Since the discovery of the ribosome as the machinery essential for protein synthesis in the mid-1950s, extensive studies have been carried out with respect to ribosomal structure, function, biosynthesis, regulation. For obvious historical reasons, these studies, especially those on the structure–function relationship, have been done mostly by using Escherichi coli, leading to enormous amounts of information on E. coli ribosomes (1). One of the most impressive recent developments is the demonstration in vitro of the peptidyl transferase activity of rRNA with few or no proteins attached, confirming the suspected essential roles of rRNAs in ribosomal functions (2–4). In parallel to the in vitro studies of E. coli ribosomes, extensive genetic and physiological studies have also been carried out, identifying all of the genes for ribosomal components and giving insight into mechanisms by which E. coli regulates the synthesis of ribosomes and their molecular components (5, 6). Genetic approaches have always been essential to test the validity of conclusions derived from in vitro experiments regarding ribosome functions or regulation of ribosome synthesis. In the article by Asai et al. published in this issue of the Proceedings (7), Squires and coworkers describe their success in constructing an E. coli strain (“Δ7 prrn”) in which each of the seven chromosomal rRNA operons is inactivated by a deletion spanning the 16S and 23S RNA-coding regions, and rRNA is transcribed from a single rRNA operon carried by a multicopy plasmid. Although this success does not provide major surprises, the newly constructed rrn deletion strains provide a powerful system for mutational analysis of the structure and function of rRNAs as well as a system for studying the significance of the presence of multiple rRNA operons in bacteria.
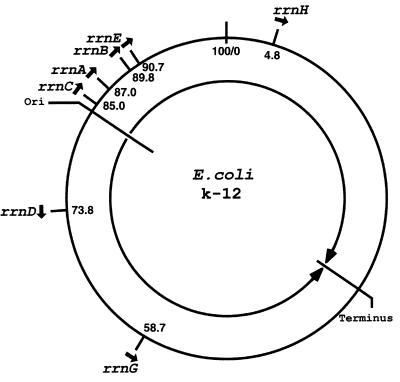
The seven rRNA operons in E. coli are located on the chromosome as shown in Fig. 1. Regarding the presence of multiple copies of rRNA genes and their chromosomal location in bacteria, two questions were repeatedly asked as soon as experimental information became available, first for Bacillus subtilis in the mid-1960s and then for E. coli around 1970. The first question concerned the heterogeneity of rRNA sequences among multiple copies of the rRNA genes and its functional significance. The second question was the significance of the chromosomal location of rRNA genes.
Figure 1.
Location of the rRNA operons (rrn) on the E. coli chromosome. Closed arrows indicate the direction of transcription. The origin (Ori) and the terminus of DNA replication also are indicated, together with two large arrows indicating the direction of DNA replication. Positions are shown in min (or centisomes) according to the new E. coli map (8).
Regarding the heterogeneity of rRNA sequences, one can now compare sequences of rRNA genes with each other based on the complete sequence of the E. coli K-12 genome elucidated recently (8), yielding, for example, sequence differences ranging from 0 to 1.3% between any two 16S rRNA genes. Such differences appear to be minor. However, it has been reported that the parasite Plasmodium has two distinct genes encoding small-subunit (18S) rRNAs with a small sequence difference (3.5%) and each is expressed in different stages of the life cycle of the organism, suggesting a functional difference between the two kinds of ribosomes containing different rRNA species (9). Thus, even though sequence heterogeneity among multiple rRNA operons is small and is probably of no functional significance, it has been difficult to establish this point convincingly. In fact, a systematic analysis of the seven E. coli rRNA operons in their native chromosomal locations demonstrated small but significant differences among the seven rRNA operons with respect to their promoter strength and their regulatory features, such as responses to heat shock or amino acid starvation (10). Such experimental results raised the possibility of functional differentiation among rRNA operons and/or the significance of the chromosomal location in relation to regulation of rRNA synthesis (10). Although in the paper by Asai et al. (7), the authors do not describe comparison of the Δ7 prrn strain with the control strain without any rrn deletion, the fact that the Δ7 prrn strain can grow reasonably well indicates that any differences among different rRNA operons, if they exist, must be small and inessential for growth under standard culture conditions. No doubt the system developed by Squires and coworkers should be useful for studying the questions related to the rRNA sequence heterogeniety as well as the significance of chromosomal locations of rRNA operons. For example, one should now be able to construct E. coli strains carrying a single rRNA operon in any number of copies located at any desired locations at either original locations or at some new locations. Such strains may be able to give definitive answers to these questions.
