Abstract
Human-aided dispersal can result in phylogeographic patterns that do not reflect natural historical processes, particularly in species prone to intentional translocations by humans. Here, we use a multiple-gene sequencing approach to assess the effects of human-aided dispersal on phylogeography of the tench Tinca tinca, a widespread Eurasian freshwater fish with a long history in aquaculture. Spatial genetic analysis applied to sequence data from four unlinked loci and 67 geographic localities (38–382 gene copies per locus) defined two groups of populations that were little structured geographically but were significantly differentiated from each other, and it identified locations of major genetic breaks, which were concordant across genes and were driven by distributions of two phylogroups. This pattern most reasonably reflects isolation in two major glacial refugia and subsequent range expansions, with the Eastern and Western phylogroups remaining largely allopatric throughout the tench range. However, this phylogeographic variation was also present in all 17 cultured breeds studied, and some populations at the western edge of the native range contained the Eastern phylogroup. Thus, natural processes have played an important role in structuring tench populations, but human-aided dispersal has also contributed significantly, with the admixed genetic composition of cultured breeds most likely contributing to the introgression.
Keywords: intron, mtDNA, secondary contact, species range, stocking, Tinca tinca
Introduction
Determining the effects of human-aided dispersal and how it overlays with natural distributional changes is essential for the effective protection of species throughout their native ranges. Translocations that occur within the limits of the natural distribution of a species do not extend its range but instead superimpose new genetic signatures on the natural diversity patterns if they involve genetically divergent populations or domestic breeds (Taylor 2004; Ferguson et al. 2007; Stone et al. 2007; Mabuchi et al. 2008; Randi 2008; Muhlfeld et al. 2009). The impacts of such translocations are therefore more difficult to detect. Molecular phylogeography offers here a powerful tool, which can also be used to resolve the ‘cryptogenic’ nature of species whose status in a given area may be either native or introduced but where clear evidence for either origin is absent (Carlton 1996).
The international trade and human-aided transport provides an effective dispersal mechanism in many aquatic organisms and freshwater fishes in particular. Up until now, phylogeographic studies of European freshwater fishes were largely focused on species that were not targets of aquaculture (e.g. Durand et al. 1999; Kotlík and Berrebi 2001; Šlechtová et al. 2004; Bohlen et al. 2007; Šedivá et al. 2008). Few economically important species have been studied phylogeographically across their ranges, but even in those cases, the focus has been primarily on putative native populations, assuming (or hoping for) negligible phylogeographic contribution of human-aided dispersal (see Nesbø et al. 1999; Triantafyllidis et al. 2002; Van Houdt et al. 2005). As a result, phylogeographic information is still lacking for many common fishes, despite their role in freshwater communities and economic importance.
One such domesticated fish (Bilio 2007) with poorly known genetic structure (Lo Presti et al. 2010; Kohlmann et al. 2010) despite the ancient history in the European aquaculture and cuisine (Giovio 1524; Lebedev 1960; Steffens 1995; García-Berthou et al. 2007) is the tench Tinca tinca (Linnaeus, 1758). The tench is widely distributed between the British Isles and Iberian Peninsula in the west to central Siberia in the east (Fig. 1), but because it has been in cultivation in Europe for a long time (Šusta 1884; Steffens 1995), its exact native range is difficult to discern: in some areas (e.g. Spain: García-Berthou et al. 2007; Italy: Gherardi et al. 2008; Turchini and De Silva 2008), it may be either native or introduced but clear evidence for either origin is absent (i.e. it is cryptogenic there). There are records of tench introduction outside its native range from as early as the 18th century (e.g. to Ireland: Kennedy and Fitzmaurice 1970), and since then, introduced populations have been established on all continents except Antarctica (Welcomme 1988; Brylińska et al. 1999). In some countries, it is even considered as an invasive, potentially harmful species due to concerns over competition with native fish (e.g. Rowe 2004; Stokes et al. 2004; Hesthagen and Sandlund 2007; Rowe et al. 2008; DeVaney et al. 2009).
Figure 1.
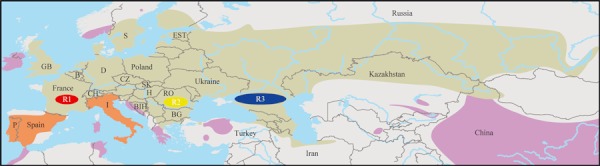
Putative native (olive) and part of non-native (violet) distribution range of the tench. Large areas where the origin is considered ambiguous are highlighted by orange. Locations of major freshwater glacial refugia in Europe, Western/Atlantic (R1), Danubian (R2), and Ponto-Caspian (R3) are indicated. Sampling countries are labeled (codes: B, Belgium; BG, Bulgaria; BIH, Bosnia and Herzegovina; CH, Switzerland; CZ, Czech Republic; D, Germany; EST, Estonia; GB, Great Britain; H, Hungary; I, Italy; P, Portugal; RO, Romania; S, Sweden; SK, Slovakia). References to the map: Urchinov 1995; Brylińska et al. 1999; Mitrofanov and Petr 1999; Savvaitova and Petr 1999, Economidis et al. 2000; Wang et al. 2004; Innal and Erk'akan 2006; Hesthagen and Sandlund 2007; Popov 2009; Mamilov et al. 2010.
Distribution of genetic diversity of freshwater fishes is largely controlled by the island-like nature of their habitats (Bernatchez and Wilson 1998), and the present-day phylogeographic patterns of temperate species have been shaped primarily by isolation in multiple glacial refugia during the last glacial maximum (18 000–23 000 years ago), followed by range expansion and drainage isolation. Many widely distributed temperate freshwater fish species therefore show deep phylogeographic subdivisions (e.g. Durand et al. 1999; Bernatchez 2001; Kotlík and Berrebi 2001; Van Houdt et al. 2005; Kotlík et al. 2008; Hänfling et al. 2009). However, some species display only a limited or shallow phylogeographic structure, which is usually interpreted as the result of a recent dispersion from only one glacial refugium (Triantafyllidis et al. 2002; Bohlen et al. 2007). Alternatively, it can point to strong effects of human-aided translocations (Hänfling et al. 2009).
The present study uses a multiple-gene sequencing approach (Brito and Edwards 2008) and barrier-detection statistics to test whether the range-wide genetic variation of the tench shows a significant phylogeographic structure that can be explained by natural processes during the last glacial–interglacial cycle. Tench occupy all major freshwater regions in Europe, so that it should be possible to identify the contribution of different refugia (Fig. 1) to its present-day distribution. However, if human-aided dispersal significantly altered recent evolutionary history of the tench, the haplotypes could have been redistributed among populations, wiping out any natural phylogeographic structure (Sanz et al. 2006). Captive breeding can produce admixed gene pools, increasing the homogenizing effect of human-aided dispersal. To assess this effect of hatchery practices, in addition to putative native populations, we also sampled various cultured strains and known introduced populations outside the native range.
Materials and methods
Sampling
Sampled populations were chosen to cover the majority of the natural range of the tench in Europe and Asia. Fin tissue samples were stored in 95% ethanol. A total of 225 individuals were collected from 76 populations and included 25 hatchery stocks and several known introductions (Fig. 2; Appendix A). A single specimen (MNHN 0000–1357) from the collection of the Museum National d'Histoire Naturelle in Paris, France, was sampled. We also analyzed 16 Czech- and foreign-cultured tench breeds maintained in the live gene bank of the Research Institute of Fish Culture and Hydrobiology in Vodňany, Czech Republic (Gela et al. 1998, 2006; Flajšhans et al. 1999), and an Italian regional breed, the Golden hump tench of Poirino highland (Gasco et al. 2010).
Figure 2.
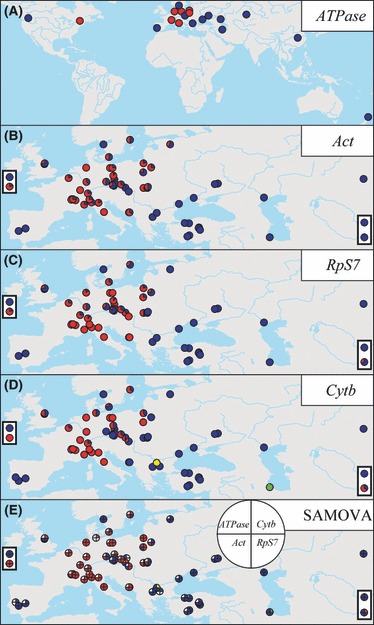
Geographic distribution of major clades and SAMOVA groups. Clade W is shown in red and clade E in blue for ATPase (A), Act (B), and RpS7 (C). For Cytb (D), clade W is in red, clade EA in blue, clade EC in green, and clade EI in yellow. The same colors are used for the SAMOVA groups (E). Boxed data points to the right and left of the maps in (B) through (E) represent identities for two sites in North America and in China and New Zealand, respectively [see (A)]. For exact haplotype distribution and frequencies, see Appendix A.
Data collection
Introns of three nuclear genes and a complete sequence of one mitochondrial gene (Table 1) were analyzed by polymerase chain reaction (PCR) amplification from genomic DNA and direct sequencing. Total genomic DNA was extracted with QIAGEN (Valencia, CA, USA) DNeasy® Tissue kit. The PCR conditions followed standard methods (Tsigenopoulos and Berrebi 2000; Machordom and Doadrio 2001). The resulting PCR products were purified using the Millipore (Bedford, MA, USA) Montage PCR centrifugal filter devices and were directly sequenced with the ABI PRISM BigDye Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, MA, USA) and purified using DyeEx Spin kit (Qiagen). The extension products were run on ABI 3730 or 3730×l automated sequencers. Sequences were assembled using SEQMAN II (DnaStar Inc., Madison, WI) with the default options. All sequence traces were inspected visually to check the accuracy of the heterozygous base calls (Hare and Palumbi 1999). Nucleotide sequences of each unique haplotype were deposited in the GenBank database under the accession numbers HM167935–HM167965.
Table 1.
