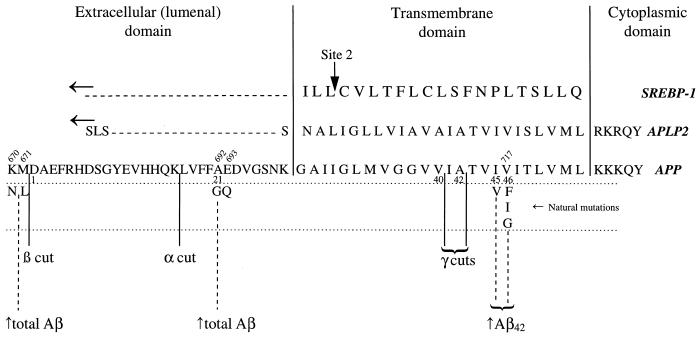
How the β-amyloid peptide (Aβ) is cleaved from its precursor, the amyloid precursor protein (APP), preoccupies a large portion of the community studying Alzheimer’s disease. And for good reason. The Aβ protein, the major component of the amyloid in senile plaques, represents a leading target in the quest for agents to combat Alzheimer’s disease. Its formation involves an unusual proteolytic event within a hydrophobic stretch of amino acids predicted to form an α-helix within the membrane. APP is a type I membrane-spanning protein of 770 residues that secretes its large ectodomain after at least two different types of cleavage. Based on the abundance of the cleavage products, the site most frequently cut is at residue 687, which lies within the Aβ peptide and thus precludes its formation. The activity of the responsible, but elusive, enzyme, dubbed the α-secretase, has the property of recognizing an optimal projection length from the membrane more strongly than any specific sequence determinants (1). The α-secretase cleavage occurs in a post-Golgi compartment, possibly including caveolae (2), and may be a glycosyl-phosphatidylinositol-linked aspartyl protease (3). A small percentage of APP is not cleaved at the α-secretase site, but instead is cleaved at residue 672 of APP (which becomes Asp-1 of Aβ) by an activity dubbed β-secretase, the first step in Alzheimer’s-type amyloidogenesis. Other Aβ species with an elongated or shortened N terminus are known (4). The β-secretase cleavage, once completed, sets the stage for the next cleavage by an activity referred to as γ-secretase. In what is now considered a critical event in the disease pathogenesis, the Aβ peptide is released after cleavage by the γ-secretase either between residues 40 and 41 or between residues 42 and 43. Normally, both forms of the Aβ peptide—Aβ42 and Aβ40—are present, with Aβ40 in great excess of Aβ42. The precise cleavage site of the γ-secretase, and hence the generation of the shorter or longer Aβ peptide, is widely believed to have important pathologic consequences. Compared with Aβ40, Aβ42 deposits earlier in the disease process (5) and assembles more rapidly into filaments in vitro (6). Explanations of the mechanism of Aβ42 vs. Aβ40 cleavage by the γ-secretase are problematic because the two proteolytic sites lie on opposite sides of the predicted α-helical transmembrane domain of APP.
By systematically mutating to phenylalanine each amino acid through the carboxyl half of the intramembranous segment of APP (or more precisely a fragment called C99 or A4CT which represents the remaining carboxyl-terminal fragment after β-secretase cleavage), Lichtenthaler et al. (7) noted a remarkable pattern among specific mutations and the exact site of γ-secretase cleavage. Based on measurements of the ratio of Aβ42/Aβ40, the authors proposed that the side of the α-helix to which the γ-secretase binds determines the relative amounts of each specific Aβ isoform. Thus, mutations can subtly shift the percentage of the γ-secretase (or γ-secretases) that binds to one or the other side of the helix. The assumption, of course, is that the APP membrane-spanning domain forms an α-helix. Although this is a reasonable assumption (8, 9), other types of helices with different periodicities or dimeric helical structures within the membrane are possible and could affect the model greatly. With aging, fatty acids are easily attacked by reactive oxygen species, resulting in alterations in membrane fluidity and deformability that could also alter the precise site of γ-secretase cleavage. In fact, free radical formation is a leading, although still sketchy, theory to explain the cause of Alzheimer’s disease in the vast majority of patients who do not carry a mutation in one of the causative genes.
A concomitant and equally intriguing observation emerging from the experimental design of Lichtenthaler et al. was the lack of a significant effect of the mutations on the level of total Aβ. Thus, an increase in Aβ42 was generally commensurate with a decrease in Aβ40, and therefore mutations within the intramembranous domain (amino acids 43–52) had little effect on the efficiency of the γ-secretase, but in some cases, had a profound effect on the relative cleavage between residues 40 and 41 versus between residues 42 and 43.