The immediate utility of the system described by Squires and coworkers regards the study of rRNA structure and function by in vivo mutational analysis. In the past, one could carry out mutational analysis of rRNAs in vitro, e.g., by in vitro transcription of rRNA genes combined with reconstitution techniques (e.g., ref. 11). However, these types of approaches were technically demanding and were not widely used. Alternative in vivo mutational analysis was also possible (reviewed in ref. 1). For example, mutational alterations were made on a plasmid-encoded rRNA gene with an antibiotic-resistance mutation and fused to an inducible strong promoter. The effects of a mutational alteration were then studied after induction of the synthesis of the rRNA from the plasmid-encoded mutant gene and in the presence of the antibiotics to inactivate the ribosomes containing the rRNA derived from the chromosomal rRNA genes, (e.g., refs. 12 and 13). The new Δ7 prrn system is no doubt simpler and cleaner, and an example of mutational analysis is mentioned in the paper (7).
Systems similar to the E. coli Δ7 prrn exist for the yeast Saccharomyces cerevisiae. In this model eukaryotic organism, approximately 150 copies of rRNA genes are tandemly repeated at a locus on chromosome XII. Yeast strains were constructed in which these genes were mostly (14) or completely (15) deleted, and rRNA synthesis was achieved by a single rRNA gene repeat cloned on a multicopy plasmid. Although the growth rates of these yeast strains were not as good as the normal yeast strains, suggesting the possible importance of the chromosomal context of rRNA genes, Liebman and coworkers used this system to carry out mutational analyses to study the structure and function of rRNA (e.g., refs. 14 and 16). However, the vast majority of biochemical, structural, and mutational studies of rRNA have been done on E. coli ribosomes, and the utility of the yeast system for structure/function studies of ribosomes has been limited. This is why the development of the new E. coli system is very exciting. Nevertheless, the yeast system has been used to analyze, in addition to the structure and function of rRNA, cis-elements controlling the expression of rRNA genes as well as cis-elements responsible for localization of the nucleolus within the nucleus (15). Of course, these sorts of analyses could also be done by using the new E. coli system. For example, all of the E. coli rRNA operons have two tandem promoters, P1 and P2. It was shown that growth rate-dependent regulation acts on the major P1 promoter and not on the minor P2 promoter, whereas both promoters are subject to stringent control. These conclusions were obtained by using a nonfunctional mini-gene on a plasmid or reporter systems (17–19), and more direct tests should now be feasible. Regarding the role of cis-elements on the localization of the nucleolus studied in the yeast system (15), it should be noted that the question has never been specifically asked as to where transcription of rRNA genes takes place in bacteria. Impressive technical advancements have been made recently for localization of a specific DNA segment by fluorescence microscopy, as exemplified by the work on the localization of the replication origin and terminus within B. subtilis cells (20). Thus, one should be able to study questions related to the location of rRNA synthesis; specifically, the question of whether each of the seven rRNA operons is localized in a different site or all seven of the genes are localized in proximity, forming a single rRNA transcription factory corresponding to the nucleolus in eukaryotes. If the latter turns out to be the case, the specific chromosomal location of the seven rRNA operons, the significance of which we would like to know (as mentioned above), could be important to achieve the formation of such a factory in a suitable location within a bacterial cell. Plasmid-encoded rRNA genes in the Δ7 prrn strain may be mobile, and could freely reach locations suitable for rRNA transcription and ribosome assembly.