Summary of polymorphism for each gene and the results of demographic analyses
| Gene | Phylogeo-graphical unit | N | Number of haplotypes | Polymorphic sites | Indels | Haplotype diversity ± SD | Nucleotide diversity ± SD (x 100) | Tajima's D | Fu's Fs | P(SSDD/R) |
|---|---|---|---|---|---|---|---|---|---|---|
| Cyt b | Clade E | 140 | 12 | 33 | 0 | 0.228 ± 0.048 | 0.181 ± 0.058 | −1.940**/**/** | −1.455 | 0.217/0.383 |
| (1141bp) | Clade EA | 130 | 8 | 7 | 0 | 0.105 ± 0.037 | 0.009 ± 0.003 | −2.065***/***/*** | −13.791***/***/*** | 0.286/0.312 |
| Clade EI | 5 | 1 | 0 | 0 | 0 | 0 | – | – | – | |
| Clade EC | 5 | 3 | 3 | 0 | 0.700 ± 0.218 | 0.105 ± 0.043 | −1.048 | −0.186 | 0.882/0.896 | |
| Clade W | 70 | 5 | 4 | 0 | 0.308 ± 0.070 | 0.029 ± 0.007 | −1.278*/−/− | −2.988*/−/* | 0.366/0.092 | |
| Total | 210 | 17 | 44 | 0 | 0.581 ± 0.029 | 0.687 ± 0.038 | 0.092 | 4.994 | 0.000/0.230 | |
| RpS7 | Clade E | 210 | 3 | 0 | 2 | 0.019 ± 0.013 | 0.002 ± 0.002 | −1.279−/*/* | −5.178−/*/*** | 0.109/0.082 |
| (868bp) | Clade W | 172 | 5 | 4 | 1 | 0.666 ± 0.018 | 0.116 ± 0.007 | 0.266 | 0.891 | 0.053/0.005 |
| Total | 382 | 8 | 15 | 5 | 0.637 ± 0.020 | 0.883 ± 0.013 | 3.669+++/+/+++ | 18.222 ++/+++/+ | 0.113/0.274 | |
| Act | Clade E | 237 | 2 | 1 | 0 | 0.008 ± 0.008 | 0.003 ± 0.003 | −0.934 | −2.952−/−/** | 0.033/0.996 |
| (289bp) | Clade W | 193 | 2 | 1 | 0 | 0.010 ± 0.010 | 0.004 ± 0.004 | −0.956 | −2.776−/−/* | 0.050/0.991 |
| Total | 430 | 4 | 6 | 0 | 0.501 ± 0.006 | 0.860 ± 0.009 | 3.240++/++/++ | 8.886 +/++/+ | 0.000/0.008 | |
| ATPase | Clade E | 26 | 1 | 0 | 0 | 0 | 0 | – | – | – |
| (100bp) | Clade W | 12 | 1 | 0 | 0 | 0 | 0 | – | – | – |
| Total | 38 | 2 | 1 | 0 | 0.444 ± 0.058 | 0.444 ± 0.058 | 1.253 | 1.538 | 0.095/0.015 |
The size of DNA fragments is given below the gene names in base pairs. The superscripts indicate probability levels that values in the neutral population can be equal or lower than observed: *P < 0.05; **P < 0.01; ***P < 0.001; equal or higher than observed: +P < 0.05; ++P < 0.01 and ‘−’ means nonsignificant result given by coalescent simulations based on number of segregating sites/the average number of nucleotide differences estimated by DNASP, version 4.50.3 (Rozas et al. 2003)/result given by ARLEQUIN version 3.11 (Excoffier et al. 2005), respectively. The value P(SSD) shows the probability of observing a less good fit between the model and observed distribution by chance under the demographic/spatial expansion scenario.
A part of nuclear DNA containing the second intron of the actin gene (Act) was amplified and sequenced using primers Act-2-R and Act-2-F described by Atarhouch et al. (2003). The intron of the gene coding for the ATP synthase beta subunit (ATPase) was amplified and sequenced using the primers described by Jarman et al. (2002). The first intron of the gene coding for the S7 ribosomal protein (RpS7) was amplified and sequenced using the primers S7RPEX1F and S7RPEX2R (Chow and Hazama 1998). Haplotypes were inferred from diploid sequence traces (Clark 1990; Won and Hey 2005) and verified by the use of fastPHASE (Scheet and Stephens 2006). The entire mitochondrial cytochrome b gene (Cytb) was amplified with the primers GluF and ThrR described by Machordom and Doadrio (2001) and sequenced with newly designed forward (5′-AAACAACCCAACAGGACT-3′) and reverse sequencing primers (5′-CAAATAGGAAATATCATTCTG-3′).
Data analyses
Sequence analysis
For each locus, we estimated the haplotype and nucleotide diversities and their variances (Nei 1987). To explore whether intragenic recombination may have affected the patterns of variation at Act, ATPase, and RpS7, we used the four-gamete test (Hudson and Kaplan 1985). McDonald and Kreitman (1991) test was performed for Cytb to test for deviation from neutrality using an outgroup species and comparing different tench clades with each other. Tinca tinca is the only species in the family Tincidae, so that a sharpbelly species, Hemiculter leucisculus, from a related family Cultridae (Chen and Mayden 2009) has been used as the outgroup (GenBank Accession no. AF095608). All the calculations were performed using DNASP, version 4.50.3 (Rozas et al. 2003).
Phylogenetic and network analyses
Rooted phylogenies were reconstructed by the maximum-likelihood criterion (ML) using PhyML version 3.0.1 (Guindon and Gascuel 2003). We used Akaike information criterion and jModelTest version 0.1 (Posada 2008) to identify the HKY+G model as the most suitable model of DNA substitution for the Cytb data and the TrN model for the RpS7 data. Sharpbelly RpS7 sequence was not available, so that a sequence (AY325789) of the rosy bitterling, Rhodeus ocellatus, from another related family Acheilognathidae was used to root the RpS7 tree. The robustness of the trees was assessed by the approximate likelihood ratio test (Anisimova and Gascuel 2006) and by bootstrap resampling (1000 replicates; Felsenstein 1985) using PhyML. A haplotype network was constructed for each gene by the statistical parsimony (Templeton et al. 1992) as implemented in TCS version 1.21 (Clement et al. 2000).
Inference of demographic history
To examine past population dynamics, we calculated two commonly used summary statistics D (Tajima 1989) and Fs (Fu 1997) with DnaSP and ARLEQUIN version 3.11 (Excoffier et al. 2005). Their significance was tested by generating random samples under constant population size using a coalescent simulation conditioned on the number of polymorphic sites (Ramírez-Soriano et al. 2008). For neutral markers, significant negative values can be expected in cases of population expansion (Tajima 1989; Fu 1997).
As another way of assessing signatures of refugial expansion, we considered the distribution of the number of pairwise nucleotide differences (mismatch distribution) by contrasting observed distributions with those expected from models of population size change. We tested whether the data fitted the sudden demographic expansion model (Rogers and Harpending 1992) or the instantaneous range expansion model (Excoffier 2004), using ARLEQUIN. The models were fitted to the data by a generalized nonlinear least-square approach, which allowed the estimation of the parameter τ = t/2 μ, the expansion time scaled by the mutation rate (Schneider and Excoffier 1999). A parametric bootstrapping approach (Schneider and Excoffier 1999) was used to obtain the probability that the observed data conform to the model using the sum of square deviations (SSD) between the observed and expected mismatch distribution as a test statistic. We considered a wide range of estimated Cytb mutation rates for fishes of about 0.005–0.125 substitutions per site per Myr, published by Dowling et al. (2002) and Burridge et al. (2008), respectively.
Spatial genetic analysis
Two complementary barrier-detection methods were applied to identify any discontinuities in the geographic distribution of genetic variation (Guillot et al. 2009). The geographic component of the phylogeographic pattern was first assessed by the spatial analysis of molecular variance using SAMOVA version 1.0 (Dupanloup et al. 2002). The advantage of SAMOVA is that it removes bias in population designation because it does not make a priori group distinction for genetic analyses. It employs a simulated annealing procedure using geographic locations of the sampling sites to cluster the sites into a user-defined number of groups (K), so that the proportion of total genetic variance between groups (FCT) is maximized and the proportion of variation among sites within groups (FSC) is minimized.
Major barriers to the distribution of genetic variation were then estimated by the Monmonier's (1973) maximum difference algorithm implemented in BARRIER version 2.2 (Manni et al. 2004), based on a matrix of the pairwise net genetic distances among sampling sites generated from DNA sequences using ARLEQUIN. The algorithm was applied to a network connecting the geographic coordinates of the sampling locations computed using Delaunay triangulation (Manni et al. 2004). Analyses were performed separately for each locus but on the same geographic network, and the results were then combined to identify barriers supported by multiple loci; the locus ATPase was excluded because of its limited geographic coverage.
Coalescent simulation
We conducted a series of simulation experiments to evaluate whether a natural population that was founded by unrelated clades at the end of the Younger Dryas, and has been isolated from other populations since then, may still carry haplotypes from different clades. This situation would correspond, for example, to tench populations inhabiting lakes in deglaciated areas of northern Europe (see Lajbner et al. 2010). In each experiment, we simulated 10 000 coalescent trees using Mesquite version 2.5 (Maddison 2008; Maddison and Maddison 2008) to estimate the distribution of the time to the most recent common ancestor (TMRCA) in such a population, and we counted the trees deeper than 3000 generations, approximately corresponding to the end of the Younger Dryas c. 11 500 years ago (Muscheler et al. 2008) and the generation time of 4 years (Monich 1953; Pekař 1965). We parameterized the simulations by female effective population size (Nef) values corresponding to known population densities of tench (c. 100–500 individuals per hectare; Lusk et al. 1998) and a lake area between 10 and 400 hectares, and assuming an equal sex ratio (Monich 1953) and the ratio of the effective population size to the adult census size, Ne/N, of 0.3 (Turner et al. 2006). We focused on the female component of population, which is represented in our data by mtDNA variation, because of its relatively shallower coalescence time depth and therefore shorter expected TMRCA compared with autosomal loci. For values of Nef yielding the number of deep trees that was <5% of all the trees simulated assuming that Nef, we considered it unlikely that a population with that effective number of females would still contain haplotypes from different clades unless the haplotypes were recently redistributed among populations through human-mediated movement. On the other hand, a high number of deep trees (i.e. more than 95%) would indicate that there is no need to invoke recent gene flow as the likely explanation for the coexistence of divergent clades in such population, which could be the result of natural postglacial contact. Although these simulation experiments make simplifying assumptions that may not be realistic, they generate ideal benchmarks for interpreting the observed data.
Results
Sequence variation
The levels of polymorphism among sequences obtained for each of the four genes (38–430 gene copies per gene) are summarized in Table 1. There were five short (<5 bp) insertion/deletion (indel) polymorphisms segregating at the RpS7 locus (Table 1) that were not associated with simple sequence repeats and could be unambiguously aligned. Of these, a two-base deletion was inferred to have occurred along the branch leading to clade W and a single-base deletion along the branch leading to clade E. Data sets from neither Act, ATPase, nor RpS7 showed evidence of homoplasy and they all passed the four-gamete test, indicating that recombination has not affected the patterns of variation at the nuclear genes in our study. The McDonald–Kreitman test provided no evidence of selection on the coding sequence of the Cytb gene (P > 0.05).
