Abstract
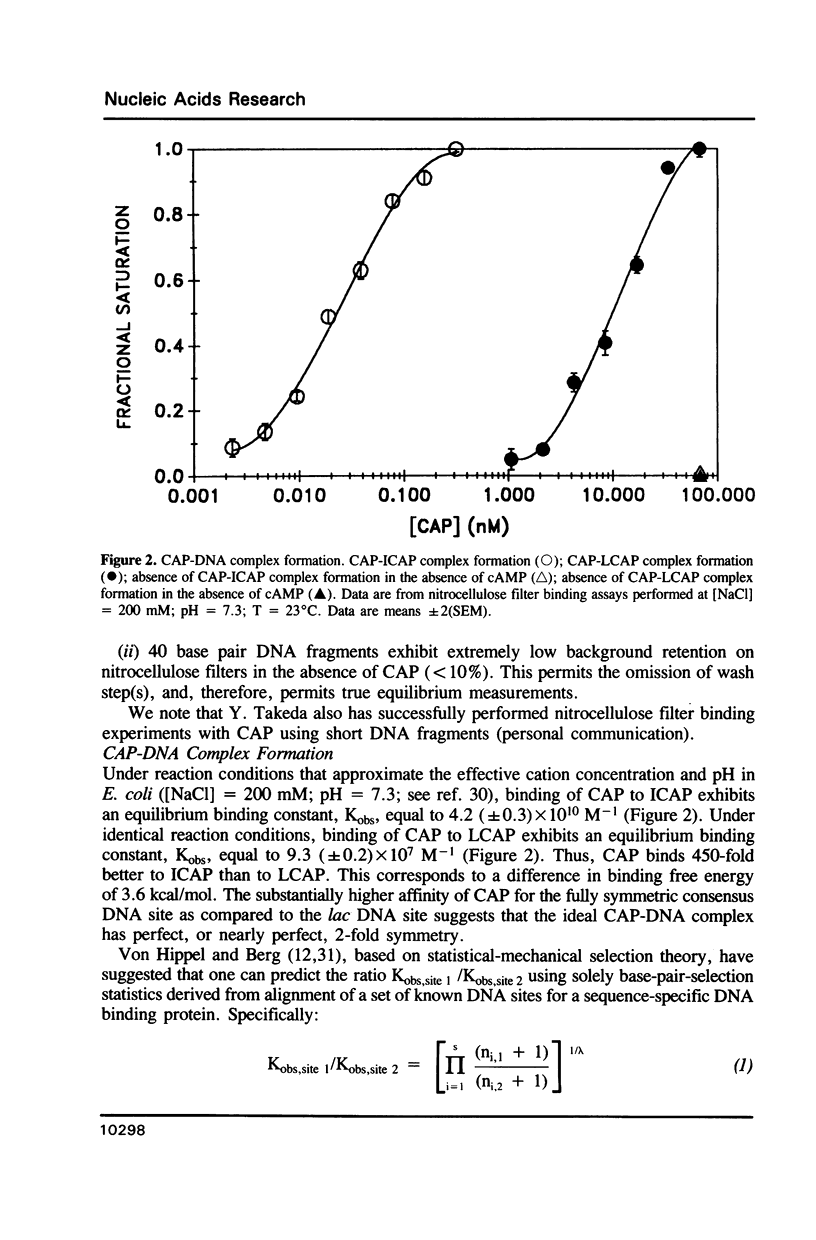
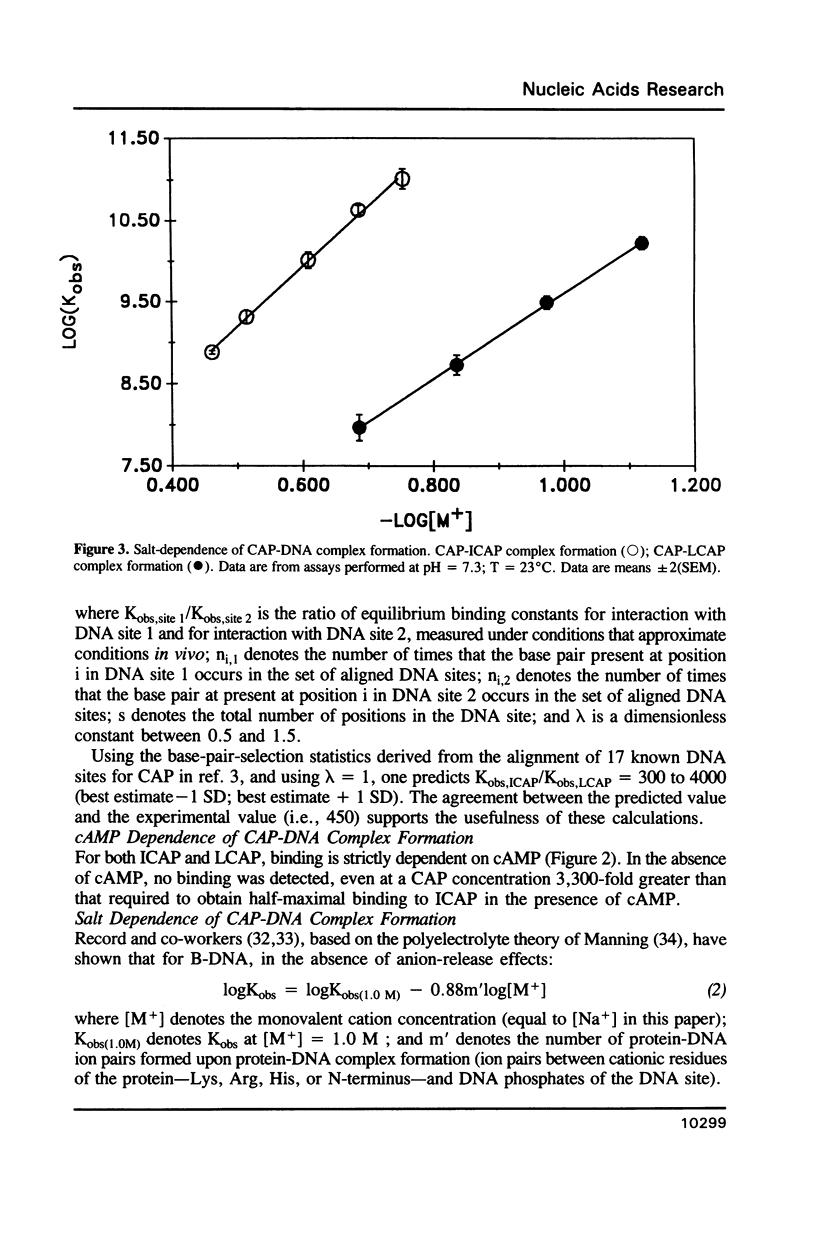
We have synthesized two 40 base pair DNA fragments; one fragment contains the consensus DNA site for CAP (fragment 'ICAP'); the other fragment contains the E. coli lac promoter DNA site for CAP (fragment 'LCAP'). We have investigated the binding of CAP to the two DNA fragments using the nitrocellulose filter binding assay. Under standard conditions [( NaCl] = 200 mM, pH = 7.3), CAP exhibits a 450-fold higher affinity for ICAP than for LCAP. The salt dependence of the binding equilibrium indicates that CAP makes eight ion pairs with ICAP, but only six ion pairs with LCAP. Approximately half of the difference in binding free energy for interaction of CAP with ICAP vs. LCAP is attributable to this difference in ion-pair formation. The pH dependence of the binding equilibrium indicates that the eight CAP-ICAP ion pairs and the six CAP-LCAP ion pairs do not involve His residues of CAP.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg O. G., von Hippel P. H. Selection of DNA binding sites by regulatory proteins. II. The binding specificity of cyclic AMP receptor protein to recognition sites. J Mol Biol. 1988 Apr 20;200(4):709–723. doi: 10.1016/0022-2836(88)90482-2. [DOI] [PubMed] [Google Scholar]
- Berg O. G., von Hippel P. H. Selection of DNA binding sites by regulatory proteins. Statistical-mechanical theory and application to operators and promoters. J Mol Biol. 1987 Feb 20;193(4):723–750. doi: 10.1016/0022-2836(87)90354-8. [DOI] [PubMed] [Google Scholar]
- Clore G. M., Gronenborn A. M. Proton nuclear magnetic resonance study of the histidine residues of the Escherichia coli adenosine cyclic 3',5'-phosphate receptor protein. pH titration behavior, deuterium exchange, and partial assignments. Biochemistry. 1982 Aug 17;21(17):4048–4053. doi: 10.1021/bi00260a021. [DOI] [PubMed] [Google Scholar]
- Ebright R. H., Cossart P., Gicquel-Sanzey B., Beckwith J. Molecular basis of DNA sequence recognition by the catabolite gene activator protein: detailed inferences from three mutations that alter DNA sequence specificity. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7274–7278. doi: 10.1073/pnas.81.23.7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright R. H., Cossart P., Gicquel-Sanzey B., Beckwith J. Mutations that alter the DNA sequence specificity of the catabolite gene activator protein of E. coli. Nature. 1984 Sep 20;311(5983):232–235. doi: 10.1038/311232a0. [DOI] [PubMed] [Google Scholar]