Starting from the Δ7 prrn strain, Squires and coworkers succeeded in replacing the plasmid carrying a single E. coli rRNA operon with a plasmid carrying an rRNA operon from different bacterial species in Enterobacteriacae, Salmonella typhimurium and Proteus vulgaris. The resultant bacterial strains did not show any obvious growth defects, demonstrating that hybrid ribosomes containing S. typhimurium (or P. vulgaris) rRNA, and E. coli ribosomal proteins are functional in vivo. This demonstration, though very striking, was not totally unexpected. Early studies demonstrated that functionally active 30S ribosomal subunits can be reconstituted in vitro from the 16S rRNA of one species of bacteria and the ribosomal proteins of a distantly related species either as a whole (21) or individually replacing each of the corresponding ribosomal (r)-proteins (22). In addition, formation of various hybrid ribosomes in vivo was demonstrated by crossing E. coli with other bacterial species belonging to Enterobacteriacae (reviewed in ref. 23). For example, Sypherd and coworkers crossed an E. coli Hfr strain and a Salmonella typhosa strain and obtained a haploid hybrid strain that carried the bulk of the S. typhosa genome, except that a segment spanning from Xyl to StrA was replaced by the corresponding region from the E. coli genome. By analyzing r-proteins of 30S subunits isolated from the hybrid strain, they demonstrated the presence of several r-proteins with an electrophoretic mobility unique to E. coli r-proteins and the absence of several S. typhosa-specific r-proteins (24). Because approximately half of the r-protein genes are now known to be localized at and adjacent to the StrA locus, the ribosomes in this hybrid strain must have contained S. typhosa rRNAs, and a significant proportion of r-proteins coming from E. coli and the remaining r-proteins from S. typhosa. Because these earlier in vivo studies were done in an effort to map genes for r-proteins (and rRNA) and analytical tools to characterize r-proteins were also limited, no attempt was made to extend these studies from evolutionary as well as structural viewpoints. The new Δ7 prrn system now gives a more powerful method to test the structural and functional compatibility of rRNA from various bacterial species with E. coli r-proteins and other components involved in the translational machinery. In addition, based on the results of hybrid ribosomes, the authors make the provocative suggestion that “… contrary to common belief, coevolution of rRNA with many other components in the translational machinery may not completely preclude the horizontal transfer of rRNA genes” (7). Horizontal transfer of r-protein genes, leading to formation of hybrid bacterial strains producing hybrid ribosomes, has been demonstrated to occur in interspecific crosses among bacterial species in Enterobactericae, as mentioned above (23, 24). Horizontal transfer of rRNA genes through interspecific crosses has also been demonstrated (25) and, although a complete replacement of the multiple host rRNA genes by the transferred rRNA gene has not been documented, it might be expected to take place perhaps with a small, but measurable, frequency if one uses effective selection methods, e.g., by the use of suitable antibiotics acting on the two rRNA species differently. The question of how significant such horizontal transfer of rRNA genes was in the evolution of rRNAs in nature may surely be a subject of future studies. Regardless of the question of evolutionary significance, the system developed by Squires and coworkers will undoubtedly stimulate the progress of ribosome research, especially research related to prokaryotic ribosomes.
Footnotes
A commentary on this article begins on page 1971.