Several implications of this work deserve comment. If cleavage at one of the two γ-secretase sites is mediated by two different enzymes, as some data suggested (see refs. in Lichtenthaler et al.), then the mutations made by Lichtenthaler and colleagues appear to affect the efficiency of the two enzymes reciprocally. Alternatively, a single γ-secretase may bind and cut on either side of the α-helix, and its efficiency at either site depends on amino acids 43–52. Given a certain topological presentation of the mutant or wild-type substrate to the enzyme, the predisposition for the cleavage site can shift, but the total amount of Aβ generated changes very little. Likewise, the Alzheimer’s disease-causing natural mutations at codon 717 increase Aβ42 and may decrease Aβ40, and therefore do not alter total Aβ (10, 11). The 717 mutations are on the same side of the predicted helix as the experimental mutations of Lichtenthaler et al. that increased the Aβ42/Aβ40 ratio.
On the other hand, shifting the ratio of Aβ42/Aβ40 is not the only way to get Alzheimer’s disease—mutations on the amino side of the Aβ peptide and its flanking sequence increase both Aβ42 and Aβ40. The Swedish double mutation at codon 670/671 and the Flemish mutation at codon 692 both have the property of increasing total Aβ (12, 13). Similarly, Down syndrome, in which there is an increased gene dosage of APP, increases Aβ42 and Aβ40 even more (14). What these conditions have in common is increased cleavage at the β-secretase site, either because of increased efficiency of the enzyme in the case of the Swedish substrate or increased amount of the substrate in Down syndrome. In either case, more of the product of β-secretase cleavage (C99) is available for the γ-secretase, which is not rate-limiting. Because γ-secretase is also active on the αCTF (the fragment remaining after α-secretase cleavage) to generate a carboxyl-terminal fragment of Aβ called p3, it is unlikely that those residues at the amino end of the Aβ peptide removed by α-secretase cleavage contribute to activity at the γ-secretase site. In fact, the γ-secretase may also cleave the amyloid precursor-like proteins. The intramembranous regions of APLPs are highly homologous to APP (15) and show absolute identity with each of the amino acids in APP shown by Lichtenthaler et al. to have an effect on the Aβ42/Aβ40 ratio.
In general, some APP-causing mutations affect the γ-secretase site to increase Aβ42 specifically, whereas others affect the β-secretase site to increase the available substrate (C99) for γ-secretase cleavage, leading to increased Aβ42 and Aβ40. In each of these cases, one can reasonably conclude that increased Aβ42 is the sine qua non for genetic forms of the disease, in which abundant amyloid deposits in the brain parenchyma. Interestingly, the mutation in the APP gene at codon 693, which causes severe amyloid deposition in the cerebral vasculature with much less in the brain parenchyma does not increase either Aβ42 or Aβ40 (16), but instead enhances the aggregation of the mutant peptide into amyloid fibrils.
For γ-cleavage to occur, one would expect some degree of helix unwinding or relaxation at the cleavage site. Ordinarily in proteolysis, the introduction of a nucleophile serves this function, but the location of the γ-secretase cleavage site deep within the bilayer makes this a costly strategy. The abundance of glycines and β-branched amino acids (Ile and Val) make the membrane-spanning region of APP unusual and may contribute to the flexibility required for alternative proteolytic sites. As APP transits from the endoplasmic reticulum to the plasma membrane and the cholesterol content of the membrane increases, there is a corresponding increase in membrane thickness. Perhaps the unusual flexibility of the membrane spanning domain permits differential access to the cleavage sites when APP is in the endoplasmic reticulum compared with the Golgi or the plasma membrane. Support for this concept stems from the differential localization of Aβ42 vs. Aβ40 within cells: the preferred site of Aβ42 is the endoplasmic reticulum/intermediate compartment and the preferred site of Aβ40 is the Golgi and beyond (17–21). A second intracellular pool of both Aβ42 and Aβ40 resides in the detergent-insoluble glycolipid fraction (22), where the membrane is rich in cholesterol and may provide a protective storage site for the toxic peptide.