- Ebright R. H., Kolb A., Buc H., Kunkel T. A., Krakow J. S., Beckwith J. Role of glutamic acid-181 in DNA-sequence recognition by the catabolite gene activator protein (CAP) of Escherichia coli: altered DNA-sequence-recognition properties of [Val181]CAP and [Leu181]CAP. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6083–6087. doi: 10.1073/pnas.84.17.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilen E., Pampeno C., Krakow J. S. Production and properties of the alpha core derived from the cyclic adenosine monophosphate receptor protein of Escherichia coli. Biochemistry. 1978 Jun 27;17(13):2469–2473. doi: 10.1021/bi00606a001. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G., Hirschman S. Z. A neighbor-interaction analysis of the hypochromism and spectra of DNA. J Mol Biol. 1965 Sep;13(2):407–427. doi: 10.1016/s0022-2836(65)80106-1. [DOI] [PubMed] [Google Scholar]
- Fried M. G., Crothers D. M. Equilibrium studies of the cyclic AMP receptor protein-DNA interaction. J Mol Biol. 1984 Jan 25;172(3):241–262. doi: 10.1016/s0022-2836(84)80025-x. [DOI] [PubMed] [Google Scholar]
- Gartenberg M. R., Crothers D. M. DNA sequence determinants of CAP-induced bending and protein binding affinity. Nature. 1988 Jun 30;333(6176):824–829. doi: 10.1038/333824a0. [DOI] [PubMed] [Google Scholar]
- Gaston K., Chan B., Kolb A., Fox J., Busby S. Alterations in the binding site of the cyclic AMP receptor protein at the Escherichia coli galactose operon regulatory region. Biochem J. 1988 Aug 1;253(3):809–818. doi: 10.1042/bj2530809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao-Huang Y., Revzin A., Butler A. P., O'Conner P., Noble D. W., von Hippel P. H. Nonspecific DNA binding of genome-regulating proteins as a biological control mechanism: measurement of DNA-bound Escherichia coli lac repressor in vivo. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4228–4232. doi: 10.1073/pnas.74.10.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb A., Spassky A., Chapon C., Blazy B., Buc H. On the different binding affinities of CRP at the lac, gal and malT promoter regions. Nucleic Acids Res. 1983 Nov 25;11(22):7833–7852. doi: 10.1093/nar/11.22.7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koop A. H., Staprans S. I., Bourgeois S. Specific binding of the cAMP receptor protein of Escherichia coli to the lactose operon promoter. Biochimie. 1985 Jan;67(1):161–175. doi: 10.1016/s0300-9084(85)80244-3. [DOI] [PubMed] [Google Scholar]
- Kotlarz D., Fritsch A., Buc H. Variations of intramolecular ligation rates allow the detection of protein-induced bends in DNA. EMBO J. 1986 Apr;5(4):799–803. doi: 10.1002/j.1460-2075.1986.tb04284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majors J. Specific binding of CAP factor to lac promoter DNA. Nature. 1975 Aug 21;256(5519):672–674. doi: 10.1038/256672a0. [DOI] [PubMed] [Google Scholar]