Genealogical and geographic relationships
The phylogenetic and network analyses split the range-wide data set for the mitochondrial Cytb into two distinct phylogroups (clades W and E) separated with 1.6% of genetic distance (Fig. 3E,F), translating to a divergence time of about 64 × 103 to 1600 × 103 years ago. The Western phylogroup was found in Europe between the British Isles and Poland, whereas the Eastern phylogroup was present from Europe throughout Asia to China, with a broad zone of overlap with the Western phylogroup in Europe (Fig. 2D). While clade W showed very little internal structure, clade E was partitioned into three subclades (Fig. 3E,F). The majority of haplotypes were in the clade EA, while the other two clades had very restricted distributions: the EC haplotypes in the Anzalee lagoon of the Caspian Sea in Iran and the EI haplotype in the Iskar River of the Danube River drainage in Bulgaria (Fig. 2D).
Figure 3.
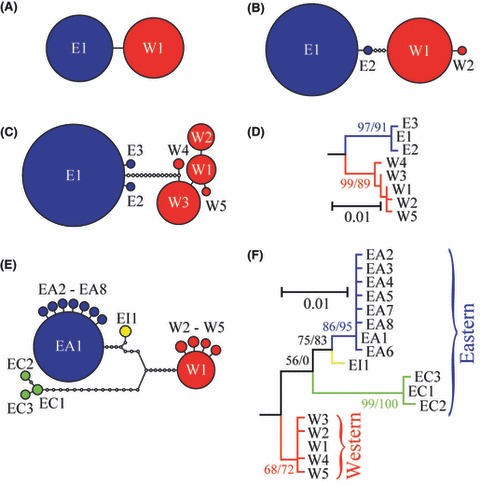
Haplotype relationships. Clade E is shown in blue and clade W in red for ATPase (A), Act (B), and RpS7 (C, D). For Cytb (E, F), clade W is in red, clade EA in blue, clade EC in green, and clade EI in yellow. The networks were constructed under the 95% maximum parsimony criterion, and the size of the circles is proportional to the haplotype frequency; small empty circles represent unobserved haplotypes. The maximum-likelihood phylograms are shown with bootstrap (from 1000 replicates)/aLRT support for major partitions in the RpS7 (D) and Cytb (F) phylogenies, with branch lengths proportional to the scale bar with the unit being a mean number of nucleotide changes per site.
We constructed a phylogenetic network for each nuclear DNA locus and a phylogenetic tree of the RpS7 haplotypes (Fig. 3A–D). The most salient feature of the inferred genealogies is the complete lineage sorting of nuclear genes between the two phylogroups in that all genes are distinguished into two clades W and E, and the divergence between the phylogroups based on sequences of the nuclear Act, ATPase, and RpS7 genes is geographically concordant with mitochondrial Cytb sequences (Fig. 2A–D). Nuclear DNA loci and mtDNA thus display striking similarities, showing a strong genealogical concordance across the distribution range of the tench. Changes in mtDNA and the three nuclear loci are concordant also across the contact zone between the two phylogroups, with only finer-scale differences being evident in phylogroup frequencies among sites (Fig. 2A–D).
The introduced populations in Turkey and China carried at all loci only clade E haplotypes, as did the overseas introduction to the state of Washington. However, the non-native populations in Bosnia and Herzegovina, in New Zealand, and in Quebec carried at one or more loci haplotypes from both clade W and clade E (Fig. 2A–D).
The phylogeographic variation observed among the tench populations was present also in the cultured breeds, with the exception of Cytb clades EC and EI that had very restricted geographic distributions. Each one of the 16 cultured breeds in the Vodňany live gene bank as well as the Italian regional breed carried haplotypes from both clades W and E at one or more loci, including the seven regional Czech breeds, three European breeds (German, Romanian, and Hungarian), three experimental breeds, and three ornamental breeds (Appendix A).
Population demographic history
The D and Fs statistics were negative for the major Cytb clades W and E as well as for clades EA and EC, reflecting the excess of rare mutations compared to the expectation under constant population size, and for clades W, E, and EA, this difference was significant (Table 1). A similar pattern was observed at the Act and RpS7 genes, with a number of D and Fs values being large and negative, and with significant results for both Act clades and the RpS7 clade E (Table 1).
For all four genes and clades W and E as well as for Cytb clades EA and EC, there was also a good fit [P (simulated SSD ≥ observed SSD) > 0.1] between the observed and the expected mismatch distribution from at least one expansion model (Table 1). The τ values obtained for Cytb clades W (0.373) and EA (3.000) translate into an expansion time of about 1308–31 134 years ago and 10 517–262 927 years ago, respectively.
Spatial genetic structure
The SAMOVA analyses identified a significant two-group spatial structure for each locus (Fig. 2E), with approximately 65% to 100% of the genetic variation proportioned between the two groups (Cytb: FCT, 0.687, P < 0.05; FSC, 0.606, P < 0.001; nuclear DNA loci: FCT, 0.667–1.000, P < 0.001; FSC, 0.000–0.080, P < 0.001). Assuming a four-group scenario for Cytb placed the Anzalee population (clade EC) and the Iskar population (clade EI) in their own separate groups (Fig. 2E), yielding higher FCT (0.791, P < 0.001) and lower FSC values (−0.095, P < 0.001) than those observed for this gene in the two-group scenario. Interestingly, one SAMOVA group was defined in the way that its distribution was clearly partitioned into distinct sets of sites, which belonged to that same group but which were not geographically adjacent (i.e. the British, one Swedish, and the Spanish and Portuguese sites were placed in the same group with sites from eastern Europe and Asia; Fig. 2E).
The BARRIER analysis overlaying five major barriers for each locus identified several discontinuities with a support from multiple loci (Fig. 4). The longest break divided the tench distribution into a western part and an eastern part and was fully supported by two loci and partially by all three loci (Fig. 4), depending on the local patterning of clades in the contact zone between the Western and Eastern phylogroups (Fig. 2B–D). Another barrier separated the Spanish and Portuguese sites from the rest of the sites with a complete support of all loci. The third barrier separated the British sites from the other sites with a support of two loci, and the fourth barrier separated the Swedish site Lake Öre sjö from the other sites in Sweden and around the Baltic Sea, with a complete support from two loci and a partial support of all loci (Fig. 4). Additional three short breaks supported by two loci were identified in central Europe (Fig. 4), following the transitions between phylogroups in that region (see Fig. 2E).
Figure 4.
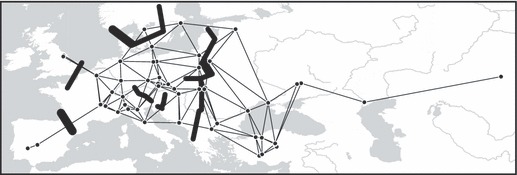
European phylogeographic breaks identified in tench data by BARRIER using the Monmonier's algorithm. Thin lines, Delaunay triangulations; thick lines, barriers supported by at least two loci. The thickness of the different barriers and their segments is proportional to the number of loci that supported them (two or three).
TMRCA distribution
The simulations of the TMRCA assuming Nef of 730 produced fewer than 5% of coalescent trees that were deeper than 3000 generations. We therefore consider it unlikely that an isolated population with this effective number of females or smaller that was founded by unrelated mtDNA clades at the end of the Younger Dryas (assuming the generation time of 4 years) would still contain haplotypes from different clades, unless the haplotypes were recently redistributed among populations by human-mediated movement. However, for any Nef larger than that, there was >5% chance that the TMRCA predated the origin of the population, and for Nef larger than 4000, more than 95% of all coalescent trees were deeper than 3000 generations. The effective number of females of 4000 would translate to an adult census size of c. 25 000 individuals assuming an equal sex ratio and the ratio Ne/N of 0.3, which would correspond to a lake area of c. 250 hectares, assuming the population density of 100 individuals per hectare.
Discussion
Pleistocene phylogeographic subdivision
The statistical method in SAMOVA detected a significant phylogeographic pattern driven by the spatial orientation of the Western and Eastern phylogroups, with high congruence between mtDNA and nuclear DNA loci (Fig. 2E). The barrier-detection method in BARRIER revealed a well-supported genetic break crossing central Europe in a north–south direction (Fig. 4), paralleling the transition between the phylogroups (Fig. 2A–D). These results together provide evidence of a strong geographic component to the present phylogeographic pattern in the tench that is highly concordant among unlinked loci.
The distribution of highly divergent, reciprocally monophyletic phylogroups is strongly reminiscent of phylogeographic discontinuities modulated by refugial isolation (Taberlet et al. 1998; Hewitt 2000). It seems thus likely that, after the last glacial maximum, the Western phylogroup originated from the western European refugium, whereas the Eastern phylogroup originated from an eastern European or western Asian refugium. This conclusion is in accordance with previous phylogeographic studies indicating putative freshwater refugia in drainages of the Atlantic tributaries and of Rhone River (Durand et al. 1999; Nesbø et al. 1999; Kotlík and Berrebi 2001) and in the Black and Caspian Sea basins (Bănărescu 1991; Kotlík and Berrebi 2001; Kotlík et al. 2004, Kotlík et al. 2008). The importance of the Ponto-Caspian refugium is supported by the findings of tench fossils from glacial deposits in the Black Sea basin (Lebedev 1960). It is interesting that a distinct Cytb clade EC occurred in the southern Caspian Sea and only there, although the widespread clade EA occurred in the northern Caspian Sea, and all tench from both sites carried the same nuclear DNA haplotypes (Fig. 2A–D). Furthermore, clade EI occurred only at one site in the Iskar River basin in the lower Danube River drainage, where again only widespread nuclear DNA haplotypes were present (Fig. 2A–D). This shows hitherto undescribed complexities in the distribution of refugia within the Ponto-Caspian region and the Danube River, and lineage sorting and/or gene flow between them.
The signatures of population expansion in both phylogroups are consistent with a history of postglacial dispersion from formerly isolated refugia. The estimates of time from population expansion are approximately consistent with an expansion following the last glacial maximum. If, on the other hand, the significant tests reflected recent introductions, the time estimates should indicate much more recent expansion. The higher age of the expansion of the Eastern phylogroup than of the Western phylogroup is congruent with phylogeographic evidence from other fishes that the geographic range occupied by the Eastern phylogroup was much less directly affected by recent glacial advances than the western European drainages (Bernatchez 2001).
Fourteen sites in central and northern Europe were assigned to one SAMOVA group by some loci and to the other SAMOVA group by the other loci (Fig. 2E), and admixed sites carrying haplotypes of both phylogroups were observed over a large area between, roughly, Belgium and Estonia (Fig. 2A–D). Changes in mtDNA and the three nuclear loci are concordant across the contact zone, supporting that this is not a matter of primary contact and selection on some of the markers but rather of a secondary contact of populations from different refugia. But can this introgression be caused entirely by human-aided dispersal? Our TMRCA simulations indicated that there is no need to invoke recent gene flow as a likely explanation for the presence of both phylogroups even in relatively small populations. Furthermore, the location of the tench contact zone matches phylogeographic subdivisions in other species where expanding populations from different refugia meet in the same area (e.g. Taberlet et al. 1998; Hewitt 2000). We therefore consider it unlikely that the overlap between the phylogroups at the sites in central Europe has been entirely caused by human transport and release. Rather, it most likely represents a region of natural postglacial contact between lineages from the eastern and western refugia.
