated coat protein were evaluated by centrifugation and the concentration of pelleted coat protein polymer and soluble coat protein monomer were determined from SDS-PAGE. (B) Micrographs of assembly products with and without scaffolding protein demonstrates the role that scaffolding protein plays in directing PC geometry.
The calculated concentration for coat protein assembly for PCs without scaffolding protein (16 μM, Figure 1) and observed experimentally for formation of aberrant coat protein complexes (8.5 to 8.9 μM) are significantly different. The assembly of non-PC polymer is below the expected critical concentration. This indicates that without the contribution of scaffolding protein, the coat protein-coat protein interaction in the non-PC polymer is actually more stable by about −0.4 kcal/mol than the conformation found in a T=7 PC. However, to form PCs the energetic difference between coat protein-coat protein interactions is overcome by the contribution of scaffolding protein to PC stability (−1.3 kcal/mol).
Affinity of scaffolding protein for different sites
To further define how each scaffolding protein contributes to PC stability, we examined the average dissociation constant of scaffolding protein for the PC lattice. This calculation was based on concentrations of free and bound scaffolding protein and the estimation of 300, 360, and 420 quasi-equivalent sites per PC (Table 1). The number of bound scaffolding protein molecules were taken from previous results (Parent, Zlotnick, and Teschke, 2006) and are based on the assumption that all molecules are productively bound to PC. Thus, the equilibrium assocation constant can be described by equation 9.
| 9) |
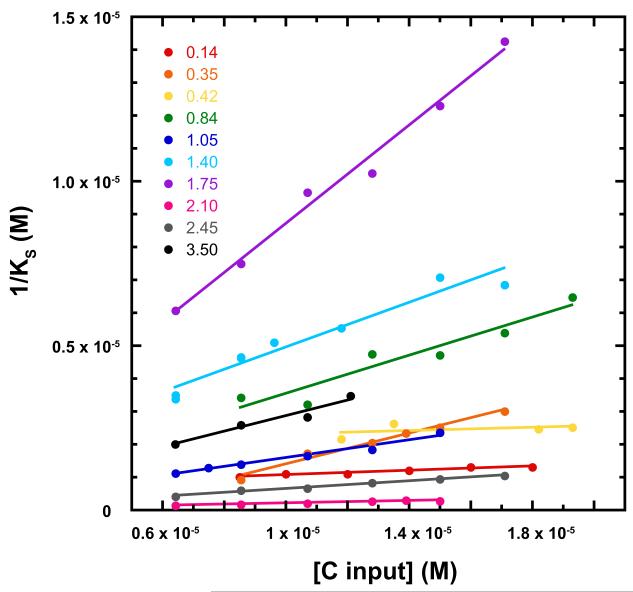
Dissociation constants (1/KS) were determined at multiple initial ratios of scaffolding/coat protein from 0.14 to 3.5, and multiple concentrations of scaffolding protein at each ratio (Figure 3). If there were a single class of binding site, KS would be expected to be constant, and thus independent of protein concentration. Conversely, if there were several classes of binding sites, we would observe the average affinity for filled sites, with the average affinity decreasing as more weak sites were filled at higher scaffolding protein concentrations. In fact, we observed a broad correlation where KS was weaker at higher protein concentrations (high [C input]). The weaker binding constant at high protein concentrations, where more binding occurs, is likely due to the use of progressively more weak sites, indicating multiple classes of independent site. Alternatively, there may be negative cooperativity between dependent sites. In either case, the association constant from equation 9 for a given set of intital conditions yields a weighted average. Our observation is consistent with previous observations that there are at least two different classes of scaffolding protein binding site within PC (Greene and King, 1994; Parker, Brouillette, and Prevelige, 2001). Extrapolated to infinite dilution of coat protein, where strong binding sites dominate the KS calculation, the average dissociation constant was 0.8 ± 0.7 μM for Snum = 300 (Table 1) Given the noise of the extrapolation, this value is consistent with the dissociation constant of approximately 0.3 μM for the 60 high affinity scaffolding protein-binding sites determined by isothermal titration calorimetry (Parker, Brouillette, and Prevelige, 2001).
Figure 3. The affinity of scaffolding protein for the PC lattice becomes weaker at higher initial concentrations of coat protein.
Each line represents data for assembly reactions with a given S/C molar ratio from 0.14 to 3.5; points are at different initial concentrations of coat protein. The monotonic change in KS is consistent with two or more classes of binding sites or negative cooperativity. The graph includes new data and data from (Parent, Zlotnick, and Teschke, 2006). The KS values in this graph were calculated for 300 scaffolding protein sites; the graph was qualitatively identical for 360 and 420 sites.