- Manning G. S. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q Rev Biophys. 1978 May;11(2):179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- Martin K. J., Schleif R. F. Equilibrium DNA-binding of AraC protein. Compensation for displaced ions. J Mol Biol. 1987 Jun 5;195(3):741–744. doi: 10.1016/0022-2836(87)90193-8. [DOI] [PubMed] [Google Scholar]
- Maurizot J. C., Grebert P. Thermodynamic parameters of the binding of the tight-binding I12X86 lac repressor to operator and non-operator DNA. FEBS Lett. 1988 Oct 24;239(1):105–108. doi: 10.1016/0014-5793(88)80554-4. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mitra S., Zubay G., Landy A. Evidence for the preferential binding of the catabolite gene activator protein (CAP) to DNA containing the lac promoter. Biochem Biophys Res Commun. 1975 Dec 1;67(3):857–863. doi: 10.1016/0006-291x(75)90755-x. [DOI] [PubMed] [Google Scholar]
- Morita T., Shigesada K., Kimizuka F., Aiba H. Regulatory effect of a synthetic CRP recognition sequence placed downstream of a promoter. Nucleic Acids Res. 1988 Aug 11;16(15):7315–7332. doi: 10.1093/nar/16.15.7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossing M. C., Record M. T., Jr Thermodynamic origins of specificity in the lac repressor-operator interaction. Adaptability in the recognition of mutant operator sites. J Mol Biol. 1985 Nov 20;186(2):295–305. doi: 10.1016/0022-2836(85)90106-8. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Pastan I., Adhya S. Cyclic adenosine 5'-monophosphate in Escherichia coli. Bacteriol Rev. 1976 Sep;40(3):527–551. doi: 10.1128/br.40.3.527-551.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porschke D., Hillen W., Takahashi M. The change of DNA structure by specific binding of the cAMP receptor protein from rotation diffusion and dichroism measurements. EMBO J. 1984 Dec 1;3(12):2873–2878. doi: 10.1002/j.1460-2075.1984.tb02223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Record M. T., Jr, Anderson C. F., Lohman T. M. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Q Rev Biophys. 1978 May;11(2):103–178. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, Lohman M. L., De Haseth P. Ion effects on ligand-nucleic acid interactions. J Mol Biol. 1976 Oct 25;107(2):145–158. doi: 10.1016/s0022-2836(76)80023-x. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Suzuki H., Bourgeois S. Lac repressor-operator interaction. I. Equilibrium studies. J Mol Biol. 1970 Feb 28;48(1):67–83. doi: 10.1016/0022-2836(70)90219-6. [DOI] [PubMed] [Google Scholar]
- Saxe S. A., Revzin A. Cooperative binding to DNA of catabolite activator protein of Escherichia coli. Biochemistry. 1979 Jan 23;18(2):255–263. doi: 10.1021/bi00569a003. [DOI] [PubMed] [Google Scholar]