Evidence for human-aided dispersal
On the other hand, the contact zone is very broad and spans across several watershed divides, and there is fairly high amount of introgression in western Europe (Fig. 2B–D). The SAMOVA analysis even placed sites from three western European regions that contained particularly high proportions of the Eastern phylogroup into the same group with the sites from eastern Europe and Asia (Fig. 2E). These sites were located in Iberian Peninsula, in Britain and in Sweden, and they were separated from the other western sites with a BARRIER support of several loci (Fig. 4). All tench from the three sites in Spain and Portugal contained exclusively the Eastern phylogroup, which strongly speaks in favor of the hypothesis that tench are not a native species on the Iberian Peninsula (García-Berthou et al. 2007; Ribeiro et al. 2009), and points to the eastern Europe or Asia as their likely source. This demonstrates the ability of detailed phylogeographic studies such as ours to resolve the status of cryptogenic species where other evidence for either native or introduced origin is absent (Carlton 1996). The lack of phylogeographic resolution means, however, that we cannot confirm or reject the native status of the populations in Italy (Gherardi et al. 2008; Turchini and De Silva 2008). The absence of strong genetic separation from more northern sites (Figs 2E and 4) suggests that tench colonization of Italy is most likely of postglacial origin.
Another site in western Europe that only contained Eastern alleles is Lake Öre sjö in southern Sweden. It may suggest that this population escaped admixture, but it may also be that the sample of only one fish (four loci) was not enough to detect the Western phylogroup if it was present in low frequency.
The British sites were separated from the other western sites by BARRIER, but they carried a mixture of the Eastern and Western phylogroups, which was reflected by their SAMOVA assignment to both groups, depending on the locus (Fig. 2E). This is probably a result of human introduction of the Eastern phylogroup to the British Isles as this phylogroup occurs in much lower frequency in western Europe. It could also be a natural colonization by both phylogroups but it would require almost complete replacement of the Eastern phylogroup in western Europe (see Searle et al. 2009).
Cultured breeds and introgression
The above evidence strongly suggests that human-aided dispersal has altered the phylogeographic structure of the tench. This implies either that tench from geographically remote populations were used for stocking, or that local source breeds carried the opposite phylogroup. Interestingly, we found that although the cultured breeds originating from different parts of Europe differed in the frequencies of the Western and Eastern phylogroups (Appendix A), all of them carried haplotypes of both phylogroups. Therefore, supplemental stocking with these or genetically related breeds would increase the probability of introgression between the phylogroups. Our recent study looked for evidence of a reproductive isolation in a postglacial lake inhabited by both phylogroups but we found no results that would point toward barriers to their interbreeding (Lajbner et al. 2010). Furthermore, at many sites within the contact zone, we observed individuals of apparently hybrid ancestry (see Fig. 2B–D). The putative hybrids were heterozygous for alternate phylogroups or were homozygous but for different phylogroups at different loci and/or carried mtDNA of the opposite phylogroup (data not shown). Finally, that both phylogroups characterized all of the examined breeds support that populations of mixed origin can persist without strong negative fitness consequences at least under cultured conditions. Therefore, the admixed genetic composition of the cultured breeds most likely contributed to the introgression between the phylogroups in natural habitats.
Phylogeography of known introductions
There is no record as to the geographic origin of tench in the Neretva River in Bosnia and Herzegovina, which is in the eastern Adriatic Sea basin where tench do not naturally occur (Glamuzina 2006). The presence of both phylogroups in the Neretva population shows that it is may have descended either from introductions from the adjacent Danube River drainage where both phylogroups occur (Fig. 2), or from genetically admixed hatchery stocks.
In Turkey, tench are probably native to some river drainages within the Black Sea basin (Brylińska et al. 1999) but it have been introduced to water systems of central and western Turkey (Korkmaz and Zencir 2005; Innal and Erk'akan 2006). The six putative non-native populations in Turkey (Appendix A) contained exclusively haplotypes of the Eastern phylogroup (Fig. 2B–D), which made them indistinguishable from the other sites in the eastern part of the range (Figs 2E and 4). This points to a local source of this introduction or to a distant source but within the range of the Eastern phylogroup.
The introduced population in China also carried only the Eastern phylogroup (Fig. 2A–D). Tench were introduced in large parts of China during the 20th century (Walker and Yang 1999; Huang et al. 2001), most probably from the Itrysh River drainage in northern China where tench naturally occur (Fig. 1). Interestingly, European cultured breeds originating from the live gene bank in Vodňany were recently imported to China to serve as a source for stocking into open waters throughout China (Wang et al. 2004). If those breeds carry both phylogroups, as did all breeds in that gene bank that we examined, this practice is likely to induce introgression of the European genes into the native populations of the tench in Asia.
The first introduction of tench from Europe to the United States occurred in 1877 (Baird 1879). By 1896, their descendants had been distributed to at least 36 states, and subsequent introductions to North America followed, including to Canada in 1986 (Quebec: Dumont et al. 2002). Both these introductions used tench from Germany (Baughman 1947; Fuller et al. 1999; Nico and Fuller 2010). Consistent with this, the population from Quebec contained both phylogroups and was placed in the same SAMOVA group with German and other western European sites (Fig. 2E). However, the Silver Lake population in the state of Washington contained only the Eastern phylogroup and it was grouped with the eastern sites by SAMOVA (Fig. 2E). This suggests that this population originated from yet another introduction to the United States that occurred in the state of Washington in 1909 (Wydoski and Whitney 2003) and which would have involved an unknown but most likely an eastern European or Asian source.
New Zealand tench were introduced several times in 19th century from Tasmania (Allport 1866; Abbott 1868; Arthur 1881; Thomson 1922; Hicks 2003), to where they had been successfully introduced from England in 1858 (Allport 1866, 1868). The North Island population contained both phylogroups (Fig. 2A–D) and it was placed in one SAMOVA group by one locus and to the other SAMOVA group by other loci (Fig. 2E). We were unable to acquire samples from Tasmania but these results suggest that England already had the Eastern phylogroup in 19th century, placing an upper limit on the time of its introduction to the British Isles.
Conclusions
The difficulty of disentangling the confounding effects of secondary dispersal from the impact of natural historical processes presents a persistent challenge for studies on the historical biogeography, particularly of species prone to intentional translocation by humans. Our study highlights that for such species, it may be useful to consider the effects of anthropogenic factors as juxtaposed with the natural phylogeographic structure rather than viewing these as mutually exclusive causes of the observed genetic and distribution patterns. We showed that natural historical processes have played an important role in genetically structuring the tench populations and that their signatures can still be detected across multiple genes. On the other hand, we demonstrated that human-aided dispersal significantly contributed to the recent evolutionary history of the tench and that the admixed genetic composition of cultured breeds most likely enhances introgression between genetically differentiated populations. It appears likely that if the current practices in open-water fisheries management continue, the human-aided migration will eventually erase the natural phylogeographic pattern for large parts of the tench range. It is also possible that, by increasing their adaptive variation, the hybridization would enhance the invasive potential of the admixed populations outside the native range, including into novel niches not occupied in the native range (Lucek et al. 2010). Within the native range, phylogroups descended from different refugia would likely show physiological adaptations to different selective environments. Stocking with individuals of the opposite phylogroup or the mixed ancestry may disrupt such adaptations, which can lead to reduction in fitness of wild populations (see Araki et al. 2008; Hutchings and Fraser 2008; Fraser et al. 2010; Marie et al. 2010; for numerous examples from salmonids). Such impacts might substantially reduce the evolutionary potential of wild populations and affect their chance of persistence (Stockwell et al. 2003; Frankham 2005).
Acknowledgments
We thank all the colleagues who assisted with sample collections, especially Adámek Z., Akbarzadeh A., Alavi M.S.H., Apostolou A., Bohlen J., Bolding B., Buras P., Cook I., Černý J., Dahlberg M., De Gelas K., Desloges C., Desloges S., Doadrio I., Dumont P., Dzuba B., Ekmekçi F.G., Flajšhans M., Gaffaroğlu M., Gallardo J.M., Gante H.F., Gasco L., Gomulka P., Gualtieri M., Henshaw A., Hertig A., Hicks B.J., Hubenova T., Kamler E., Kohlmann K., Korte E., Korwin-Kossakowski M., Koščo J., Lopatin O., Mamilov N., Memiş D., Navodaru I., Nico L.G., Paaver T., Pekárik L., Persat H., Petr T., Piačková V., Polli B., Rossi S., Sudakova N., Sychrová O., Švátora M., Vandeputte M., Vassilev M., and Wang J. We thank Choleva L. for his assistance with nuclear markers selection and Marková S., PelikánováŠ., Šedivá A., Bohlen-Šlechtová V., Janko K., Ráb P., and many others for their advice. The work was supported by the Ministry of Education, Youth and Sports of the Czech Republic (LC06073, MSM6007665809), by the Academy of Sciences of the Czech Republic (IRP IAPG AV0Z50450515 and IGA UZFG/05/22), and by the Czech Science Foundation (206/09/1154).