The presence of Aβ42 in the ER fits well with another important observation in Alzheimer’s disease—the localization of presenilin to the endoplasmic reticulum. Mutations in presenilin cause an inherited early-onset form of the disease and effect the Aβ42/Aβ40 ratio. Over 50 different mutations in presenilin have been described (23), and all of those studied to date increase Aβ42 production about 1.5- to 3.0-fold. Further support for a functional relationship between APP metabolism and presenilin comes from PS1-deficient mice, which have reduced γ-secretase activity (24). Several recently presented abstracts suggested that presenilin itself can cleave at the γ-secretase site (25) and that presenilin-binding molecules may regulate the cleavage of APP (26).
The intramembranous cleavage of APP is so unusual that it is often compared with the only other well established example of an intramembranous cleavage—that of the sterol-regulatory element-binding proteins (SREBP-1 and SREBP-2), a pair of membrane-bound transcription factors that regulate cholesterol homeostasis (27). A metalloprotease called S2P that is itself a polytopic membrane protein is responsible for the intramembranous cleavage of SREBP (28). Processing of SREBP, which forms a hairpin in the endoplasmic reticulum membrane occurs in two steps—the first cleavage in a hydrophilic loop within the lumen (site 1) and the second at the Leu-484–Cys-485 bond within the membrane (site 2). Importantly, the positions of the cleavage sites relative to the membrane in SREBP and APP differ (29). The SREBP site is located at the outer margin near the polar head groups of the phospholipids, whereas the APP site lies deep within the membrane. Also unlike APP, mutagenesis of the conserved amino acids around site 2, including those residues at the cleavage site itself, does not affect cleavage (29). What APP and SREBP do have in common is the hint of a novel and little-known chemistry that goes on within the membrane.
Figure 1.
Comparison of amino acid sequences of three membrane-spanning proteins: Sterol-regulatory element-binding protein 1 (SREBP-1), amyloid precursor-like protein 2 (APLP2), and the amyloid precursor protein (APP). The numbering above the APP sequence refers to the codon corresponding to the position in the holoprotein. The numbering below the APP sequence refers to the amino acid position beginning with Asp-1 of the Aβ peptide. Known disease-causing mutations of APP are shown below the sequence and their tendency to increase (↑) either total Aβ or Aβ42 is noted. Intramembranous cleavage sites are noted as site 2 in SREBP and as the γ cuts in APP.
ABBREVIATIONS
- Aβ
β-amyloid
- APP
amyloid precursor protein
- SREBP
sterol-regulatory element-binding protein
Footnotes
A commentary on this article begins on page 3053.
References
- 1.Sisodia S S. Proc Natl Acad Sci USA. 1992;89:6075–6079. doi: 10.1073/pnas.89.13.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ikezu T, Trapp B D, Song K S, Schlegel A, Lisanti M P, Okamoto T. J Biol Chem. 1998;273:10485–10495. doi: 10.1074/jbc.273.17.10485. [DOI] [PubMed] [Google Scholar]
- 3.Komano H, Seeger M, Gandy S, Wang G T, Krafft G A, Fuller R S. J Biol Chem. 1998;273:31648–31651. doi: 10.1074/jbc.273.48.31648. [DOI] [PubMed] [Google Scholar]
- 4.Evin G, Beyreuther K, Masters C L. Amyloid. 1994;1:263–280. [Google Scholar]
- 5.Saido T C, Iwatsubo T, Mann D M A, Shimada H, Ihara Y, Kawashima S. Neuron. 1995;14:457–466. doi: 10.1016/0896-6273(95)90301-1. [DOI] [PubMed] [Google Scholar]
- 6.Jarrett J T, Berger E P, Lansbury P T., Jr Biochemistry. 1993;32:4693–4697. doi: 10.1021/bi00069a001. [DOI] [PubMed] [Google Scholar]