Discussion
Our new analysis of PC formation yields results that are consistent with experiments and expands our understanding of the role of scaffolding protein. It accommodates varying occupancy of scaffolding protein, provides descriptions of scaffolding protein site heterogeneity, intrinsic coat-coat protein interaction energy, and the critical concentration for scaffolding protein-independent assembly at high coat protein concentrations. The observed critical concentration for uncontrolled coat protein assembly significantly undershoots the value estimated from ΔGPC,C,S=0, suggesting an energetic compromise is required to complete a PC resulting in a large number of less than ideal coat-coat protein interactions. While it is well established that scaffolding protein controls assembly geometry (Lenk et al., 1975; Prevelige, Thomas, and King, 1988; Suhanovsky et al., 2010), scaffolding protein also increases the yield, i.e. it stabilizes PC (Parent, Zlotnick, and Teschke, 2006), indicating that scaffolding protein provides the basis for initiating and stabilizing assembly of T=7 particles.
Based on our data, we estimate 300 to 350 scaffolding protein sites per PC (this paper and (Parent, Zlotnick, and Teschke, 2006). These estimates are consistent with the 300-360 sites determined from biochemical measurements (Casjens et al., 1985; Parent et al., 2005; Prevelige, Thomas, and King, 1993). A maximum of approximately 300 scaffolding protein molecules per procapsid were found in vivo (King, Hall, and Casjens, 1978). The concentration dependent changes in average binding constant for scaffolding protein indicates that there are mutliple classes of scaffolding protein sites, with a sub-micromolar Kd for the highest affinity sites. This heterogeneity, and a plethora of low affinity sites, may also explain the differences in estimates of the number of scaffolding protein sites in a procapsid.
A scaffolding protein molecule makes a surprisingly small contribution to PC stability, substantially smaller than its binding energy for PCs. This indicates that scaffolding protein binding is only partly linked to PC stability, contrary to the assumption of our previous analysis (Parent, Zlotnick, and Teschke, 2006). The simplest interpretation (Wyman and Gill, 1990) is that scaffolding protein can bind to a given site on a PC in two states, with or without stabilizing the complex. The average difference in binding energy for these two states is ΔGPC,S, −1.3 kcal/mol (Table 1). The consistent value for ΔGPC,S over a broad range of scaffolding protein – coat protein concentrations (demonstrated by the hyperbolic fit in Figure 1) suggests that both low and high affinity scaffolding protein binding events contribute to PC assembly and stability. Complexes of scaffolding protein and coat protein, out of the context of a PC, may exist but at low concentration due to weak affinity.
Novel implications resulting from this mathematical analysis
There are several implications that can be derived from this analysis. Consider:
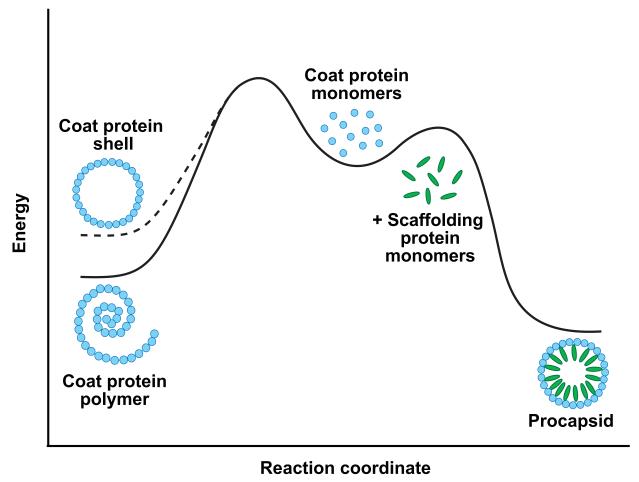
The difference between the 16 μM Kd estimated for a coat-coat protein interaction in a PC and the ~8.5 μM critical concentration observed for coat protein self-assembly suggests that interactions between subunits must be strained to obtain a PC. While the coat-coat protein interaction is stronger for the non-PC polymer than calculated for PC without scaffolding protein (shell), the scaffolding protein-mediated coat-coat protein interaction of PCs is much stronger (Figure 4). This higher energy state of coat-coat protein interaction in an empty coat protien shell may be functionally important for the lattice to undergo the transition to produce the stable, mature head (Galisteo, Gordon, and King, 1995). Similar conformational instability may be an energy source for the transitions associated with maturation in other bacteriophages like HK97 (Johnson, 2010) and the eukaryotic Herpesviruses (Brown and Newcomb, 2011).