Appendix A
Origin of tench specimens with haplotype codes and frequencies.
| Coordinates | Haplotype codes (counts) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Locality* | Basin | Water body | Country† | Latitude | Longitude | CytB | Act | RpS7 | ATPase | N | Year |
| Open waters (nonfarm sites) | |||||||||||
| Linkebeek | Scheldt/North Sea | Artificial pond | B | 50.77 | 4.33 | EA1(1), W1(3) | E1(2), W1(8) | E1(2), W2(1), W3(5) | – | 5 | 2005 |
| Osikovica | Danube/Black Sea | Iskar tributary | BG | 42.94 | 24.00 | EI1(5) | E1(10) | E1(10) | E1(2) | 5 | 2005 |
| Blagoevgrad | Struma/Aegean Sea | Struma | BG | 42.02 | 23.09 | EA1(1) | E1(2) | E1(1), E2(1) | – | 1 | 2005 |
| Karaotok Park Prirode | Neretva/Adriatic Sea | Canal Sunca‡ | BIH | 43.05 | 17.80 | W1(1) | E1(1), W1(1) | W3(1), W4(1) | – | 1 | 2005 |
| Stolac | Neretva/Adriatic Sea | Bregava‡ | BIH | 43.08 | 17.96 | EA1(3) | E1(2), W1(4) | W2(3), W3(1), W4(2) | – | 3 | 2005 |
| Noyan | Saint Lawrence River/Atlantic Ocean | Richelieu River‡ | CDN | 45.12 | −73.26 | W1(3) | E1(1), W1(5) | E1(1), W2(4), W3(1) | – | 3 | 2005 |
| Zurich | Rhine/North Sea | Zurich | CH | 47.30 | 8.62 | EA1(1), W1(4) | E1(2), W1(8) | E1(1), W2(6), W3(1) | – | 5 | 2005 |
| Lugano | Po/Adriatic Sea | Lugano | CH | 45.98 | 8.97 | W1(2) | E1(2), W1(6) | W2(2), W3(1), W5(1) | – | 4 | 2006 |
| Olomouc | Danube/Black Sea | Morava | CZ | 49.61 | 17.25 | – | – | – | E1(2) | 1 | 1863 |
| Kokořín | Elbe/North Sea | Pšovka | CZ | 50.44 | 14.58 | EA1(1), W1(1) | E1(1), W1(3) | W1(1), W2(1) | – | 2 | 2003 |
| Felchow | Oder/Baltic Sea | Grosser Felchowsee | D | 53.06 | 14.13 | W1(1) | E1(2), W1(6) | W1(3), W2(1) | W1(2) | 5 | 1997 |
| Haaven | Wesser/North Sea | Hunte | D | 53.09 | 8.21 | W1(3), W5(1) | W1(8) | E1(1), W1(3), W2(4) | W1(2) | 4 | 2004 |
| Hessen | Rhine/North Sea | Rhine | D | 49.92 | 8.32 | W3(4) | E1(3), W1(5) | E1(4), W2(2), W3(2) | – | 4 | 2005 |
| Plön | Schwentine/Baltic Sea | Vierer see | D | 54.13 | 10.47 | EA1(1), W1(1) | – | – | – | 2 | 2007 |
| Döllner Heide | Oder/Baltic Sea | Kleiner Döllnsee | D | 52.98 | 13.57 | EA1(2), W1(2) | E1(1), W1(7) | W3(6) | – | 5 | 1996 |
| Guadalupe | Guadiana/Atlantic Ocean | Guadalupejo | E | 39.44 | −5.31 | EA1(1) | E1(2) | E1(2) | – | 1 | 2006 |
| Võnnu | Narva/Baltic Sea | Emajõgi | EST | 58.83 | 27.00 | EA1(5) | E1(6), W1(4) | E1(10) | – | 5 | 2005 |
| Priay | Rhône/Mediterranean Sea | Ain | F | 46.00 | 5.27 | W1(2) | E1(1), W1(3) | W1(1), W3(1) | – | 2 | 2005 |
| Belley | Rhône/Mediterranean Sea | Rhône | F | 45.78 | 5.81 | W1(2) | W1(4) | W1(4) | W1(2) | 2 | 2005 |
| Gérardmer | Rhine/North Sea | Gérardmer | F | 48.07 | 6.87 | W1(2) | W1(4) | E1(1), W3(3) | – | 2 | 2005 |
| Warbutts | Ouse/North Sea | Artificial pond | GB | 54.05 | −1.01 | EA3(1), W1(1) | E1(3), W1(1) | E1(3), W1(1) | – | 2 | 2005 |
| Stillingfleet | Ouse/North Sea | Artificial pond | GB | 53.86 | −1.09 | EA1(2) | E1(3), W1(1) | E1(2), W2(1), W4(1) | – | 2 | 2005 |
| Cascina Belgiardino | Po/Adriatic Sea | Adda (Cavo Roggione) | I | 45.28 | 9.48 | W1(3) | E1(2), W1(4) | W1(2), W2(1), W3(2), W4(1) | – | 3 | 2005 |
| Ghazian | Caspian Sea | Anzalee lagoon | IR | 37.47 | 49.33 | EC1(3), EC2(1), EC3(1) | E1(10) | E1(3), E3(1) | E1(2) | 5 | 2005 |
| Sadyrbay | Tengiz – Korgalzhyn | Korgalzhyn | KZ | 50.59 | 70.29 | EA1(3) | E1(6) | E1(6) | E1(2) | 3 | 2005 |
| Hamilton | Waikato/Tasman Sea | Hamilton Lake‡ | NZ | −37.80 | 175.28 | EA1(1), W1(2) | E1(8) | E1(4), W3(2) | E1(2) | 4 | 2003–2005 |
| Lentiscais | Tejo/Atlantic Ocean | Tejo | P | 39.73 | −7.49 | EA1(1) | – | – | – | 1 | 2007 |
| Sątopy-Samulewo | Pregel/Baltic Sea | Sajna | PL | 54.08 | 21.06 | EA6(1), EA7(1),W1(3) | E1(1), E2(1), W1(8) | E1(2), W1(2), W2(1), W3(5) | W1(2) | 5 | 2006 |
| Kurowo | Vistula/Baltic Sea | Narew | PL | 53.12 | 22.80 | EA1(2) | E1(1), W1(3) | E1(3), W1(1) | – | 2 | 2005 |
| Tulcea | Danube/Black Sea | Danube delta | RO | 45.00 | 29.00 | EA1(3), EA4(1) | E1(8) | E1(8) | – | 4 | 2004 |
| Astrakhan | Volga/Caspian Sea | Volga | RUS | 46.41 | 48.00 | EA1(4), EA8(1) | E1(10) | E1(10) | E1(2) | 5 | 2006 |
| Vabacken | Bäveå/North Sea | Öre sjö | S | 58.31 | 12.13 | EA1(1) | E1(2) | E1(2) | E1(2) | 1 | 2007 |
| Stockholm | Mälaren/Baltic Sea | Mälaren | S | 59.33 | 18.07 | EA1(1), W1(2) | E1(3), W1(3) | E1(4), W1(1), W3(1) | – | 3 | 2007 |
| Börringe | Segeå/Baltic Sea | Havgårdssjön | S | 55.49 | 13.36 | EA1(3), W2(1) | E1(4), W1(4) | E1(2), W1(2), W2(1), W3(1) | – | 4 | 2007 |
| Moravský Svätý Ján | Danube/Black Sea | Dlhé lúky | SK | 48.59 | 17.00 | EA1(1), W1(1) | E1(3), W1(1) | E1(1), W2(1), W3(2) | – | 2 | 2006 |
| Buzica | Danube/Black Sea | Ida | SK | 48.55 | 21.08 | EA1(2) | W1(4) | E1(2), W3(2) | – | 2 | 2006 |
| Michalovce | Danube/Black Sea | ZemplínskáŠírava | SK | 48.76 | 22.07 | EA1(1) | E1(1), W1(1) | E1(2) | – | 1 | 2006 |
| Gabčíkovo | Danube/Black Sea | Starý les | SK | 47.77 | 17.73 | EA1(2), W3(1) | E1(4) | W1(1), W3(3) | – | 3 | 2004–2005 |
| Oborín | Danube/Black Sea | Laborec | SK | 48.54 | 21.90 | EA1(2) | E1(4) | E1(4) | – | 2 | 2006 |
| Sapanca | Sakarya/Black Sea | Sapanca gölü‡ | TR | 40.71 | 30.28 | EA1(4), EA5(1) | E1(10) | E1(10) | – | 5 | 2006 |
| Örencik | Yenice Irmaği/Black Sea | Abant gölü‡ | TR | 40.60 | 31.28 | EA1(2) | E1(4) | E1(4) | – | 2 | 2006 |
| Gedikli | Göksu/Mediterraean Sea | Beyşehir gölü‡ | TR | 37.91 | 31.33 | EA1(3) | E1(6) | E1(6) | – | 3 | 2006 |
| Köprüköy | Kızıl Irmak/Black Sea | Köprüköy baraji‡ | TR | 39.57 | 33.43 | EA1(2) | E1(4) | E1(4) | – | 2 | 2006 |
| Kirikkale | Kızıl Irmak/Black Sea | Kapulukaya baraji‡ | TR | 39.69 | 33.46 | EA1(2) | E1(4) | E1(4) | – | 2 | 2004 |
| Toklumen | Kızıl Irmak/Black Sea | Hirfanlı baraji‡ | TR | 39.13 | 33.71 | EA1(2) | E1(4) | E1(4) | – | 2 | 2005 |
| Kırıntı | Aksu Çayi/Mediterraean Sea | Kovada gölü‡ | TR | 37.65 | 30.87 | EA1(3) | E1(6) | E1(6) | – | 3 | 2006 |
| Savincy | Donets/Azov Sea | Siverskyj Donets | UA | 49.38 | 37.02 | EA1(4) | E1(8) | E1(8) | – | 4 | 2006 |
| Gola Pristan | Dnipro/Black Sea | Dnipro delta | UA | 46.31 | 32.31 | EA1(4) | E1(8) | E1(8) | E1(4) | 4 | 2006 |
| Senkove | Donets/Azov Sea | Krasnyj Oskol | UA | 49.51 | 37.69 | EA1(2) | E1(4) | E1(4) | E1(2) | 2 | 2006 |
| Medical Lake | Columbia River/Pacific Ocean | Silver lake‡ | USA | 47.54 | −117.65 | EA1(5) | E1(10) | E1(10) | E1(2) | 5 | 2005 |
| Fish farms | |||||||||||
| Plovdiv | Maritsa/Aegean Sea | Fish pond | BG | 42.15 | 24.72 | EA1(2) | – | – | – | 2 | 2007 |
| Vegas del Guadiana | Guadiana/Atlantic Ocean | Fish pond | E | 38.89 | −6.88 | EA1(5) | E1(10) | E1(10) | E1(2) | 5 | 2006 |
| Mionnay | Rhône/Mediterranean Sea | Fish pond | F | 45.90 | 4.92 | W1(2) | W1(4) | W1(1), W2(1), W3(2) | – | 2 | 2005 |
| Bouligneux | Rhône/Mediterranean Sea | Fish pond | F | 46.02 | 4.99 | W1(1), W2(1) | E1(2), W1(2) | W3(2) | – | 2 | 2005 |
| Perugia | Tiber/Tyrrhenian Sea | Trasimeno Lake§ | I | 43.15§ | 12.10§ | W1(2) | W1(4) | W3(4) | W1(2) | 2 | 2005 |
| Mincio, Bonferraro di Sorga | Po/Adriatic Sea | Garda Lake§ | I | 45.55§ | 10.70§ | W1(3) | E1(1), W1(4), W2(1) | W1(2), W2(2), W3(2) | – | 3 | 2005 |
| Żabieniec | Vistula/Baltic Sea | Fish pond | PL | 52.05 | 21.03 | W1(1), W5(1) | E1(3), W1(1) | E1(1), W3(3) | W1(2) | 2 | 2005 |
| Wuhan | Yangtze River/East China Sea | Fish pond | PRC | 30.56 | 114.37 | EA1(3), EA2(1) | E1(8) | E1(2) | E1(2) | 4 | 2004 |
| Italian regional breed | |||||||||||
| Ceresole d′Alba | Po/Adriatic Sea | Fish pond | I | 44.80 | 7.82 | W1(2) | E1(1), W1(3) | W1(3), W3(1) | – | 2 | 2005 |
| Live gene bank in Vodňany | |||||||||||
| Regional breeds | |||||||||||
| Hluboká, new stock | Elbe/North Sea | Fish pond | CZ | 49.05 | 14.43 | EA1(2), W1(1) | E1(1), W1(5) | E1(1), W1(2), W3(3) | – | 3 | 2004 |
| Hluboká, old stock | Elbe/North Sea | Fish pond | CZ | 49.05 | 14.43 | EA1(3) | E1(2), W1(4) | W2(2), W3(4) | – | 3 | 2004 |
| Mariánské Lázně | Elbe/North Sea | Fish pond | CZ | 49.97 | 12.70 | EA1(3) | E1(3), W1(3) | E1(4), W3(2) | – | 3 | 2005 |
| Tábor (Milevsko), new stock | Elbe/North Sea | Fish pond | CZ | 49.45 | 14.36 | EA1(2), W1(2) | E1(2), W1(4) | E1(2), W3(2) | – | 4 | 2004 |