- 7.Lichtenthaler S F, Wang R, Grimm K, Uljon S N, Masters C L, Beyreuther K. Proc Natl Acad Sci USA. 1999;96:3053–3058. doi: 10.1073/pnas.96.6.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lemmon M A, Flanagan J M, Hunt J F, Adair B D, Bormann B J, Dempsey C E, Engelman D M. J Biol Chem. 1992;267:7683–7689. [PubMed] [Google Scholar]
- 9.Lee N H, Weinstock K G, Kirkness E F, Earle-Hughes J A, Fuldner R A, Marmaros S, Glodek A, Gocayne J D, Adams M D, Kerlavage A R, et al. Proc Natl Acad Sci USA. 1995;92:8303–8307. doi: 10.1073/pnas.92.18.8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maruyama K, Tomita T, Shinozaki K, Kume H, Asada H, Saido T C, Ishiura S, Iwatsubo T, Obata K. Biochem Biophys Res Commun. 1996;227:730–735. doi: 10.1006/bbrc.1996.1577. [DOI] [PubMed] [Google Scholar]
- 11.Tamaoka A, Odaka A, Ishibashi Y, Usami M, Sahara N, Suzuki N, Nukima N, Mizusawa H, Shoji S, Kanazawa I, Mori H. J Biol Chem. 1994;269:32721–32724. [PubMed] [Google Scholar]
- 12.Citron M, Vigo-Pelfrey C, Teplow D B, Miller C, Schenk D, Johnston J, Winblad B, Venizelos N, Lannfelt L, Selkoe D J. Proc Natl Acad Sci USA. 1994;91:11993–11997. doi: 10.1073/pnas.91.25.11993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haass C, Hung A Y, Selkoe D J, Teplow D B. J Biol Chem. 1994;269:17741–17748. [PubMed] [Google Scholar]
- 14.Tokuda T, Fukushima T, Ikeda S, Sekijima Y, Shoji S, Yanagisawa N, Tamaoka A. Ann Neurol. 1997;41:271–273. doi: 10.1002/ana.410410220. [DOI] [PubMed] [Google Scholar]
- 15.Wasco W, Gurubhagavatula S, Paradis M D, Romano D M, Sisodia S S, Hyman B T, Neve R L, Tanzi R E. Nat Genet. 1993;5:95–99. doi: 10.1038/ng0993-95. [DOI] [PubMed] [Google Scholar]
- 16.De Jonghe C, Zehr C, Yager D, Prada C M, Younkin S, Hendriks L, Van Broeckhoven C, Eckman C B. Neurobiol Dis. 1998;5:281–286. doi: 10.1006/nbdi.1998.0202. [DOI] [PubMed] [Google Scholar]
- 17.Xu H, Sweeney D, Wang R, Thinakaran G, Lo A C Y, Sisodia S S, Greenberg P, Gandy S. Proc Natl Acad Sci USA. 1997;94:3748–3752. doi: 10.1073/pnas.94.8.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wild-Bode C, Yamazaki T, Capell A, Leimer U, Steiner H, Ihara Y, Haass C. J Biol Chem. 1997;272:16085–16088. doi: 10.1074/jbc.272.26.16085. [DOI] [PubMed] [Google Scholar]
- 19.Hartmann T, Bieger S C, Bruhl B, Tienari P J, Ida N, Allsop D, Roberts G W, Masters C L, Dotti C G, Unsicker K, Beyreuther K. Nat Med. 1997;3:1016–1018. doi: 10.1038/nm0997-1016. [DOI] [PubMed] [Google Scholar]
- 20.Cook D G, Forman M S, Sung J C, Leight S, Kolson D L, Iwatsubo T, Lee V M, Doms R W. Nat Med. 1997;3:1021–1023. doi: 10.1038/nm0997-1021. [DOI] [PubMed] [Google Scholar]
- 21.Xia W, Zhang J, Ostaszewski B L, Kimberly W T, Seubert P, Koo E H, Shen J, Selkoe D J. Biochemistry. 1998;47:16465–16471. doi: 10.1021/bi9816195. [DOI] [PubMed] [Google Scholar]
- 22.Lee S J, Liyanage U, Bickel P E, Xia W, Lansbury P T, Kosik K S. Nat Med. 1998;4:730–734. doi: 10.1038/nm0698-730. [DOI] [PubMed] [Google Scholar]
- 23.Cruts M, Van Broeckhoven C. Hum Mutat. 1998;11:183–190. doi: 10.1002/(SICI)1098-1004(1998)11:3<183::AID-HUMU1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 24.DeStrooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F. Nature (London) 1998;391:387–390. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- 25.Wolfe M S, Moore C L, Xia W, Leatherwood D D, Donkor I O, Selkoe D J. Soc Neurosci Abstr. 1998;24:1005. [Google Scholar]
- 26.Morin P, Medina M, Brown A M C, Kosik K S. Neurobiol Aging. 1998;19:S38. (abstr.). [Google Scholar]
- 27.Brown M S, Goldstein J L. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 28.Rawson R B, Zelenski N G, Nijhawan D, Ye J, Sakai J, Hasan M T, Chang T Y, Brown M S, Goldstein J L. Mol Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- 29.Duncan E A, Dave U P, Sakai J, Goldstein J L, Brown M S. J Biol Chem. 1998;273:17801–17809. doi: 10.1074/jbc.273.28.17801. [DOI] [PubMed] [Google Scholar]