The observed concentration dependence for the dissociation constant of scaffolding protein for PC sites indicates heterogeneity in binding. This is inconsistent with a single value for the contribution of scaffolding protein to PC stability, the ΔGPC,S term of equation 7. However, heterogeneity of scaffolding protein sites is consistent with the data in Figure 3, which shows that not all scaffolding protein sites are equal in interaction energy. These data suggest that scaffolding protein binding early in the assembly of a PC may face and impose far different forces than an scaffolding protein bound toward the latter stages of PC closure. The variation of binding energies calculated from the equilibrium of scaffolding protein with PC suggests that the values ΔGPC,C, ΔGPC,S, and ΔGS all vary with the saturation of scaffolding protein sites on the PC lattice.
Low concentrations of scaffolding protein are sufficient to direct assembly of T=7 particles, though such particles are less thermodynamically stable than PCs that are nearly saturated with scaffolding protein. For example, assembly at about 100 scaffolding protein per PC (i.e., ΔGPC = 420*(−6.45 kcal/mol) + 100*(−1.30 kcal/mol)) is less stable than aberrant uncontrolled assembly of the same 420 coat proteins (i.e., 420*(−6.77 kcal/mol)). To explain this result, either high affinity scaffolding protein sites contribute more to stability or the initial scaffolding protein molecules help overcome a kinetic barrier to nucleating assembly with PC geometry. Either possiblity suggests the hypothesis that the effect of scaffolding protein on coat-coat protein interaction geometry propagates throughout the process of P22 assembly, a path not postulated in most previously defined models of assembly (Zlotnick and Mukhopadhyay, 2011). It is likely that rigorously characterizing assembly will require development of new approaches to observing assembly of single particles and modeling reactions with coarse grained molecular dynamics simulations (Elrad and Hagan, 2008; Nguyen, Reddy, and Brooks, 2007; Rapaport, 2010).
Figure 4. Scaffolding protein stabilizes procapsids.
In the absence of scaffolding protein (left half of diagram), an empty coat protein shell is less stable than a non-icosahedral coat protein polymer. The interaction of scaffolding protein with coat protein (right half of diagram) stabilizes and leads to formation of PCs. Experimentally, a PC stripped of scaffolding protein does not redistribute to the polymer form because of the high activation energy barrier to dissociation (Singh and Zlotnick, 2003). The thermodynamic stability of the empty coat protein shell (dashed line) was determined by extrapolation of the data in Figure 1 to S = 0. The observation that even a small amount of scaffolding protein is sufficient to induce PC formation indicates that scaffolding protein also lowers the kinetic barrier to PC formation. Figure not drawn to scale.
In summary, our analysis supports the hypothesis that bacteriophage P22 assembly is a highly dynamic process. Scaffolding protein contributes binding energy and provides geometric constraints, particularly in the early stages of assembly. These contributions continue as assembly proceeds, but it appears that the requirement for scaffolding protein, based on structural and geometric arguments (Lander et al., 2006; Prevelige et al., 1990), becomes progressively smaller and may be dispensible during the latter stages of assembly.
Materials and Methods
Refolded coat protein monomers
Coat protein monomers were obtained as described previously (Anderson and Teschke, 2003; Fuller and King, 1982; Parent, Zlotnick, and Teschke, 2006). Briefly, urea-unfolded coat protein monomers were refolded by extensive dialysis against 20 mM phosphate buffer, pH 7.6 at 4 °C, and clarified by centrifugation at 175,000 × g at 4 °C for 20 min.