| Tábor, old stock | Elbe/North Sea | Fish pond | CZ | 49.40 | 14.69 | EA1(3) | E1(2), W1(4) | E1(2), W1(1), W3(3) | – | 3 | 2004 |
| Velké Meziříčí | Elbe/North Sea | Fish pond | CZ | 49.35 | 16.02 | EA1(1), W1(1), W4(1) | E1(2), W1(4) | E1(2), W1(4) | – | 3 | 2004 |
| Vodňany | Elbe/North Sea | Fish pond | CZ | 49.15 | 14.18 | EA1(3) | E1(4), W1(2) | E1(5), W2(1) | – | 3 | 2004 |
| European breeds | |||||||||||
| Königswartha (Germany) | Elbe/North Sea | Fish pond | D | 51.31 | 14.33 | EA1(1), W1(1) | E1(1), W1(9) | E1(1), W1(5), W2(2) | – | 5 | 2004 |
| Romania | Danube/Black Sea | Fish pond | RO | EA1(1), B1(3) | E1(5), W1(3) | E1(2), W1(1), W3(3) | – | 4 | 2004 | ||
| Hungaria | Danube/Black Sea | Fish pond | H | EA1(5) | E1(2), W1(8) | E1(1), W3(3) | – | 5 | 2004 | ||
| Experimental breeds | |||||||||||
| Leather ‘92 | Fish pond | CZ | W2(3) | E1(1), W1(5) | E1(6) | – | 3 | 2004 | |||
| Synthetic | Fish pond | CZ | EA1(3) | E1(3), W1(3) | E1(2), W2(2), W3(2) | – | 3 | 2004 | |||
| Gynogenetic | Fish pond | CZ | EA1(3) | W1(6) | E1(4), W3(1), W4(1) | – | 3 | 2004 | |||
| Ornamental breeds | |||||||||||
| Golden | Fish pond | CZ | EA1(2) | E1(2), W1(2) | E1(1), W1(3) | – | 2 | 2004 | |||
| Blue | Fish pond | CZ | EA1(2) | E1(1), W1(3) | E1(1), W1(1), W3(2) | – | 2 | 2004 | |||
| Alampic | Fish pond | CZ | EA1(2) | E1(1), W1(3) | E1(1), W1(2), W3(1) | – | 2 | 2004 | |||
For the geographic breeds in the live gene bank, locality identifies the original source of the breed, while for the experimental and ornamental breeds, only the breed name is given.
The countries are coded as follows: Belgium (B), Bulgaria (BG), Bosnia (BIH), Canada (CDN), Switzerland (CH), Czech Republic (CZ), Germany (D), Spain (E), Estonia (EST), France (F), Great Britain (GB), Hungary (H), Italy (I), Iran (IR), Kazakhstan (KZ), New Zealand (NZ), Portugal (P), Poland (PL), China (PRC), Romania (RO), Russia (RUS), Sweden (S), Slovakia (SK), Turkey (TR), Ukraine (UA), United States of America (USA).
Known introduced population.
Information about the source population.
Literature cited
- Abbott F. 1868. Tench fish supplied from the Royal Society's gardens during the year 1868. Monthly Notices of Papers and Proceedings of the Royal Society of Tasmania for 1868. Page83.
- Allport M. 1866. pp. 61–64. Report on the present state of the fry of the salmon and salmon trout at the Plenty; and of the taking of the first spawn from the brown trout. Monthly Notices of Papers and Proceedings of the Royal Society of Tasmania for 1866. Pages.
- Allport M. 1868. pp. 33–36. Remark's on Mr Krefft's Notes on the fauna of Tasmania. Monthly Notices of Papers and Proceedings of the Royal Society of Tasmania for 1868. Pages.
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Systematic Biology. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- Araki H, Berejikian BA, Ford MJ, Blouin MS. Fitness of hatchery-reared salmonids in the wild. Evolutionary Applications. 2008;1:342–355. doi: 10.1111/j.1752-4571.2008.00026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur W. History of fish culture in New Zealand. Transactions and Proceedings of the Royal Society of New Zealand. 1881;14:180–210. [Google Scholar]
- Atarhouch T, Rami M, Cattaneo-Berrebi G, Ibanez C, Augros S, Boissin E, Dakkak A, et al. Primers for EPIC amplification of intron sequences for fish and other vertebrate population genetic studies. BioTechniques. 2003;35:676–678. doi: 10.2144/03354bm02. 680, 682. [DOI] [PubMed] [Google Scholar]
- Baird SF. United States Commission of Fish and Fisheries, Part V., Report of the Commissioner for 1877. Washington, DC: U.S. Government Printing Office; 1879. [Google Scholar]
- Bănărescu P. Zoogeography of Fresh Waters 2: Distribution and Dispersal of Fresh Water Animals in North America and Eurasia. Wiesbaden: AULA-Verlag; 1991. [Google Scholar]
- Baughman JL. The tench in America. Journal of Wildlife Management. 1947;11:197–204. [Google Scholar]
- Bernatchez L. The evolutionary history of brown trout (Salmo trutta L.) inferred from phylogeographic, nested clade, and mismatch analyses of mitochondrial DNA variation. Evolution. 2001;55:351–379. doi: 10.1111/j.0014-3820.2001.tb01300.x. [DOI] [PubMed] [Google Scholar]
- Bernatchez L, Wilson C. Comparative phylogeography of nearctic and palearctic fishes. Molecular Ecology. 1998;7:431–452. [Google Scholar]
- Bilio M. Controlled reproduction and domestication in aquaculture – the current state of the art, Part I. Aquaculture Europe. 2007;32(1):5–14. [Google Scholar]
- Bohlen J, Šlechtová V, Doadrio I, Ráb P. Low mitochondrial divergence indicates a rapid expansion across Europe in the weather loach, Misgurnus fossilis (L.) Journal of Fish Biology. 2007;71(Suppl B):186–194. [Google Scholar]
- Brito PH, Edwards SV. Multilocus phylogeography and phylogenetics using sequence-based markers. Genetica. 2008;135:439–455. doi: 10.1007/s10709-008-9293-3. [DOI] [PubMed] [Google Scholar]
- Brylińska M, Bryliński E, Bnińska M. Tinca tinca (Linnaeus 1758) In: Bănărescu PM, editor. The Freshwater Fishes of Europe, 5/I: Cyprinidae 2/I. Wiesbaden: AULA-Verlag; 1999. pp. 229–302. [Google Scholar]
- Burridge CP, Craw D, Fletcher D, Waters JM. Geological dates and molecular rates: fish DNA sheds lights on time dependency. Molecular Biology and Evolution. 2008;25:624–633. doi: 10.1093/molbev/msm271. [DOI] [PubMed] [Google Scholar]
- Carlton JT. Biological invasions and cryptogenic species. Ecology. 1996;77:1653–1655. [Google Scholar]
- Chen WJ, Mayden RL. Molecular systematics of the Cyprinoidea (Teleostei: Cypriniformes), the world's largest clade of freshwater fishes: further evidence from six nuclear genes. Molecular Phylogenetics and Evolution. 2009;52:544–549. doi: 10.1016/j.ympev.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Chow S, Hazama K. Universal PCR primers for S7 ribosomal protein gene introns in fish. Molecular Ecology. 1998;7:1255–1256. [PubMed] [Google Scholar]
- Clark AG. Inference of haplotypes from PCR-amplified samples of diploid populations. Molecular Biology and Evolution. 1990;7:111–122. doi: 10.1093/oxfordjournals.molbev.a040591. [DOI] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Molecular Ecology. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- DeVaney SC, McNyset KM, Williams JB, Peterson AT, Wiley EO. A tale of four “Carp”: invasion potential and ecological niche modeling. Public Library of Science ONE. 2009;4:e5451. doi: 10.1371/journal.pone.0005451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling TE, Tibbets CA, Minckley WL, Smith GR. Evolutionary relationships of the plagopterins (Teleostei: Cyprinidae) from cytochrome b sequences. Copeia. 2002;2002:665–678. [Google Scholar]
- Dumont P, Vachon N, Leclerc J, Guibert A. Intentional introduction of Tench in Southern Quebec. In: Claudi R, Nantel P, Muckle-Jeffs E, editors. Alien Invaders in Canada's Waters, Wetlands and Forests. Ottawa: Canadian Forest Service, Natural Resources Canada; 2002. pp. 169–177. [Google Scholar]
- Dupanloup I, Schneider S, Excoffier L. A simulated annealing approach to define the genetic structure of populations. Molecular Ecology. 2002;11:2571–2581. doi: 10.1046/j.1365-294x.2002.01650.x. [DOI] [PubMed] [Google Scholar]
- Durand JD, Persat H, Bouvet Y. Phylogeography and postglacial dispersion of the chub (Leuciscus cephalus) in Europe. Molecular Ecology. 1999;8:989–997. doi: 10.1046/j.1365-294x.1999.00654.x. [DOI] [PubMed] [Google Scholar]
- Economidis PS, Dimitriou E, Pagoni R, Michaloudi E, Natsis L. Introduced and translocated fish species in the inland waters of Greece. Fisheries Management and Ecology. 2000;7:239–250. [Google Scholar]
- Excoffier L. Patterns of DNA sequence diversity and genetic structure after a range expansion: lessons from the infinite-island model. Molecular Ecology. 2004;13:853–864. doi: 10.1046/j.1365-294x.2003.02004.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Ferguson A, Fleming I, Hindar K, Skaala Ř, McGinnity P, Cross TF, Prodöhl P. Farm escapes. In: Verspoor E, Stradmeyer L, Nielsen J, editors. The Atlantic Salmon: Genetics, Conservation and Management. Oxford: Blackwell; 2007. pp. 357–398. [Google Scholar]