References
- 1.Green R, Noller H. Annu Rev Biochem. 1997;66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- 2.Noller H, Hoffarth V, Zimniak L. Science. 1992;256:1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- 3.Nitta I, Ueda T, Watanabe K. RNA. 1998;4:257–267. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Nitta I, Kamada Y, Noda H, Ueda T, Watanabe K. Science. 1998;281:666–669. doi: 10.1126/science.281.5377.666. [DOI] [PubMed] [Google Scholar]
- 5.Keener J, Nomura M. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1417–1431. [Google Scholar]
- 6.Gourse R L, Gaal T, Bartlett M S, Appleman J A, Ross W. Annu Rev Micriobiol. 1996;50:645–677. doi: 10.1146/annurev.micro.50.1.645. [DOI] [PubMed] [Google Scholar]
- 7.Asai T, Zaporojets D, Squires C, Squires C L. Proc Natl Acad Sci USA. 1999;96:1971–1976. doi: 10.1073/pnas.96.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blattner F R, Plunkett G, III, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 9.Gunderson J H, Sogin M L, Wollett G, Hollingdale M, de la Cruz V F, Waters A P, McCutchan T F. Science. 1987;238:933–937. doi: 10.1126/science.3672135. [DOI] [PubMed] [Google Scholar]
- 10.Condon C, Philips J, Fu Z-Y, Squires C, Squires C L. EMBO J. 1992;11:4175–4185. doi: 10.1002/j.1460-2075.1992.tb05511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denman R, Weitzmann C, Cunningham P R, Negre D, Nurse K, Colgan J, Pan Y-C, Miedel M, Ofengand J. Biochemistry. 1989;28:1002–1011. doi: 10.1021/bi00429a013. [DOI] [PubMed] [Google Scholar]
- 12.Hui A, de Boer H A. Proc Natl Acad Sci USA. 1987;84:4762–4766. doi: 10.1073/pnas.84.14.4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacob W F, Santer M, Dahlberg A E. Proc Natl Acad Sci USA. 1987;84:4757–4761. doi: 10.1073/pnas.84.14.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chernoff Y O, Vincent A, Liebman S W. EMBO J. 1994;13:906–913. doi: 10.1002/j.1460-2075.1994.tb06334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oakes M, Aris J P, Brockenbrough J S, Wai H, Vu L, Nomura M. J Cell Biol. 1998;143:23–34. doi: 10.1083/jcb.143.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chernoff Y O, Newnam G P, Liebman S W. Proc Natl Acad Sci USA. 1996;93:2517–2522. doi: 10.1073/pnas.93.6.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gourse R L, de Boer H A, Nomura M. Cell. 1986;44:197–205. doi: 10.1016/0092-8674(86)90498-8. [DOI] [PubMed] [Google Scholar]
- 18.Gafny R, Cohen S, Nachaliel N, Glaser G. J Mol Biol. 1994;243:152–156. doi: 10.1006/jmbi.1994.1641. [DOI] [PubMed] [Google Scholar]
- 19.Josaitis C A, Gaal T, Gourse R L. Proc Natl Acad Sci USA. 1995;92:1117–1121. doi: 10.1073/pnas.92.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teleman A A, Graumann P L, Lin D C-H, Grossman A D, Losick R. Curr Biol. 1998;8:1102–1109. doi: 10.1016/s0960-9822(98)70464-6. [DOI] [PubMed] [Google Scholar]
- 21.Nomura M, Traub P, Bechmann H. Nature (London) 1968;219:793–799. doi: 10.1038/219793b0. [DOI] [PubMed] [Google Scholar]
- 22.Higo K, Held W, Kahan L, Nomura M. Proc Natl Acad Sci USA. 1973;70:944–948. doi: 10.1073/pnas.70.3.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sypherd P S, Osawa S. In: Ribosomes. Nomura M, Tissieres A, Lengyel P, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1974. pp. 669–678. [Google Scholar]
- 24.O’Neil D M, Baron L S, Sypherd P S. J Bacteriol. 1969;99:242–247. doi: 10.1128/jb.99.1.242-247.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sypherd P, O’Neil D, Taylor M. Cold Spring Harbor Symp Quant Biol. 1969;34:77–84. doi: 10.1101/sqb.1969.034.01.012. [DOI] [PubMed] [Google Scholar]