Assembly and analysis of PCs
PCs were assembled as previously described (Parent, Zlotnick, and Teschke, 2006). Briefly, refolded coat protein monomers at a final concentration of 0.3-0.9 mg/mL (6.5-19 μM) were mixed with scaffolding protein at concentrations corresponding to scaffolding/coat protein molar ratios ranging from 0.14 to 3.5. Several reactions were performed at each scaffolding/coat protein molar ratio by varying both the coat protein and scaffolding protein concentrations. Assembly reactions were done in 20 mM sodium phosphate (pH 7.6), 50 mM NaCl and incubated at 20 °C for >20 h in a total volume of 125 μl. Assembly reactions were applied to a 15 mL Sepharose 4B column run at a flow rate of 0.5 ml/min at room temperature in 20 mM sodium phosphate, pH 7.6, 50 mM NaCl buffer. Samples of each fraction were then run on 10% SDS polyacrylamide gels. The gels were silver stained (Rabilloud, Carpentier, and Tarroux, 1988) and the coat and scaffolding protein bands were quantified by densitometry using BioRad Quantity One software. The fraction of scaffolding or coat protein in the peaks corresponding to PC or to the unreacted subunits was simply calculated from the total of each protein applied to the column. Data from Parent, Zlotnick and Teschke, 2006 and new data corresponding to molar ratios of 0.14, 1.75 and 3.5 were analyzed to determine the concentration of coat and scaffolding proteins in PCs and as soluble subunits.
The average number of scaffolding protein per PC was plotted against the input S/C ratio and fit with a rectangular hyperbola:
| 10) |
where N is a unitary association constant of scaffolding protein for procapsids that scales the ratio of bound to the ratio of input scaffolding protein. Snum is the maximum number of binding sites for scaffolding protein. Smin is the minumum number of bound scaffolding proteins.
Uncontrolled assembly
Coat protein was assembled in the absence of scaffolding protein by concentrating aliquots of assembly-competent coat protein monomers (prepared as described above) by dialysis against a solution of 20% PEG 20K in 20 mM sodium phosphate (pH 7.6), 50 mM NaCl. The total concentration of coat protein in each sample after concentration was determined by the absorbance at 280 nm after denaturation with 6 M guanidine hydrochloride. Assembled coat protein polymers were sedimented by centrifugation through a 20% sucrose cushion at 107,000 g for 90 minutes at 20 °C in an Sorvall RP55-S rotor. The concentrations of pelleted coat protein polymer and coat protein monomer remaining in solution were determined by densitometry of Coomassie-stained SDS-PAGE gels using BioRad Quantity One software.
Negative stain electron microscopy
Aliquots (3 μL) of the uncontrolled assembly reactions were applied to carbon-coated, 300-mesh copper grids, allowed to absorb for 1 minute, and the grids were then washed with 2-3 drops of water followed by staining with 1% aqueous uranyl acetate for 30 seconds. Excess stain was blotted off with filter paper and the grids were air-dried. The images were acquired using an AMT XR-40 (2048×2048 pixel) camera side mounted on a Technai Biotwin G2 Spirit transmission electron microscope (nominal magnification of 68,000) operated at 80 kV.
Acknowledgements
We thank Dr. Marie Cantino for use of the University of Connecticut Electron Microscopy Center and her expert guidance in electron microscopy. This work was supported by NIH grants R01 AI077688 to AZ and R01 GM076661 to CMT.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson E, Teschke CM. Folding of phage P22 coat protein monomers: kinetic and thermodynamic properties. Virology. 2003;313(1):184–97. doi: 10.1016/s0042-6822(03)00240-x. [DOI] [PubMed] [Google Scholar]
- Brown JC, Newcomb WW. Herpesvirus Capsid Assembly: Insights from Structural Analysis. Current opinion in virology. 2011;1(2):142–149. doi: 10.1016/j.coviro.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, Adams MB, Hall C, King J. Assembly-controlled autogenous modulation of bacteriophage P22 scaffolding protein gene expression. J Virol. 1985;53(1):174–9. doi: 10.1128/jvi.53.1.174-179.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, King J. P22 morphogenesis. I: Catalytic scaffolding protein in capsid assembly. J Supramol Struct. 1974;2(2-4):202–24. doi: 10.1002/jss.400020215. [DOI] [PubMed] [Google Scholar]