- Flajšhans M, Linhart O, Šlechtová V, Šlechta V. Genetic resources of commercially important fish species in the Czech Republic: present state and future strategy. Aquaculture. 1999;173:469–481. [Google Scholar]
- Frankham R. Stress and adaptation in conservation genetics. Journal of Evolutionary Biology. 2005;18:750–755. doi: 10.1111/j.1420-9101.2005.00885.x. [DOI] [PubMed] [Google Scholar]
- Fraser DJ, Houde ALS, Debes PV, O'Reilly P, Eddington JD, Hutchings JA. Consequences of farmed–wild hybridization across divergent wild populations and multiple traits in salmon. Ecological Applications. 2010;20:935–953. doi: 10.1890/09-0694.1. [DOI] [PubMed] [Google Scholar]
- Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller PL, Nico LG, Williams JD. Nonindigenous Fishes Introduced into Inland Waters of the United States. Bethesda, Maryland: American Fisheries Society Special Publication 27; 1999. [Google Scholar]
- García-Berthou E, Boix D, Clavero M. Non-indigenous animal species naturalized in Iberian inland waters. In: Gherardi F, editor. Biological Invaders in Inland Waters: Profiles, Distribution, and Threats. Dordrecht: Invading Nature: Springer Series in Invasion Ecology, Springer; 2007. pp. 123–140. [Google Scholar]
- Gasco L, Gai F, Lussiana C, Lo Presti R, Malfatto V, Daprà F, Zoccarato I. Morphometry, slaughtering performances, chemical and fatty acid composition of the protected designation of origin “Golden hump tench of Poirino highland” product. Reviews in Fish Biology and Fisheries. 2010;20:357–365. [Google Scholar]
- Gela D, Linhart O, Flajšhans M, Duda P. A live gene bank of tench, Tinca tinca (L.) strains in the Czech Republic. Polish Archives of Hydrobiology. 1998;45:311–314. [Google Scholar]
- Gela D, Flajšhans M, Kocour M, Rodina M, Linhart O. Tench broodstock management in breeding station under conditions of pond culture. Aquaculture International. 2006;14:195–203. [Google Scholar]
- Gherardi F, Bertolino S, Bodon M, Casellato S, Cianfanelli S, Ferraguti M, Lori E, et al. Animal xenodiversity in Italian inland waters: distribution, modes of arrival, and pathways. Biological Invasions. 2008;10:435–454. [Google Scholar]
- Giovio P. De Romanis Piscibus Libellus. Rome. (in Latin): F. Minitius Caluus; 1524. [Google Scholar]
- Glamuzina B. České Budějovice: University of South Bohemia; 2006. Status of introduced tench, Tinca tinca in Hutovo Blato wetlands, Adriatic Sea drainage. In IX. česká ichtyologická konference: sborník příspěvků z IX. konference s mezinárodníúčastí, Vodňany, 4.–5.5. 2006, p14. [Google Scholar]
- Guillot G, Leblois R, Coulon A, Frantz AC. Statistical methods in spatial genetics. Molecular Ecology. 2009;18:4734–4756. doi: 10.1111/j.1365-294X.2009.04410.x. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hänfling B, Dümpelmann C, Bogutskaya NG, Brandl R, Brändle M. Shallow phylogeographic structuring of Vimba vimba across Europe suggests two distinct refugia during the last glaciation. Journal of Fish Biology. 2009;75:2269–2286. doi: 10.1111/j.1095-8649.2009.02415.x. [DOI] [PubMed] [Google Scholar]
- Hare MP, Palumbi SR. The accuracy of heterozygous base calling from diploid sequence and resolution of haplotypes using allele-specific sequencing. Molecular Ecology. 1999;8:1749–1752. doi: 10.1046/j.1365-294x.1999.00738-1.x. [DOI] [PubMed] [Google Scholar]
- Hesthagen T, Sandlund OT. Non-native freshwater fishes in Norway: history, consequences and perspectives. Journal of Fish Biology. 2007;71(Suppl D):173–183. [Google Scholar]
- Hewitt GM. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- Hicks BJ. Managing Invasive Freshwater Fish in New Zealand. Wellington, New Zealand: Department of Conservation; 2003. Biology and potential impacts of rudd (Scardinius erythrophthalmus L.) in New Zealand; pp. 49–58. Proceedings of a workshop hosted by Department of Conservation 10–12 May 2001, Hamilton. [Google Scholar]
- Huang D, Liu J, Hu C. Fish resources in Chinese reservoirs and their utilization. In: De Silva SS, editor. Reservoir and Culture-based Fisheries: Biology and Management. Canberra: ACIAR Proceedings No. 98; 2001. pp. 16–21. Proceedings of an International Workshop held in Bangkok, Thailand from 15–18 February 2000. [Google Scholar]
- Hudson RR, Kaplan NL. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchings JA, Fraser DJ. The nature of fisheries- and farming-induced evolution. Molecular Ecology. 2008;17:294–313. doi: 10.1111/j.1365-294X.2007.03485.x. [DOI] [PubMed] [Google Scholar]
- Innal D, Erk'akan F. Effects of exotic and translocated fish species in the inland waters of Turkey. Reviews in Fish Biology and Fisheries. 2006;16:39–50. [Google Scholar]
- Jarman SN, Ward RD, Elliott NG. Oligonucleotide primers for PCR amplification of coelomate introns. Marine Biotechnology. 2002;4:347–355. doi: 10.1007/s10126-002-0029-6. [DOI] [PubMed] [Google Scholar]
- Kennedy M, Fitzmaurice P. The biology of the tench Tinca tinca (L.) in Irish waters. Proceedings of the Royal Irish Academy. 1970;69B:31–82. [PubMed] [Google Scholar]
- Kohlmann K, Kersten P, Panicz R, Memiş D, Flajšhans M. Genetic variability and differentiation of wild and cultured tench populations inferred from microsatellite loci. Reviews in Fish Biology and Fisheries. 2010;20:279–288. [Google Scholar]
- Korkmaz AŞ, Zencir Ö. Tench Invasion in Turkish freshwater. 2005. Abstracts of the international workshop on biological invasions in Inland waters, p42. Florence.
- Kotlík P, Berrebi P. Phylogeography of the barbel (Barbus barbus) assessed by mitochondrial DNA variation. Molecular Ecology. 2001;10:2177–2185. doi: 10.1046/j.0962-1083.2001.01344.x. [DOI] [PubMed] [Google Scholar]
- Kotlík P, Bogutskaya NG, Ekmekçi FG. Circum Black Sea phylogeography of Barbus freshwater fishes: divergence in the Pontic glacial refugium. Molecular Ecology. 2004;13:87–95. doi: 10.1046/j.1365-294x.2003.02021.x. [DOI] [PubMed] [Google Scholar]
- Kotlík P, Marková S, Choleva L, Bogutskaya NG, Ekmekçi FG, Ivanova PP. Divergence with gene flow between Ponto-Caspian refugia in an anadromous cyprinid Rutilus frisii revealed by multiple gene phylogeography. Molecular Ecology. 2008;17:1076–1088. doi: 10.1111/j.1365-294X.2007.03638.x. [DOI] [PubMed] [Google Scholar]
- Lajbner Z, Kohlmann K, Linhart O, Kotlík P. Lack of reproductive isolation between the Western and Eastern phylogroups of the tench. Reviews in Fish Biology and Fisheries. 2010;20:289–300. [Google Scholar]
- Lebedev VD. Quaternary Freshwater Fish Fauna of European Part of USSR. Leningrad. (in Russian): Izdatelstvo Moskovskovo universiteta; 1960. [Google Scholar]
- Lo Presti R, Gasco L, Lisa C, Zoccarato I, Di Stasio L. PCR-RFLP analysis of mitochondrial DNA in tench Tinca tinca. Journal of Fish Biology. 2010;76:401–407. doi: 10.1111/j.1095-8649.2009.02495.x. [DOI] [PubMed] [Google Scholar]
- Lucek K, Roy D, Bezault E, Sivasundar A, Seehausen O. Hybridization between distant lineages increases adaptive variation during a biological invasion: stickleback in Switzerland. Molecular Ecology. 2010;19:3995–4011. doi: 10.1111/j.1365-294X.2010.04781.x. [DOI] [PubMed] [Google Scholar]
- Lusk S, Lusková V, Halačka K. The status of tench (Tinca tinca (L.)) in aquatic habitats of the floodplain along the lower reaches of the River Dyje (Czech Republic) Polish Archives of Hydrobiology. 1998;45:407–414. [Google Scholar]
- Mabuchi K, Senou H, Nishida M. Mitochondrial DNA analysis reveals cryptic large-scale invasion of non-native genotypes of common carp (Cyprinus carpio) in Japan. Molecular Ecology. 2008;17:796–809. doi: 10.1111/j.1365-294X.2007.03626.x. [DOI] [PubMed] [Google Scholar]
- Machordom A, Doadrio I. Evidence of a Cenozoic Betic-Kabilian connection based on freshwater fish phylogeography (Luciobarbus, Cyprinidae) Molecular Phylogenetics and Evolution. 2001;18:252–263. doi: 10.1006/mpev.2000.0876. [DOI] [PubMed] [Google Scholar]
- Maddison WP. 2008. Coalescence Package for Mesquite. Version 2.5 http://mesquiteproject.org.
- Maddison WP, Maddison DR. 2008. Mesquite: a modular system for evolutionary analysis. Version 2.5 http://mesquiteproject.org.