- Shanblatt S. H., Revzin A. The binding of catabolite activator protein and RNA polymerase to the Escherichia coli galactose and lactose promoters probed by alkylation interference studies. J Biol Chem. 1986 Aug 15;261(23):10885–10890. [PubMed] [Google Scholar]
- Stormo G. D., Hartzell G. W., 3rd Identifying protein-binding sites from unaligned DNA fragments. Proc Natl Acad Sci U S A. 1989 Feb;86(4):1183–1187. doi: 10.1073/pnas.86.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Blazy B., Baudras A., Hillen W. On the origin of selectivity in recognition by cyclic adenosine 3',5'-monophosphate receptor protein of its specific binding site of the lactose promoter region. J Mol Biol. 1983 Jul 15;167(4):895–899. doi: 10.1016/s0022-2836(83)80118-1. [DOI] [PubMed] [Google Scholar]
- Takeda Y., Ohlendorf D. H., Anderson W. F., Matthews B. W. DNA-binding proteins. Science. 1983 Sep 9;221(4615):1020–1026. doi: 10.1126/science.6308768. [DOI] [PubMed] [Google Scholar]
- Terry B. J., Jack W. E., Rubin R. A., Modrich P. Thermodynamic parameters governing interaction of EcoRI endonuclease with specific and nonspecific DNA sequences. J Biol Chem. 1983 Aug 25;258(16):9820–9825. [PubMed] [Google Scholar]
- Vershon A. K., Liao S. M., McClure W. R., Sauer R. T. Bacteriophage P22 Mnt repressor. DNA binding and effects on transcription in vitro. J Mol Biol. 1987 May 20;195(2):311–322. doi: 10.1016/0022-2836(87)90652-8. [DOI] [PubMed] [Google Scholar]
- Warwicker J., Engelman B. P., Steitz T. A. Electrostatic calculations and model-building suggest that DNA bound to CAP is sharply bent. Proteins. 1987;2(4):283–289. doi: 10.1002/prot.340020404. [DOI] [PubMed] [Google Scholar]
- Weber I. T., Steitz T. A. A model for the non-specific binding of catabolite gene activator protein to DNA. Nucleic Acids Res. 1984 Nov 26;12(22):8475–8487. doi: 10.1093/nar/12.22.8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber I. T., Steitz T. A. Model of specific complex between catabolite gene activator protein and B-DNA suggested by electrostatic complementarity. Proc Natl Acad Sci U S A. 1984 Jul;81(13):3973–3977. doi: 10.1073/pnas.81.13.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber I. T., Steitz T. A. Structure of a complex of catabolite gene activator protein and cyclic AMP refined at 2.5 A resolution. J Mol Biol. 1987 Nov 20;198(2):311–326. doi: 10.1016/0022-2836(87)90315-9. [DOI] [PubMed] [Google Scholar]
- Winter R. B., von Hippel P. H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 2. The Escherichia coli repressor--operator interaction: equilibrium measurements. Biochemistry. 1981 Nov 24;20(24):6948–6960. doi: 10.1021/bi00527a029. [DOI] [PubMed] [Google Scholar]
- de Crombrugghe B., Busby S., Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984 May 25;224(4651):831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]