- Chen DH, Baker ML, Hryc CF, DiMaio F, Jakana J, Wu W, Dougherty M, Haase-Pettingell C, Schmid MF, Jiang W, Baker D, King JA, Chiu W. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(4):1355–60. doi: 10.1073/pnas.1015739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortines JR, Weigele PR, Gilcrease EB, Casjens SR, Teschke CM. Decoding bacteriophage P22 assembly: identification of two charged residues in scaffolding protein responsible for coat protein interaction. Virology. 2011;421(1):1–11. doi: 10.1016/j.virol.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrad OM, Hagan MF. Mechanisms of size control and polymorphism in viral capsid assembly. Nano Lett. 2008;8(11):3850–7. doi: 10.1021/nl802269a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fane BA, Prevelige PE., Jr. Mechanism of scaffolding-assisted viral assembly. In: Chiu W, Johnson JE, editors. Virus Structure. Vol. 64. Academic Press; San Diego: 2003. pp. 259–99. [DOI] [PubMed] [Google Scholar]
- Fuller MT, King J. Assembly in vitro of bacteriophage P22 procapsids from purified coat and scaffolding subunits. Journal of molecular biology. 1982;156(3):633–65. doi: 10.1016/0022-2836(82)90270-4. [DOI] [PubMed] [Google Scholar]
- Galisteo ML, Gordon CL, King J. Stability of wild-type and temperature-sensitive protein subunits of the phage P22 capsid. J.Biol.Chem. 1995;270:16595–16601. doi: 10.1074/jbc.270.28.16595. [DOI] [PubMed] [Google Scholar]
- Greene B, King J. Binding of scaffolding subunits within the P22 procapsid lattice. Virology. 1994;205:188–197. doi: 10.1006/viro.1994.1634. [DOI] [PubMed] [Google Scholar]
- Johnson JE. Virus particle maturation: insights into elegantly programmed nanomachines. Current Opinion in Structural Biology. 2010;20(2):210–6. doi: 10.1016/j.sbi.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Uchida M, O’Neil A, Li R, Prevelige PE, Douglas T. Implementation of p22 viral capsids as nanoplatforms. Biomacromolecules. 2010;11(10):2804–9. doi: 10.1021/bm100877q. [DOI] [PubMed] [Google Scholar]
- Katen SP, Zlotnick A. Thermodynamics of Virus Capsid Assembly. Methods in Enz. 2009;455:395–417. doi: 10.1016/S0076-6879(08)04214-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J, Botstein D, Casjens S, Earnshaw W, Harrison S, Lenk E. Structure and assembly of the capsid of bacteriophage P22. Philos Trans R Soc Lond B Biol Sci. 1976;276(943):37–49. doi: 10.1098/rstb.1976.0096. [DOI] [PubMed] [Google Scholar]
- King J, Hall C, Casjens S. Control of the synthesis of phage P22 scaffolding protein is coupled to capsid assembly. Cell. 1978;15(2):551–60. doi: 10.1016/0092-8674(78)90023-5. [DOI] [PubMed] [Google Scholar]
- Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312(5781):1791–5. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- Lenk E, Casjens S, Weeks J, King J. Intracellular visualization of precursor capsids in phage P22 mutant infected cells. Virology. 1975;68(1):182–99. doi: 10.1016/0042-6822(75)90160-9. [DOI] [PubMed] [Google Scholar]
- Nguyen HD, Reddy VS, Brooks CL., 3rd Deciphering the kinetic mechanism of spontaneous self-assembly of icosahedral capsids. Nano Lett. 2007;7(2):338–44. doi: 10.1021/nl062449h. [DOI] [PubMed] [Google Scholar]
- O’Neil A, Reichhardt C, Johnson B, Prevelige PE, Douglas T. Genetically programmed in vivo packaging of protein cargo and its controlled release from bacteriophage P22. Angewandte Chemie. 2011;50(32):7425–8. doi: 10.1002/anie.201102036. [DOI] [PubMed] [Google Scholar]
- Padilla-Meier GP, Teschke CM. Conformational changes in bacteriophage P22 scaffolding protein induced by interaction with coat protein. Journal of molecular biology. 2011;410(2):226–40. doi: 10.1016/j.jmb.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Doyle SM, Anderson E, Teschke CM. Electrostatic interactions govern both nucleation and elongation during phage P22 procapsid assembly. Virology. 2005;340(1):33–45. doi: 10.1016/j.virol.2005.06.018. [DOI] [PubMed] [Google Scholar]
- Parent KN, Suhanovsky MM, Teschke CM. Phage P22 procapsids equilibrate with free coat protein subunits. Journal of molecular biology. 2007;365(2):513–22. doi: 10.1016/j.jmb.2006.09.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Zlotnick A, Teschke CM. Quantitative Analysis of Multi-component Spherical Virus Assembly: Scaffolding Protein Contributes to the Global Stability of Phage P22 Procapsids. J Mol Biol. 2006;359(4):1097–106. doi: 10.1016/j.jmb.2006.03.068. [DOI] [PubMed] [Google Scholar]