- Mamilov NSh, Balabieva GK, Koishybaeva GS. Distribution of alien fish species in small waterbodies of the Balkhash basin. Russian Journal of Biological Invasions. 2010;1(3):181–186. [Google Scholar]
- Manni F, Guérard E, Heyer E. Geographic patterns of (genetic, morphologic, linguistic) variation: how barriers can be detected by “Monmonier's algorithm”. Human Biology. 2004;76:173–190. doi: 10.1353/hub.2004.0034. [DOI] [PubMed] [Google Scholar]
- Marie AD, Bernatchez L, Garant D. Loss of genetic integrity correlates with stocking intensity in brook charr (Salvelinus fontinalis. Molecular Ecology. 2010;19:2025–2037. doi: 10.1111/j.1365-294X.2010.04628.x. [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Mitrofanov VP, Petr T. Fish and fisheries in the Altai, Northern Tien Shan and Lake Balkhash (Kazakhstan) In: Petr T, editor. Fish and Fisheries at Higher Altitudes: Asia, FAO Fisheries Technical Paper 385. Rome, Italy: Food and Agriculture Organization of the United Nations; 1999. pp. 149–167. [Google Scholar]
- Monich JK. Rozmnozhenie i razvitie linja (Tinca tinca L.) v Zapadnoj Syberii (Reproduction and ontogeny of the tench (Tinca tinca L.) in Western Siberia) Trudy Tomskogo Gosudarstvennogo Universiteta. 1953;125:106–115. (in Russian) [Google Scholar]
- Monmonier M. Maximum-difference barriers: an alternative numerical regionalization method. Geographic Analysis. 1973;3:245–261. [Google Scholar]
- Muhlfeld CC, Kalinowski ST, McMahon TE, Taper ML, Painter S, Leary RF, Allendorf FW. Hybridization rapidly reduces fitness of a native trout in the wild. Biology Letters. 2009;5:328–331. doi: 10.1098/rsbl.2009.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscheler R, Kromer B, Björck S, Svensson A, Friedrich M, Kaiser KF, Southon J. Tree rings and ice cores reveal 14C calibration uncertainties during the Younger Dryas. Nature Geoscience. 2008;1:63–267. [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Nesbø CL, Fossheim T, Vøllestad LA, Jakobsen KS. Genetic divergence and phylogeographic relationships among European perch (Perca fluviatilis) populations reflect glacial refugia and postglacial colonization. Molecular Ecology. 1999;8:1387–1404. doi: 10.1046/j.1365-294x.1999.00699.x. [DOI] [PubMed] [Google Scholar]
- Nico LG, Fuller PL. >Tinca tinca. Gainesville, FL: USGS Nonindigenous Aquatic Species Database; 2010. http://nas.er.usgs.gov/queries/FactSheet.asp?speciesID=652 Revision Date: 4/24/2006. [Google Scholar]
- Pekař Č. Pozorování průběhu výtěru lína obecného (Tinca tinca L.) v údolní nádrži Lipno (Observation of the tench (Tinca tinca L.) spawning in Lipno dam lake.) Bulletin VÚRH Vodňany. 1965;1(2):14–18. (in Czech) [Google Scholar]
- Popov PA. Species composition and pattern of fish distribution in Siberia. Journal of Ichthyology. 2009;49:483–495. [Google Scholar]
- Posada D. jModelTest: phylogenetic Model Averaging. Molecular Biology and Evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Ramírez-Soriano A, Ramos-Onsins SE, Rozas J, Calafell F, Navarro A. Statistical power analysis of neutrality tests under demographic expansions, contractions and bottlenecks with recombination. Genetics. 2008;179:555–567. doi: 10.1534/genetics.107.083006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randi E. Detecting hybridization between wild species and their domesticated relatives. Molecular Ecology. 2008;17:285–293. doi: 10.1111/j.1365-294X.2007.03417.x. [DOI] [PubMed] [Google Scholar]
- Ribeiro F, Collares-Pereira MJ, Moyle PB. Non-native fish in the fresh waters of Portugal, Azores and Madeira Islands: a growing threat to aquatic biodiversity. Fisheries Management and Ecology. 2009;16:255–264. [Google Scholar]
- Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology and Evolution. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- Rowe DK. Potential effects of tench (Tinca tinca) in New Zealand freshwater ecosystems. NIWA Client Report HAM2004–005. Hamilton, New Zealand: National Institute of Water and Atmospheric Research; 2004. [Google Scholar]
- Rowe DK, Moore A, Giorgetti A, Maclean AC, Grace P, Wadhwa S, Cooke J. Review of the Impacts of Gambusia, Redfin Perch, Tench, Roach, Yellowfin Goby and Streaked Goby in Australia. Canberra, Australia: Prepared for the Australian Government Department of the Environment, Water, Heritage and the Arts; 2008. http://www.environment.gov.au/biodiversity/invasive/publications/introduce-fish.html. [Google Scholar]
- Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Sanz N, Cortey M, Pla C, García-Marín JL. Hatchery introgression blurs ancient hybridization between brown trout (Salmo trutta) lineages as indicated by complementary allozymes and mtDNA markers. Biological Conservation. 2006;130:278–289. [Google Scholar]
- Savvaitova KA, Petr T. Fish and fisheries in Lake Issyk-Kul (Tien Shan), River Chu and Pamir Lakes. In: Petr T, editor. Fish and Fisheries at Higher Altitudes: Asia, FAO Fisheries Technical Paper 385. Rome, Italy: Food and Agriculture Organization of the United Nations; 1999. pp. 168–187. [Google Scholar]
- Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. American Journal of Human Genetics. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S, Excoffier L. Estimation of demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: application to human mitochondrial DNA. Genetics. 1999;152:1079–1089. doi: 10.1093/genetics/152.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle JB, Kotlík P, Rambau RV, Marková S, Herman JS, McDevitt AD. The Celtic fringe of Britain: insights from small mammal phylogeography. Proceedings of the Royal Society of London. Series B: Biological Sciences. 2009;276:4287–4429. doi: 10.1098/rspb.2009.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šedivá A, Janko K, Šlechtová V, Kotlík P, Simonović P, Delic A, Vassilev M. Around or across the Carpathians: colonization model of the Danube basin inferred from genetic diversification of stone loach (Barbatula barbatula) populations. Molecular Ecology. 2008;17:1277–1292. doi: 10.1111/j.1365-294X.2007.03656.x. [DOI] [PubMed] [Google Scholar]
- Šlechtová V, Bohlen J, Freyhof J, Persat H, Delmastro GB. The Alps as barrier to dispersal in cold-adapted freshwater fishes? Phylogeographic history and taxonomic status of the bullhead in the Adriatic freshwater drainage. Molecular Phylogenetics and Evolution. 2004;33:225–239. doi: 10.1016/j.ympev.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Steffens W. The tench (Tinca tinca L.), a neglected pond fish species. Polish Archives of Hydrobiology. 1995;42:161–180. [Google Scholar]
- Stockwell CA, Hendry AP, Kinnison MT. Contemporary evolution meets conservation biology. Trends in Ecology & Evolution. 2003;18:94–101. [Google Scholar]
- Stokes K, O'Neill K, McDonald RA. Invasive Species in Ireland. Belfast: Queens University Belfast; 2004. Unpublished report to Environment and Heritage Service and National Parks and Wildlife Service. Quercus. [Google Scholar]
- Stone GN, Challis RJ, Atkinson RJ, Csoka G, Hayward A, Mutun S, Preuss S, et al. The phylogeographic clade trade: tracing the impact of human-mediated dispersal on the colonisation of northern Europe by the oak gallwasp Andricus kollari. Molecular Ecology. 2007;16:2768–2781. doi: 10.1111/j.1365-294X.2007.03348.x. [DOI] [PubMed] [Google Scholar]
- Šusta J. 1884. Výživa kapra a jeho družiny rybničné (Nutrition of carp and other pond species). ČSAZ, Praha (1884) Reviewed edition 1938, Prague. (in Czech)
- Taberlet P, Fumagalli L, Wustsaucy AG, Cosson JF. Comparative phylogeography and postglacial colonization routes in Europe. Molecular Ecology. 1998;7:453–464. doi: 10.1046/j.1365-294x.1998.00289.x. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor EB. Evolution in mixed company: evolutionary inferences from studies of natural hybridization in Salmonidae. In: Hendry AP, Stearns S, editors. Evolution Illuminated. Salmon and their Relatives. Oxford: Oxford University Press; 2004. pp. 232–263. [Google Scholar]
- Templeton AR, Crandall KA, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics. 1992;132:619–638. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson GM. The Naturalisation of Animals and Plants in New Zealand. Cambridge: Cambridge University Press; 1922. [Google Scholar]
- Triantafyllidis A, Krieg F, Cottin C, Abatzopoulos TJ, Triantaphyllidis C, Guyomard R. Genetic structure and phylogeography of European catfish (Silurus glanis) populations. Molecular Ecology. 2002;11:1039–1055. doi: 10.1046/j.1365-294x.2002.01501.x. [DOI] [PubMed] [Google Scholar]
- Tsigenopoulos CS, Berrebi P. Molecular phylogeny of North Mediterranean freshwater barbs (genus Barbus: Cyprinidae) inferred from cytochrome b sequences: biogeographic and systematic implications. Molecular Phylogenetics and Evolution. 2000;14:165–179. doi: 10.1006/mpev.1999.0702. [DOI] [PubMed] [Google Scholar]
- Turchini G, De Silva S. Bio-economical and ethical impacts of alien finfish culture in European Inland waters. Aquaculture International. 2008;16:243–272. [Google Scholar]
- Turner TF, Osborne MJ, Moyer GR, Benavides MA, Alo D. Life history and environmental variation interact to determine effective population to census size ratio. Proceedings of the Royal Society of London. Series B: Biological Sciences. 2006;273:3065–3073. doi: 10.1098/rspb.2006.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urchinov ZhU. Fisheries in the Zarafshan River basin (Uzbekistan) In: Petr T, editor. Inland Fisheries Under the Impact of Irrigated Agriculture: Central Asia, FAO Fisheries Circular 894. Rome, Italy: Food and Agriculture Organization of the United Nations; 1995. pp. 58–62. [Google Scholar]
- Van Houdt J, De Cleyn L, Perretti A, Volckaert F. A mitogenic view on the evolutionary history of the Holarctic freshwater gadoid, burbot (Lota lota. Molecular Ecology. 2005;14:2445–2457. doi: 10.1111/j.1365-294X.2005.02590.x. [DOI] [PubMed] [Google Scholar]
- Walker KF, Yang HZ. Fish and fisheries in western China. In: Petr T, editor. Fish and Fisheries at Higher Altitudes. Food and Agriculture Organization of the United Nations, Rome, Italy: FAO Fisheries Technical Paper; 1999. pp. 231–271. [Google Scholar]
- Wang J, Min W, Guan M, Hu S. Tench farming in China: present status and future prospects. In: Sakowicz S IVth, editor. International Workshop on Biology and Culture of the Tench, Tinca tinca (L.) Olsztyn: Inland Fisheries Institute in Olsztyn; 2004. p. 32. Wierzba, September 20–23 2004. Programme and Abstracts. [Google Scholar]
- Welcomme RL. International Introductions of Inland Aquatic Species. Rome, Italy: Food and Agriculture Organization of the United Nations; 1988. FAO Fisheries Technical Paper 294. [Google Scholar]
- Won YJ, Hey J. Divergence population genetics of chimpanzees. Molecular Biology and Evolution. 2005;22:297–307. doi: 10.1093/molbev/msi017. [DOI] [PubMed] [Google Scholar]
- Wydoski RS, Whitney RR. Inland Fishes of Washington. 2nd Edn. Seattle: American Fisheries Society, Bethesda, Maryland and University of Washington Press; 2003. Revised and Expanded. [Google Scholar]