- Parker MH, Brouillette CG, Prevelige PE., Jr. Kinetic and calorimetric evidence for two distinct scaffolding protein binding populations within the bacteriophage P22 procapsid. Biochemistry. 2001;40(30):8962–70. doi: 10.1021/bi0026167. [DOI] [PubMed] [Google Scholar]
- Parker MH, Prevelige PE., Jr. Electrostatic interactions drive scaffolding/coat protein binding and procapsid maturation in bacteriophage P22. Virology. 1998;250(2):337–49. doi: 10.1006/viro.1998.9386. [DOI] [PubMed] [Google Scholar]
- Prevelige J, Thomas D, King J. Scaffolding protein regulates the polymerization of P22 coat subunits into icosahedral shells in vitro. J.Mol.Biol. 1988;202:743–757. doi: 10.1016/0022-2836(88)90555-4. [DOI] [PubMed] [Google Scholar]
- Prevelige PE, Fane BA. Building the machines: scaffolding protein functions during bacteriophage morphogenesis. Advances in experimental medicine and biology. 2012;726:325–50. doi: 10.1007/978-1-4614-0980-9_14. [DOI] [PubMed] [Google Scholar]
- Prevelige PE, Thomas D, King J. Nucleation and growth phases in the polymerization of coat and scaffolding subunits into icosahedral procapsid shells. Biophys.J. 1993;64:824–835. doi: 10.1016/S0006-3495(93)81443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevelige PEJ, Thomas D, King J, Towse SA, Thomas G. J. j. Conformational states of the bacteriophage P22 capsid subunit in relation to self-assembly. Biochemistry. 1990;29:5626–5633. doi: 10.1021/bi00475a030. [DOI] [PubMed] [Google Scholar]
- Rabilloud T, Carpentier G, Tarroux P. Improvement and simplification of low-background silver staining of proteins by using sodium dithionite. Electrophoresis. 1988;9(6):288–91. doi: 10.1002/elps.1150090608. [DOI] [PubMed] [Google Scholar]
- Rapaport DC. Studies of reversible capsid shell growth. Journal of physics. Condensed matter : an Institute of Physics journal. 2010;22(10):104115. doi: 10.1088/0953-8984/22/10/104115. [DOI] [PubMed] [Google Scholar]
- Singh S, Zlotnick A. Observed hysteresis of virus capsid disassembly is implicit in kinetic models of assembly. J Biol Chem. 2003;278(20):18249–55. doi: 10.1074/jbc.M211408200. [DOI] [PubMed] [Google Scholar]
- Suhanovsky MM, Parent KN, Dunn SE, Baker TS, Teschke CM. Determinants of bacteriophage P22 polyhead formation: the role of coat protein flexibility in conformational switching. Molecular microbiology. 2010;77(6):1568–82. doi: 10.1111/j.1365-2958.2010.07311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhanovsky MM, Teschke CM. Bacteriophage P22 capsid size determination: roles for the coat protein telokin-like domain and the scaffolding protein amino-terminus. Virology. 2011;417(2):418–29. doi: 10.1016/j.virol.2011.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanford C. The Hydrophobic Effect: Formation of Micelles and Biological Membranes. 2nd ed John Wiley and Sons, Inc.; New York: 1980. [Google Scholar]
- Teschke CM, Fong DG. Interactions between coat and scaffolding proteins of phage P22 are altered in vitro by amino acid substitutions in coat protein that cause a cold-sensitive phenotype. Biochemistry. 1996;35(47):14831–40. doi: 10.1021/bi960860l. [DOI] [PubMed] [Google Scholar]
- Teschke CM, Parent KN. ‘Let the phage do the work’: using the phage P22 coat protein structures as a framework to understand its folding and assembly mutants. Virology. 2010;401(2):119–30. doi: 10.1016/j.virol.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuman-Commike PA, Greene B, Jakana J, McGough A, Prevelige PE, Chiu W. Identification of additional coat-scaffolding interactions in a bacteriophage P22 mutant defective in maturation. J Virol. 2000;74(8):3871–3. doi: 10.1128/jvi.74.8.3871-3873.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman J, Gill SJ. Binding and Linkage: Functional Chemistry of Biological Macromolecules. University Science Books; Herndon: 1990. [Google Scholar]
- Zlotnick A. To build a virus capsid. An equilibrium model of the self assembly of polyhedral protein complexes. J Mol Biol. 1994;241(1):59–67. doi: 10.1006/jmbi.1994.1473. [DOI] [PubMed] [Google Scholar]
- Zlotnick A, Mukhopadhyay S. Virus assembly, allostery and antivirals. Trends Microbiol. 2011;19(1):14–23. doi: 10.1016/j.tim.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]